Abstract
Coronavirus (COVID-19) is a pandemic, which caused suddenly unexplained pneumonia cases and caused a devastating effect on global public health. Computerized tomography (CT) is one of the most effective tools for COVID-19 screening. Since some specific patterns such as bilateral, peripheral, and basal predominant ground-glass opacity, multifocal patchy consolidation, crazy-paving pattern with a peripheral distribution can be observed in CT images and these patterns have been declared as the findings of COVID-19 infection. For patient monitoring, diagnosis and segmentation of COVID-19, which spreads into the lung, expeditiously and accurately from CT, will provide vital information about the stage of the disease. In this work, we proposed a SegNet-based network using the attention gate (AG) mechanism for the automatic segmentation of COVID-19 regions in CT images. AGs can be easily integrated into standard convolutional neural network (CNN) architectures with a minimum computing load as well as increasing model precision and predictive accuracy. Besides, the success of the proposed network has been evaluated based on dice, Tversky, and focal Tversky loss functions to deal with low sensitivity arising from the small lesions. The experiments were carried out using a fivefold cross-validation technique on a COVID-19 CT segmentation database containing 473 CT images. The obtained sensitivity, specificity, and dice scores were reported as 92.73%, 99.51%, and 89.61%, respectively. The superiority of the proposed method has been highlighted by comparing with the results reported in previous studies and it is thought that it will be an auxiliary tool that accurately detects automatic COVID-19 regions from CT images.
Keywords: COVID-19 segmentation, Convolutional neural network, Attention-based SegNet
Introduction
Since December 2019, a large and increasing outbreak of a new coronavirus has emerged in Wuhan, Hubei province of China [1]. While the International Committee on Taxonomy of Viruses (ICTV) called this virus as SARS-CoV-2, the World Health Organization (WHO) has named it Coronavirus Disease 2019 (COVID-19) [2]. It has been confirmed that the new coronavirus is transmitted from human to human. The WHO has reported 78,811 laboratory-confirmed cases covering more than 2200 cases outside of China [3]. Due to the increasing population mobility in the globalizing world, the virus spreads rapidly all over the world and its destructive effects have been seen on the routine daily life, global economy, and general public health [4]. Therefore, the WHO has declared this infectious disease as a pandemic.
COVID-19 infection leads to acute respiratory illness and even fatal acute respiratory distress syndrome (ARDS) with approximately 17% to 29% rates. The fatality rate is predicted to be around 2.3% [5]. Fever, dyspnea, cough, myalgia, and headache have been defined as the symptoms of this disease. The real-time reverse transcriptase-polymerase chain reaction (RT-PCR) assay has been adopted as the gold standard for the SARS-CoV-2 diagnosis. Although it is believed that this test has high specificity for COVID-19 infection diagnosis, its sensitivity has been reported to be as low as 60–70%. Besides, one of the medical imaging techniques, computed tomography (CT), has been adopted as a vital method to support the diagnosis and management of patients with COVID-19 infection [3]. Several specific patterns such as bilateral, peripheral, and basal predominant ground-glass opacity (GGO), multifocal patchy consolidation, and crazy-paving pattern with a peripheral distribution observed at chest CT images have been declared as the findings of COVID-19 infection [1]. These definitions have increased the role of radiography and chest CT for COVID-19 infection diagnosis. It is believed that chest CT findings are of a key role in the assessment of COVID-19 infection [6]. In clinical practice, the GGO accompanied by the interlobular septa thickening or crazy-paving pattern, consolidation, and air bronchogram sign have been observed commonly. Besides, chest CT manifestations in patients with COVID-19 have been related to patient’s age [7].
A specific drug or vaccine is still not recommended for the treatment of this infection. Laboratory conditions must be appropriate and sufficient for the implementation of RT-PCR assay. It is also a time-consuming process taking hours, or even days before the results are available. Therefore, due to the lack of a sufficient number of test kits and inadequate hospital equipment, the diagnosis period of suspicious patients is significantly delayed and the disease has not been prevented from concentrating in certain centers. At this point, quantitative CT analysis can be an alternative diagnostic tool and its role is constantly evolving with modest scientific evidence [6, 8–10].
When the literature is examined, it is seen that many different algorithms have been used to classify or segment COVID-19 infection by using CT images. For the classification task, a deep learning model based on the Darknet-19 was suggested to detect COVID-19 infection using CT images. The number of filters was gradually increased in architecture. Binary and multi-class classification were realized. The model achieved 98.08% and 87.02% classification achievements for binary and multi-class classification tasks, respectively [11]. COVIDiagnosis-Net covering SqueezeNet and Bayesian optimization was introduced for COVID-19 infection detection. The overall classification score was reported as 98.26% [4]. The deep features, which were extracted a novel convolutional neural network (CNN) model using Bayesian optimization method, were applied as the input to k-nearest neighbor ( NN), support vector machine (SVM), and decision-tree (DT) machine learning models. The most efficient results yielded by SVM with 98.97% accuracy [12]. In another study, the generalization abilities of pretrained deep CNNs were investigated for the same purpose. The most efficient results were ensured by ResNet-101 and Xception models with 99.02% accuracy scores [13]. A deep learning-based methodology was offered using X-ray images for COVID-19 diagnosis. In the study, SVM was utilized as a classifier and this machine learning technique was fed with the deep features, which were extracted from the fully connected layer of ResNet-50 model. The model achieved a 95.38% classification score [14]. Three pretrained deep CNN-based models, which are ResNet-50, InceptionV3, and InceptionResNetV2, were adopted for COVID-19 infection diagnosis. Fivefold cross-validation method was used in the experiment and the best scores were obtained using the ResNet-50 model with 98% classification success [15]. COVIDX-Net, which is a framework of deep learning classifiers to diagnose COVID-19 using X-ray images, was come up with an end-to-end learning schema. This study pointed out that the VGG-19 and DenseNet showed good and similar performances whereas the performance of the Inception V3 model was not enough successful [16]. The fuzzy color and image stacking techniques were embedded into MobileNetV2 and SqueezeNet with the social mimic optimization algorithm to detect COVID-19 infection from Chest CT images. The model ensured 99.27% classification achievement [17].
The automatic segmentation of anatomical structures is an important step for many medical image analysis tasks [18–22]. This approach depicts the regions of interest (ROIs) such as lung, lobes, bronchopulmonary segments, and infected regions or lesions, in the chest X-ray or CT images [23]. The recently proposed segmentation networks for COVID-19 employs U-Net [2, 24–28], U-Net++ [29, 30], and VB-Net [31]. A hybrid COVID-19 detection model that relies on an improved marine predator algorithm (IMPA) was proposed for X-ray image segmentation. The ranking-based diversity reduction (RDR) strategy was utilized to enhance the performance of the model. Thanks to the model, the similar small regions were extracted on X-ray images [32]. Since the segmentation of CT images is of great significance to help patient monitoring as well as diagnosis of COVID-19, U-Net-based segmentation network model based on attention mechanism was proposed. This model explores the spatial and channel attention features to capture rich contextual relationships. As a result, the dice score was reported as 83.1% in the study [20]. An AI system, which automatically analyzes CT images to identify COVID-19 pneumonia features, was proposed. The segmentation procedure was embedded in the proposed model. The lung region extraction, lesion segmentation, and lesion classification were realized to ease the burden of radiologists [30]. A new deep CNN model using a fully CNN in conjunction with an adversarial critic model was offered for lung segmentation based on the chest X-ray. The model was called Attention U-Net Based Adversarial Architectures. The dice score was reported as 97.5% on the JSRT dataset [33].
As seen in the reviewed literature, the deep CNN models have been used for both classification and segmentation of COVID-19 infection. U-shape architecture with symmetric encoding and decoding blocks has been adopted to learn better visual semantics and detailed contextures. In this study, a SegNet-based network using the attention gate (AG) mechanism for the automatic segmentation of COVID-19 regions from CT images is proposed. AGs have been embedded in the proposed model with a minimum computing load. In this way, the precision and predictive capabilities of the proposed model are enhanced. As a result, we achieved promising results.
The rest of this study is organized as follows: the methodology is given in “Methodology”. Experimental works and results are presented in “Description of Database and Experimental Work”. Concluding remarks are presented in “Conclusions”.
Methodology
Attention-Based Network Architecture (A-SegNet)
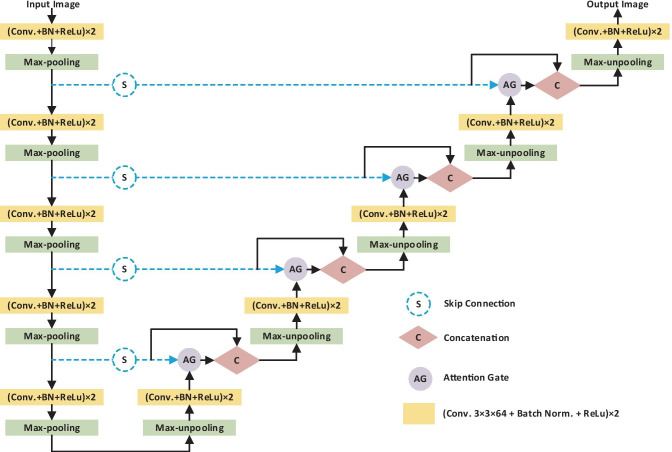
CNNs are one of the most popular method that was used in many tasks such as classification, localization, and segmentation [18, 19, 34–40]. CNNs outperform conventional methods in medical image analysis. Further, in a study proposed by Wolz et al. [41], it was even claimed that CNNs were faster than graph-cut and multi-atlas segmentation techniques. The success of CNNs can be associated with learning each activation map with stochastic gradient descent (SGD) optimization, sharing the learned convolution filters in all pixels, and using the structural information by the convolution operator most appropriately. Even so, it is difficult to reduce the false positive value for small objects with large variability in shapes, such as retinal vessel and organ segmentation [42, 43]. In such cases, existing CNNs localize the task separately and require previous additional object localization models to simplify subsequent classification/segmentation steps or to guide localization using weak tags to improve the accuracy [20, 44]. Here, we present that the segmentation of COVID-19 regions can be provided by integrating attention gates (AG) into a standard SegNet model (cf. Fig. 1) [21].
Fig. 1.
Proposed network architecture (A-SegNet), equipped with AG modules
SegNet is a fully connected neural network architecture proposed for pixel-based image segmentation. SegNet architecture uses feature maps that are produced by encoder-decoder pairs to learn different resolutions. Each encoder layer sequence applies the maximum pooling operation to the result obtained after applying the convolution, batch normalization, and ReLu processes. Then, the output of the maximum pooling is fed to the input of both the next encoder and the corresponding decoder. Decoders are generally similar to encoders, and the main difference between the two is that they do not have a nonlinear effect. The decoders sample the input they receive upstream using the indexes from the encoding step. The result obtained from the last decoder is given to the softmax layer and the final output is obtained.
In the proposed method, we incorporated AGs to the standard SegNet architecture to emphasize the salient features that pass through the skip connections. An AG has two input signals. The first one takes the map of the feature carried by the skip connection. The other input uses the coarse feature map obtained from the output of the previous decoder layer. The irrelevant and noisy feature responses extracted from the coarse-scale are clarified in skip connections. The output of the AG is fed into the next decoder. This process is performed for each skip connection. In this way, attention weights increase the resolution of features and provide better segmentation performance.
Attention Gate Module
The dataset used in this study contains different mass sizes and shapes. Also, in many CT slices, the mass size is quite small compared with the background region. Therefore, segmentation-based classic CNNs produce low sensitivity results. Concerning these features, an encoder-decoder-based SegNet architecture in which AGs [45] are integrated is proposed to segment the COVID-19 regions. The general structure of an AG is shown in Fig. 2.
Fig. 2.
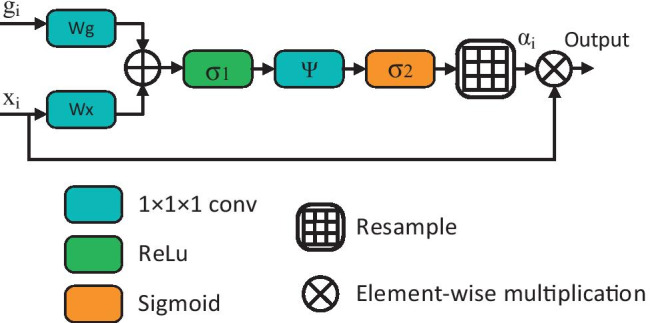
Schematic of the AG
The , shown in Fig. 2, is the feature map of the layer output and refers to a gating signal vector that is collected from a coarser scale and decides the focus region for each pixel. On the other hand, the coefficient provides distinct activations related to the target task by suppressing the noise in irrelevant property responses. Finally, the final output of the AGs is produced by element-wise multiplication of and maps:
| 1 |
In general, attention modules are used in two ways, multiplicative [46] or additive [47]. The multiplicative attention modules have faster computing capability and use less memory in practice as they can be applied as matrix multiplication. However, it has been suggested that additive attention modules perform better performance in experiments due to their large input features [48]. In our experiments, we preferred the first in terms of faster calculation, and it is formulated as follows:
| 2 |
where is the ReLu function: , and also expressed as the sigmoid function: . Finally, and are the linear transformations using the 1 × 1 × 1 dimensional convolution operator, respectively, and are bias terms. All AG weights are initialized randomly and updated according to the principle of back-propagation.
Loss Function
Cross-Entropy Loss
Cross-entropy is a commonly used loss function in deep learning practices to cope with binary classification problems that compute the probability of being a particular class or not [49]. Let and represent the input CT slice and the corresponding segmented ground-truth COVID-19 map, respectively. The main purpose in segmentation problems is to learn mapping during the training of the network [50]. The baseline cross-entropy loss has been expressed as follows:
| 3 |
where and represent the pixel index and the total pixels, respectively. According to the statement in Eq. (3), we can see that the cross-entropy loss gives equal weight to the loss of different pixels, which do not take into account unbalanced pixel distributions.
Dice Loss
The Dice Score Coefficient (DSC) [33] is the most common and simple approach that compares and evaluates segmentation results to calculate the overlap rate of predicted masks and ground-truth, and so, for the given and images, DSC is calculated as follows:
| 4 |
where, , and represent true-positives, false-negatives, and false-positive indices, respectively. Let be the class tag corresponding to any set, and represent ground-truth and predicted results, respectively. Hence, the dice score can be rearranged as follows:
| 5 |
where ε is the constant coefficient added to prevent numerical instability due to the division of the equation to zero and its value is very small. Thus, the linear Dice Loss (DL) of the segmentation prediction can be calculated as follows:
| 6 |
Tversky Loss
One of the biggest handicaps of DL is that it considers false negative and false positive predictions uniformly. This will cause low recall alongside high precision. If so the estimated region of interest (ROI) is small, the weights of false-negative pixels should be higher than the weights of false-positive pixels. It can be easily overcome this deficiency by adding weights and , which are tunable parameters. Thus, Eq. (5) is rearranged and the mathematical definition of the Tversky Similarity score can be expressed as follows [51]:
| 7 |
where and , that is, the overline of was simply used to express the complement of the class. As a result, Tversky loss can be easily obtained from Eq. (4) and is expressed as follows:
| 8 |
Focal Tversky Loss
Another disadvantage of the dice score is that it is difficult to segment small ROIs as they do not contribute to the loss sufficiently. To struggle with this handicap, Abraham et al. [52] recommended the Focal Tversky Loss function (FTL):
| 9 |
where is a real number ranging from 1 to 3. In practice, if a pixel is misclassified with a high Tversky score, the FTL is unaffected. However, if the Tversky score is small and the pixel is misclassified, the FTL will decrease significantly. We have trained our network for different , , and values of FTL to help segment small COVID-19 regions.
Description of Database and Experimental Work
Database and Pre-Processing
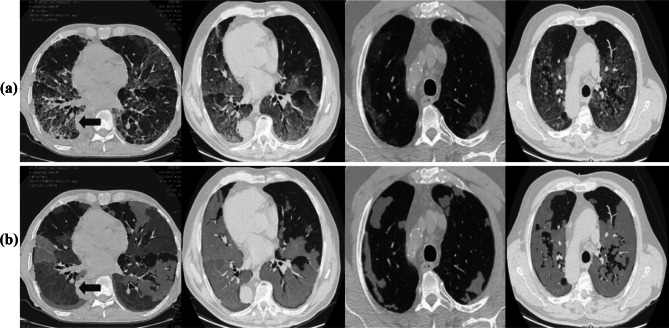
The COVID-19 CT segmentation databases used in the experiments were collected by the Italian Society of Medical and Interventional Radiology and consists of two parts. The first database contains 100 axial CT images of 60 patients with COVID-19, as well as 100 ground-truth images labeled by the radiologist in 3 categories such as ground-glass, consolidation, and pleural effusion. The images are 512 × 512 pixels in size and presented to the researchers in grayscale NIFTI file format. The second database contains a total of 829 axial CT slices and is recorded as 9 CT volumes. The radiologists reported that only 373 of the sections in this database had COVID-19 cases and were segmented. Also, unlike the first database, the resolutions of these slices are 630 × 630 pixels. Also, it is seen in Fig. 3 that only 233 slices contain the consolidation regions and these regions occupy a very small area in the second database. Therefore, we segmented all lesions in ground-truth as a single case of COVID-19 lesion. Also, by combining the images of both data sets containing the relevant case, we have taken into account 473 CT images in total for use in experiments.
Fig. 3.
Sample images of COVID-19 CT segmentation database. a Raw images, b ground-truths corresponding to a. Ground-glass, consolidation, and pleural effusion cases are shown in orange, blue, and green, respectively
We aim to produce the correct COVID-19 segmentation masks from CT images. Hence to avoid including undesired parts/organs in CT images, Hounsfield units were mapped to the intensity window as [−1024, 600]. Then, the mapped images were normalized to the 0–255 range. Finally, all images were rescaled to a resolution of 256 × 256 pixels to reduce the computation cost during training, and the segmentation results were interpreted with a fivefold cross-validation technique.
Experimental Works and Results
The network we specified in our proposal is implemented in MATLAB environment and trained with a workstation that is equipped with a single Nvidia GPU Quadro P5000. During the training, the network’s performances of different loss functions such as dice, Tversky, and focal Tversky were evaluated in the aforementioned database, considering the fivefold cross-validation technique. In all of the experiments, maximum epoch and mini-batch size values were chosen as 50 and 16, respectively. The initial learning rate was gradually reduced by 0.5 per 10 epoch, initializing from 0.1 the loss functions. The loss functions were also optimized by employing a stochastic gradient descent optimizer with momentum.
The segmentation performance of the proposed work was evaluated by three metrics namely dice, sensitivity, and specificity, respectively. Evaluation metrics are defined as follows:
| 10 |
| 11 |
| 12 |
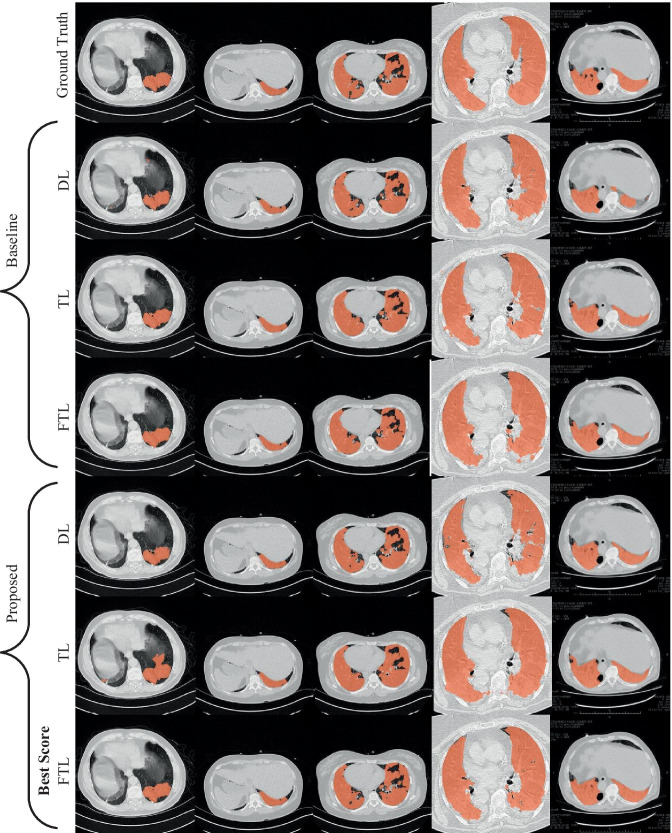
where TP, FP, TN, and FN show the true positive, false positive, true negative, and false negative samples, respectively [53]. Figure 4 shows sample ground-truths and obtained segmentation results with different loss functions. While the first row of Fig. 4 showed the ground truth segmentations, the rows 2–4 showed the baseline segmentations. The proposed segmentation achievements were given in the last three rows of Fig. 4.
Fig. 4.
Sample ground-truths and related segmentation results obtained with different loss functions. “for TL, α = 0.3, β = 0.7” and “for FTL, α = 0.3, β = 0.7, γ = 4/3”
From Fig. 4, it was observed that the proposed methods’ achievements (A-SegNet-FTL) were better than the baseline segmentations. Generally speaking, the boundary of the segmentation was quite regular and there were no small holes inside the segmentations. The region of interest was correctly segmented. For the sample image in the first column of Fig. 4, it was seen that almost all approaches produced similar segmentations. Only, A-SegNet-TL produced over-segmentation for this sample. For the second sample image, as all approaches almost produced identical segmentations, baseline approaches produced segmentations that contain holes inside the region of interest. The proposed approaches produced identical results with ground truth results. For the other samples, as the obtained segmentations were quite similar, a close look shows the quality segmentations of the proposed method.
Table 1 also shows the quantitative evaluation scores for both the proposed method and baseline methods, respectively. As observed in Table 1, various parameters were used during experiments. For the baseline results, it was seen that the best dice (87.56%) and sensitivity (92.13%) scores were obtained with the SegNet + FTL (α = 0.3, β = 0.7, γ = 4/3) and the best specificity (99.71%) score was obtained with SegNet + FTL (α = 0.7, β = 0.3, γ = 4/3), respectively. For the proposed method, it was seen that while highest dice (89.61%) and sensitivity (92.73%) scores were obtained with A-SegNet + FTL (α = 0.3, β = 0.7, γ = 4/3), the best specificity score (99.75%) was obtained by the A-SegNet + FTL (α = 0.7, β = 0.3, γ = 4/3), respectively.
Table 1.
Quantitative evaluation of COVID-19 segmentation results
| Model | Parameters | Dice (%) | Sensitivity (%) | Specificity (%) | |
|---|---|---|---|---|---|
| Baseline | SegNet + DL | 86.06 | 86.30 | 99.45 | |
| SegNet + TL | α = 0.7, β = 0.3 | 71.02 | 67.54 | 99.48 | |
| SegNet + TL | α = 0.5, β = 0.5 | 68.97 | 66.70 | 99.49 | |
| SegNet + TL | α = 0.3, β = 0.7 | 74.85 | 86.38 | 98.70 | |
| SegNet + FTL | α = 0.7, β = 0.3, γ = 4/3 | 86.58 | 82.47 | 99.71 | |
| SegNet + FTL | α = 0.5, β = 0.5, γ = 4/3 | 87.44 | 88.27 | 99.54 | |
| SegNet + FTL | α = 0.3, β = 0.7, γ = 4/3 | 87.56 | 92.13 | 99.39 | |
| Ours | A-SegNet + DL | 88.52 | 88.90 | 99.56 | |
| A-SegNet + TL | α = 0.7, β = 0.3 | 74.65 | 71.07 | 99.44 | |
| A-SegNet + TL | α = 0.5, β = 0.5 | 88.52 | 88.31 | 99.62 | |
| A-SegNet + TL | α = 0.3, β = 0.7 | 74.61 | 79.03 | 99.28 | |
| A-SegNet + FTL | α = 0.7, β = 0.3, γ = 4/3 | 86.96 | 82.22 | 99.75 | |
| A-SegNet + FTL | α = 0.5, β = 0.5, γ = 4/3 | 89.16 | 87.99 | 99.66 | |
| A-SegNet + FTL | α = 0.3, β = 0.7, γ = 4/3 | 89.61 | 92.73 | 99.51 |
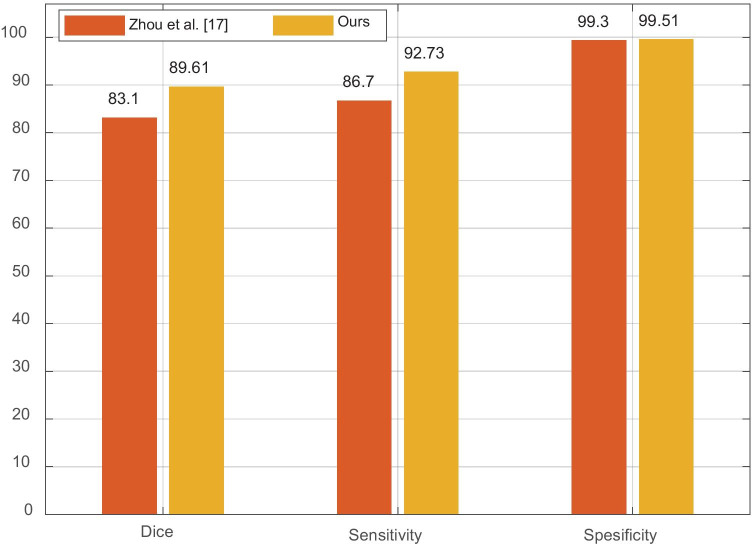
A further comparison of the proposed method with Zhou et al. [20] is given in Fig. 5. The bar illustration was used to show the comparisons. As seen in Fig. 5, our dice, sensitivity, and specificity scores were 6.51%, 6.03%, and 0.21% better than the compared methods evaluation scores.
Fig. 5.
Comparison of the proposed method with a state-of-the-art method [20]
Because of variable infection regions in the lungs, similar imaging, and large inter-case variations, accurate evaluation of CT images is still a challenging task in clinical practice. In the early stages of the disease, the infections caused by COVID-19 are seen in small regions of the lung and it is really hard to detect these patterns in CT images with a visual examination. Besides, imaging biomarkers of COVID-19 such as GGO, mosaic sign, air bronchogram, and interlobular septal thickening are similar in some severe and non-severe cases. In this scope, the proposed CNN model supports the decision-making process of the field experts. The results of this study point out that understanding chest CT imaging of COVID-19 helps detect infection early as well as has the potential to support clinical management processes.
Conclusions
In this paper, SegNet-based network using the attention gate (AG) mechanism was proposed for the automatic segmentation of COVID-19 regions from CT images. The proposed network was trained with various loss functions namely, dice, Tversky, and focal Tversky, respectively. A CT dataset was used in experiments and successful segmentation was obtained. The following conclusions were acquired from the experiments:
The attention gate mechanism improved the segmentation performance.
Training of the proposed network with the focal Tversky loss produced the better segmentations than the others.
Adding the γ parameter improved the segmentation results.
Low α and high β parameters produced better performance.
In future works, it is planning to apply optimization for obtaining α, β, and γ parameters. Besides, dense network structures with AG will be investigated for COVID-19 segmentation in CT images.
Acknowledgements
The authors would like to thank Dr. Omer C. Kuloglu (MD), who is a radiologist in Lokman Hekim Hayat Hospital, for his valuable comments on the CT images.
Declarations
Ethical Approval
This article does not contain any data, or other information from studies or experimentation, with the involvement of human or animal subjects.
Conflict of Interest
The authors declare that they have no conflict to interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Liu P, Tan X. 2019 Novel Coronavirus (2019-nCoV) Pneumonia. Radiology. 2020;295:19. doi: 10.1148/radiol.2020200257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng C, Deng X, Fu Q, Zhou Q, Feng J, Ma H, et al. Deep learning-based detection for COVID-19 from chest CT using weak label. MedRxiv. 2020 doi: 10.1101/2020.03.12.20027185. [DOI] [Google Scholar]
- 3.Kanne JP, Little BP, Chung JH, Elicker BM, Ketai LH. Essentials for Radiologists on COVID-19: An Update—Radiology Scientific Expert Panel. Radiology n.d.;0:200527. 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed]
- 4.Ucar F, Korkmaz D. COVIDiagnosis-Net: deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med Hypotheses. 2020;140:109761. doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang T-H, Wu J-L, Chang L-Y. Clinical characteristics and diagnostic challenges of pediatric COVID-19: a systematic review and meta-analysis. J Formos Med Assoc. 2020 doi: 10.1016/j.jfma.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prokop M, van Everdingen W, van Rees Vellinga T, van Ufford J, Stöger L, Beenen L, et al. CO-RADS – A categorical CT assessment scheme for patients with suspected COVID-19: definition and evaluation. Radiology n.d.;0:201473. 10.1148/radiol.2020201473. [DOI] [PMC free article] [PubMed]
- 7.Chen Z, Fan H, Cai J, Li Y, Wu B, Hou Y, et al. High-resolution computed tomography manifestations of COVID-19 infections in patients of different ages. Eur J Radiol. 2020;126:108972. doi: 10.1016/j.ejrad.2020.108972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng Z, Qin L, Cao Q, Dai J, Pan A, Yang W, et al. Quantitative computed tomography of the coronavirus disease 2019 (COVID-19) pneumonia. Radiol Infect Dis 2020. 10.1016/j.jrid.2020.04.004. [DOI] [PMC free article] [PubMed]
- 9.Shen C, Yu N, Cai S, Zhou J, Sheng J, Liu K, et al. Quantitative computed tomography analysis for stratifying the severity of Coronavirus Disease 2019. J Pharm Anal. 2020;10:123–129. doi: 10.1016/j.jpha.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toğaçar M, Ergen B, Cömert Z. A deep feature learning model for pneumonia detection applying a combination of mRMR feature selection and machine learning models. IRBM. 2019 doi: 10.1016/j.irbm.2019.10.006. [DOI] [Google Scholar]
- 11.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya] U [Rajendra. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med 2020:103792. 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed]
- 12.Nour M, Cömert Z, Polat K. A Novel Medical Diagnosis model for COVID-19 infection detection based on Deep Features and Bayesian Optimization. Appl Soft Comput 2020:106580. 10.1016/j.asoc.2020.106580. [DOI] [PMC free article] [PubMed]
- 13.Ardakani AA, Kanafi AR, Acharya UR, Khadem N, Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput Biol Med 2020:103795. 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed]
- 14.Sethy PK, Behera SK. Detection of Coronavirus Disease (COVID-19) based on deep features. Preprints. 2020 doi: 10.20944/preprints202003.0300.v1. [DOI] [Google Scholar]
- 15.Narin A, Kaya C, Pamuk Z. Automatic Detection of Coronavirus Disease (COVID-19) Using X-ray Images and Deep Convolutional Neural Networks 2020. [DOI] [PMC free article] [PubMed]
- 16.Hemdan EE-D, Shouman MA, Karar ME. COVIDX-Net: A Framework of Deep Learning Classifiers to Diagnose COVID-19 in X-Ray Images 2020.
- 17.Toğaçar M, Ergen B, Cömert Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput Biol Med. 2020;121:103805. doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, editors. Med. Image Comput. Comput. Interv. -- MICCAI 2015, Cham: Springer International Publishing; 2015, p. 234–41.
- 19.Budak Ü, Guo Y, Tanyildizi E, Şengür A. Cascaded deep convolutional encoder-decoder neural networks for efficient liver tumor segmentation. Med Hypotheses. 2020;134:109431. doi: 10.1016/j.mehy.2019.109431. [DOI] [PubMed] [Google Scholar]
- 20.Zhou T, Canu S, Ruan S. An automatic COVID-19 CT segmentation network using spatial and channel attention mechanism 2020. [DOI] [PMC free article] [PubMed]
- 21.Badrinarayanan V, Kendall A, Cipolla R. SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39:2481–2495. doi: 10.1109/TPAMI.2016.2644615. [DOI] [PubMed] [Google Scholar]
- 22.Başaran E, Cömert Z, Çelik Y. Convolutional neural network approach for automatic tympanic membrane detection and classification. Biomed Signal Process Control. 2020;56:101734. doi: 10.1016/j.bspc.2019.101734. [DOI] [Google Scholar]
- 23.Shi F, Wang J, Shi J, Wu Z, Wang Q, Tang Z, et al. Review of Artificial Intelligence Techniques in Imaging Data Acquisition, Segmentation and Diagnosis for COVID-19. IEEE Rev Biomed Eng 2020:1. 10.1109/RBME.2020.2987975. [DOI] [PubMed]
- 24.Cao Y, Xu Z, Feng J, Jin C, Han X, Wu H, et al. Longitudinal assessment of COVID-19 using a deep learning–based quantitative CT pipeline: illustration of two cases. Radiol Cardiothorac Imaging. 2020;2:e200082. doi: 10.1148/ryct.2020200082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang L, Han R, Ai T, Yu P, Kang H, Tao Q, et al. Serial quantitative chest CT assessment of COVID-19: deep-learning approach. Radiol Cardiothorac Imaging. 2020;2:e200075. doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi X, Jiang Z, YU Q, Shao C, Zhang H, Yue H, et al. Machine learning-based CT radiomics model for predicting hospital stay in patients with pneumonia associated with SARS-CoV-2 infection: A multicenter study. MedRxiv 2020. 10.1101/2020.02.29.20029603. [DOI] [PMC free article] [PubMed]
- 27.Gozes O, Frid-Adar M, Greenspan H, Browning PD, Zhang H, Ji W, et al. Rapid AI Development Cycle for the Coronavirus (COVID-19) Pandemic: Initial Results for Automated Detection & Patient Monitoring using Deep Learning CT Image Analysis 2020.
- 28.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, et al. Artificial Intelligence Distinguishes COVID-19 from Community Acquired Pneumonia on Chest CT. Radiology n.d.;0:200905. 10.1148/radiol.2020200905.
- 29.Chen J, Wu L, Zhang J, Zhang L, Gong D, Zhao Y, et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study. MedRxiv 2020. 10.1101/2020.02.25.20021568
- 30.Jin S, Wang B, Xu H, Luo C, Wei L, Zhao W, et al. AI-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system in four weeks. MedRxiv. 2020 doi: 10.1101/2020.03.19.20039354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shan F, Gao Y, Wang J, Shi W, Shi N, Han M, et al. Lung Infection Quantification of COVID-19 in CT Images with Deep Learning 2020.
- 32.Abdel-Basset M, Mohamed R, Elhoseny M, Chakrabortty RK, Ryan M. A hybrid COVID-19 detection model using an improved marine predators algorithm and a ranking-based diversity reduction strategy. IEEE Access 2020:1. 10.1109/ACCESS.2020.2990893.
- 33.Gaál G, Maga B, Lukács A. Attention U-Net Based Adversarial Architectures for Chest X-ray Lung Segmentation 2020.
- 34.Bai W, Sinclair M, Tarroni G, Oktay O, Rajchl M, Vaillant G, et al. Human-level CMR image analysis with deep fully convolutional networks 2017.
- 35.Budak Ü, Cömert Z, Çıbuk M, Şengür A. DCCMED-Net: densely connected and concatenated multi Encoder-Decoder CNNs for retinal vessel extraction from fundus images. Med Hypotheses. 2020;134:109426. doi: 10.1016/j.mehy.2019.109426. [DOI] [PubMed] [Google Scholar]
- 36.Budak Ü, Cömert Z, Rashid ZN, Şengür A, Çıbuk M. Computer-aided diagnosis system combining FCN and Bi-LSTM model for efficient breast cancer detection from histopathological images. Appl Soft Comput. 2019;85:105765. doi: 10.1016/j.asoc.2019.105765. [DOI] [Google Scholar]
- 37.Zaharchuk G, Gong E, Wintermark M, Rubin D, Langlotz CP. Deep learning in neuroradiology. Am J Neuroradiol. 2018;39:1776–1784. doi: 10.3174/ajnr.A5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee C-Y, Xie S, Gallagher P, Zhang Z, Tu Z. Deeply-Supervised Nets. In: Lebanon G, Vishwanathan SVN, editors. Proc. Eighteenth Int. Conf. Artif. Intell. Stat., vol. 38, San Diego, California, USA: PMLR; 2015, p. 562–70.
- 39.Deniz E, Sengür A, Kadiroglu Z, Guo Y, Bajaj V, Budak Ü. Transfer learning based histopathologic image classification for breast cancer detection. Heal Inf Sci Syst. 2018;6:18. doi: 10.1007/s13755-018-0057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Şengür D. Investigation of the relationships of the students’ academic level and gender with Covid-19 based anxiety and protective behaviors: A data mining approach 2020;15:93–9.
- 41.Wolz R, Chu C, Misawa K, Fujiwara M, Mori K, Rueckert D. Automated abdominal multi-organ segmentation with subject-specific atlas generation. IEEE Trans Med Imaging. 2013;32:1723–1730. doi: 10.1109/TMI.2013.2265805. [DOI] [PubMed] [Google Scholar]
- 42.Guo Y, Budak Ü, Şengür A. A novel retinal vessel detection approach based on multiple deep convolution neural networks. Comput Methods Programs Biomed. 2018;167:43–48. doi: 10.1016/j.cmpb.2018.10.021. [DOI] [PubMed] [Google Scholar]
- 43.Guo Y, Budak Ü, Vespa LJ, Khorasani E, Şengür A. A retinal vessel detection approach using convolution neural network with reinforcement sample learning strategy. Measurement. 2018;125:586–591. doi: 10.1016/j.measurement.2018.05.003. [DOI] [Google Scholar]
- 44.Pesce E, Withey] S [Joseph, Ypsilantis P-P, Bakewell R, Goh V, Montana G. Learning to detect chest radiographs containing pulmonary lesions using visual attention networks. Med Image Anal 2019;53:26–38. 10.1016/j.media.2018.12.007. [DOI] [PubMed]
- 45.Oktay O, Schlemper J, Folgoc L Le, Lee M, Heinrich M, Misawa K, et al. Attention U-Net: Learning Where to Look for the Pancreas 2018.
- 46.Luong T, Pham H, Manning CD. Effective Approaches to Attention-based Neural Machine Translation. Proc. 2015 Conf. Empir. Methods Nat. Lang. Process., Lisbon, Portugal: Association for Computational Linguistics; 2015, p. 1412–21. 10.18653/v1/D15-1166.
- 47.Bahdanau D, Cho K, Bengio Y. Neural Machine Translation by Jointly Learning to Align and Translate 2014.
- 48.Britz D, Goldie A, Luong M-T, Le Q. Massive Exploration of Neural Machine Translation Architectures. Proc. 2017 Conf. Empir. Methods Nat. Lang. Process., Copenhagen, Denmark: Association for Computational Linguistics; 2017, p. 1442–51. 10.18653/v1/D17-1151.
- 49.Li R, Li M, Li J, Zhou Y. Connection Sensitive Attention U-NET for Accurate Retinal Vessel Segmentation 2019.
- 50.Shankaranarayana SM, Ram K, Mitra K, Sivaprakasam M. Joint Optic Disc and Cup Segmentation Using Fully Convolutional and Adversarial Networks. In: Cardoso MJ, Arbel T, Melbourne A, Bogunovic H, Moeskops P, Chen X, et al., editors. Fetal, Infant Ophthalmic Med. Image Anal., Cham: Springer International Publishing; 2017, p. 168–76.
- 51.Tversky A. Features of similarity. Psychol Rev. 1977;84:327–352. doi: 10.1037/0033-295X.84.4.327. [DOI] [Google Scholar]
- 52.Abraham N, Khan NM. A Novel Focal Tversky Loss Function With Improved Attention U-Net for Lesion Segmentation. 2019 IEEE 16th Int. Symp. Biomed. Imaging (ISBI 2019), 2019, p. 683–7. 10.1109/ISBI.2019.8759329.
- 53.Şengür D, Turhan M. Prediction Of The Action Identification Levels Of Teachers Based On Organizational Commitment And Job Satisfaction By Using K-Nearest Neighbors Method. 2018;13:61–68. [Google Scholar]