Abstract
Every class of RNA forms base-paired structures that impact biological functions. Chemical probing of RNA structure, especially with the advent of strategies such as SHAPE-MaP, vastly expands the scale and quantitative accuracy over which RNA structure can be examined. These methods have enabled large-scale structural studies of mRNAs and lncRNAs, but the length and complexity of these RNAs makes interpretation of the data challenging. We have created modules available through the open-source Integrative Genomics Viewer (IGV) for straightforward visualization of RNA structures and associated and complementary experimental data. Here we present detailed and stepwise strategies for exploring and visualizing complex RNA structures in IGV. Individuals can use these instructions and supplied sample data to become adept at using IGV to visualize RNA structure models in conjunction with useful allied information.
Keywords: RNA structure, lncRNA, mRNA, Integrative Genomics Viewer, SHAPE-MaP, base pairing
1. Introduction
RNA, like DNA, forms base-paired structures, as was first conclusively demonstrated for a simple synthetic helix in 1956 [1] and for tRNA in 1974 [2]. The secondary structures of RNAs of every class (including tRNA, rRNA, sRNA, miRNA, mRNA, and lncRNA) have been implicated in the biological functions of these RNAs [3–5]. Chemical or enzymatic probing of RNA in conjunction with thermodynamically-informed structure modeling has a long and successful history of defining structure models for RNAs not amenable to crystallization and for RNAs under biologically relevant or experimentally varied solution and cellular conditions [6]. Advances in RNA structure probing technologies such as SHAPE-MaP have facilitated studies of complex mRNAs [7], lncRNAs [8], and viral RNAs [9, 10]. These RNA molecules are often thousands of nucleotides in length, presenting notable challenges in data visualization and interpretability.
To enable efficient examination of the structures of long RNAs, we have created visualization modules for the Integrative Genomics Viewer (IGV). IGV is cross-platform and open-source software that supports visualization of diverse experimental data, especially from studies using arrays and high-throughput sequencing readout strategies [11]. IGV was developed to flexibly display genomic, clinical, and experimental data with an emphasis on integrative and interactive analyses. The software allows visual comparison of many types of experiments and is quite responsive to user interaction.
We previously added several functionalities to IGV that enable exploration of RNA structure models and base-pairing probabilities, scaling easily from visualization of the entirety of long RNAs to focused examination of individual helices [12]. Base pairs are conveniently rendered as arcs. Here we present stepwise instructions and general recommendations for visualizing RNA structure models and associated data in IGV (see Note 3.1). We provide three example datasets derived from recent studies in the Supporting Information. Researchers can use these instructions and the sample data to quickly and efficiently become adept at interrogating RNA structural information in conjunction with a variety of complementary information. Due to the wide availability of next-generation sequencing, SHAPE-MaP probing strategies can be readily employed by diverse non-expert laboratories. The visualization tools described here facilitate making RNA structure analysis a routine component of examining diverse biological systems.
2. Method
2.1. Collect files in appropriate formats
Download and extract provided “busan_rna_igv_vis_2019_SI.zip” to use sample data files with this tutorial or prepare files from your own sources.
- Transcript sequence (required)
- Text file with a .fa extension in FASTA format.
- The first line of the file should begin with the ‘>‘ (greater-than) character, followed directly by a sequence name or ID with no special characters or spaces.
- Remaining lines should contain the nucleotide sequence without spaces.
- Chemical reactivity profiles (use either of the following two file formats)
- .shape file: This is a tab-delimited text file with two columns. First column is nucleotide position, starting with 1. Second column is normalized SHAPE reactivity data values; no-data positions are set to −999.
- .map file:
- Same format as .shape, but with two additional columns. Third column is standard error, fourth column is nucleotide sequence.
- ShapeMapper2 software outputs .map files containing chemical probing reactivities calculated using mutational profiling. ShapeMapper2 and associated documentation is available at: https://github.com/Weeks-UNC/shapemapper2/blob/master/README.md.
- Base-pairing (secondary) structure model and/or estimated base-pairing probabilities
- A commonly used format for defining a base-pairing model has the extension .ct. These files are produced by the Fold module of RNAstructure.
- Dot-bracket (.db, .dbn) files are also supported, most commonly used for small hand-edited structures or used alongside multiple sequence alignments in other software packages.
- Pairing probabilities are calculated by the partition and ProbabilityPlot modules of RNAstructure. These files have the extension .dp.
- For file format reference, see https://software.broadinstitute.org/software/igv/RNAsecStructure
- Generation of RNA structure model files is not covered in detail here, since this method is focused on graphical exploration. For long RNAs, we recommend using Superfold (available at https://weeks.chem.unc.edu/software.html), which automates the process of performing structure modeling over computationally manageable windows and merging the resulting structures. Superfold accepts a .map file as input and produces both a .ct file, containing a single predicted minimum free energy structure, and a .dp file, containing estimated base pairing probabilities.
- Annotations such as gene coding regions, repeat sequences, or sites of known function (optional)
- The .gff3 file format is convenient for most uses and is readily hand-edited. See the included examples in “busan_rna_igv_vis_2019_SI.zip” and further documentation at https://software.broadinstitute.org/software/igv/GFF.
- Important: Ensure that the names listed in the first column of the .gff3 file match the name given in the first line of the FASTA file and not the FASTA filename or other text.
- One or more linear profiles from complementary experiments or computational analyses (this list is not exhaustive, see Note 3.1). These data are not required, but can substantially enrich RNA structure analyses.
- Protein-binding enrichment data (for example CLIP or RIP data)
- GC-content median over fixed windows
- SHAPE reactivity median over fixed windows
- Estimated per-nucleotide or median Shannon entropy over fixed windows
- Common file formats are .wig, .bedgraph, and .tdf (see https://software.broadinstitute.org/software/igv/FileFormats)
- Important: Ensure that the sequence names listed in the first column of a .wig or .bedgraph file match the name given in the first line of the FASTA file (not including the ‘>‘ character) and not the FASTA filename.
2.2. Load and import files into IGV
Download IGV (available at https://software.broadinstitute.org/software/igv/download) and launch
- Load nucleotide sequence
- Click “Genomes” in menu bar and select “Load Genome from File”
- Select FASTA file and click “Open”
- Select “E_coli/sequence.fa” if using the example dataset or select your own .fa file
- Load SHAPE reactivity profiles, base-pairing probabilities, and annotations
- Click “File” in menu bar and select “Load from File”
- Select “E_coli/SHAPE_reactivity.map”, “E_coli/base_pairing_probability.dp”, and “E_coli/gene_annotations.gff3” if following along with example dataset or select your own files, and click “Open”
- Click “Continue” in any popup dialog boxes that appear (see Note 3.2)
2.3. Adjust track display
Steps listed here are optional and specific values given are suggestions. Users should adjust settings for comfortable display for their particular screen size, platform, and dataset.
- Set view preferences
- Click “View”, “Preferences”, “General”
- Check “Display all tracks in a single panel”
- Uncheck “Show attributes panel”
- Rename tracks
- Right-click track, select “Rename track”, and enter a descriptive name
- For the example dataset, replace “SHAPE_reactivity.shape.wig” with “SHAPE reactivity” and “base_pairing_probability.dp.bp” with “Pairing probability”.
- If working with a higher resolution display, consider adjusting track name size settings
- Shift-left-click and drag track names to select all tracks. Right click track or track names, select “Change font size”, and increase the value to 16. See Note 3.3.
- If track names are cut off or abbreviated: click “View”, “Set Name Panel Width” and set to a larger value.
- The default font size can be changed by clicking “View”, “Preferences”, “General”, then clicking “Change” next to “Default font”. This will only affect tracks or files loaded in the future.
- Reorder tracks by left-clicking and dragging track names
- Drag gene annotations track directly above other tracks
- Drag SHAPE reactivity profile above base pairing probability arcs
- If zoomed in far enough to see individual nucleotide identities, it can be useful to move the sequence track directly above base pairing arcs to visualize complementary pairs and the sequences of unpaired regions.
- Adjust SHAPE profile track range
- Right click reactivity profile track, and select “Set Data Range”
- Set “Min” and “Mid” values to 0 and “Max” value to 3
- Widen pairing probability arc track
- Right click on the arc track, select “Change Track Height”
- Set to a larger value such as 100
2.4. Examine functional sites in example biologically important RNAs
2.4.1. E. coli mRNA gene translation start sites
The provided example of an E. coli transcript is notable in that it contains two non-ribosomal protein genes rimM and trmD (encoding a ribosome maturation factor and a tRNA methyltransferase, respectively) that are located between two ribosomal protein-coding genes rpsP and rplS (encoding S16 and L19, respectively) [13] (Fig. 1a). The ribosomal proteins encoded by rpsP and rplS are translated at high levels; in contrast, the rimM and trmD gene products are translated at lower levels. In addition, the translation rates of rimM and trmS are largely uncoupled from those of the surrounding genes [14]. Examining the structures around the translation start sites of each gene provides clues to explain these differences.
- Zoom in on the start codon regions of rplS and rimM. Use any of the following:
- Click and drag to select range in ruler (as shown in Fig. 1a)
- Click the ‘+’ button in the upper right area of the toolbar several times and drag track window to scroll
- Double-click on an annotation graphic several times
- Enter a gene, annotation name, or numeric range in text box
Figure 1. Visualization of E. coli mRNA translation start sites.
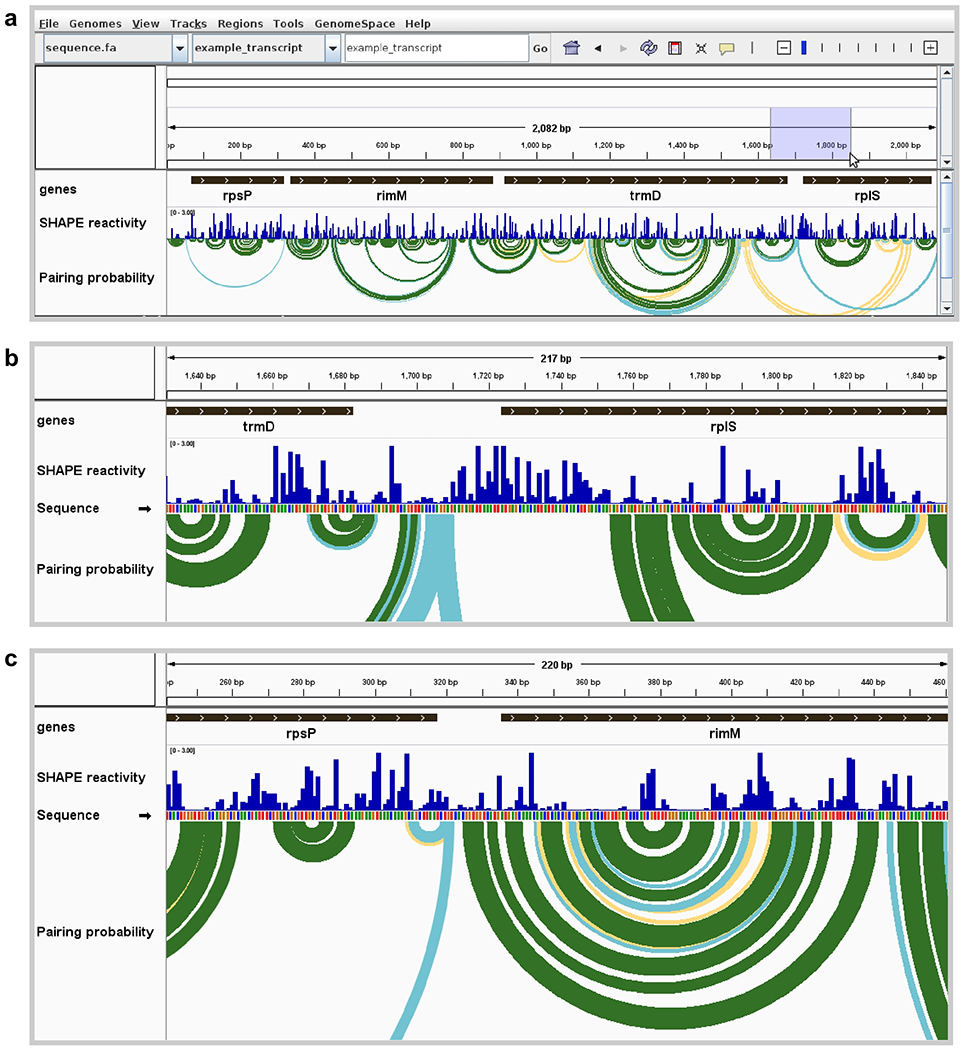
(a) Full view of an E. coli polycistronic transcript showing gene boundaries, SHAPE reactivity profile, and modeled base pairing probabilities. Pairing probability is indicated by arc color: green, >80% probability; blue, 30–80%; yellow, 10–30%. High SHAPE reactivities in blue shaded region are indicative of an unstructured region around the rplS translation start site. (b) Zoomed view of unstructured region surrounding the start codon of rplS, showing highly SHAPE-reactive positions modeled as unpaired (corresponding to no arcs indicative of base pairing). (c) Zoomed view of the start codon and surrounding region of rimM, showing positions with low SHAPE reactivities and corresponding well-determined base-pairing structure model. Data from ref. (13).
Note the differing structural contexts of the translation start sites of rplS and rimM. In particular, the region surrounding the AUG codon in rplS is unstructured, evidenced by high SHAPE reactivities and lack of highly probable base pair arcs in structure models (Fig. 1b). This lack of structure near the start codon likely provides a high ribosome accessibility, allowing translation initiation in the absence of a Shine-Dalgarno sequence [15].
2.4.2. Murine LHR mRNA sequence motifs
In mice, ZFP36L2, a zinc finger protein, regulates expression of the luteinizing hormone receptor (LHR) mRNA during oocyte maturation [16]. ZFP36L2 is a member of a class of zinc-finger-containing proteins that bind RNA targets containing the sequence motif “AUUUA”, termed adenine-uridine-rich elements (AREs) [17]. Surprisingly, gel-shift assays revealed that ZFP36L2 bound only one of the three AREs present within the LHR 3’ untranslated region [16], raising the intriguing possibility that the RNA structural context of these sequence motifs influences protein binding.
- Examine the nucleotide sequence of a structure model (data from ref. [18])
- Load the provided LHR sequence, SHAPE reactivity data, structure model, and annotations from supporting files folder “LHR”, as in step 2.2 (see Fig. 2a).
- Zoom in on the region of the annotated AREs until the nucleotide sequence becomes visible (see Fig. 2b).
Visualize the highly structured element in Fig. 2b as a traditional planar graph (optional). This requires third-party software such as the StructureEditor component of RNAstructure available at https://rna.urmc.rochester.edu/RNAstructure.html; see Note 3.4.
Figure 2. Visualization of structures around AREs in LHR mRNA.
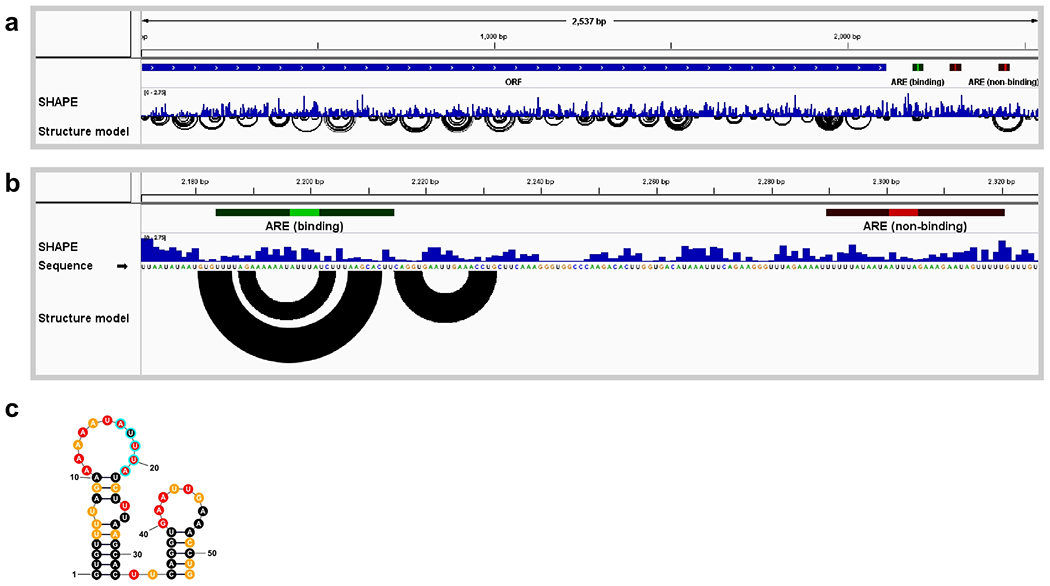
(a) Full view of the LHR transcript showing SHAPE reactivity profile and base-pairing structure model. Annotations include an open reading frame (blue bar labeled ORF) and three AREs (highlighted in green and red). (b) Zoomed view of the region of two AREs, one functional and one non-functional based on binding assays with ZFP36L2 protein(16). The arcs in the Structure model track, which indicate base pairs, suggest that the upstream ARE is highly structured, whereas the downstream ARE is not. (c) ARE structure rendered as a planar graph using the StructureEditor component of RNAstructure. SHAPE reactivities are indicated by color: red, reactivity >0.85; orange, 0.4-0.84; black, <0.4. Core ARE sequence highlighted with cyan outline. Data from ref. (18).
SHAPE reactivities and structure modeling suggested that the functional motif is located in the context of a hairpin loop (Fig. 3c) and that RNA structure influences the binding affinity for ZFP36L2. These RNA structure-based hypotheses were supported through extensive mutagenesis studies [18].
Figure 3. Murine Xist long non-coding RNA repeat region.
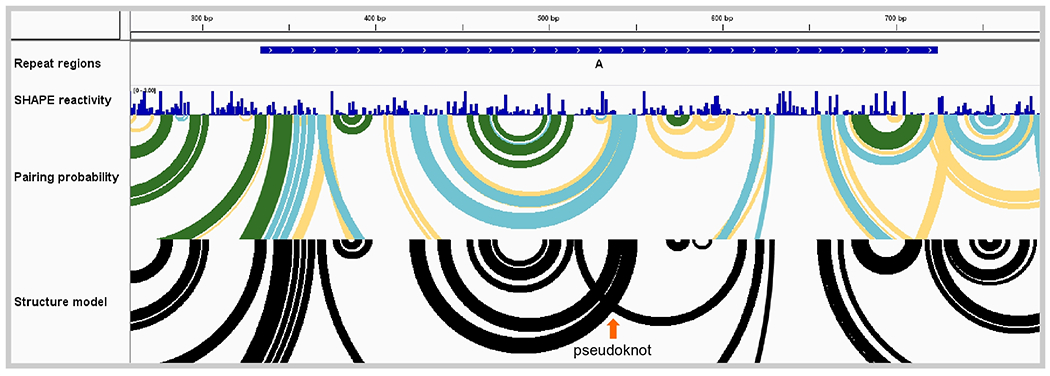
View of one Xist repeat region (repeat A) showing SHAPE reactivity profile, base-pairing probabilities, and base-pairing structure model. Pairing probability is indicated by arc color: green, >80% probability; blue, 30-80%; yellow, 10-30%. The abundance of medium- and low-probability base pairing arcs (in blue and yellow) suggests that no single structure (including the minimum free energy structure shown with black arcs) predominates in this region and that, instead, an ensemble of RNA structural states is present. Data from ref. (8).
2.4.3. Murine Xist lncRNA repeat elements
The Xist long non-coding RNA localizes to the nucleus and mediates the formation of a chromatin-modifying protein complex that silences gene expression on the female X chromosome [19]. Xist contains multiple repetitive sequence regions, some of which are implicated in Xist localization, interaction with members of the protein complex, and X-chromosome inactivation [20–22].
For functionally important regions of an RNA, there is often a single, thermodynamically stable secondary structure. Examples of such well-defined secondary structures include ligand-bound riboswitches, the bacterial 16S rRNA, or the stable base paired structures overlapping the rimM gene (Fig. 1; green arcs). In contrast, some RNAs instead adopt a family (or ensemble) of structures. Modeling RNA structural ensembles remains an important experimental and computational frontier, but the visualization of estimated pairing probabilities is a useful approach that begins to address variability within populations of folded RNA molecules.
- Examine base-pairing probabilities (data from ref. [8])
- Load the provided murine Xist sequence, SHAPE reactivity data, structure model, base pairing probabilities, and repeat region annotations from supporting files folder “Xist”, as in step 2.2.
- Zoom in on repeat A as in Fig. 3.
Although the minimum free energy structure model, by definition, displays a single secondary structure for Xist repeat region A (Fig. 3; black arcs), the pairing probability arcs show multiple overlapping low- and medium-probability helices (Fig. 3; blue and yellow arcs). These data support a model in which this region of the Xist RNA does not have a well-defined secondary structure overall. A possible pseudoknotted structure is evident in the “Structure model” track as overlapping arcs, highlighted with an orange arrow in Fig. 3.
Supplementary Material
Footnotes
This brief report is focused on the visualization of RNA structure probing data and structure models and is not a comprehensive guide to IGV. For general guides and documentation to IGV, see: https://software.broadinstitute.org/software/igv/UserGuide and https://software.broadinstitute.org/software/igv/FileFormats.
- .ct, .map., and .shape file formats do not contain sequence name, strand, or nucleotide offset position. Therefore, upon import, IGV converts these files into file formats that contain this information, without overwriting the original input files.
- The default settings in the file conversion popup dialog cover the most common RNA structure exploration scenario, that is, one transcript sequence and chemical reactivity profiles corresponding to this sequence. If examining a short gene-specific primer amplicon (see ref. [23]) within a larger sequence, users may need to manually adjust the beginning and end positions of reactivity profiles or structures.
The IGV user interface may appear unusably small on some newer high-resolution displays. Recent Java 11 builds of IGV provide support for high-resolution displays (see https://software.broadinstitute.org/software/igv/download).
The IGV modules discussed here visualize secondary structures as arc diagrams. Rendering traditional RNA secondary structure figures (sometimes referred to as airport, planar, or tree diagrams) requires additional software. Commonly used packages include VARNA, Ribosketch, RNAstructure StructureEditor, and XRNA. See informal discussion at https://github.com/Weeks-UNC/shapemapper2/blob/master/docs/other_software.md.
References
- 1.Rich A, Davies DR (1956) A new two stranded helical structure: polyadenylic acid and polyuridylic acid. J Am Chem Soc 78:3548–3549. 10.1021/ja01595a086 [DOI] [Google Scholar]
- 2.Kim SH, Suddath FL, Quigley GJ, et al. (1974) Three-Dimensional Tertiary Structure of Yeast Phenylalanine Transfer RNA. Science 185:435–440. 10.1126/science.185.4149.435 [DOI] [PubMed] [Google Scholar]
- 3.Eddy SR (2001) Non–coding RNA genes and the modern RNA world. Nat Rev Genet 2:919–929. 10.1038/35103511 [DOI] [PubMed] [Google Scholar]
- 4.Parker BJ, Moltke I, Roth A, et al. (2011) New families of human regulatory RNA structures identified by comparative analysis of vertebrate genomes. Genome Res 21:1929–1943. 10.1101/gr.112516.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonenberg N, Hinnebusch AG (2009) Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell 136:731–745. 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mailler E, Paillart J-C, Marquet R, et al. (2018) The evolution of RNA structural probing methods: From gels to next-generation sequencing. Wiley Interdiscip Rev RNA 10:e1518. 10.1002/wrna.1518 [DOI] [PubMed] [Google Scholar]
- 7.Corley M, Solem A, Phillips G, et al. (2017) An RNA structure-mediated, posttranscriptional model of human α−1-antitrypsin expression. Proc Natl Acad Sci 114:E10244–E10253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smola MJ, Christy TW, Inoue K, et al. (2016) SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc Natl Acad Sci 113:10322–10327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dethoff EA, Boerneke MA, Gokhale NS, et al. (2018) Pervasive tertiary structure in the dengue virus RNA genome. Proc Natl Acad Sci 115:11513–11518. 10.1073/pnas.1716689115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dadonaite B, Barilaite E, Fodor E, et al. (2017) The structure of the influenza A virus genome. 10.1101/236620 [DOI] [Google Scholar]
- 11.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Busan S, Weeks KM (2017) Visualization of RNA structure models within the Integrative Genomics Viewer. RNA 23:1012–1018. 10.1261/rna.060194.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mustoe AM, Busan S, Rice GM, et al. (2018) Pervasive Regulatory Functions of mRNA Structure Revealed by High-Resolution SHAPE Probing. Cell 173:181–195.e18. 10.1016/j.cell.2018.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wikström PM, Björk GR (1988) Noncoordinate translation-level regulation of ribosomal and nonribosomal protein genes in the Escherichia coli trmD operon. J Bacteriol 170:3025–3031. 10.1128/jb.170.7.3025-3031.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scharff LB, Childs L, Walther D, Bock R (2011) Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding Sites. PLoS Genet 7:e1002155. 10.1371/journal.pgen.1002155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ball CB, Rodriguez KF, Stumpo DJ, et al. (2014) The RNA-Binding Protein, ZFP36L2, Influences Ovulation and Oocyte Maturation. PLoS ONE 9:e97324. 10.1371/journal.pone.0097324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai WS, Carballo E, Thorn JM, et al. (2000) Interactions of CCCH Zinc Finger Proteins with mRNA. J Biol Chem 275:17827–17837. 10.1074/jbc.m001696200 [DOI] [PubMed] [Google Scholar]
- 18.Ball CB, Solem AC, Meganck RM, et al. (2017) Impact of RNA structure on ZFP36L2 interaction with luteinizing hormone receptor mRNA. RNA 23:1209–1223. 10.1261/rna.060467.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gendrel A-V, Heard E (2014) Noncoding RNAs and Epigenetic Mechanisms During X-Chromosome Inactivation. Annu Rev Cell Dev Biol 30:561–580. 10.1146/annurev-cellbio-101512-122415 [DOI] [PubMed] [Google Scholar]
- 20.Chu C, Zhang QC, da Rocha ST, et al. (2015) Systematic Discovery of Xist RNA Binding Proteins. Cell 161:404–416. 10.1016/j.cell.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sunwoo H, Colognori D, Froberg JE, et al. (2017) Repeat E anchors Xist RNA to the inactive X chromosomal compartment through CDKN1A-interacting protein (CIZ1). Proc Natl Acad Sci 114:10654–10659. 10.1073/pnas.1711206114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarma K, Levasseur P, Aristarkhov A, Lee JT (2010) Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci 107:22196–22201. 10.1073/pnas.1009785107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smola MJ, Rice GM, Busan S, et al. (2015) Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat Protoc 10:1643–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


