Abstract
Background.
Donor-derived cell-free DNA (dd-cfDNA) is a noninvasive biomarker for the early detection of organ transplant rejection and other causes of graft injury. For nonrejection renal injuries, there is little information about the performance characteristics of this biomarker. We highlight some of the possible causes of kidney injury that may arise in patients with normal dd-cfDNA levels.
Methods.
We performed a retrospective analysis of solitary renal transplant cases between January 2017 and November 2019. Those who had an abnormal laboratory or pathological finding within 1 mo of a normal dd-cfDNA test were selected. Subgroups were stratified for those who had normal or abnormal/rising serum creatinine, and differences between the groups were analyzed.
Results.
Of 414 individuals who received a kidney transplant, 24 (7.5%) had a total of 41 normal dd-cfDNA values and 51 abnormal laboratory tests or histologic findings. The most common graft-injuring event was BK virus viremia (24 of 51). Other abnormal findings included urinary traction infections (n = 4), CMV viremia (n = 4), and biopsies demonstrating antibody-mediated rejection (AMR) (n = 2), T cell–mediated rejection (n = 1), focal segmental glomerulosclerosis (n = 2), nondonor-specific antibody chronic AMR (n = 1), and interstitial fibrosis and tubular atrophy (n = 7). Subgroup analysis of those with normal dd-cfDNA and normal/stable versus abnormal/rising creatinine showed that BK virus viremia was the most common abnormal finding in both groups at 53% and 38% respectively. On biopsy, 1 case of acute T cell–mediated rejection (1B and 2B) was seen with normal/stable creatinine, whereas 1 of nonspecific C4d focally positive and 1 of nondonor-specific antibody AMR were seen with abnormal/rising creatinine.
Conclusions.
Low levels of serum dd-cfDNA do not preclude detection of active graft-injuring events and that subclinical injuries may be developing. Context is important in the interpretation of dd-cfDNA, so renal biopsy remains a part of the diagnostic pathway for allograft dysfunction and maintenance of allograft health.
INTRODUCTION
Allograft rejection is a major cause of graft failure.1 Thus, timely and accurate diagnosis allows a significant number of transplanted kidneys to be salvaged. However, other etiologies of allograft dysfunction, such as acute tubular necrosis (ATN), medication-induced nephrotoxicity, BK virus (BKV) and cytomegalovirus (CMV) infections, and recurrent glomerular kidney disease can present with similar biochemical patterns, such as elevated creatinine, proteinuria, and new-onset or worsening hypertension.2,3 To prevent allograft loss and minimize long-term graft injury, discriminating between allograft rejection and other causes of graft injury is critical so appropriate treatment can be administered.
Subclinical graft injury may occur despite normal serum creatinine measurements, so improved methods to detect treatable causes of kidney injury could prevent irreversible graft damage. Donor-derived cell-free DNA (dd-cfDNA) has been developed as a noninvasive biomarker for the early detection of organ transplant rejection.3 Using targeted amplification and sequencing, the test can quantify DNA released by the donor organ as a measure of graft injury without the variability, inconsistency, or risks associated with graft biopsy. It may also identify graft injury before there are measurable changes in the serum creatinine. Studies have demonstrated that serum dd-cfDNA >1% may identify acute antibody-mediated rejection (AMR) especially in the setting of positive donor-specific antibodies (DSAs).4 dd-cfDNA may also help identify patients with borderline T cell–mediated rejection (TCMR) 1A histology who are likely to develop worsening kidney function if the level is in the high-normal range (>0.5%).5 Recent data suggest that elevations in dd-cfDNA are also observed in patients with DSA, BKV nephropathy, urinary traction infections (UTIs), and ATN.6,7 Although an abnormal dd-cfDNA result does not replace renal biopsy yet, it offers a noninvasive method to inform possible etiologies of allograft dysfunction in specific recipients. Current indications for obtaining dd-cfDNA tests include protocol and surveillance testing, abnormal creatinine, and other abnormalities, such as increasing proteinuria or newly elevated DSA.
Given dd-cfDNA’s potential in detecting other, no-rejection kidney-injuring events, the determination of the sensitivity and specificity of dd-cfDNA for these events is relevant both to broadening and focusing its clinical applications. Thus, identifying the histopathology and diagnosis in recipients with normal dd-cfDNA levels will allow for a better understanding of this biomarker’s performance in nonrejection scenarios. Here, we present a case series of kidney transplant recipients who had normal follow-up dd-cfDNA <1% in the presence of abnormal histopathology findings found on screening investigations and graft biopsies. We highlight some of the possible causes of kidney injury that may arise in patients with normal dd-cfDNA levels.
MATERIALS AND METHODS
This retrospective case series describes a group of adult kidney transplant recipients who had undergone dd-cfDNA testing (Allosure, CareDx, Brisbane, CA) between 0- and 31-mo posttransplant at a single center. Both for-cause and surveillance dd-cfDNA tests were performed. After obtaining institutional review board approval, all solitary renal transplant recipients, between January 2017 and November 2019, who underwent at least 1 dd-cfDNA test were reviewed. Correlation between organ injury events and biomarkers is described in the context of normal dd-cfDNA results. Exclusion criteria included abnormal dd-cfDNA values, a history of previous transplantation or multiorgan transplantation. An abnormal dd-cfDNA result was defined as >1% and a high-normal dd-cfDNA result was defined as >0.5%. Posttransplant follow-up was scheduled as per institutional protocol, which included routine dd-cfDNA, DSA, and BK viremia testing at 6 wk, then 3-, 6-, 9-, and 12-mo posttransplant. Additional investigations for allograft dysfunction were performed depending on the clinical context.
We investigated kidney recipients who had an abnormal routine laboratory value within 1 mo of a previous normal dd-cfDNA test. Abnormal test results done before dd-cfDNA testing were excluded to avoid treatment effects. Routine labs used to investigate graft-injuring events included serum creatinine, urine albumin:creatinine ratio, blood and urine cultures, serum BKV and CMV loads, DSA specificity and mean fluorescent index measured by Luminex, transplant doppler ultrasound, and renal biopsy results. Serum creatinine >2.0 mg/dL or an increase in creatinine >20% from baseline was considered abnormal. Serum tacrolimus levels were used to assess for calcineurin inhibitor (CNI) nephrotoxicity. Bacterial UTI was defined by the presence of >105 colony forming units based on culture. Serum BKV and CMV loads are reported as copies/mL and were considered abnormal whenever viral loads were detectable. Abnormal ultrasound findings included hydronephrosis, urothelial thickening, significant debris within the collecting system, or abnormality in renal blood flow suspicious for inflammatory events or vascular complications. Renal biopsy identifying AMR or TCMR, BKV nephropathy, recurrence of primary disease (such as focal segmental glomerulosclerosis [FSGS]), ATN, and chronic graft fibrosis were all considered abnormal.
Statistics
Descriptive statistics were compared between those with normal and abnormal creatinine levels in the setting of normal dd-cfDNA levels. Nominal variables were compared using Chi-square tests and continuous variables compared using Student’s t-test. All statistical analyses were performed using GraphPad Prism 8.
RESULTS
Between January 2017 and November 2019, a total of 414 individuals received a kidney transplant at our center. Forty-five were excluded because of a history of multiorgan transplant or retransplantation, and another 50 were excluded for a positive dd-cfDNA result. Out of the remaining 319 recipients, 24 individuals (7.5%) had a total of 41 normal dd-cfDNA tests associated with 51 laboratory tests that identified lesions and pathologies that could be associated with graft injury (Figure 1). Mean dd-cfDNA level was 0.28% ± 0.79% and mean creatinine was 1.92 ± 0.72 mg/dL. The mean number of days between a normal dd-cfDNA level and a biopsy or biochemical measure revealing a cause for graft injury was 10.53 ± 9 d. The mean number of days between a serum creatinine level and another test revealing an abnormal result was 8.84 ± 9.22 d. Of the 51 investigations with abnormal results, there were 4 pairs of blood tests and biopsies that demonstrated BKV viremia and nephropathy. BKV viremia was the most common graft-injuring event observed (24 of 51); however, half (12 of 24) of these tests detected only low viral loads (<1500 copies) (Table 1). Other abnormal findings included UTIs (n = 4), CMV viremia (n = 4), and biopsies demonstrating chronic AMR (n = 4), TCMR (n = 1), recurrent FSGS (n = 2), BKV nephropathy (n = 4), non-DSA AMR (n = 1), focal C4d positivity (n = 1), and interstitial fibrosis and tubular atrophy (IF/TA) (n = 6). Interestingly, there was no DSA detected in any of the assays included in this analysis, even if there was evidence of chronic AMR on biopsy. There were also no histologic changes associated with CMV viremia seen in biopsies. There were a total of 4 dd-cfDNA levels in the high-normal range (>0.5%) associated with 6 graft-injuring events, as 2 of the high-normal dd-cfDNA were associated with 2 graft-injuring events each (Figure 2). Graft-injuring events in this group include: BKV viremia (n = 2, <1500 copies and 7 400 000 copies), BKV nephropathy (n = 1), chronic active antibody-mediated allograft rejection (n = 2), and mild IF/TA (n = 1). Further assessment of the etiologies of IF/TA revealed a variety of causes including resolving TCMR, chronic HIV infection, and chronic thrombotic microangiopathy.
FIGURE 1.
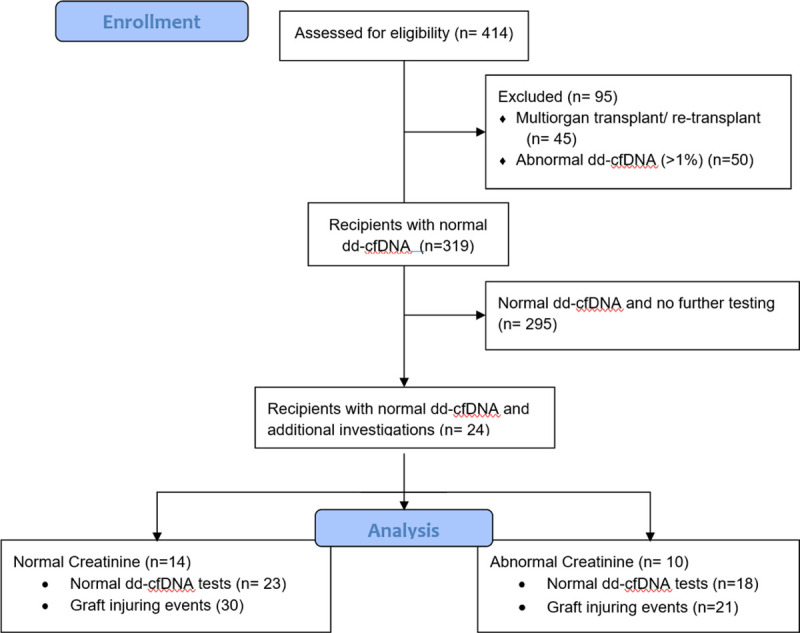
CONSORT flow diagram of the study cohort. dd-cfDNA, donor-derived cell-free DNA.
TABLE 1.
Abnormal investigations in patients with normal dd-cfDNA
| BK viremia | Number of abnormal results (n = 51) |
|---|---|
| Low-viral load | 12 (23) |
| High-viral load | 12 (23) |
| CMV viremia | 4 (8) |
| UTI | 4 (8) |
| Biopsy | |
| TCMR | 1 (2) |
| AMR | 4 (8) |
| Recurrent FSGS | 2 (4) |
| BK nephropathy | 4 (8) |
| IF/TA | 6 (12) |
| Non-DSA AMR | 1 (2) |
| Focal C4d deposition | 1 (2) |
Data presented as n (%).
AMR, antibody-mediated rejection; CMV, cytomegalovirus; dd-cfDNA, donor-derived cell-free DNA; DSA, donor-specific antibody; FSGS, focal segmental glomerulosclerosis; IF/TA, interstitial fibrosis and tubular atrophy; TCMR, T cell–mediated rejection; UTI, urinary tract infection.
FIGURE 2.
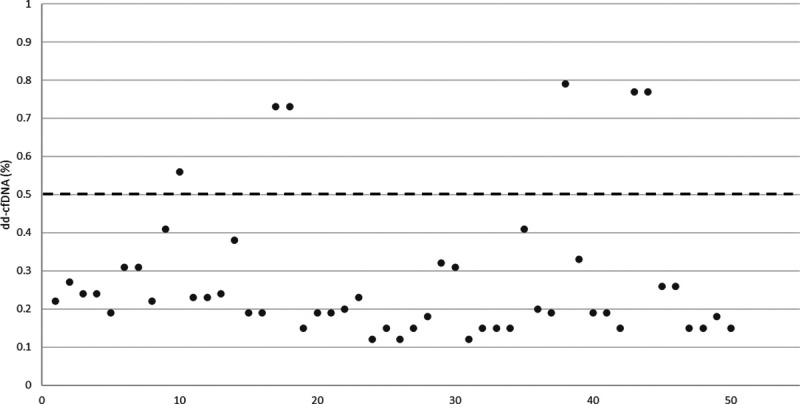
dd-cfDNA level (%) associated with graft-injuring event. The dashed line (- -) represents the threshold between low-normal and high-normal dd-cfDNA levels. dd-cfDNA, donor-derived cell-free DNA.
Subgroup analysis of graft-injuring events was performed with the cohort divided into events occurring with normal (n = 30) versus abnormal creatinine (n = 21) and normal dd-cfDNA level. Donor and recipient characteristics of the 2 groups were similar (Table 2). Creatinine drawn at the time dd-cfDNA level was significantly different between the 2 groups (1.40 versus 2.58 mg/dL, P < 0.001), whereas there was no difference in serum FK levels (6.88 versus 6.62 ng/mL, P = 0.70) (Table 3). There was no significant difference in dd-cfDNA levels (0.26 versus 0.27%, P = 0.88) when patients were stratified by creatinine.
TABLE 2.
Baseline characteristics of recipients with normal dd-cfDNA and normal creatinine vs abnormal creatinine
| Normal creatinine (n = 14) | Abnormal creatinine (n = 10) | P | ||
|---|---|---|---|---|
| Sex (n=) | Male (%) | 8 (57) | 7 (70) | |
| Female (%) | 6 (43) | 3 (30) | ||
| Race (n=) | Caucasian (%) | 7 (50) | 1 (10) | 0.05 |
| African American (%) | 5 (36) | 8 (80) | ||
| Asian (%) | 2 (14) | 0 | ||
| Other (%) | 0 | 1 (10) | ||
| Recipient age (y, mean) | 61.7 | 52.8 | 0.06 | |
| Donor age (y, mean) | 46.5 | 44.4 | 0.81 | |
| KDPI (%, mean) | 69 | 73 | 0.76 | |
| Type of dialysis (n=) | Preemptive (%) | 4 (29) | 0 | 0.10 |
| HD (%) | 9 (64) | 10 (100) | ||
| PD (%) | 1 (7) | 0 | ||
| Duration of dialysis (y, mean) | 5.1 | 6 | 0.59 | |
| Induction immunosuppression (n=) | Alemtuzumab | 5 (36) | 6 (60) | 0.32 |
| Antithymocyte globulin | 2 (14) | 2 (20) | ||
| Basiliximab | 7 (50) | 2 (20) |
Values expressed as mean when applicable. Data are presented as mean or n (%). Student t-test for continuous variables; χ2 test for binary or categorical variables (global P value).
dd-cfDNA, donor-derived cell-free DNA; HD, hemodialysis; KDPI, kidney donor profile index; PD, peritoneal dialysis.
TABLE 3.
Characteristics of graft-injuring events with normal dd-cfDNA and normal creatinine vs abnormal creatinine
| Normal creatinine (n = 30) | Abnormal creatinine (n = 21) | P | |
|---|---|---|---|
| Serum creatinine (mg/dL) | 1.41 | 2.58 | <0.001 |
| Allosure (%) | 0.26 | 0.27 | 0.88 |
| Mean time from transplant to event (y) | 0.81 | 0.81 | 0.99 |
| Tacrolimus level (ng/mL) | 6.88 | 6.62 | 0.70 |
| Maintenance immunosuppression (n=) | 0.31 | ||
| Tacro + MPA + steroid | 13 | 9 | |
| Tacro + MPA | 13 | 7 | |
| Tacro + steroid | 2 | 4 | |
| Sirolimus + leflunomide | 2 | 0 | |
| Sirolimus + leflunomide + steroid | 0 | 1 |
Values expressed as mean when applicable. Student t-test for continuous variables; χ2 test for binary or categorical variables (global P value).
dd-cfDNA, donor-derived cell-free DNA; MPA, mycophenolic acid; Tacro, tacrolimus.
In those with normal dd-cfDNA and normal/stable creatinine, 53% of abnormal test results were BKV viremia (16 of 30), of which 44% (7 of 16) had low viral loads (<1500 copies) (Figure 3). Serum BKV loads otherwise ranged between 1600 and 679 950 copies. There were 2 cases of low viral load CMV infection and 3 cases of bacterial UTI. Among the 9 abnormal biopsy results in this group, 1 was TCMR, 1 was chronic active AMR along with low level BKV replication (4100 copies), and 1 was recurrent FSGS. Minimally active BKV nephropathy was seen in 2 of the biopsies (520 and 75 000 copies), and mild IF/TA without a specific etiology was observed in 4 biopsies.
FIGURE 3.
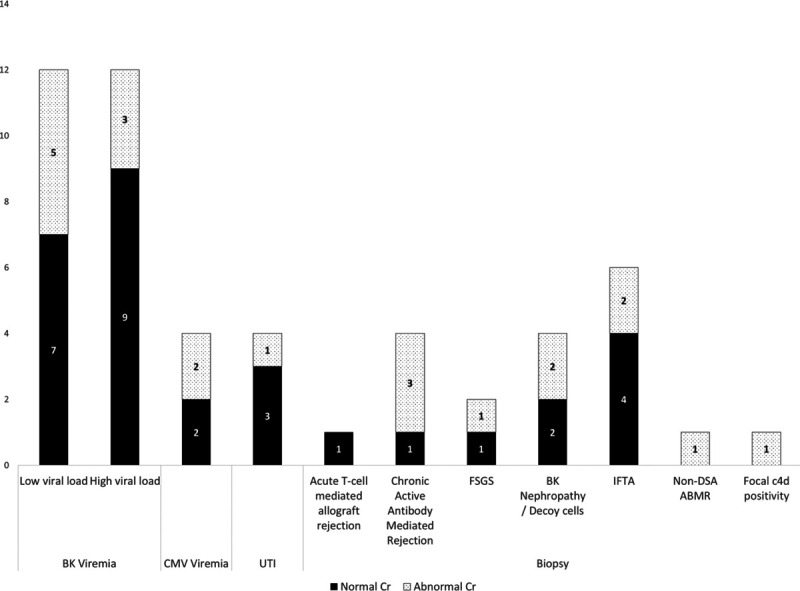
Graft-injuring events seen on laboratory investigations and histology in patients with normal dd-cfDNA broken down by normal and abnormal creatinine. ABMR, antibody-mediated rejection; CMV, cytomegalovirus; dd-cfDNA, donor-derived cell-free DNA; DSA, donor-specific antibody; FSGS, focal segmental glomerulosclerosis; IFTA, interstitial fibrosis and tubular atrophy; UTI, urinary tract infection.
In patients with normal dd-cfDNA and abnormal creatinine, BKV viremia made up 38% (8 of 21) of the abnormal investigation results, whereas abnormal biopsies consisted of nearly half of the results (10 of 21). BKV loads ranged from 660 000 to 7 400 000 copies in 4 of 9 patients, whereas the remainder demonstrated low viral loads (<1500 copies). Two recipients had CMV infections with low viral loads, and 1 had a bacterial UTI. On histology, 2 biopsies revealed active or pattern B BKV nephropathy. Three biopsies demonstrated chronic AMR, 1 had thrombotic microangiopathy that was attributed to non-DSA chronic AMR, and 1 had recurrent FSGS. The remaining abnormal biopsy findings did not have clear clinical diagnoses or etiologies, including 1 with focal C4d deposition without microvascular inflammation, and 2 with IF/TA.
DISCUSSION
Allograft biopsy remains the current standard for diagnosis of graft dysfunction. However, substantial graft injury may occur before clinical evidence that would prompt a for-cause graft biopsy. For this reason, some centers utilize surveillance kidney biopsy for the early identification of allograft rejection. The opportunity to develop a sensitive and reproducible measure of graft injury before the development of graft dysfunction could improve the opportunity to prolong graft life.8,9 dd-cfDNA was developed as a noninvasive assay to identify donor genetic material released into serum from graft-injury events such as rejection. Data from the DART study showed that a dd-cfDNA threshold of 1% yielded 59% sensitivity and 85% specificity in detection of rejection. However, dd-cfDNA has been noted to be elevated in numerous other conditions10-12 including nonrejection acute kidney injury.13 Although the DART study determined 1% dd-cfDNA to be a threshold of reasonable sensitivity and specificity for detection of rejection, more recent assessment has shown that TCMR and borderline rejection can be identified at a lower threshold of dd-cfDNA of 0.5%.5 The utility and interpretation of dd-cfDNA in detecting other graft-injuring events have not been fully defined.
In this series, we described graft-injuring events in the setting of dd-cfDNA level <1%. Most of these dd-cfDNA levels were low, at <0.5%, with only 6 of 51 between 0.5% and 1.0%. The graft-injuring events included TCMR and chronic AMR, BKV, and CMV infections, recurrent primary renal disease, and bacterial UTIs. These observations demonstrate that even with low levels of dd-cfDNA released into the serum, there may be an underlying pathologic process that could cause long-term injury to the graft.14,15 Even though the mechanisms of cfDNA release into blood have not been fully elucidated, the most common proposed pathways are through apoptosis, necrosis, and active DNA release.16 These pathways are all initiated by an injury that prompts a cascade of DNA fragmentation and subsequent release into the blood. The half-life of cfDNA is short at approximately 30 min, so detection of dd-cfDNA is consistent with ongoing cellular injury and death. The risk of irreversible damage from these pathologies reinforces the need to continue to perform for-cause and surveillance tests posttransplant. A single assay or biomarker alone, such as dd-cfDNA, is not sufficient to detect all cell-injuring events. Hence, there is need for surveillance with DSA, BKV, and CMV, especially during the first-year posttransplant.
The sensitivity of dd-cfDNA can be augmented when used in combination with other assays. Jordan et al demonstrated that detection of AMR was improved when used in combination with DSA.17 In our recipients with normal dd-cfDNA, we assessed if serum creatinine could help adequately differentiate the presence or absence of graft-injuring events. The 2 groups stratified by serum creatinine had similar serum tacrolimus levels at the time of serum creatinine sampling, indicating those with elevated creatinine had acute kidney injury due to a cause other than CNI toxicity. In the group with normal dd-cfDNA and normal creatinine, there was only a slightly higher incidence of UTIs (3 of 30 events) compared with those with abnormal creatinine (1 of 21 events). Rejection was seen on biopsy equally in both groups. Both TCMR and chronic active AMR were seen in the group with normal serum creatinine, and only AMR in the group with abnormal creatinine. Although BKV viremia was present in both groups (normal creatinine n = 16, abnormal creatinine n = 8), normal creatinine was less likely to be associated with high BKV loads of >105 copies (n = 1) versus abnormal creatinine (n = 3). Active or pattern B BKV nephropathy was only seen with an abnormal creatinine. This suggests that serum creatinine along with dd-cfDNA could differentiate early versus advanced BKV infection. Low BKV viral load was not expected to cause an elevation of dd-cfDNA levels, unlike high BKV viral load, which could result in graft injury. Interestingly, 9 of 16 participants with high BKV viral load still had a normal dd-cfDNA and serum creatinine levels, which could reflect low degree of graft injury. Given the small sample size, no statistical conclusions can be made; however, we demonstrated that all types of graft-injury events were seen in both groups.
Although the effect of rejection on dd-cfDNA has been studied, there is little understanding of how other etiologies of graft injury affect the amount of dd-cfDNA. Graft-injuring events have been shown to cause changes in gene expression and impact long-term graft outcomes, even if histologic changes found on biopsy do not correlate with a specific diagnosis.18 Transplantation and posttransplant infections alter recipients’ gut and urinary microbiota,19,20 and studies show that changes in the microbiota can affect immunosuppression dosing and modulate immune response to graft injury.21,22 In this study cohort, there were several biopsies that demonstrated only IF/TA without a specific etiology. Normal levels of dd-cfDNA suggested minimal, if any, cellular injury, however, alterations in epigenetic or microbiota activities could influence long-term changes. Unfortunately, without a specific cause for graft dysfunction and in the absence of a defined etiology, the management would remain continued surveillance. The future of diagnosis of graft health or graft dysfunction could involve a combination of tests, including dd-cfDNA, gene expression profiling of pathways involved in rejection,23 and microarray analysis of tissue samples to better diagnose graft injury, predict outcomes, and provide early treatment.
This report is limited by the small sample size and its retrospective nature. Although more recipients during the study period were diagnosed with graft-injuring events, there were many cases in which the dd-cfDNA assay was not done simultaneously or within the 30-d window we defined for our analysis. The time interval between dd-cfDNA testing and detection of cell-injuring events was set relatively long as there was no protocol for repeating dd-cfDNA when a cell-injuring event was identified. Thus, some of the events that were identified with low dd-cfDNA levels could have been low because the injury occurred after the dd-cfDNA test and repeat testing was simply not done. Conversely, out of the 295 recipients with normal dd-cfDNA levels without additional testing, there were likely recipients with subclinical graft-injuring events that were never detected because of a lack of other indications for investigation. The follow-up period varied between a few months to 2.5 y, which increased the heterogeneity of the data, and during that time period clinical practice for testing for graft-injuring events may have changed. The variety of abnormal findings with normal dd-cfDNA levels, both acute and chronic, further supports the nonspecific nature of serum testing and that no single test can provide a definitive diagnosis. To eliminate treatment effects that would alter the dd-cfDNA level, our study only included abnormal assays performed after the normal dd-cfDNA test. Because of this time interval, it is possible the graft injury started after the dd-cfDNA assay was obtained. The results do indicate that even in the setting of a normal dd-cfDNA level and normal serum creatinine, there can be significant histologic changes from allograft injury. Until there is improvement in interpretation of biomarkers, renal biopsy along with other testing will need to remain a part of the diagnostic pathway for allograft dysfunction and maintenance of allograft health.
Footnotes
Published online 5 March, 2021.
J.S.B. and M.R.W. are recipients of research grants from CareDx and Natera.
The authors declare no conflicts of interest.
W.Y.X. participated in data collection, analysis, interpretation, and article preparation. K.K. and N.G. participated in data collection and article preparation. C.B.D., J.R.S., and M.R.W. participated in article preparation. J.S.B. participated in data collection, analysis, interpretation, and article preparation.
REFERENCES
- 1.Siddiqi N, McBride MA, Hariharan S. Similar risk profiles for post-transplant renal dysfunction and long-term graft failure: UNOS/OPTN database analysis. Kidney Int. 2004; 65:1906–1913 [DOI] [PubMed] [Google Scholar]
- 2.Hart A, Smith JM, Skeans MA, et al. OPTN/SRTR 2018 annual data report: kidney. Am J Transplant. 2020; 20Suppl 120–130 [DOI] [PubMed] [Google Scholar]
- 3.Joosten SA, Sijpkens YW, van Kooten C, et al. Chronic renal allograft rejection: pathophysiologic considerations. Kidney Int. 2005; 68:1–13 [DOI] [PubMed] [Google Scholar]
- 4.Cornell LD, Smith RN, Colvin RB. Kidney transplantation: mechanisms of rejection and acceptance. Annu Rev Pathol. 2008; 3:189–220 [DOI] [PubMed] [Google Scholar]
- 5.Stites E, Kumar D, Olaitan O, et al. High levels of dd-cfDNA identify patients with TCMR 1A and borderline allograft rejection at elevated risk of graft injury. Am J Transplant. 2020; 20:2491–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solez K, Colvin RB, Racusen LC, et al. Banff 07 classification of renal allograft pathology: updates and future directions. Am J Transplant. 2008; 8:753–760 [DOI] [PubMed] [Google Scholar]
- 7.Bloom RD, Bromberg JS, Poggio ED, et al. ; Circulating Donor-Derived Cell-Free DNA in Blood for Diagnosing Active Rejection in Kidney Transplant Recipients (DART) Study Investigators. Cell-free DNA and active rejection in kidney allografts. J Am Soc Nephrol. 2017; 28:2221–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gielis EM, Ledeganck KJ, De Winter BY, et al. Cell-free DNA: an upcoming biomarker in transplantation. Am J Transplant. 2015; 15:2541–2551 [DOI] [PubMed] [Google Scholar]
- 9.Friedewald JJ, Kurian SM, Heilman RL, et al. ; Clinical Trials in Organ Transplantation 08 (CTOT-08). Development and clinical validity of a novel blood-based molecular biomarker for subclinical acute rejection following kidney transplant. Am J Transplant. 2019; 19:98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimony A, Zahger D, Gilutz H, et al. Cell free DNA detected by a novel method in acute ST-elevation myocardial infarction patients. Acute Card Care. 2010; 12:109–111 [DOI] [PubMed] [Google Scholar]
- 11.Chiu TW, Young R, Chan LY, et al. Plasma cell-free DNA as an indicator of severity of injury in burn patients. Clin Chem Lab Med. 2006; 44:13–17 [DOI] [PubMed] [Google Scholar]
- 12.Ren B, Liu F, Xu F, et al. Is plasma cell-free DNA really a useful marker for diagnosis and treatment of trauma patients? Clin Chim Acta. 2013; 424:109–113 [DOI] [PubMed] [Google Scholar]
- 13.Peer V, Abu Hamad R, Berman S, et al. Renoprotective effects of DNAse-I treatment in a rat model of ischemia/reperfusion-induced acute kidney injury. Am J Nephrol. 2016; 43:195–205 [DOI] [PubMed] [Google Scholar]
- 14.Al Midani A, Elands S, Collier S, et al. Impact of urinary tract infections in kidney transplant recipients: a 4-year single-center experience. Transplant Proc. 2018; 50:3351–3355 [DOI] [PubMed] [Google Scholar]
- 15.Binet I, Nickeleit V, Hirsch HH, et al. Polyomavirus disease under new immunosuppressive drugs: a cause of renal graft dysfunction and graft loss. Transplantation. 1999; 67:918–922 [DOI] [PubMed] [Google Scholar]
- 16.Sherwood K, Weimer ET. Characteristics, properties, and potential applications of circulating cell-free DNA in clinical diagnostics: a focus on transplantation. J Immunol Methods. 2018; 463:27–38 [DOI] [PubMed] [Google Scholar]
- 17.Jordan SC, Bunnapradist S, Bromberg JS, et al. Donor-derived cell-free DNA identifies antibody-mediated rejection in donor specific antibody positive kidney transplant recipients. Transplant Direct. 2018; 4:e379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Einecke G, Reeve J, Sis B, et al. A molecular classifier for predicting future graft loss in late kidney transplant biopsies. J Clin Invest. 2010; 120:1862–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JR, Muthukumar T, Dadhania D, et al. Gut microbial community structure and complications after kidney transplantation: a pilot study. Transplantation. 2014; 98:697–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu JF, Muthusamy A, Al-Ghalith GA, et al. Urinary microbiome associated with chronic allograft dysfunction in kidney transplant recipients. Clin Transplant. 2018; 32:e13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrade-Oliveira V, Amano MT, Correa-Costa M, et al. Gut bacteria products prevent AKI induced by ischemia-reperfusion. J Am Soc Nephrol. 2015; 26:1877–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JR, Muthukumar T, Dadhania D, et al. Gut microbiota and tacrolimus dosing in kidney transplantation. PLoS One. 2015; 10:e0122399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng MC. The AlloMapTM genomic biomarker story: 10 years after. Clin Transplant. 2017; 31 [DOI] [PubMed] [Google Scholar]


