Abstract
Objective
Trauma quality improvement programs and registries improve care and outcomes for injured patients. Designated trauma centers calculate injury scores using dedicated trauma registrars; however, many injuries arrive at nontrauma centers, leaving a substantial amount of data uncaptured. We propose automated methods to identify severe chest injury using machine learning (ML) and natural language processing (NLP) methods from the electronic health record (EHR) for quality reporting.
Materials and Methods
A level I trauma center was queried for patients presenting after injury between 2014 and 2018. Prediction modeling was performed to classify severe chest injury using a reference dataset labeled by certified registrars. Clinical documents from trauma encounters were processed into concept unique identifiers for inputs to ML models: logistic regression with elastic net (EN) regularization, extreme gradient boosted (XGB) machines, and convolutional neural networks (CNN). The optimal model was identified by examining predictive and face validity metrics using global explanations.
Results
Of 8952 encounters, 542 (6.1%) had a severe chest injury. CNN and EN had the highest discrimination, with an area under the receiver operating characteristic curve of 0.93 and calibration slopes between 0.88 and 0.97. CNN had better performance across risk thresholds with fewer discordant cases. Examination of global explanations demonstrated the CNN model had better face validity, with top features including “contusion of lung” and “hemopneumothorax.”
Discussion
The CNN model featured optimal discrimination, calibration, and clinically relevant features selected.
Conclusion
NLP and ML methods to populate trauma registries for quality analyses are feasible.
Keywords: trauma surgery, machine learning, interpretability
INTRODUCTION
Trauma is the fourth leading cause of death in the United States (US) across all age groups and accounted for an estimated 136 billion dollars in healthcare costs in 2010.1 The development of statewide trauma systems and registries have increased the amount of data available to researchers to better understand the epidemiology of trauma in the US, informing trauma quality programs to develop practice changes and improve health outcomes.2 Trauma registries rely on certified trauma coders to manually abstract relevant information from the electronic health record (EHR) after discharge and summarize patient injuries in the form of Abbreviated Injury Scores (AIS). The manual calculation of injury scores is a requirement to receive state designation and the American College of Surgeons Committee on Trauma verification but it is time- and resource-intensive.3 In addition, an estimated 30%–50% of patients with major injuries receive care at nontrauma centers, which may not have the same formalized programs or resources as state-designated trauma centers to track pertinent epidemiologic data for quality improvement, research, and planning.4–6 As a result, despite the accumulation of information in trauma registries across the US, a significant portion of care after injury remains uncaptured.
Methods in machine learning (ML) and natural language processing (NLP) have the potential to automate data capture for clinical registries. Information on mechanism and severity of the injury and patient functional status and outcomes are embedded in the unstructured free text that makes up the majority of the EHR.7 With the use of NLP techniques, this information can be incorporated into supervised ML algorithms that can learn from reference standards, such as the manually generated AIS, to potentially automate the collection of data from centers without the need to manually input data into a trauma registry. NLP can mine and analyze these data sources to populate clinical registries, providing standardized, and comprehensive injury scoring for patients to accompany the conventional discrete structured elements that make up a trauma registry. With refinement, these automated tools may also allow injury scores to become available at point-of-care for risk prognostication or better allocation of hospital resources.8
In this study, we aim to determine the optimal ML algorithm for a classifier using NLP methods to discriminate between cases of severe and nonsevere chest injury using clinical documents from trauma encounters. We have previously studied the optimal text preprocessing and time after presentation to generate accurate predictions from a single modeling approach; we expand upon this work by examining multiple modeling approaches with a focus on model interpretability and clinical relevance. We view this as initial steps and proof-of-concept toward development of an automated classifier for nontrauma centers. We hypothesize that similar predictive validity metrics will be achieved across different ML approaches but model interpretability and clinical face validity of the most important text features will differ.
MATERIALS AND METHODS
Data source and study setting
The Loyola University Medical Center (LUMC) Level I trauma registry was queried for adult patients presenting between January 1, 2014 and October 22, 2018 as a trauma activation, trauma transfer, or direct transfer after a mechanism attributed to trauma or burn injury; the registry is manually generated by trauma registrars certified by the Trauma Quality Improvement Program and serves as the gold-standard reference label. Patients were linked between the trauma registry and the EHR by medical record number. The AIS for the chest was used to develop the outcome of interest for supervised ML; AIS is graded on a 6-point ordinal scale from minor to unsurvivable. An AIS chest score greater than 2 served as the outcome of interest, as it is considered a severe chest injury and is associated with an increase in likelihood of mortality.9 The final analytic cohort is depicted in Supplementary Figure S1. Comparisons in patient characteristics were made between those with and without severe chest injury using the Wilcoxon rank-sum tests for continuous variables and the chi-square test for categorical variables.
Text preprocessing from clinical documents
Clinical documents from the EHR were organized by entry time into the EHR. Only clinical documents from the first 8 hours after presentation to the emergency department were included in the analysis; no structured data elements (demographics, vitals, laboratory values, and so on) were used in the analysis to minimize the amount of feature engineering required. Prior work with a logistic regression model demonstrated 8 hours as the minimum amount of data needed to achieve optimal performance metrics for classification of severe injury. Inclusion of additional data across the entire encounter only increased computational requirements without further improvement in model discrimination.10
Sensitivity analysis was performed to remove the potential heterogeneity of clinical documentation by trauma care providers by only including routinely collected radiology reports during an ED evaluation. The routine documentation that is common to nontrauma centers include chest radiographs and chest computed tomography (CT) radiographs with standardized reporting by board-certified radiologists. The collection of these reports during routine ED care are time-sensitive, so we utilized the data from the first 8 hours after presentation to the ED, similar to our primary analysis. In addition, we also examined all chest radiology reports during the encounter in a second sensitivity analysis.
Documents were processed using the Apache clinical Text Analysis and Knowledge Extraction System (cTAKES) to map free text from the EHR to clinical concepts in the National Library of Medicine Unified Medical Language System (UMLS) in the form of concept unique identifiers (CUI).11 Mapping documents to CUIs provides a method of working with text without protected health information (PHI) for reporting to state and national registries, and condenses the feature space by mapping similar free-text phrases to a single CUI (e.g., hematoma, hematomas, and blood clot all map to CUI C0018944).
Development of ML models
The overall data corpus was randomly divided into an 80% (n = 7033) training dataset and a 20% dataset (n = 1758) holdout testing dataset. Due to the overall low rate of severe chest injury in the cohort, the training dataset was downsampled to a 1:1 distribution of cases to controls. All models were trained on the downsampled training dataset with hyperparameters (Supplementary Table S2) tuned by a random grid search to maximize the area under the receiver operating characteristic curve (AUROC) using 5-fold cross-validation; results were reported from the independent 20% holdout test dataset. The prevalence of cases in the test dataset was not adjusted and the results reported reflect the true prevalence of severe chest injury in our cohort. The final analytic cohort was downsampled to have a 50% case-rate (n = 429), and the case-rate in the holdout test dataset still reflected the true prevalence of the trauma cohort at 6.4% (n = 113).
Models utilized in the analysis were logistic regression with elastic net (EN) regularization, extreme gradient boosted (XGB) machines, and convolutional neural network (CNN). For EN and XGB, binary CUI values (presence vs. absence) served as inputs for supervised ML; a 300-dimension CUI embedding layer was used for the input into the CNN. For the embeddings, CUIs were ordered by note entry time and tokenized across the encounter, preserving the architecture and context of the clinical documents within each encounter.12
The nonparametric DeLong method was used to test for the statistical significance of differences in AUROC between models.13 Sensitivity and specificity with 95% confidence intervals (95% CI) were compared at thresholds set to hold specificity and sensitivity at 80%, respectively. Discrimination was also assessed with classification plots, which allow for a global assessment of model discrimination by depicting the variation in true positive and false positive rates with varying thresholds. The classification plot also allows for comparisons of models where the AUROC is numerically similar but the shape of the curve may provide insights on optimal model discrimination.14 To account for the differences in prevalence between the downsampled training dataset and the holdout testing dataset, model predictions were calibrated using isotonic calibration. Next, calibration was assessed by visual plots, calibration slope, and calibration intercept.15 The concordance of predictions across models was also evaluated for the test dataset.
Global model interpretability
To examine the clinical face validity of the three models, we applied global model interpretability metrics. For the interpretable models (EN and XBG), interpretation was made by directly examining feature importance. The beta coefficients from the EN model were extracted and ranked. For the XGB model, permutation feature importance was extracted by averaging the improvement in squared error risk across all boosted trees.16–18 For the CNN model, a surrogate local interpretable model-agnostic explanations (LIME) model was applied to approximate the predictions to explain individual predictions locally and then average the feature weights from the local explanations to derive a global measure.19,20 The global LIME measure had a median R2 (variance explained) of 0.69 (IQR 0.46–0.93), which was an acceptable approximation for the CNN model. The extracted features from all three ML models were rescaled to a 100-point scale to facilitate comparisons of feature importance from the training dataset.
Error analysis
The false positive and false negative cases predicted by the model were compared against the reference labels generated by the trauma registrars. This comparison was performed to better understand the model’s shortcomings when compared to the certified trauma registrars. Local LIME explanations for a random sampling of false positive and false negative cases predicted by the CNN model were generated, and we subsequently conducted a manual chart review (SK) to determine the source of the error by the false prediction.
This study was considered exempt by the Institutional Review Board of Loyola University Chicago. All data acquisition, processing, and analysis were conducted in the R programming language (version 3.6.0) using RStudio (version 1.2.1335).21,22
RESULTS
Patient and data characteristics
Between January 1, 2014 and October 22, 2018, there were 9084 encounters manually annotated by certified trauma registrars with linkage to the EHR. Of these, 293 patients (3.2%) were excluded due to a lack of CUI data; details of these patients are presented in Supplementary Table S1. Of the remaining 8 791 encounters, 542 (6.2%) had a severe chest injury. The characteristics of patients with and without severe chest injury are presented in Table 1. Patients with severe chest injury had higher rates of operative intervention, higher Elixhauser readmission and mortality scores,23 and higher rates of in-hospital death (P < .01 for all comparisons). The data corpus consisted of clinical documents filed into the EHR within the first 8 hours after presentation to the ED with a total of 102 493 reports and 15068 unique CUIs.
Table 1.
Patient characteristics and outcomes between severe and nonsevere injury
| Nonsevere chest injury | Severe chest injuryc | P-value | |
|---|---|---|---|
| n | 8249 | 542 | |
| Age, median (IQR) | 47 (30–65) | 42 (27–60) | <.001 |
| Sex, n (%) | <.001 | ||
| Male | 5459 (66.2) | 406 (74.9) | |
| Female | 2790 (33.8) | 136 (25.1) | |
| Race, n (%) | .026 | ||
| White | 4683 (56.8) | 282 (52.0) | |
| Black | 1922 (23.3) | 153 (28.2) | |
| Othera | 1644 (19.9) | 107 (19.7) | |
| Admitting service, n (%) | <.001 | ||
| Trauma | 3992 (48.3) | 483 (89.1) | |
| Burns | 1499 (18.2) | 29 (5.4) | |
| Orthopedic surgery | 834 (10.1) | 0 (0.0) | |
| Other | 1924 (23.3) | 30 (5.5) | |
| Operative intervention, n (%) | 2101 (25.5) | 194 (35.8) | <.001 |
| OR time (mins), median (IQR) | 185 (116–270) | 167 (104–274) | .31 |
| Comorbidities, n (%) | |||
| CHF | 213 (2.6) | 12 (2.2) | .70 |
| Hypertension | 1428 (17.3) | 111 (20.5) | .068 |
| Pulmonary disease | 468 (5.7) | 74 (18.2) | <.001 |
| Diabetes | 523 (6.3) | 44 (8.1) | .12 |
| Renal disease | 210 (2.5) | 19 (3.5) | .22 |
| Liver disease | 192 (2.3) | 25 (4.6) | .001 |
| Coagulopathy | 257 (3.1) | 57 (10.5) | <.001 |
| Alcohol misuse | 592 (7.2) | 65 (12.0) | <.001 |
| Drug misuse | 441 (5.3) | 55 (10.1) | <.001 |
| Elixhauser scores, median (IQR)c | |||
| Readmission score | 8 (0–21) | 13 (4–22) | .002 |
| Mortality score | 0 (−1–10) | 4 (0–13) | <.001 |
| Length of stay, median (IQR) | 2.3 (0.7–5.9) | 5.4 (1.6–13.4) | <.001 |
| Disposition, n (%) | <.001 | ||
| Home | 5831 (70.7) | 246 (45.4) | |
| Discharge to HC facility | 1822 (22.1) | 146 (26.9) | |
| AMA | 163 (2.0) | 3 (0.6) | |
| In-hospital death | 342 (4.1) | 145 (26.8) | |
| Otherb | 91 (1.1) | 2 (0.4) |
AMA: against medical advice; HC: healthcare; IQR: interquartile range; OR: operating room.
Other Race = American Indian, Asian, Hispanic, Multiracial, Hawaiian, Pacific Islander, Unknown.
Other Disposition = Hospice, law enforcement, unknown.
Elixhauser Scores calculated using diagnosis codes from the entire encounter.23
ML model parameters
The EN and CNN classifiers had the highest AUROC, at 0.93 (95% CI [0.91, 0.94]) and 0.93 (95% CI [0.91, 0.95]), respectively, as compared with the XGB classifier, which had an AUROC of 0.91 (95% CI [0.89, 0.94], DeLong test p < 0.05). At a specificity of 80%, the EN classifier demonstrated the highest sensitivity (0.95, 95% CI [0.89, 0.98]), followed by the CNN (0.93, 95% CI [0.87, 0.97]) and XGB (0.91, 95% CI [0.84, 0.96]) classifiers. Conversely, at a sensitivity of 80%, the CNN classifier demonstrated the highest specificity (0.91, 95% CI [0.89, 0.92]), followed by the EN (0.88, 95% CI [0.86, 0.90]) and XGB classifiers (0.85, 95% CI [0.84, 0.87]).
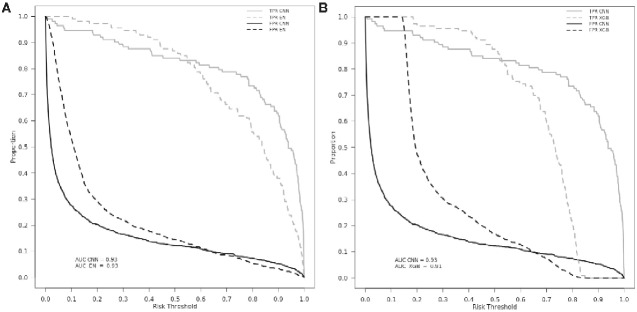
Classification plots comparing the CNN with both the EN and XGB models are shown in Figure 1. Although EN demonstrated the highest sensitivity, examination of the classification plot reveals that the CNN had a higher true-positive rate across most risk thresholds. Similarly, the false-positive rate was lower across most thresholds in the CNN model in comparison with EN and XGB. Concordance of the three models’ predictions from the holdout test dataset can be found in Table 2. The CNN model had a higher overall percentage of accurate predictions and a lower number of false-positive results.
Figure 1.
Classification plots comparing (A) CNN and EN and (B) CNN and XGB models. CNN model is indicated with the solid lines in each figure. TPR = true positive rate (grey); FPR = false positive rate (black); AUC = area under curve. X-axis represents threshold at which TPR/FPR are measured.
Table 2.
Concordance of model predictions across test dataset
| EN (n, %) |
XGB (n, %) |
CNN (n, %) |
||||
|---|---|---|---|---|---|---|
| Model correct | 1435 | 81.6 | 1452 | 82.6 | 1528 | 86.9 |
| All models correct | 1357 | 77.2 | 1357 | 77.2 | 1357 | 77.2 |
| Model correct, 1 or both other models wrong | 78 | 4.4 | 95 | 5.4 | 171 | 9.7 |
| Positive case | 10 | 0.6 | 5 | 0.3 | 4 | 0.2 |
| Negative case | 68 | 3.9 | 90 | 5.1 | 167 | 9.5 |
Total number in holdout test dataset = 1758.
All three models had good calibration. The calibrated EN model had a slope of 0.97 (95% CI [0.83, 1.14]) and an intercept of 0.13 (95% CI [−0.19, 0.46]), the calibrated XGB model had a slope of 1.02 (95% CI [0.86, 1.18]) and an intercept of 0.15 (95% CI [−0.18, 0.49]), and the calibrated CNN model with slope of 0.88 (95% CI [0.75, 1.02]) and an intercept of 0.05 (95% CI [−0.25, 0.36]).
Sensitivity analysis
In the first sensitivity analysis, including only the radiology reports (chest radiographs and CT) from the first 8 hours of ED presentation, we observed a small decrease in AUROC to 0.88 (95% CI [0.84, 0.92]) and sensitivity and specificity of 0.84 (95% CI [0.76, 0.91]) and 0.83 (95% CI [0.81, 0.86]), respectively. Expanding the corpus of radiology reports to the entire hospitalization demonstrated minimal gains with an AUROC of 0.89 (95% CI [0.86, 0.92]), The sensitivity and specificity of this classifier were 0.87 (95% CI [0.78, 0.93]) and 0.79 (95% CI [0.76, 0.81]), respectively.
Global model interpretability
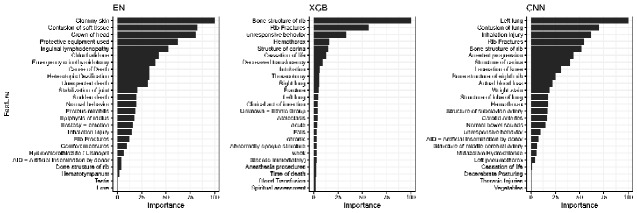
To assess the variation in clinical face validity of the three models, we examined the global model feature importance. Each model had selections of CUIs with high clinical face validity, consistent with features identified in the official trauma registrar AIS dictionary as indicative of severe chest injury, such as “C0035522—Rib Fractures” or “C0035561—Bone structure of rib” (Figure 2).24,25 However, the examination of other highly ranked features for each model identified clinically irrelevant features that do not align with formal clinical classifications for a severe chest injury. The EN model had the most extraneous features for severe chest injury, including CUIs such as “C0578736—Inguinal Lymphadenopathy” or “C0029396—Heterotopic ossification.” Overall, the XGB and CNN had more clinically relevant features selected and better face validity.
Figure 2.
Top global model explanations from ML models to predict severe chest injury. X-axis represents rescaled variable importance from (1) EN beta coefficients, (2) XGB permuted feature importance, and (3) CNN training dataset averaged LIME explanations. Y-axis represents the preferred text definition of CUIs; CUI codes omitted here for clarity. CNN LIME interpretation median r2 = 0.69 (IQR 0.46–0.93).
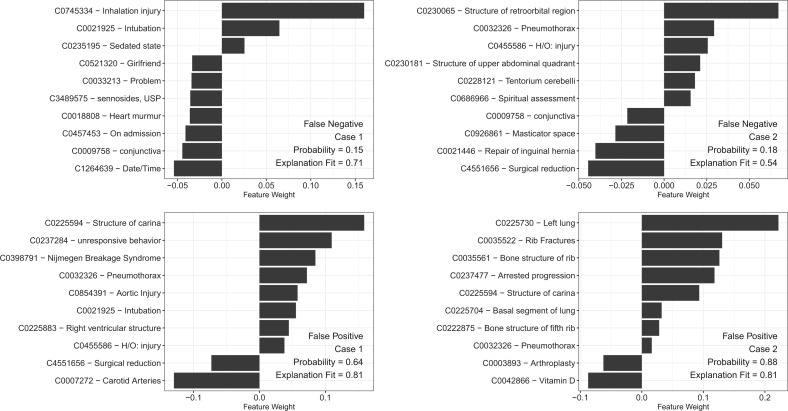
Error analysis
To assess the shortcomings of our model we generated LIME explanations for the false negative and false positive cases predicted by our CNN model. Two representative examples of each type are depicted in Figure 3A–D. In Figure 3A, the patient was a 27-year-old male with a grade II inhalation injury and 6% partial thickness burns to the face and upper extremities. The model identified inhalation injury and mechanical ventilation as important predictors, but a lack of anatomical injury and lower prevalence of severe chest injuries from our burn unit (5.4%) likely contributed to a false negative label. Figure 3B depicts the case of a 70-year-old male patient who presented as outside hospital transfer after fall from standing and was found to lack a complete workup at the outside facility, undergoing a CT chest on hospital day 4. He subsequently was found to have a pneumothorax, but the imaging report fell outside of the 8-hour time frame that the model was trained on and thus was labeled as negative.
Figure 3.
Local model interpretations generated by LIME explainer for CNN model for false negative cases (A, B) and false positive cases (C, D). X-axis represents feature weight for top ten concept unique identifiers for each case. Probability represents probability of positive case; explanation fit represents r2 of LIME classifier for the selected case.
For the false positive cases, such as Figure 3C and D, we find that negation prevents the correct labeling of the case. In the first case, the patient is a 75-year-old female found unresponsive and intubated in the trauma bay; this patient was then found to have no chest injury, with the radiology report reading “no evidence for acute aortic or solid organ injury or fracture.” The second case features a patient presenting as an outside hospital transfer with only a single fracture of the fifth rib, but a similar interpretation in the radiology read prevented accurate representation of this patient’s injury pattern. In both situations, the misinterpretation of negated terms in the radiology report led to a false positive label by the classifier, which did not accurately identify the negation.
DISCUSSION
In this study, we developed ML classifiers for prediction of severity of chest injury using only clinical documents from the EHR. We found that the best discrimination, as measured by AUROC, was achieved by the EN and CNN models. The examination of classification plots for these two models showed the CNN had an overall higher true positive rate across thresholds. Discordant predictions between the three models revealed that the CNN classifier did better at minimizing false positives and false negatives. Global feature importance demonstrated a more balanced selection of clinically relevant CUIs in the CNN model than for either EN or XGB. The CNN’s performance largely derives from routinely collected chest radiology reports with small loss in performance when other clinical documents were excluded in sensitivity analysis. Examination of local explanations for false negative and false-positive results shed light on the shortcomings of the model with respect to subpopulations and negation extraction.
Predictive validity metrics demonstrated that the EN and CNN models had the best balance of discrimination and calibration. Both classifiers had similar AUROC as well as slope-intercept values for calibration. Classification plots were more evident in displaying better performance across regions of the discrimination curves for the CNN classifier over the other models. The CNN model had more accurate predictions in the borderline cases with fewer false negative cases and more true-positive cases at higher thresholds than the other models, suggesting that it better handled the distribution and weight of CUI features in class assignment.
In examining model interpretability, the EN and CNN models showed stark differences for clinical face validity, possibly due to the differing CUI inputs and the use of an embedding approach for the CNN model.12,26 While all three models identified anatomical injuries as important to prediction, the EN and, to a lesser extent, the XGB models had a considerable amount of noise in their top-ranked features. We used binary one-hot encoded CUI data to train both the EN and XGB classifiers which simplified the value of textual data. While it is possible that term frequency-inverse document frequency or similar weighting might have enhanced model interpretability, our previous work demonstrated no difference in classification as compared with binary CUIs.10 Examination of the top CNN features indicated a more balanced ranking of global features to develop an accurate prediction. The CUI embedding for the CNN has the ability to account for repeated mentions and identify similar CUIs that may have translated into a more cohesive ranking of top features.27 The embeddings may have better represented the temporal manner of a trauma encounter, with relevant information being repeated and carried forward in documentation.27
The averaged LIME explanations used for global feature importance further supported CNN as the optimal model. To our knowledge, this study represents the first use of averaging local LIME explanations to provide a global explanation for a neural network with CUI embeddings. By acquiring a good average explanation fit across local LIME explanations, we inferred that the top global features utilized by the CNN were uniformly predictive of severe chest injury.28
Examining the local explanations for the cases that were incorrectly predicted by the CNN reveals patterns about the issues limiting prediction accuracy. We found that subpopulations and unusual presentations of severe chest injury, as found in patients presenting after burns, or logistical issues, such as discovery of injury after the 8-hour time point, were significant enough to cause false-negative results. Conversely, our false-positive LIME explanations suffered due to challenges with negation in text. cTAKES contains a rule-based negation module that is known to have issues with complex patterns of negation, which.is well described by other authors.29,30 In our examples, the model failed to identify language about the nonpresence of injury despite employing the negation features of cTAKES. The use of LIME explanations for global level explanations and local error analysis may help target areas for classifier improvement to gain the trust of trauma registrars and administrators seeking to implement these tools for quality improvement programs and reporting in their health systems.
The CNN classifier is an initial step toward an automated trauma registry for quality control, internal evaluation, and reporting to state and federal entities, though the model requires further refinement prior to implementation, given the need for highly accurate registry data for quality reporting and research. A brief survey of our institution’s trauma registrar revealed that manual chart reviews range between one and 2 hours; therefore, NLP algorithms can save substantial time and effort for documentation. To our knowledge, this is the first work that focuses on automating the coding of injury severity using methods in NLP. Prior work in this domain has largely focused on NLP methods for patient identification, modeling using structured patient data, or conversion of patient information to billing diagnosis codes for summative reports.31–33 Several authors in other clinical domains have noted the importance of using NLP methods to capture information sequestered in clinical text.34 NLP and supervised ML methods have previously been used to build clinical registries in oncology and neurology, though the target tasks were narrow in focus and domain specific.35–37 These studies focused largely on mining of data and less on interpretability. Furthermore, the use of CUIs as coded structured data from the free text allows for portability of classifiers by sharing the CUI vocabulary of trained models, enabling centers to aggregate data without leakage of PHI.38 The multiple facets described have broad implications for building accurate and interpretable ML models to populate complex data fields within clinical registries to identify practice gaps and inform improvements in patient care.
Several limitations are present in our study. As a single-center study, our results are biased toward the prevalence and pattern of trauma seen at our level I trauma center. Our CNN classifier requires external validation. This validation would need to be performed in multiple stages, both at trauma centers with registrars performing manual coding as well as at centers without trauma centers, the latter being necessary to determine the generalizability of the clinical documentation used as the source data. These data are obtained from a level I trauma center and are likely more complete and standardized to comply with rigorous reporting requirements. However, many of the top features identified in variable importance analysis were radiographic features, leading us to perform a sensitivity analysis using only chest radiology reports. Chest imaging is an essential component of the trauma survey and common to even nontrauma EDs. Sensitivity analysis showed a model with only the chest radiograph reports in the first 8 hours had minimal loss in performance compared to our full model.
For our global variable importance analysis for the CNN model, we used a LIME explainer on a local level to obtain a sense of global variable importance that is susceptible to the error of each local prediction. We attempted to use other well-described methods such as obtaining Shapley values, but the dense feature space included in analysis made development of the interactions between the features computationally infeasible.
For our analysis, we utilized clinical documents in the form of a CUI output from the cTAKES NLP engine, which may not be available at all health systems due to lack of expertise or sufficient hardware for processing large quantities of clinical documents. Lastly, the clinical documents at our center used in analyses have specific provider and regional differences in documentation that may bias our results and lead to correlations of features not necessarily directly associated with our outcome, but may have indirect clinical consequences of severe chest injury. In addition, CUIs can account for lexical variations as multiple terms map to the same CUI.
CONCLUSION
The CNN classifier demonstrated good discrimination, calibration, and interpretability for identifying cases of severe chest injury. Similar classifiers may be refined to eventually define AIS across all nine anatomical regions in an automated manner leveraging methods in ML and NLP. This study is a first step toward automating the capture and reporting of injury scores for trauma centers and emergency departments across the US.
FUNDING
Drs. Afshar, Churpek, Dligach and Kulshrestha received support for article research from the National Institutes of Health (NIH). Dr. Afshar received support from the NIH (K23 AA024503 and R01 DA051464). Dr. Churpek received funding from an R01 from National Institute of General Medical Sciences (R01 GM123193), research support from EarlySense (Tel Aviv, Israel), and he has a patent pending (ARCD. P0535US.P2) for risk stratification algorithms for hospitalized patients. Dr. Dligach is supported by the National Library of Medicine of the National Institutes of Health (R01LM012973 and R01LM010090). Dr. Kulshrestha is supported by NIH National Institute on Alcohol Abuse and Alcoholism T32 AA1352719. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The remaining authors have disclosed that they do not have any potential conflicts of interest
AUTHOR CONTRIBUTIONS
Literature search and study design: SK, DD, CJ, MMC, MA. Data collection and analysis: SK, MA. Data interpretation: SK, MA, CJ, DD, MMC, APO, RG. Writing: SK, MA, MMC, CJ, DD, AS, JMG, JG, JMK. Critical Revision: APO, AS, JMK, JG, JMG, RG.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank the Loyola University Medical Center trauma registrars Holly Molloy and Kathleen Dodaro for their invaluable help in preparing the source data that supported this manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at JAMIA Open online.
DATA AVAILABILITY STATEMENT
The data underlying this article cannot be shared publicly to protect the privacy of the patients included in this study. The data will be shared on reasonable request to the corresponding author. A representative code script in the R programming language has made available online: doi:10.5061/dryad.1c59zw3tw.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. National Center for Injury P, Control. Web-based Injury Statistics Query and Reporting System (WISQARS): Centers for Disease Control and Prevention, 2005.
- 2. DiMaggio C, Ayoung-Chee P, Shinseki M, et al. Traumatic injury in the United States: In-patient epidemiology 2000-2011. Injury 2016; 47 (7): 1393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Trauma A. Resources for Optimal Care of the Injured Patient. 6th ed. Chicago, IL: American College of Surgeons, 2014. [Google Scholar]
- 4. MacKenzie EJ, Hoyt DB, Sacra JC, et al. National inventory of hospital trauma centers. JAMA 2003; 289 (12): 1515–22. [DOI] [PubMed] [Google Scholar]
- 5. Ciesla DJ, Pracht EE, Cha JY, Langland-Orban B.. Geographic distribution of severely injured patients: implications for trauma system development. J Trauma Acute Care Surg 2012; 73 (3): 618–24. [DOI] [PubMed] [Google Scholar]
- 6. Hsia RY, Wang E, Torres H, Saynina O, Wise PH.. Disparities in trauma center access despite increasing utilization: data from California, 1999 to 2006. J Trauma 2010; 68 (1): 217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ford E, Carroll JA, Smith HE, Scott D, Cassell JA.. Extracting information from the text of electronic medical records to improve case detection: a systematic review. J Am Med Inform Assoc 2016; 23 (5): 1007–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kreimeyer K, Foster M, Pandey A, et al. Natural language processing systems for capturing and standardizing unstructured clinical information: a systematic review. J Biomed Inform 2017; 73: 14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Copes WS, Champion HR, Sacco WJ, et al. Progress in characterizing anatomic injury. J Trauma 1990; 30 (10): 1200–7. [DOI] [PubMed] [Google Scholar]
- 10. Kulshrestha S, Dligach D, Joyce C, et al. Prediction of severe chest injury using natural language processing from the electronic health record. Injury 2021; 52 (2): 205–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010; 17 (5): 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Si Y, Wang J, Xu H, Roberts K.. Enhancing clinical concept extraction with contextual embeddings. J Am Med Inform Assoc 2019; 26 (11): 1297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. DeLong ER, DeLong DM, Clarke-Pearson DL.. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988; 44 (3): 837–45 [PubMed] [Google Scholar]
- 14. Verbakel JY, Steyerberg EW, Uno H, et al. ROC plots showed no added value above the AUC when evaluating the performance of clinical prediction models. J Clin Epidemiol 2020; 126: 207–16. [DOI] [PubMed] [Google Scholar]
- 15. Huang Y, Li W, Macheret F, Gabriel RA, Ohno-Machado L.. A tutorial on calibration measurements and calibration models for clinical prediction models. J Am Med Inform Assoc 2020; 27 (4): 621–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining: Association for Computing Machinery, 2016:785–94.
- 17. Kuhn M. Building Predictive Models in R Using the caret Package. J Stat Soft 2008; 28 (5): 1–26. [Google Scholar]
- 18. Hastie T, Tibshirani R, Friedman J.. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2nd ed. New York, NY: Springer Science & Business Media, 2009. [Google Scholar]
- 19. Ribeiro MT, Singh S, Guestrin C. Why Should I Trust You?: Explaining the Predictions of Any Classifier. arXiv [cs.LG] 2016
- 20.lime: Local Interpretable Model-Agnostic Explanations [program]. 0.5.1 version, 2019.
- 21. RStudio: Integrated Development for R [program]. 1.2.1335 version. Boston, MA, 2019.
- 22.R: A Language and Environment for Statistical Computing [program]. 3.6.1 version. Vienna, Austria: R Foundation for Statistical Computing, 2019.
- 23. Elixhauser A, Steiner C, Harris DR, Coffey RM.. Comorbidity measures for use with administrative data. Med Care 1998; 36 (1): 8–27. [DOI] [PubMed] [Google Scholar]
- 24. Moore EE, Cogbill TH, Jurkovich GJ, et al. Organ injury scaling. III: chest wall, abdominal vascular, ureter, bladder, and urethra. J Trauma 1992; 33 (3): 337–9. [PubMed] [Google Scholar]
- 25. Moore EE, Malangoni MA, Cogbill TH, et al. Organ injury scaling. IV: thoracic vascular, lung, cardiac, and diaphragm. J Trauma 1994; 36 (3): 299–300. [PubMed] [Google Scholar]
- 26. Topaz M, Murga L, Gaddis KM, et al. Mining fall-related information in clinical notes: Comparison of rule-based and novel word embedding-based machine learning approaches. J Biomed Inform 2019; 90: 103. [DOI] [PubMed] [Google Scholar]
- 27. Beam AL, Kompa B, Schmaltz A, et al. Clinical Concept Embeddings Learned from Massive Sources of Multimodal Medical Data. arXiv [cs.CL] 2018 [PMC free article] [PubMed]
- 28. Wang F, Kaushal R, Khullar D.. Should health care demand interpretable artificial intelligence or accept Black Box. Ann Intern Med 2020; 172 (1): 59–60. [DOI] [PubMed] [Google Scholar]
- 29. Sohn S, Wu S, Chute CG.. Dependency parser-based negation detection in clinical narratives. AMIA Summits Transl Sci Proc 2012; 2012: 1– 8. [PMC free article] [PubMed] [Google Scholar]
- 30. Wu S, Miller T, Masanz J, et al. Negation’s not solved: generalizability versus optimizability in clinical natural language processing. PLoS ONE 2014; 9 (11): e112774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Day S, Christensen LM, Dalto J, Haug P.. Identification of trauma patients at a level 1 trauma center utilizing natural language processing. J Trauma Nurs 2007; 14 (2): 79–83. [DOI] [PubMed] [Google Scholar]
- 32. Riddick L, Long WB, Copes WS, Dove DM, Sacco WJ.. Automated coding of injuries from autopsy reports. Am J Forensic Med Pathol. 1998; 19 (3): 269–74. [DOI] [PubMed] [Google Scholar]
- 33. Hagiwara S, Oshima K, Murata M, et al. Model for predicting the injury severity score. Acute Med Surg 2015; 2 (3): 158–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Friedman C, Rindflesch TC, Corn M.. Natural language processing: state of the art and prospects for significant progress, a workshop sponsored by the National Library of Medicine. J Biomed Inform 2013; 46 (5): 765–73. [DOI] [PubMed] [Google Scholar]
- 35. Senders JT, Cho LD, Calvachi P, et al. Automating clinical chart review: an open-source natural language processing pipeline developed on free-text radiology reports from patients with glioblastoma. JCO Clin Cancer Inform 2020; 4 (4): 25–34. [DOI] [PubMed] [Google Scholar]
- 36. Garg R, Oh E, Naidech A, Kording K, Prabhakaran S.. Automating ischemic stroke subtype classification using machine learning and natural language processing. J Stroke Cerebrovasc Dis 2019; 28 (7): 2045–51. [DOI] [PubMed] [Google Scholar]
- 37. Al-Haddad MA, Friedlin J, Kesterson J, Waters JA, Aguilar-Saavedra JR, Schmidt CM.. Natural language processing for the development of a clinical registry: a validation study in intraductal papillary mucinous neoplasms. HPB 2010; 12 (10): 688–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sheller MJ, Edwards B, Reina GA, et al. Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Sci Rep 2020; 10 (1): 12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article cannot be shared publicly to protect the privacy of the patients included in this study. The data will be shared on reasonable request to the corresponding author. A representative code script in the R programming language has made available online: doi:10.5061/dryad.1c59zw3tw.