Abstract
Purpose
Gastric adenocarcinoma is one of the most important causes of cancer death and lacks effective treatment. Eighty-four gastric adenocarcinoma tissue samples along with the clinical information were collected. After analyzing the expression of HOXC11 and LSH in the gastric adenocarcinoma tissues, we explored the prognosis of patients and its correlation with clinical characteristics. Both HOXC11 and LSH were over-expressed in MKN-45 cell lines to verify the effect of high expression of HOXC11 and LSH on GAC.
Methods
The expression of HOXC11 and LSH in 84 cases with gastric adenocarcinoma (GAC) was detected via immunohistochemistry, including 17 cases in stage I, 7 cases in stage II, 27 cases in stage III and 33 cases in stage IV. The expression levels of HOXC11 and LSH, and the clinicopathological characteristics of the samples, were also studied. Cell proliferation, migration, cell cycle and apoptosis assays were utilized for demonstrating malignancy of HOXC11 and LSH over-expressed cells.
Results
Among 84 GAC pathological samples, 12 high HOXC11 expression, and 72 showed low expression; 54.8% (46/84) high LSH expression, and 45.2% (38/84) exhibited low expression. Survival analysis of the Kaplan-Meier plotter gastric cancer datasets showed that subjects with low expression of HOXC11 and LSH had a longer survival time, with a median survival time of 40.2 and 36.4 months, while the subjects with high HOXC11 and LSH expression were only 20.5 and 10 months, respectively. Meanwhile, HOXC11 and LSH over-expressed cells showed a stronger proliferous and migratory ability, and a sped up cell cycle.
Conclusion
The high expression level of HOXC11 and LSH both manifested the poor survival prognosis of GAC patients, and more pronounced malignant phenotype in GAC cells indicated that HOXC11 and LSH can be a strong predictive factor of inferior disease-free survival. From this, we can consider that HOXC11 and LSH both have significant status in GAC stage and survival prediction.
Keywords: GAC, homeobox C11, lymphoid-specific helicase, survival, biomarker
Introduction
According to the Global Cancer Observatory (GCO) cancer database (www.iarc.fr.), the incidence of gastric carcinoma (GC) ranks fifth among all cancers, but ranks third in mortality, because of the high risk of stomach cancer and the lack of effective treatment. Gastric adenocarcinoma (GAC) is the most common histological type (~95%) among all malignancies of gastric origin,1 which can be classified as intestinal or diffuse based on Lauren classification, and the intestinal type cancers typically have a better prognosis than diffuse cancers.2 Although GAC has a high cure rate in the early stages, patients with advanced (clinical stage IV) GAC have a median survival of just 9–10 months.3 The progress in GAC treatment is lagging compared with other tumors, and more research is needed to overcome the obstacles of GAC to prolong survival.4 Growing evidence suggests intricate interplay among epigenetic regulators and mechanisms in gastric cancer.5
Homeobox (HOX) was initially discovered in Drosophila melanogaster as a shared sequence element of approximately 180 bp in homeotic genes and encodes the homeodomain (HD) that normally functions as a DNA-binding domain.6,7 HD proteins control many cellular processes by regulating the expression of downstream target genes. The HOX genes can be divided into four chromosomal clusters, HOXA, HOXB, HOXC, and HOXD that positioned at 7p15.3, 17q21.3, 12q13.3 and 2q31, respectively, each containing between 9 and 11 genes.8 Recently, HOXC11 has been found to be highly expressed in breast cancer9 and clear cell renal cell carcinoma.10 Compared with normal tissues, cervical cancer also has HOXC11 high expression level due to gene activation.11 In human renal cell carcinoma (RCC) and the HK-2 human epithelial cell line, the expression of HOXC11 was still found to be significantly higher and the higher expression promoted proliferation, indicating that HOXC11 may be an important determinant of RCC patient prognosis.10 The roles of HOXC11 in gastric adenocarcinoma require further identification.
Lymphoid-specific helicase (LSH, also known as PASG, SMARCA6 or HELLS) is a major epigenetic regulator that controls DNA methylation patterns.12,13 The LSH gene encodes a protein related to the SNF2 family of chromatin-remodeling ATPases14,15 and is an essential factor for establishing an appropriate DNA, CpG islands and histones methylation level and efficiently repairing DNA double-strand breaks.16,17 Deletion of LSH affects phosphorylation of the histone variant H2AX during DNA damage, resulting in DNA double-strand breaks that cannot be effectively repaired.18 Such genomic instability promotes the generation of tumors. LSH can also activate the expression of metabolic genes at the transcriptional level19 that are considered proliferation drivers for their overexpression in several cancer types.20 Changes in DNA methylation often occur in tumor cells and are primarily manifested as hypomethylation of oncogenes and hypermethylation of tumor suppressor genes, resulting in instability of the tumor cell genome.21 Previous research has confirmed that LSH is not only abnormally expressed in gastric cancer, but is also highly expressed in several types of tumors, such as nasopharyngeal carcinoma,22,23 hepatocellular carcinoma,24 lung cancer,25,26 clear cell renal cell carcinoma,27 gliomas28 and osteosarcoma,29 and correlated with progression and poor prognosis of glioma patients. It also reported that LSH associated with the HOX gene families, which regulating DNA methylation and HOX gene silencing during development.30
In the present study, the expression of HOXC11 and LSH in 84 clinical samples and samples from two databases, The Cancer Genome Atlas database and Kaplan-Meier plotter gastric cancer datasets, were analyzed, as well as the relationship between HOXC11 and LSH expression and clinicopathological features and survival times. After that, HOXC11 and LSH were over-expressed in MKN-45 gastric signet ring cell carcinoma cell line for cell proliferation, migration, cell cycle, and apoptosis assays.
Materials and Methods
Patients in the Study
To analyze the impact of HOXC11 and LSH expression levels on the patient, and their relationship with surgery, 84 patients with GAC who underwent a pathological examination at Xiangya Hospital were included in the study, which including 73 subjects with R0 resection (complete resection rate 86.9%), 10 subjects with only exploratory laparotomy and 1 subject with gastric perforation repair as a palliative therapy. 18 of 84 patients are still alive, however, 15 patients have passed away and 51 of 84 patients we have lost contact. Among the 84 subjects, 4 had well-differentiated tumors, 24 had tumors with intermediate differentiation, and 56 had tumors with poor differentiation. Of the 84 GAC patients, 53 were males, and 31 were females, aged 31–74 years (average age of 57 years), with stage I (n=21), II (n=23), III (n=20), and IV (n=20) GAC; the tumor and regional lymph node and metastasis (TNM) classification system for malignant tumors (American Joint Committee on Cancer and Union for International Cancer Control, 2018)31 was used. The data used in this study were obtained after the patient’s personal information was removed, which met the ethical requirements of protecting patient privacy. The ethics committee of Cancer Research Institute of Central South University approved the study.
Databases’ Messages
We used data associated with 373 samples from The Cancer Genome Atlas database, including 30 normal and 343 GAC samples, and the differences in HOXC11 and LSH expression levels between normal and GAC tissues were evaluated. We also assessed data of 876 GC patients from Kaplan-Meier plotter gastric cancer datasets, and the relationship between HOXC11 and LSH expression levels and survival time was analyzed in patients with different clinicopathological characteristics, gender, TNM staging, differentiation, Lauren classification, and stages.
Immunohistochemical Staining
Gastric adenocarcinoma tissues were obtained from the Department of Pathology of Xiangya Hospital. After formalin fixation and paraffin embedding, the sections were prepared and then incubated with a mouse monoclonal antibody against human HOXC11 (dilution, 1:150, Novus#NBP2-00499) or LSH (dilution, 1:200, Santa Cruz#sc-46665) at 4°C overnight, followed by incubation with a biotinylated goat antibody against mouse immunoglobulin (Beijing Zhongshan Jinqiao Corporation#SAP-9100). Images captured by CX41 microscope (OLYMPUS, Tokyo, Japan) with a DP-72 Microscope Digital Camera System (OLYMPUS, Tokyo, Japan), and the results were differentially quantified by two pathologists from Second Xiangya Hospital, Changsha, China. To evaluate HOXC11 and LSH expression, immunohistochemical staining results were classified into four groups on the basis of intensity and extent. The proportion of cell with positive protein expression was categorized as showed: 0, <10% immunopositive cells; 1, 10%-30% positive cells; 2, 30%-60% positive cells; 3, >60% positive cells. Relative intensity categorized the staining intensity as follows: 1, negative; 2, weak; 3, moderate; and 4, strong. The proportion and intensity scores were then multiplied to obtain a total score. Methods references from Du YB et al.32 To get the final statistical results, we detected 10 normal gastric mucosa as a control experiment. HOXC11 and LSH average scores of these 10 samples are 4.8 and 4.5. Therefore, scores 0, 3, 6 were considered low-level expression, whereas scores 9 and 12 were considered high-level expression.
Cell Culture and Plasmids
MKN-45 (Procell#CL-0292), a gastric signet ring cell carcinoma cell line, was used in this paper. The cell line was cultured by RPMI1640 (Gibco) and 10% (v/v) FBS added in the medium under 37°C with 5% CO2 in cell incubator. The HOXC11 and LSH overexpression plasmid was established by inserting the HOXC11 and LSH cDNA into the pLVX-EF1α-IRES-Puro vector (Clontech#631988). Plasmids were transfected into HEK293T (Procell#CL-0005) cell by Neofect® DNA transfection reagent (Neofect#TF201201) for lentivirus. After 2 days of lentivirus infection, puromycin (1μg/mL) (Sangon Biotech Corporation#A610593) was used for screening positive clones.
Western Blot and Antibodies
After trypsin digested, cells were washed twice with 1×PBS and lysed on ice with IP lysis buffer added protease inhibitor cocktail for 2 hours. Protein electrophoresis was conducted in SDS-polyacrylamide gel and then transferred to a polyvinylidene fluoride membrane. After immunoblotted with primary antibody and followed by incubation with a secondary antibody, we used an enhanced chemiluminescence system for detecting the antibody signals. The HOXC11 antibody was purchased on Novus (#NBP2-00499, 1:500), LSH antibody was purchased on Santa Cruz (#sc-46665, 1:1000), and β-actin antibody was purchased on Proteintech (#66009-1-IG, 1:2000).
Cell Proliferation and Migration Assays
The proliferation viability assays were conducted under a density of 1000 cells/well in 100μL medium in 96-well plates and detected by Cell Counting Kit-8 (CCK-8) (biomake#B34302). In migration assay, 1×105 cells in 200μL 1% FBS medium were seeded in transwell inserts. 800μL 10% FBS medium was added in 24-well plates. After 24 hours, cells were fixed with methanol for 15 min and then stained with crystal violet for 15 min. Took pictures and counted cell under microscope.
Cell Cycle and Apoptosis Assays
In cell cycle assays, 1×106 cells were seeded in 6-well plates. After 24 hours of serum-free culture medium, 10% FBS culture medium was used for 20h. Collected cells and washed twice by 1×PBS, immobilizes cells by 70% ethanol in −20°C overnight. After 1×PBS washed, the cells were resuspended with 100μL 1×PBS and added with a final concentration of 100μg/mL RNase (Sangon Biotech Corporation#B500474) and 50μg/mL Propidium iodide (Sangon Biotech Corporation#A601112). BD FACSDivaTM flow cytometer detects the cell cycle. In cell apoptosis assays, 10μmol Cisplatin (Selleck#S1166) was used for cell apoptosis induction. Annexin V-FITC/PI apoptosis detection kit (KeyGEN BioTECH#KGA108) and BD FACSDivaTM flow cytometer were used for cell apoptosis detection.
Statistical Methods
The criteria of the TNM classification system for malignant tumors (American Joint Committee on Cancer and Union for International Cancer Control, 2018) were used in our study. An unpaired t-test and Pearson correlation coefficients were carried out by Prism 8.0 GraphPad software, which were used to conduct a statistical analysis of TCGA data. The relationship between the protein expression levels of HOXC11 and LSH and clinicopathologic characteristics were tested using a chi-square test, which was performed with SPSS 23.0 statistical software (SPSS Inc, Chicago, IL). Statistical analysis of Kaplan-Meier plotter gastric cancer datasets was carried out using the Kaplan-Meier method and Log rank test by Prism 8.0 GraphPad software. All the cell experiments were repeated at least three times, t-test was used to analyze the difference between two groups by Prism 8.0 GraphPad software. A two-tailed P value of less than 0.05 was considered statistically significant, and the confidence interval is 95%.
Results
HOXC11 and LSH are Highly Expressed in GAC Samples in TCGA Database
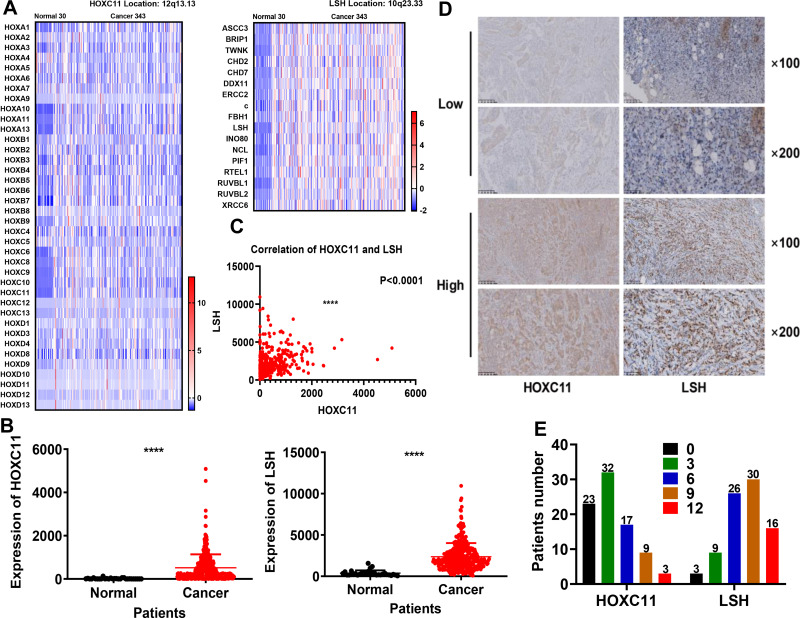
After analysis of 373 cases of GAC samples in TCGA database, we compared related genes of the HOX gene family and DNA helicase family (Figure 1A) and found that the expression of HOXC11 and LSH in GAC tissues was higher than in normal (Figure 1B), which was especially obvious in their gene family. Meanwhile, the expression level between HOXC11 and LSH shows a high degree of correlation (Figure 1C).
Figure 1.
Expression of HOXC11 and LSH in 373 TCGA and 84 clinical GAC samples. (A) Heat map of HOXC11, LSH and related gene expression levels in 373 TCGA samples. (B) Expression level of HOXC11 and LSH in 373 TCGA samples. The expression of HOXC11 and LSH in GAC tissues is higher than that in normal tissues. Shown is the mean ± SD of experiments, ****P < 0.0001. (C) The correlation of HOXC11 and LSH expression in TCGA samples. Shown is the mean ± SD of experiments, ****P < 0.0001. (D) Immunohistochemical staining was used to examine the HOXC11 and LSH protein expression levels in GAC tissues from 84 clinical patients. (E) Distribution of the number of patients in each score for HOXC11 and LSH in 84 clinical GAC samples.
The Expression of HOXC11 and LSH in Clinical Pathological Tissues and Its Relationship with Clinicopathological Characteristics
After scoring the 84 pathological samples, we counted some relevant information such as age, gender and tumor stage. We showed some immunohistochemical images to illustrate the scoring situation (Figure 1D). In 84 GAC clinical pathological samples, the expression of HOXC11 was primarily concentrated in the nucleus. Similarly, HOXC11 staining was focused on the remaining glandular structures in the lamina propria and was in the nucleus of acinar cells. Besides, the surrounding connective tissue showed little staining. And positive LSH immunohistochemical staining was mainly concentrated in the nucleus and primarily observed in the parenchyma; staining in the stroma was rarely seen. Meanwhile, nuclear staining gradually deepened with an increase in expression level. Overall, 14.3% (12/84) of samples had high HOXC11 expression and 85.7% (72/84) had low expression (Table 1). At the same time, 54.8% (46/84) of samples had high LSH expression and 45.2% (38/84) had low expression. In addition, the number of patients with high expression of both HOXC11 and LSH was 9 (10.7%), and 35 (41.7) patients showed low expression of both proteins. The score distribution of HOXC11 and LSH is shown in figure (Figure 1E).
Table 1.
HOXC11 and LSH Expression in 84 GAC Tissues
| Item | LSH High Expression | LSH Low Expression | Total |
|---|---|---|---|
| HOXC11 high expression | 9 (10.7) | 3 (3.6) | 12 (14.3) |
| HOXC11 low expression | 37 (44.0) | 35 (41.7) | 72 (85.7) |
| Total | 46 (54.8) | 38 (45.2) | 84 (100) |
Then, the relationship between the expression level and the clinicopathological characteristics was analyzed (Table 2). A chi-square test showed the expression level of LSH was related to tumor invasion (T) and lymph node metastasis (N) stage and was also associated with clinical staging, indicated that the expression level of LSH might be associated with tumor invasion and lymphatic metastasis thus affected the tumor stage. However, there was no significant association between the HOXC11 expression level and different clinicopathologic characteristics.
Table 2.
Association Between HOXC11 and LSH Expression and the Clinical Characteristics of 84 Patients with GAC
| Clinicopathologic Characteristics | No.(%) | HOXC11 Expression No.(%) | P | LSH Expression No.(%) | P | ||
|---|---|---|---|---|---|---|---|
| High | Low | High | Low | ||||
| Age | |||||||
| ≥58 | 44 (52.4) | 6 (13.6) | 38 (86.4) | 1.000 | 25 (56.8) | 19 (43.2) | 0.827 |
| <58 | 40 (47.6) | 6 (15.0) | 34 (85.0) | 21 (52.5) | 19 (47.5) | ||
| Gender | |||||||
| Male | 53 (63.1) | 7 (13.2) | 46 (86.8) | 0.753 | 31 (58.5) | 22 (41.5) | 0.496 |
| Female | 31 (36.9) | 5 (16.1) | 26 (83.9) | 15 (48.4) | 16 (51.6) | ||
| Tumor invasion(T) | |||||||
| T1 | 17 (20.2) | 2 (11.8) | 15 (88.2) | 0.439 | 3 (17.6) | 14 (82.4) | 0.005 |
| T2 | 7 (8.4) | 2 (28.6) | 5 (71.4) | 4 (57.1) | 3 (42.9) | ||
| T3 | 27 (32.1) | 2 (7.41) | 25 (92.6) | 16 (59.3) | 11 (40.7) | ||
| T4 | 33 (39.3) | 6 (18.2) | 27 (81.8) | 23 (69.7) | 10 (30.3) | ||
| Lymph node metastasis(N) | |||||||
| N0 | 37 (44.0) | 4 (10.8) | 33 (89.2) | 0.279 | 12 (32.4) | 25 (67.6) | 0.000 |
| N1 | 11 (13.1) | 1 (9.09) | 10 (90.9) | 4 (36.4) | 7 (63.6) | ||
| N2 | 5 (6.0) | 2 (40.0) | 3 (60.0) | 3 (60.0) | 2 (40.0) | ||
| N3 | 14 (16.7) | 1 (7.1) | 13 (92.9) | 14 (100.0) | 0 (0.0) | ||
| Nx | 17 (20.2) | 4 (23.5) | 13 (76.5) | 13 (76.5) | 4 (23.5) | ||
| Metastasis(M) | |||||||
| M0 | 64 (76.2) | 8 (12.5) | 56 (87.5) | 0.467 | 32 (50.0) | 32 (50.0) | 0.132 |
| M1 | 20 (23.8) | 4 (20.0) | 16 (80.0) | 14 (70.0) | 6 (30.0) | ||
| Stage | |||||||
| I | 21 (25.0) | 3 (14.3) | 18 (85.7) | 0.770 | 6 (28.6) | 15 (71.4) | 0.003 |
| II | 23 (27.4) | 2 (8.7) | 21 (91.3) | 10 (43.5) | 13 (56.5) | ||
| III | 20 (23.8) | 3 (15.0) | 17 (85.0) | 16 (80.0) | 4 (20.0) | ||
| IV | 20 (23.8) | 4 (20.0) | 16 (80.0) | 14 (70.0) | 6 (30.0) | ||
| R0 resection | |||||||
| Yes | 73 (86.9) | 10 (13.7) | 63 (86.3) | 0.654 | 37 (50.7) | 36 (49.3) | 0.101 |
| NO | 11 (13.1) | 2 (18.2) | 9 (81.8) | 9 (81.8) | 2 (18.2) | ||
| Postoperative chemotherapy | |||||||
| Yes | 36 (42.9) | 3 (8.33) | 33 (91.7) | 0.314 | 21 (58.3) | 15 (41.7) | 0.668 |
| No | 17 (20.2) | 4 (23.5) | 13 (76.5) | 10 (58.8) | 7 (41.2) | ||
| Uncertain | 31 (36.9) | 5 (16.1) | 26 (83.9) | 15 (48.4) | 16 (51.6) | ||
| Differentiation | |||||||
| Well | 4 (4.7) | 0 (0.0) | 4 (100) | 0.378 | 0 (0.0) | 4 (100.0) | 0.071 |
| Middle | 24 (28.6) | 2 (8.33) | 22 (91.7) | 13 (54.2) | 11 (45.8) | ||
| Poor | 56 (66.7) | 10 (17.9) | 46 (82.1) | 33 (58.9) | 23 (41.1) | ||
| HP | |||||||
| (-) | 52 (61.9) | 6 (11.5) | 46 (88.5) | 0.220 | 28 (53.8) | 24 (46.2) | 0.055 |
| (+) | 13 (15.5) | 1 (7.69) | 12 (92.3) | 4 (30.8) | 9 (69.2) | ||
| Uncertain | 19 (22.6) | 5 (26.3) | 14 (73.7) | 14 (73.7) | 5 (26.3) | ||
HOXC11 and LSH Expression Level Influence Overall Survival
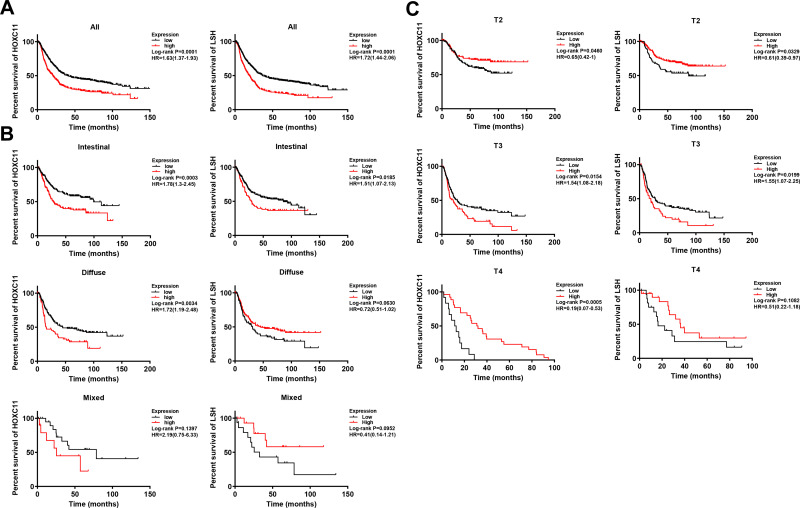
Survival analysis of the Kaplan-Meier plotter gastric cancer datasets showed that patients with low expression of HOXC11 and LSH had longer survival times (Figure 2A), with a median survival time of 40.2 and 36.4 months (Table 3). The median survival time of patients with high HOXC11 and LSH expression was only 20.5 and 10 months, respectively. Notably, the survival time of patients with low expression of both HOXC11 and LSH was significantly longer than that of those with high expression in Lauren classification intestinal gastric cancer (Figure 2B). The median survival time of patients with low and high HOXC11 expression was 99.4 and 28 months, and that of patients with low and high LSH expression was 85.8 and 28.7 months, but the survival time of LSH in diffuse and mixed gastric cancer has no difference. Meanwhile, the survival time of patients with low HOXC11 and LSH expression in T3 gastric cancer was longer than that of those with high expression (Figure 2C). In stage T2 and T4, a similar trend of survival time was shown between HOXC11 and LSH.
Figure 2.
Survival analysis of HOXC11 and LSH expression in GC tissues from Kaplan-Meier plotter gastric cancer datasets. (A) The survival of patients with high HOXC11 or LSH expression was significantly shorter than that of those with low expression. (B) Survival analysis of HOXC11 or LSH expression in GC tissues under Lauren classification. (C) Survival analysis of HOXC11 or LSH expression in GC tissues under tumor invasion (T) classification.
Table 3.
HOXC11 and LSH Expression in Kaplan–Meier Plotter Gastric Cancer Datasets
| Items | No.(%) | Median Survival Time of HOXC11, Month | Median Survival Time of LSH, Month | ||||
|---|---|---|---|---|---|---|---|
| High | Low | P | High | Low | P | ||
| All | 876 | 20.5 | 40.2 | 0.0001 | 10.0 | 36.4 | 0.0001 |
| Gender | |||||||
| Male | 69.8% | 20.3 | 35.8 | 0.0003 | 14.5 | 34.7 | 0.0001 |
| Female | 30.2% | 17.4 | 77.2 | 0.0003 | 28.7 | 45.2 | 0.0999 |
| Tumor invasion(T) | |||||||
| T1 | 2.8% | – | – | – | – | – | – |
| T2 | 48.5% | – | – | 0.0460 | – | 87.0 | 0.0329 |
| T3 | 41.0% | 20.3 | 27.5 | 0.0154 | 20.0 | 27.8 | 0.0199 |
| T4 | 7.6% | 31.6 | 12.0 | 0.0005 | 36.4 | 16.4 | 0.1082 |
| Lymph node metastasis(N) | |||||||
| N0 | 14.9% | 123.8 | 113.2 | 0.0547 | – | 123.6 | 0.3063 |
| N1+2+3 | 85.1% | 32.1 | 45.8 | 0.1236 | 53.0 | 33.3 | 0.0639 |
| N1 | 45.4% | 57.8 | – | 0.0980 | – | 70.2 | 0.0569 |
| N2 | 24.4% | 24.4 | 30.4 | 0.3104 | 30.2 | 21.2 | 0.0812 |
| N3 | 15.3% | 16.2 | 12.3 | 0.0182 | 17.4 | 12.3 | 0.2606 |
| Metastasis(M) | |||||||
| M0 | 88.8% | 46.0 | 113.2 | 0.0240 | 113.2 | 56.9 | 0.0624 |
| M1 | 11.2% | 11.0 | 11.8 | 0.4910 | 11.4 | 11.0 | 0.0521 |
| Stage | |||||||
| I | 10.2% | – | – | 0.1355 | – | 113.2 | 0.1344 |
| II | 21.2% | 123.8 | 123.6 | 0.0539 | – | 123.6 | 0.1597 |
| III | 46.2% | 25.5 | 44.0 | 0.0013 | 25.9 | 34.4 | 0.0479 |
| IV | 22.4% | 17.0 | 15.9 | 0.0029 | 17.4 | 14.3 | 0.0790 |
| Differentiation | |||||||
| Well | 12.1% | 27.4 | – | 0.0046 | 26.6 | – | 0.0054 |
| Middle | 25.4% | 22.3 | 113.2 | 0.0866 | 41.2 | 22.3 | 0.2249 |
| Poor | 62.5% | 32.6 | 24.4 | 0.2540 | 15.4 | 40.0 | 0.0009 |
| Lauren classification | |||||||
| Intestinal | 54.0% | 28.0 | 99.4 | 0.0003 | 28.7 | 85.8 | 0.0185 |
| Diffuse | 40.6% | 15.7 | 44.7 | 0.0034 | 44.7 | 31.1 | 0.0630 |
| Mixed | 5.4% | 25.5 | 78.6 | 0.1397 | – | 32.6 | 0.0952 |
| HER2 | |||||||
| Negative | 60.7% | 26.7 | 46.8 | 0.0007 | 26.7 | 44.6 | 0.0160 |
| Positive | 39.3% | 17.5 | 30.4 | 0.0005 | 14.6 | 29.1 | 0.0001 |
Abbreviations: GAC, gastric adenocarcinoma; GC, gastric carcinoma; GCO, Global Cancer Observatory; HOX, homeobox; HD, homeodomain; RCC, renal cell carcinoma; LSH, lymphoid-specific helicase; PRC, polycomb repressive complex.
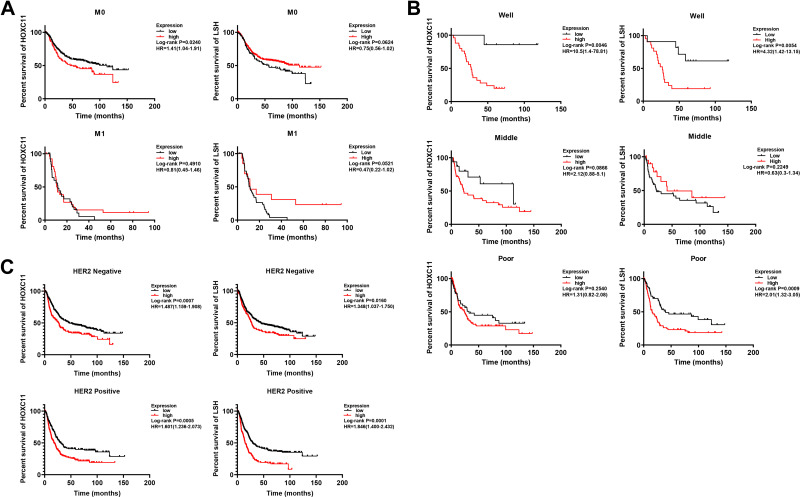
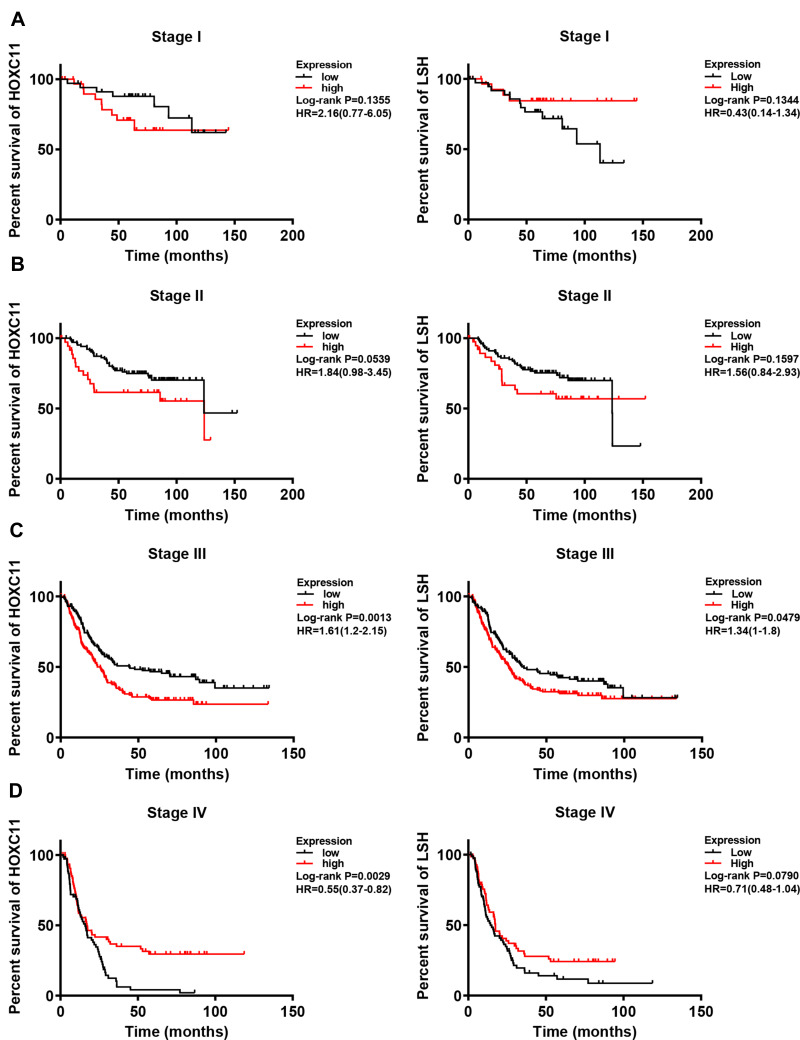
Intriguingly, in patients with initially well-differentiated, non-metastatic gastric cancer, high HOXC11 expression often indicated a poor outcome (Figure 3A and B). Moreover, the survival time of patients with high LSH expression with good differentiation or poor differentiation was significantly shorter than that of patients with low expression. As a marker associated with tumor invasion and metastasis, an important status likewise belongs to human epidermal growth factor receptor 2 (HER2). The high expression of HOXC11 and LSH in both HER2 positive and negative patients suggested a worse prognosis (Figure 3C). In different stages of gastric cancer patients, the relationship between overall survival and expression of HOXC11 and LSH is shown in Figure 4.
Figure 3.
Survival analysis of HOXC11 and LSH expression in GC tissues from Kaplan-Meier plotter gastric cancer datasets. (A) Survival analysis of HOXC11 or LSH expression in GC tissues under metastasis (M) classification. (B) Survival analysis of HOXC11 or LSH expression in GC tissues under diverse differentiation. (C) Survival analysis of HOXC11 or LSH expression in HER2 negative or positive GC tissues.
Figure 4.
Survival analysis of HOXC11 or LSH expression in GC tissues under different stage. (A) Survival analysis of HOXC11 or LSH expression in GC tissues under stage I. (B) Survival analysis of HOXC11 or LSH expression in GC tissues under stage II. (C) Survival analysis of HOXC11 or LSH expression in GC tissues under stage III. (D) Survival analysis of HOXC11 or LSH expression in GC tissues under stage IV.
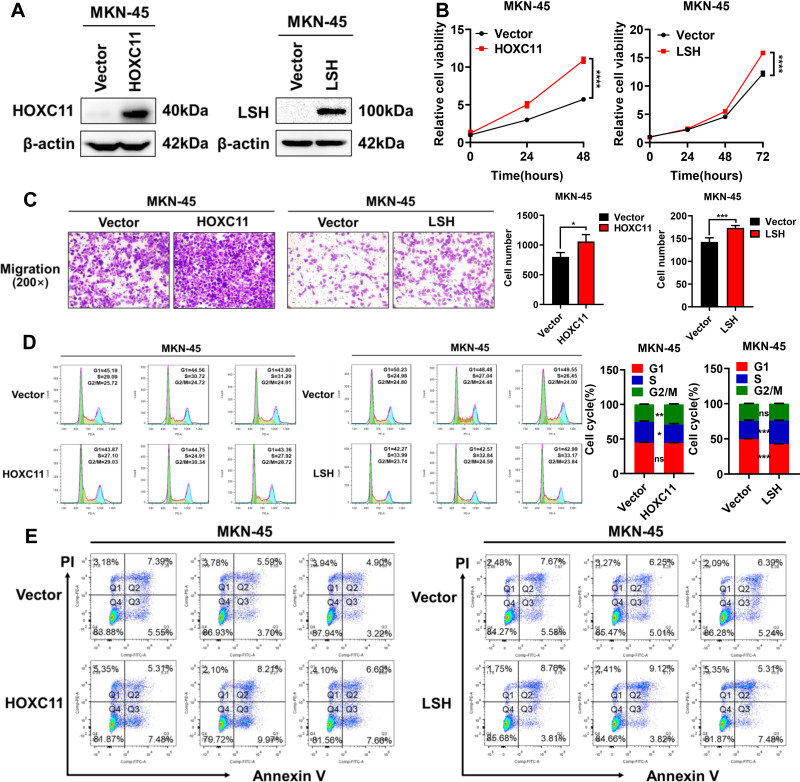
HOXC11 and LSH Overexpression Accelerates GAC Cell Malignant Progression
To verify the effect of HOXC11 and LSH high expression on gastric cancer, HOXC11 and LSH over-expressed plasmids were stably transferred into the MKN-45 cell line, and the protein expressions of HOXC11 and LSH were successfully upregulated (Figure 5A). Obviously, HOXC11 and LSH overexpression facilitate the GAC cell proliferation (Figure 5B) and migration (Figure 5C). Flow cytometry detected the cell cycle, and apoptosis induced by 10μmol Cisplatin. HOXC11 overexpression made more cells enter G2/M phase from S phase. At the same time, LSH overexpression caused cells to enter S phase from G1 phase (Figure 5D). Both genes have an ability to speed up the cell cycle process. But the results of apoptosis assays show that HOXC11 and LSH can not affect the cell apoptosis which induced by Cisplatin (Figure 5E).
Figure 5.
Overexpression of HOXC11 and LSH promotes GAC progression. (A) The establishment of HOXC11 and LSH over-expressed cell line detected by Western blot. (B) Cell viability was detected by CCK-8. Shown is the mean ± SD of experiments (n=5), ****, P < 0.0001. (C) HOXC11 and LSH overexpression enhanced migration of MKN-45 cells. Shown is the mean ± SD of experiments (n=3), *P < 0.05, ***P < 0.001. (D) HOXC11 and LSH accelerates cell cycle detected by flow cytometry. Shown is the mean ± SD of experiments (n=3), *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant. (E) Flow cytometry detected the cell apoptosis of HOXC11 and LSH.
Discussion
The HOX protein encoded by the HOX gene functions as a transcription factor and usually acts as a monomer or homodimers and directly targeted the transcription of downstream targets or interacted with protein cofactors.33 Though the downstream targets of the HOX gene remain uncertain, it does not restrict the function of HOX protein in some domain such as apoptosis, differentiation, angiogenesis, development, or motility.34 As a member of HOX family, HOXC11 has an important status in tumor progression. In our 84 pathological samples, the expression level of HOXC11 related to the tumor invasion, lymph node metastasis stage and clinical staging of the patients. It mentioned us that HOXC11 has potential as a biomarker in GAC. Meanwhile, in Kaplan-Meier plotter gastric cancer datasets, the expression level of HOXC11 is an obvious prognosis factor because the higher expression level showed a worse prognosis.
LSH maintains genome stability of cells through establishing correct DNA methylation levels and patterns, and DNA methylation is a crucial factor in the development of several malignancies. In primary non-small cell lung cancer, it was found that many deletions of LSH loci and some tumor-specific exons appeared simultaneously.35 With LSH deficiency bring about seriously absent of DNA methylation, the expression of protein-coding genes also can be partially regulated through compensatory mechanisms such as H2K27me3 redistribution.36,37 High expression of LSH protein also occurs in tumors and is believed to be closely related to the hypermethylation state. These findings suggest LSH plays an important role in the occurrence and development of tumors.
In Kaplan-Meier plotter gastric cancer datasets, the influence of HOXC11 and LSH expression level to the survival time of patients appears a similar trend in some patient types. However, there is no direct evidence of interaction between HOXC11 and LSH. As a regulator of DNA methylation, LSH can bind to PRC1, which is part of the polycomb repressive complex (PRC), affecting PRC-mediated histone modification,30,38,39 and the HOX gene is affected by PRC as one of its targets. Epigenetic control is a common mechanism for controlling HOX gene expression, and the CpG island in the silenced HOX gene promoter is typically methylated.40 However, the effect of LSH on transcriptional activation or silencing of the HOX gene in tumors is related to the tumor site,30,41,42 and cannot be simply summarized as an up-regulation or down-regulation of the expression level. The regulation of LSH on some genes of HOX family cannot be generalized and believed that LSH can directly affect HOXC11.
Here, analyses of clinical sample and various databases show that patients with high expression of HOXC11 and LSH have a shorter survival time, which indicates that the expression levels of HOXC11 and LSH are most likely important factors affecting patient survival time. This is especially true in intestinal gastric cancer. GC patients who showed well-differentiated tumors in surgery, without metastasis, but who had high HOXC11 expression had significantly shorter survival times. This suggests that HOXC11 has significance in GC malignant progression. HOXC11 expression in metastatic melanoma cells is higher than in primary melanoma cells, which also suggests the significance of HOXC11 in metastasis and invasion.43 However, because of the limited sample size, the sample in this study cannot fully reflect the overall characteristics. Based on this, the validation of the malignant evolution of HOXC11 and LSH in MKN-45 cell line also confirmed the effect of high expression of HOXC11 and LSH on GAC.
Overall, our results suggest that both HOXC11 and LSH are predictive of survival in patients with GAC. However, how the up-regulation or down-regulation of HOXC11 and LSH expression promote GAC progression requires further discussion. It is worth mentioning we believe that HOXC11 and LSH are key genes in the progression of gastric adenocarcinoma and may be biomarkers and therapeutic targets for prediction of future prognosis.
Acknowledgments
We would like to thank all laboratory members for their critical discussion and precious suggestions of this manuscript and to apologize to those members not mentioned due to space limitations.
Funding Statement
This work was supported by the National Natural Science Foundation of China (82073097 [SL], 81874139 [SL], 81672991 [SL], 82072594 [YT], 81672787 [YT]), Shenzhen Science and Technology Program (KQTD20170810160226082 [YT]), Shenzhen Municipal Government of China [JCYJ20180507184647104 [YT]), and the Hunan Provincial Key Area R&D Program (2019SK2253 [YT]).
Ethics Approval and Informed Consent
All procedures performed in studies involving human participants were under the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The ethics committee of Cancer Research Institute of Central South University has approved this study, and all samples were obtained with informed consent.
Consent for Publication
All the authors: The authors confirm that all the figures, tables, and content are publically available.
Author Contributions
All authors contributed significantly to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests. This manuscript has been read and approved by all authors and is not under consideration for publication elsewhere.
References
- 1.Ajani JA, Lee J, Sano T, Janjigian YY, Fan D, Song S. Gastric adenocarcinoma. Nat Rev Dis Primers. 2017;3:17036. [DOI] [PubMed] [Google Scholar]
- 2.Olsen S, Jin L, Fields RC, Yan Y, Nalbantoglu I. Tumor budding in intestinal-type gastric adenocarcinoma is associated with nodal metastasis and recurrence. Hum Pathol. 2017;68:26–33. doi: 10.1016/j.humpath.2017.03.021 [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends—an update. Cancer Epidemiol Biomarkers Prevent. 2016;25(1):16–27. doi: 10.1158/1055-9965.EPI-15-0578 [DOI] [PubMed] [Google Scholar]
- 4.Corso S, Giordano S. How can gastric cancer molecular profiling guide future therapies? Trends Mol Med. 2016;22(7):534–544. doi: 10.1016/j.molmed.2016.05.004 [DOI] [PubMed] [Google Scholar]
- 5.Sung H, Yang HH, Zhang H, et al. Common genetic variants in epigenetic machinery genes and risk of upper gastrointestinal cancers. Int J Epidemiol. 2015;44(4):1341–1352. doi: 10.1093/ije/dyv050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burglin TR, Affolter M. Homeodomain proteins: an update. Chromosoma. 2016;125(3):497–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fongang B, Kong F, Negi S, Braun W, Kudlicki A. A conserved structural signature of the homeobox coding DNA in HOX genes. Sci Rep. 2016;6(1):35415. doi: 10.1038/srep35415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Procino A, Cillo C. The HOX genes network in metabolic diseases.. Cell Biol Int. 2013;37(11):1145–1148. doi: 10.1002/cbin.10145 [DOI] [PubMed] [Google Scholar]
- 9.McIlroy M, McCartan D, Early S, et al. Interaction of developmental transcription factor HOXC11 with steroid receptor coactivator SRC-1 mediates resistance to endocrine therapy in breast cancer [corrected]. Cancer Res. 2010;70(4):1585–1594. doi: 10.1158/0008-5472.CAN-09-3713 [DOI] [PubMed] [Google Scholar]
- 10.Liu Y-J, Zhu Y, Yuan H-X, Zhang J-P, Guo J-M, Lin Z-M. Overexpression of HOXC11 homeobox gene in clear cell renal cell carcinoma induces cellular proliferation and is associated with poor prognosis. Tumour Biol. 2015;36(4):2821–2829. doi: 10.1007/s13277-014-2909-6 [DOI] [PubMed] [Google Scholar]
- 11.Saha SS, Chowdhury RR, Mondal NR, Roy S, Sengupta S. Expression signatures of HOX cluster genes in cervical cancer pathogenesis: impact of human papillomavirus type 16 oncoprotein E7. Oncotarget. 2017;8(22):36591–36602. doi: 10.18632/oncotarget.16619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lungu C, Muegge K, Jeltsch A, Jurkowska RZ. An ATPase-deficient variant of the SNF2 family member HELLS shows altered dynamics at pericentromeric heterochromatin. J Mol Biol. 2015;427(10):1903–1915. doi: 10.1016/j.jmb.2015.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, et al. Lsh, an epigenetic guardian of repetitive elements. Nucleic Acids Res. 2004;32(17):5019–5028. doi: 10.1093/nar/gkh821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi YJ, Yoo NJ, Lee SH. Mutation of HELLS, a chromatin remodeling gene, gastric and colorectal cancers. Pathol Oncol Res. 2015;21(3):851–852. doi: 10.1007/s12253-014-9862-y [DOI] [PubMed] [Google Scholar]
- 15.Termanis A, Torrea N, Culley J, Kerr A, Ramsahoye B, Stancheva I. The SNF2 family ATPase LSH promotes cell-autonomous de novo DNA methylation in somatic cells. Nucleic Acids Res. 2016;44(16):7592–7604. doi: 10.1093/nar/gkw424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geiman TM, Durum SK, Muegge K. Characterization of gene expression, genomic structure, and chromosomal localization ofhells (Lsh). Genomics. 1998;54(3):477–483. doi: 10.1006/geno.1998.5557 [DOI] [PubMed] [Google Scholar]
- 17.Mjelle R, Hegre SA, Aas PA, et al. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair (Amst). 2015;30:53–67. doi: 10.1016/j.dnarep.2015.03.007 [DOI] [PubMed] [Google Scholar]
- 18.Burrage J, Termanis A, Geissner A, Myant K, Gordon K, Stancheva I. The SNF2 family ATPase LSH promotes phosphorylation of H2AX and efficient repair of DNA double-strand breaks in mammalian cells. J Cell Sci. 2012;125(22):5524–5534. doi: 10.1242/jcs.111252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Y, Mao C, Yang R, et al. EGLN1/c-Myc induced lymphoid-specific helicase inhibits ferroptosis through lipid metabolic gene expression changes. Theranostics. 2017;7(13):3293–3305. doi: 10.7150/thno.19988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Yuen ST, Xu J, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46(6):573–582. doi: 10.1038/ng.2983 [DOI] [PubMed] [Google Scholar]
- 21.Jia J, Shi Y, Chen L, et al. Decrease in lymphoid specific helicase and 5-hydroxymethylcytosine is associated with metastasis and genome instability. Theranostics. 2017;7(16):3920–3932. doi: 10.7150/thno.21389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S, Tao Y-G. Chromatin remodeling factor LSH affects fumarate hydratase as a cancer driver. Chin J Cancer. 2016;35(1):72. doi: 10.1186/s40880-016-0138-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He X, Yan B, Liu S, et al. Chromatin remodeling factor lsh drives cancer progression by suppressing the activity of fumarate hydratase. Cancer Res. 2016;76(19):5743–5755. doi: 10.1158/0008-5472.CAN-16-0268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Law C-T, Wei L, Tsang FH-C, et al. HELLS regulates chromatin remodeling and epigenetic silencing of multiple tumor suppressor genes in human hepatocellular carcinoma. Hepatology. 2019;69(5):2013–2030. doi: 10.1002/hep.30414 [DOI] [PubMed] [Google Scholar]
- 25.Wang R, Shi Y, Chen L, et al. The ratio of FoxA1 to FoxA2 in lung adenocarcinoma is regulated by LncRNA HOTAIR and chromatin remodeling factor LSH. Sci Rep. 2016;5(1):17826. doi: 10.1038/srep17826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Y, He Y, Liu S, Tao Y. Chromatin remodeling factor lymphoid-specific helicase inhibits ferroptosis through lipid metabolic genes in lung cancer progression. Chin J Cancer. 2017;36(1):82. doi: 10.1186/s40880-017-0248-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen D, Maruschke M, Hakenberg O, Zimmermann W, Stief CG, Buchner A. TOP2A, HELLS, ATAD2, and TET3 are novel prognostic markers in renal cell carcinoma. Urology. 2017;102:265.e261-265.e267. doi: 10.1016/j.urology.2016.12.050 [DOI] [PubMed] [Google Scholar]
- 28.Xiao D, Huang J, Pan Y, et al. Chromatin remodeling factor LSH is upregulated by the LRP6-GSK3β-E2F1 axis linking reversely with survival in gliomas. Theranostics. 2017;7(1):132–143. doi: 10.7150/thno.17032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu SC, Benavente CA. Chromatin remodeling protein HELLS is upregulated by inactivation of the RB-E2F pathway and is nonessential for osteosarcoma tumorigenesis. Oncotarget. 2018;9(66):32580–32592. doi: 10.18632/oncotarget.25953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xi S, Zhu H, Xu H, Schmidtmann A, Geiman TM, Muegge K. Lsh controls Hox gene silencing during development. Proc Natl Acad Sci U S A. 2007;104(36):14366–14371. doi: 10.1073/pnas.0703669104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng J, Zhang R, Pan Y, et al. Comparison of the staging of regional lymph nodes using the sixth and seventh editions of the tumor-node-metastasis (TNM) classification system for the evaluation of overall survival in gastric cancer patients: findings of a case-control analysis involving a single institution in China.. Surgery. 2014;156(1):64–74. doi: 10.1016/j.surg.2014.03.020 [DOI] [PubMed] [Google Scholar]
- 32.Du Y-B, Dong B, Shen L-Y, et al. The survival predictive significance of HOXC6 and HOXC8 in esophageal squamous cell carcinoma. J Surg Res. 2014;188(2):442–450. doi: 10.1016/j.jss.2014.01.017 [DOI] [PubMed] [Google Scholar]
- 33.Holland PWH. Evolution of homeobox genes. Wiley Interdisciplinary Rev. 2013;2(1):31–45. doi: 10.1002/wdev.78 [DOI] [PubMed] [Google Scholar]
- 34.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10(5):361–371. doi: 10.1038/nrc2826 [DOI] [PubMed] [Google Scholar]
- 35.Yano M, Ouchida M, Shigematsu H, et al. Tumor-specific exon creation of the HELLS/SMARCA6 gene in non-small cell lung cancer. Int J Cancer. 2004;112(1):8–13. doi: 10.1002/ijc.20407 [DOI] [PubMed] [Google Scholar]
- 36.Ren J, Finney R, Ni K, Cam M, Muegge K. The chromatin remodeling protein Lsh alters nucleosome occupancy at putative enhancers and modulates binding of lineage specific transcription factors. Epigenetics. 2019;14(3):277–293. doi: 10.1080/15592294.2019.1582275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu W, McIntosh C, Lister R, et al. Genome-wide DNA methylation patterns in LSH mutant reveals de-repression of repeat elements and redundant epigenetic silencing pathways. Genome Res. 2014;24(10):1613–1623. doi: 10.1101/gr.172015.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh CA, Bolger JC, Byrne C, et al. Global gene repression by the steroid receptor coactivator SRC-1 promotes oncogenesis. Cancer Res. 2014;74(9):2533–2544. doi: 10.1158/0008-5472.CAN-13-2133 [DOI] [PubMed] [Google Scholar]
- 39.Bantignies F, Roure V, Comet I, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144(2):214–226. doi: 10.1016/j.cell.2010.12.026 [DOI] [PubMed] [Google Scholar]
- 40.Tao Y, Xi S, Briones V, Muegge K, Freitag M. Lsh mediated RNA polymerase II stalling at HoxC6 and HoxC8 involves DNA methylation. PLoS One. 2010;5(2):e9163. doi: 10.1371/journal.pone.0009163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joo MK, Park -J-J, Chun HJ. Impact of homeobox genes in gastrointestinal cancer. World J Gastroenterol. 2016;22(37):8247–8256. doi: 10.3748/wjg.v22.i37.8247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhatlekar S, Fields JZ, Boman BM. HOX genes and their role in the development of human cancers. J Mol Med. 2014;92(8):811–823. doi: 10.1007/s00109-014-1181-y [DOI] [PubMed] [Google Scholar]
- 43.deBlacam C, Byrne C, Hughes E, et al. HOXC11–SRC-1 regulation of S100beta in cutaneous melanoma: new targets for the kinase inhibitor dasatinib. Br J Cancer. 2011;105(1):118–123. doi: 10.1038/bjc.2011.193 [DOI] [PMC free article] [PubMed] [Google Scholar]