Abstract
The deubiquitinating enzyme (DUB)–mediated cleavage of ubiquitin plays a critical role in balancing protein synthesis and degradation. Ubiquitin-specific protease 4 (USP4), a member of the largest subfamily of cysteine protease DUBs, removes monoubiquitinated and polyubiquitinated chains from its target proteins. USP4 contains a DUSP (domain in USP)–UBL (ubiquitin-like) domain and a UBL-insert catalytic domain, sharing a common domain organization with its paralogs USP11 and USP15. USP4 plays a critical role in multiple cellular and biological processes and is tightly regulated under normal physiological conditions. When its expression or activity is aberrant, USP4 is implicated in the progression of a wide range of pathologies, especially cancers. In this review, we comprehensively summarize the current knowledge of USP4 structure, biological functions, pathological roles, and cellular regulation, highlighting the importance of exploring effective therapeutic interventions to target USP4.
Keywords: USP4, deubiquitinase, NF-κB, cancer, inflammation
Introduction
Normal cellular homeostasis depends on the balance of protein synthesis and degradation. In eukaryotes, protein degradation is mainly carried out by the ubiquitin–proteasome system (UPS). Through a cascade between the ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3), the substrate proteins are decorated with ubiquitin and transferred to the 26S proteasome where they undergo degradation (Hershko et al., 1983; Hershko and Ciechanover, 1998). Ubiquitin is a 76-amino-acid protein and contains seven lysine (Lys) residues (K6, K11, K27, K29, K33, K48, and K63). These different Lys residues serve as substrates to form ubiquitin chains with distinct structures and functions. Therefore, in addition to degradation, ubiquitin modifications can also direct proteasome-independent fates, such as alteration of protein subcellular localization and activity, or involvement in histone function, DNA repair and replication, signal transduction, and other cellular processes (Hicke, 2001; Pickart and Fushman, 2004; Kulathu and Komander, 2012).
Like many other posttranslational modifications (PTMs), ubiquitination is a dynamic and reversible process. Deubiquitinating enzymes (DUBs) antagonize ubiquitination by removing ubiquitin moieties from substrate proteins (Amerik and Hochstrasser, 2004). Approximately 100 DUBs have been identified in humans, which are classified into six families consisting of the ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian-tumor proteases (OTUs), Machado–Joseph disease protein domain proteases (MJDs), JAMM/MPN domain-associated metallopeptidases (JAMMs) (Amerik and Hochstrasser, 2004), and monocyte chemotactic protein-induced proteases (MCPIPs) (Liang et al., 2010). Among them, the USP family is the largest, containing almost 60 members with each displaying diverse substrate specificity, modification preference, and regulatory mechanism.
USP4, also known as ubiquitous nuclear protein (UNP), is one of the best-studied USPs. USP4 was initially discovered, cloned, and characterized by Gupta et al. (1993) as a nuclear oncoprotein related to tre-2/tre-17/USP6. Later, it was found to contain the USP cysteine (Cys) and histidine (His) boxes and deubiquitinating activity and then proved to be USPs (Gray et al., 1995; Gilchrist et al., 1997; Frederick et al., 1998; Gilchrist and Baker, 2000). Tumor suppressor pRb is the first characterized substrate of USP4 (Blanchette et al., 2001; DeSalle et al., 2001), and later studies revealed many others. USP4 is now known to deubiquitinate many well-known target proteins associated with key processes involved in both cellular homeostasis and disease, especially cancer where USP4 is frequently dysregulated (Wang Y. et al., 2020).
This review comprehensively describes the current knowledge concerning all aspects of USP4. First, we provide an introduction to the USP4 gene and its structure, including domain architecture, catalytic activity, and substrate-binding sites. Second, we discuss the biological functions of USP4 in normal states and its pathological roles in diseases including a summary of its protein substrates. Third, we focus on the many different mechanisms involved in the cellular regulation of USP4. Finally, the developmental status of pharmacological interventions against USP4 is described. Altogether, this analysis provides fundamental insights into the potential of USP4 as a therapeutic target in multiple disease states.
Chromosomal Location, Isoforms, and Subcellular Localization of USP4
The human USP4 gene is located on chromosome band 3p21.3 and comprised 22 exons (Gray et al., 1995). The gene encodes two dominant protein isoforms designated long and short that possess 963 and 916 amino acids, respectively (Frederick et al., 1998). The mRNA of USP4 is ubiquitously expressed in most normal tissues and cell lines at constant levels except in skeletal muscle, heart, pancreas, and testis where higher mRNA levels are detected, and in embryonic stem cells where expression is considerably lower (Gupta et al., 1994; Frederick et al., 1998). However, the subcellular localization of the USP4 protein often differs (Di Fruscio et al., 1998; Frederick et al., 1998; Soboleva et al., 2005). For example, USP4 is only expressed in the cytoplasm of HepG2 [human hepatocellular carcinoma (HCC)] cells, while being exclusively nuclear in Saos-2 (human osteosarcoma) cells (Soboleva et al., 2005). This discrepancy is partially due to nuclear–cytoplasmic shuttling where USP4 possesses both nuclear export signal and nuclear localization signal (NLS) motifs (Soboleva et al., 2005). Other mechanisms involving phosphorylation and associations with some regulators are also involved in modulating USP4 subcellular localization (Song et al., 2010; Zhang et al., 2012). Moreover, two isoforms of USP4 also have distinct subcellular distributions. While the short isoform is distributed throughout the cell, the long isoform tends to be cytoplasmic, but the regulatory mechanism remains unknown (Frederick et al., 1998). It is noteworthy that the expression ratio of the two isoforms seems to be pathological relevant as it was found to be altered explicitly in Paget disease of the bone (Klinck et al., 2014).
Structure of USP4
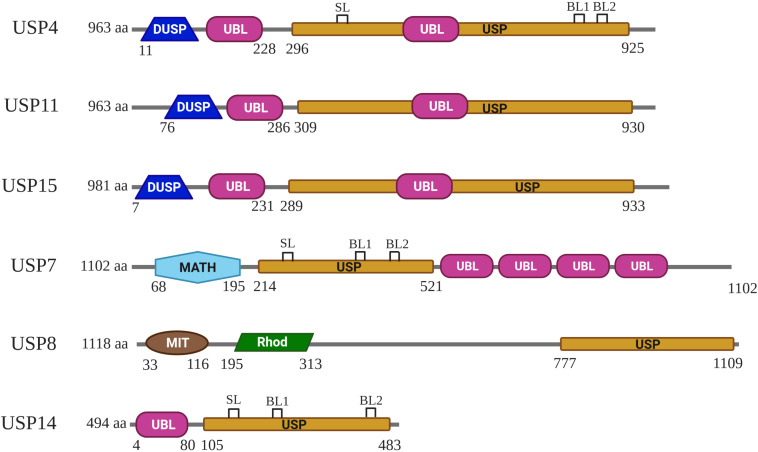
Like all other USPs, USP4 is a multiple domain protein. USP4 is identified to be a paralog of USP15 and USP11, sharing a common domain organization, which consists of a DUSP (domain in USP), two UBL (ubiquitin-like), and a bi-part catalytic domain (also known as USP domain) (Angelats et al., 2003; Komander et al., 2009; Elliott et al., 2011; Vlasschaert et al., 2015) (Figure 1). UBL1 is located in USP4 N-terminal, whereas UBL2 is embedded in the C-terminal catalytic domain as a part of the insert (Ye et al., 2009; Clerici et al., 2014) (Figure 2A). Both UBL1 and UBL2 are crucial for the catalytic activity of USP4.
FIGURE 1.
Schematic representation of the domain composition for USP4, USP11, USP15, USP7, USP8, and USP14. DUSP, UBL, USP, MATH, MIT, and Rhod domains are represented in different colors. The domain architecture of these USPs shown is based on Komander et al. (2009). SL (switching loop), BL1 (blocking loops 1), and BL2 (blocking loops 2) of USP4, USP7, and USP14 shown are based on Clerici et al. (2014), and Wertz and Murray (2019).
FIGURE 2.
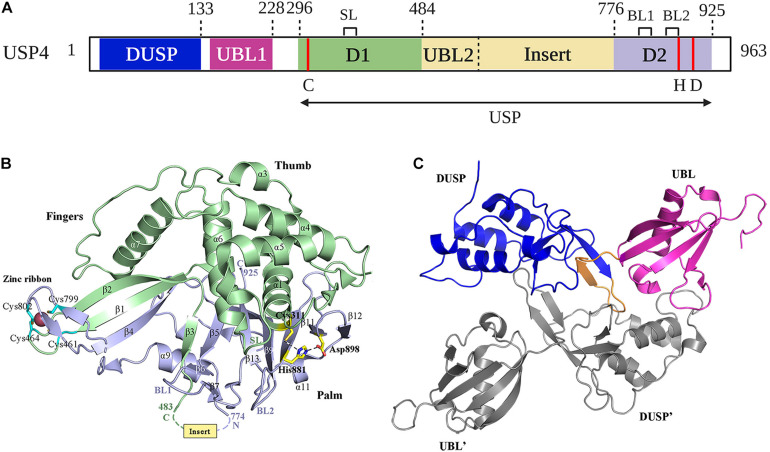
Structure of USP4. (A) Schematic illustration of the domain organization of USP4 in detail. The position of the catalytic residues (C, H, D) is indicated in red. SL spans residues 385–392; BL1 and BL2 span residues 831–834 and 874–880, respectively. Cartoon representation of (B) USP4-D1D2 domain crystal structure and (C) USP4 DUSP-UBL dimer structure are colored as in (A). The catalytic triad and zinc-coordinating residues are represented as sticks. Dotted lines indicate the position of the USP4 insert.
C-Terminal Catalytic Domain
The catalytic core of USP4 is a bi-part domain (residues 296–925) located in C-terminal, which contains the domain 1 (D1), the domain 2 (D2), and a UBL-containing insert (Clerici et al., 2014; Vlasschaert et al., 2015) (Figure 2A). The crystal structure of USP4-D1D2 shows six molecules of USP4-D1D2 per asymmetric unit with equivalent conformation, while the insert, located between beta-strand β3 and helix α9, has an asymmetric distribution (Clerici et al., 2014). USP4–D1D2 adopts the characteristic fold seen for all USPs, which resembles an extended right hand that comprised three domains: fingers, thumb, and palm (Hu et al., 2002) (Figure 2B). The D1 fragment contains the thumb and part of the fingers, including the Cys box (residues 303–320) and QQD box (residues 390–403) of the active site, whereas the D2 fragment completes the active site with the His box (residues 864–885, 894–903, and 915–922) and the rest of the fingers and the palm (Quesada et al., 2004). Like other USPs with the exceptions of USP7 (Hu et al., 2002) and USP15 (Ward et al., 2018), the catalytic triad of USP4 is in a catalytically competent configuration (Hu et al., 2005; Avvakumov et al., 2006; Renatus et al., 2006; Komander et al., 2008; Samara et al., 2010).
USP4 and other Cys proteases have a catalytic mechanism like that of the well-studied plant enzyme papains (Storer and Ménard, 1994). First, the His residue deprotonates the thiol group of the Cys residue. Second, the Cys residue launches a nucleophilic attack on the isopeptide bond, releasing the ε-amine of the target Lys residue and producing a covalent acylenzyme intermediate with ubiquitin. Lastly, with the help of water molecules, USP4 undergoes diacylation. Consequently, free ubiquitin is released. The aspartic acid (Asp) residue is required to polarize the His residue to stabilize the enzyme’s catalytic activity (Komander et al., 2009). Several structures have a role in USP4 activation. In the fingers, the Zn2+ ion brings together the D1 and D2, tetrahedrally coordinated by Cys on anti–parallel β-strands β1 and β2 in D1, and β4 in D2 (Clerici et al., 2014) (Figure 2B). This zinc-finger ribbon seems to be in the contracted “closed-hand” configuration seen in USP8 that blocks ubiquitin access (Avvakumov et al., 2006) (Figure 1). A similar role was assigned to three ubiquitin-binding surface loops (BLs) observed in USP4, of which the BL1 and BL2 were reported in USP14, where they block the active site and relocate after ubiquitin binding, and the third loop corresponding to the SL plays a role in USP7 activation (Hu et al., 2002; Faesen et al., 2011) (Figures 1, 2A,B). Superposition of the six non-crystallographic symmetry-related molecules of USP4–D1D2 shows that both the zinc-finger ribbon and the three BLs have flexibility, which is required for the dynamic exchange of ubiquitin in and out of the active site (Clerici et al., 2014).
N-Terminal DUSP-UBL Domain
The N-terminal DUSP-UBL domain spans residues 1–228, separated from the catalytic domain by a connecting linker (residues 229–295) (Clerici et al., 2014) (Figure 2A). DUSP-UBL domain exists not only in USP4 but also in USP15 and USP11 (Elliott et al., 2011) (Figure 1). The crystal structure of USP4 DUSP-UBL shows a dimer formation where the subunits are packed against each other in an antiparallel orientation (Harper et al., 2011) (Figure 2C). This packing creates an extensive interface formed through the DUSP domains, but not UBL domains from the different subunits, suggesting that the N-terminal UBL domain has an independent motion (Elliott et al., 2011).
The primary function of the DUSP-UBL domain is to activate USP4 by promoting catalytic domain turnover. A study conducted by Clerici et al. (2014) revealed that the DUSP-UBL domain could not only improve the ubiquitin off-rates but also promote faster on-rates through directly binding to the insert. In addition, deletion of the DUSP, the insert, or the linker region that separates the DUSP-UBL to the catalytic domain from USP4 showed that losing only one of these three domains can reduce the catalytic activity of USP4. Thus, the DUSP-UBL, the linker, and the inserted domain are essential for USP4 activation. Two isoforms of USP4 arise from the inclusion or skipping of exon 7 (E7), which is serine-rich with several phosphorylation sites and forms the major part of the flexible linker region (Vlasschaert et al., 2016). Given that there are no differences in USP4 deubiquitinating activity between long and short forms, the part of the linker region formed by E7 may not be crucial for USP4 activation because the short linker retains sufficient flexibility to enable comparable domain interactions (Clerici et al., 2014; Vlasschaert et al., 2016).
Substrate-Binding Domains
All USP4 subdomains have been found to play a role in substrate recognition (Table 1). The majority of substrates bind to the C-terminal catalytic domain, and few interact with the N-terminal DUSP-UBL domain. In addition, the binding sites may contain part of two subdomains or even three or more subdomains. For example, histone deacetylase 2 (HDAC2) associates with amino acids 188–302 of USP4 (Li Z. et al., 2016), ARF-binding protein 1 (ARF-BP1) interacts with residue 188 to C-terminus of the enzyme (Zhang et al., 2011), and RNA-binding protein with serine-rich domain 1 (RNPS1) binds to the full-length USP4 composed of DUSP, UBL1, UBL2, and UCH domains (Kwon et al., 2017).
TABLE 1.
Substrate-binding domains in USP4.
| Binding domains | Substrates | References | Binding domains | Substrates | References |
| The catalytic (USP) domain | TβRI | Zhang et al., 2012 | DUSP domain | RIG-I | Wang et al., 2013 |
| CtIP | Liu et al., 2015; Wijnhoven et al., 2015 | Rheb | Deng et al., 2019 | ||
| RORγt | Yang et al., 2015 | UBL1 domain | S9 | Zhao et al., 2012 | |
| IRF8 | Lin et al., 2017 | DUSP-UBL domain | BRAP | Hayes et al., 2012 | |
| β-Catenin | Yun et al., 2015 | Amino acids 188–302 | HDAC2 | Li Z. et al., 2016 | |
| TAK1 | Fan et al., 2011 | Residue 188 to C-terminus | ARF-BP1 | Zhang et al., 2011 | |
| TRAF2 | Xiao et al., 2012 | Amino acids 613–923 | TAK1 | Zhao et al., 2018 | |
| TRAF6 | Xiao et al., 2012; Xu et al., 2018 | DUSP, UBL1, UBL2, and UCH domains | RNPS1 | Kwon et al., 2017 | |
| RIP1 | Hou et al., 2013 |
Cellular and Biological Function of USP4
USP4 functions to remove monoubiquitination and polyubiquitination, including K48 and K63 conjugated ubiquitin chains, from target proteins (Zhao et al., 2009; Zhou et al., 2012). Given that numerous substrates are influenced by USP4, USP4 has been found to play an indispensable role in a wide range of cellular and biological processes, as discussed below and summarized in Table 2.
TABLE 2.
Substrates of USP4 in cellular and biological processes.
| Substrates | Type of ubiquitination removed by USP4 | References | Substrates | Type of ubiquitination removed by USP4 | References |
| Pre-mRNA splicing | Wnt/β-catenin signaling pathway | ||||
| PRP3 | K63-linked ubiquitin chains | Song et al., 2010 | β-Catenin | K48-linked ubiquitin chains | Yun et al., 2015 |
| RNP1 | K63-linked ubiquitin chains | Kwon et al., 2017 | NLK | None reported | Zhao et al., 2009 |
| DNA repair | TCF4 | None reported | Zhao et al., 2009 | ||
| CtIP | Non-deubiquitination | Liu et al., 2015; Wijnhoven et al., 2015 | NF-κB signaling pathway | ||
| Cell growth and differentiation | TRAF2 | K63-linked ubiquitin chains | Xiao et al., 2012 | ||
| Dvl | K63-linked ubiquitin chains | Zhou et al., 2016 | TRAF6 | K48- and K63-linked ubiquitin chains | Xiao et al., 2012; Zhou et al., 2012; Xu et al., 2018 |
| MyoD | None reported | Yun and Kim, 2017 | TAK1 | K63-linked ubiquitin chains | Fan et al., 2011; Liang et al., 2013 |
| SMAD4 | Monoubiquitination | Zhou et al., 2017 | RIP1 | K63-linked ubiquitin chains | Hou et al., 2013 |
| Immune response | RIG-I | K48-linked ubiquitin chains | Wang et al., 2013 | ||
| RORγt | K48-linked ubiquitin chains | Yang et al., 2015 | Hyaluronan synthesis | ||
| IRF4 | K48- and K63-linked ubiquitin chains | Guo et al., 2017 | HAS2 | Monoubiquitination | Mehić et al., 2017 |
| IRF8 | K48-linked ubiquitin chains | Lin et al., 2017 | Other biological functions | ||
| P53 signaling pathway | AQP2 | None reported | Murali et al., 2019 | ||
| HDAC2 | None reported | Li Z. et al., 2016 | A2a receptor | None reported | Milojevic et al., 2006 |
| ARF-BP1 | None reported | Zhang et al., 2011 | BRAP | None reported | Uras et al., 2012 |
| TGF-β signaling pathway | PDK1 | Monoubiquitination | Hayes et al., 2012 | ||
| TβRI | None reported | Zhang et al., 2012 | S9 | Non-deubiquitination | Zhao et al., 2012 |
Pre-mRNA Splicing
The activity and composition of the spliceosome, which is critical for pre-mRNA splicing, are regulated by reversible ubiquitination (Nottrott et al., 2002; Jurica and Moore, 2003; Maeder and Guthrie, 2008). Song et al. (2010) showed that USP4 could deubiquitinate K63-linked polyubiquitin chains of pre-mRNA-processing factor 3 (PRP3), a component of the U4 snRNP, to promote ejection of U4 proteins from the spliceosome, thereby ensuring the correct pre-mRNA splicing. Moreover, loss of USP4 disturbs efficient pre-mRNA splicing of several cell cycle regulators, such as α-tubulin and budding uninhibited by benzimidazoles 1(BUB1), resulting in the impairment of mitotic progression (Song et al., 2010). USP4 was also reported to deubiquitinate RNPS1, which aids in selecting splicing sites and activating the pre-mRNA splicing process (Kwon et al., 2017).
DNA Repair
USP4 has been shown to participate in DNA double-strand break resection and homologous recombination (HR) repair upon DNA damage. By directly interacting with CtIP, a DNA-end resection factor controlling the DNA damage-induced G2/M checkpoint, USP4 promotes CtIP foci formation at DNA damage sites. Although USP4 does not directly deubiquitinate CtIP, its interaction with CtIP is essential for HR as blocking their binding reduces HR efficiency (Liu et al., 2015; Wijnhoven et al., 2015).
Cell Growth and Differentiation
Anckar and Bonni (2015) initially reported that USP4 had opposing effects on cell morphogenesis in the mammalian cerebellum where it promoted axon growth in granule neurons but concomitantly restricted the growth and elaboration of dendrites. Later studies confirmed roles for USP4 in cell growth and differentiation (Zhou et al., 2016, 2017; Yun and Kim, 2017). USP4 suppresses Wnt3a-induced osteoblast differentiation and mineralization by cleaving K63-linked polyubiquitin chains from disheveled (Dvl) and inhibiting the accumulation of β-catenin and activation of its downstream cascades (Zhou et al., 2016). Through downregulating myoblast determination protein (MyoD) activity, USP4 also exerts an antagonistic function in myoblast differentiation (Yun and Kim, 2017). Recently, SMAD4, an intracellular effector for transforming growth factor β (TGF-β) family cytokines, was shown to be a novel substrate of USP4. The binding of USP4 to SMAD4 prevents its monoubiquitination in the cytoplasm and subsequently enhances SMAD4-mediated activin and bone morphogenetic protein (BMP) pathways. In mice, this leads to the suppression of embryonic stem cell differentiation to sustain their self-renewal potential (Zhou et al., 2017), whereas, in early zebrafish embryos, this process can maintain morphogenic events (Tse et al., 2009; Zhou et al., 2017). However, a single knockout of either USP4 or USP15 does not impair viability and normal development in mice, whereas a double knockout of USP4 and USP15 leads to embryonic lethality (Vlasschaert et al., 2015). These results suggest for embryonic development that USP4 and USP15 are functionally redundant.
Immune Response
Conflicting functions have been reported where USP4 can act as an enhancer or an inhibitor of immune responses. For example, by stabilizing retinoic acid receptor–related orphan receptor γt (RORγt), USP4 facilitates interleukin-17A (IL-17A) transcription and T helper type 17 (TH17) cell differentiation (Yang et al., 2015). USP4 also promotes the development of TH2 cells and upregulation of TH2-related cytokines through physically interacting with and deubiquitinating interferon regulatory factor 4 (IRF4) (Guo et al., 2017). In contrast to these activation responses, IRF8, another immune system-restricted IRF required for T-helper cell differentiation, has been recognized as a substrate of USP4. Deconjugation of polyubiquitin chains from IRF8 mediated by USP4 is vital for maintaining the function of regulatory T cells, which negatively regulate many different immune reactions (Lin et al., 2017).
P53, TGF-β, Wnt/β-Catenin, and Nuclear Factor κB Signaling Pathways
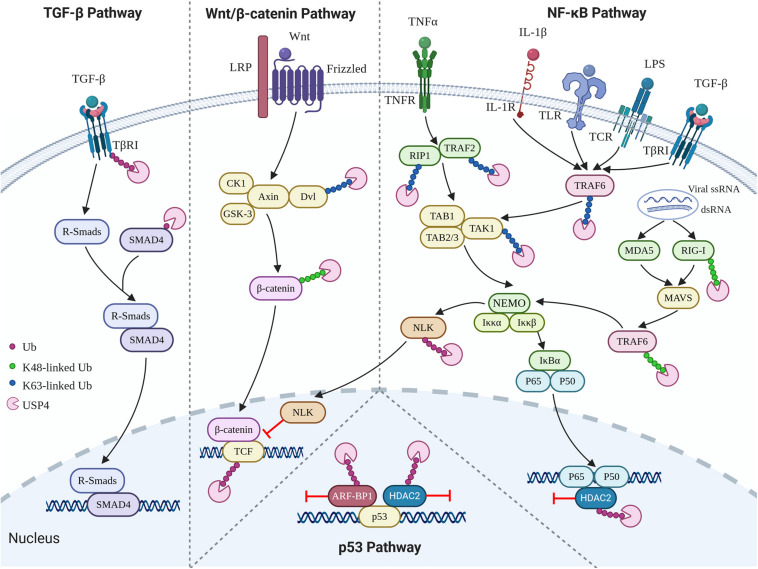
In addition to the biological contexts mentioned above, USP4 is also involved in the regulation of p53, TGF-β, Wnt/β-catenin, and nuclear factor κB (NF-κB) signaling pathways (Figure 3). USP4 inhibits p53 transcriptional activation via stabilizing HDAC2 and ARF-BP1, which acts as a deacetylase and E3 ubiquitin ligase of p53, respectively (Zhang et al., 2011; Li Z. et al., 2016). Loss of UPS4 leads to the activation of several p53-directed pathways, including upregulation of apoptosis, premature cell senescence, and reduced oncogene-associated transformation (Zhang et al., 2011). USP4 can also deubiquitinate TGF-β type I receptor (TβRI) and sustain its plasma membrane expression in a SMAD7-independent fashion, leading to hyperactivation of the TGF-β pathway (Zhang et al., 2012). The Wnt/β-catenin signaling pathway is an important pathway governing developmental, homeostatic, and pathological processes (Clevers and Nusse, 2012). USP4 was initially identified as a repressor of the Wnt/β-catenin signaling pathway, given that USP4 can mediate deubiquitination and stabilization of Dvl, a key molecule involved in the turnover of cytosolic β-catenin (Zhao et al., 2009; Zhou et al., 2016). However, later studies demonstrated that USP4 enhanced Wnt signaling through stabilizing and facilitating nuclear localization of β-catenin, especially in cancer (Yun et al., 2015; Hwang et al., 2016). Furthermore, two other Wnt signaling components, transcription factor 4 (TCF4) and Nemo-like kinase (NLK) have also been identified as substrates of USP4 (Zhao et al., 2009).
FIGURE 3.
Role of USP4 in p53, TGF-β, Wnt/β-catenin, and NF-κB signaling pathways.
NF-κB, a transcription factor involved in a broad range of cellular responses such as inflammation, immunity, and apoptosis, has been extensively reported to be negatively regulated by USP4 (Fan et al., 2011; Xiao et al., 2012; Zhou et al., 2012; Hou et al., 2013; Wang et al., 2013; Li Z. et al., 2016; Wang X. D. et al., 2020). Tumor necrosis factor receptor–associated factor 2 (TRAF2), TRAF6, and TGF-β activated kinase 1 (TAK1) are three crucial NF-κB signaling components. K63-linked polyubiquitination of TAK1 mediated by K63-linked polyubiquitination of TRAF2 or TRAF6 is an essential step in IκB phosphorylation, degradation, and further translocation of NF-κB to the nucleus and upregulation of the subsequent proinflammatory responses (Deng et al., 2000; Wu et al., 2005; Yamazaki et al., 2009; Fan et al., 2010). Several studies have indicated that USP4 acts as a deubiquitinase for TRAF2, TRAF6, and TAK1 to prevent tumor necrosis factor α (TNF-α), IL-1β-, TGF-β-, T-cell receptor (TCR)-, and lipopolysaccharide (LPS)-induced NF-κB activation (Fan et al., 2011; Xiao et al., 2012; Zhou et al., 2012; Wang X. D. et al., 2020). Besides, the suppressing role of USP4 in NF-κB signaling is also seen in its deubiquitinating activity toward receptor-interacting protein 1 (RIP1), whose K63-linked polyubiquitin chains function as scaffolds for the assembly of TAK1 kinase, and also HDAC2, which is a deacetylase associating with HDAC1 to target the p65 (RelA) subunit (Hou et al., 2013; Li Z. et al., 2016). Retinoic acid–inducible gene I–like receptors (RLRs), including retinoic acid–inducible gene I (RIG-I), are other inducible factors of the NF-κB signaling pathway (Gürtler and Bowie, 2013). Wang et al. (2013) discovered that USP4 interferes with the RIG-I-mediated activation of NF-κB. However, Xu et al. (2018) demonstrated that USP4 could positively regulate the RLR-induced NF-κB signaling pathway by removing K48-linked ubiquitin chains from TRAF6. Therefore, deubiquitinating K63- or K48-linked chains in TRAF6, respectively, may trigger anti-inflammatory or proinflammatory responses.
Other Biological Functions
USP4 is highly expressed in the kidney where it functions to regulate whole-body water homeostasis through deubiquitinating arginine vasopressin-induced membrane accumulation of aquaporin 2 (AQP2), the water channel of the kidney collecting duct (Murali et al., 2019). USP4 also influences the expression of the Gs-coupled receptor A2a adenosine receptor in cultured hippocampal neurons, acting to relax its quality control in the endoplasmic reticulum and enhancing its cell surface expression (Milojevic et al., 2006). Moreover, other studies have described BRCA1-associated protein, phosphoinositide-dependent kinase 1, and hyaluronan synthase 2 (HAS2) as substrates of USP4 (Hayes et al., 2012; Uras et al., 2012; Mehić et al., 2017). For the latter, USP4 removes monoubiquitination from HAS2, which leads to decreased hyaluronan synthesis (Mehić et al., 2017). USP4 may also take part in proteasome assembly via binding to the S9 subunit of the proteasome, although its catalytic activity is not necessary for this process (Zhao et al., 2012).
Dysregulation of USP4 in Disease
USP4 is essential for normal physiological functions, although its dysregulation can lead to different pathologies. Frederick et al. (1998) obtained the first evidence of the link between USP4 and cancer when they demonstrated the lower expression of USP4 in small cell lung cancer and squamous cell lung cancer compared with normal samples. Since then, numerous studies have documented USP4 to function as an oncoprotein or a tumor repressor in a range of different cancer types. USP4 was also reported to have impacts on many other pathological contexts, such as viral and bacterial infections, acute injury and inflammation, chronic fibrosis-related diseases, metabolic disorders, and immune disease (Table 3). Together, these findings implicate USP4 as a suitable candidate to be targeted in a wide variety of diseases.
TABLE 3.
Multifaceted role of USP4 in diseases.
| Diseases | Substrates | Substrate-related pathways | Expression of USP4 (cells or tissues) | Role of USP4 | References | |
| Cancer | Breast cancer | TβRI | TGF-β pathway | Upregulated (human cells and clinical samples) | Promoter | Zhang et al., 2012; Cao et al., 2016 |
| HDAC2 | Several cell proliferation-associated genes expression | None reported | Promoter | Wang et al., 2018 | ||
| PCD4 | None reported | Downregulated (clinical samples) | Suppressor | Li Y. et al., 2016 | ||
| Colorectal cancer | β-Catenin | Wnt/β-catenin pathway | Upregulated (clinical samples) | Promoter | Yun et al., 2015 | |
| Rheb | mTORC1 pathway | None reported | Promoter | Deng et al., 2019 | ||
| PRL-3 | AKT pathway | Upregulated (clinical samples) | Promoter | Xing et al., 2016 | ||
| Brain metastatic lung adenocarcinoma | β-Catenin | Wnt/β-catenin pathway | Upregulated (human cells) | Promoter | Hwang et al., 2016 | |
| Lung cancer | TRAF2, TRAF6 | NF-κB pathway | None reported | Suppressor | Xiao et al., 2012 | |
| Twist1 | Oct4/Sox2 expression | Upregulated (clinical samples) | Promoter | Li et al., 2020 | ||
| None reported | NF-κB pathway | Downregulated (stemness-enriched human lung cancer cells and advanced lung cancer clinical samples) | Suppressor | Lai et al., 2020 | ||
| HNSCC | RIP1 | NF-κB pathway | Upregulated (clinical samples) | Suppressor | Hou et al., 2013 | |
| GBM | TβRI | TGF-β/ERK pathway | Upregulated (clinical samples) | Promoter | Zhou Y. et al., 2019 | |
| None reported | P53 pathway | Upregulated (human cells and clinical samples) | Promoter | Qin et al., 2019 | ||
| Melanoma | None reported | P53 pathway | Upregulated (human cells and clinical samples) | Promoter | Guo et al., 2018 | |
| HCC | CypA | MAPK pathway | Upregulated (clinical samples) | Promoter | Li et al., 2018 | |
| TβRI | TGF-β pathway | Upregulated (mesenchymal-phenotype human HCC cells and clinical samples) | Promoter | Heo et al., 2014; Qiu et al., 2018 | ||
| Viral infections | VSV | RIG-I | IFN-β pathway | Downregulated (virus-infected human cells) | Suppressor | Wang et al., 2013 |
| EV71 | TRAF6 | NF-κB pathway | Downregulated (virus-infected human cells) | Suppressor | Xu et al., 2018 | |
| Acute injury and inflammation | Spinal cord injury | TRAF6 | NF-κB pathway | Downregulated (rat microglial cells) | Suppressor | Jiang et al., 2017 |
| Intracerebral hemorrhage | None reported | Caspase-3/Bax/Bcl-2 pathway | Upregulated (rat neurons adjacent to the hematoma) | Promoter | Liu et al., 2017 | |
| Hepatic (I/R) injury | TAK1 | TAK1/JNK1/2/p38 pathway | Upregulated (mouse liver tissues suffering I/R injury) | Suppressor | Zhou J. et al., 2019 | |
| Chronic fibrosis-related diseases | Cardiac hypertrophy and fibrosis | TAK1 | TAK1/JNK1/2/p38 pathway | Downregulated (clinical samples) | Suppressor | He et al., 2016 |
| Liver fibrosis and cirrhosis | TβRI | TGF-β pathway | Upregulated (mouse fibrous liver tissues) | Promoter | Wu et al., 2019; Zhu et al., 2019 | |
| Pathological scarring | TβRI | TGF-β pathway | Upregulated (clinical samples) | Promoter | Zhang J. et al., 2019; Huang et al., 2020 | |
| MMT during peritoneal dialysis | TβRI | TGF-β pathway | None reported | Promoter | Xiao et al., 2015 | |
| Metabolic disorders | NAFLD | TAK1 | TAK1/JNK/IRS/AKT/GSK3 pathway | Downregulated (clinical samples) | Suppressor | Zhao et al., 2018; Zhang Y. et al., 2019 |
| Immune disease | RHD | RORγt | IL-17A transcription | Upregulated (clinical samples) | Promoter | Yang et al., 2015 |
| IRF4 | IL-4 transcription | None reported | Promoter | Guo et al., 2017 | ||
Cancer
Examining 14 types of human cancer, Zhang et al. (2011) reported that the expression level of USP4 was upregulated in the majority of cases. A series of tumors, including breast cancer, colorectal cancer, lung cancer, brain metastatic lung adenocarcinoma, glioblastoma (GBM), melanoma, HCC, and adrenocortical carcinoma, overexpress USP4. Moreover, USP4 overexpression contributes to the initiation, progression, and/or the recurrence of tumors and is correlated with poor prognosis (Velázquez-Fernández et al., 2005; Laurell et al., 2009; Zhang et al., 2012; Yun et al., 2015; Cao et al., 2016; Hwang et al., 2016; Xing et al., 2016; Guo et al., 2018; Li et al., 2018, 2020; Qiu et al., 2018; Wang et al., 2018; Deng et al., 2019; Qin et al., 2019; Zhou Y. et al., 2019; Geng et al., 2020). In breast cancer, higher expression of USP4 is associated with distant metastasis (Geng et al., 2020). By promoting activation of the relaxin/TGF-β1/Smad2/matrix metalloproteinase 9 axis and transcriptional repression activity of HDAC2, USP4 induces breast cell invasion, migration, and proliferation both in vitro and in vivo (Cao et al., 2016; Wang et al., 2018). Phosphatase of regenerating liver-3 (PRL-3), a member of the protein tyrosine phosphatase family, plays a significant role in tumorigenesis (Liang et al., 2007; Al-Aidaroos and Zeng, 2010). Deubiquitinating PRL-3 by USP4 leads to AKT activation and E-cadherin reduction in colorectal cancer, where an elevated level of USP4 is associated with tumor size, differentiation, distant metastasis, and poor survival (Xing et al., 2016). This procancer effect is also seen in the USP4-induced deubiquitination of Rheb, which can promote mTOR complex 1–mediated cellular activities, including the control of cellular autophagy, proliferation, and cell size (Deng et al., 2019). As mentioned previously, β-catenin is a substrate of USP4, and this interaction affects the tumorigenicity of colorectal cancer, as well as brain metastasis in lung adenocarcinoma (Yun et al., 2015; Hwang et al., 2016).
Recently, USP4 was found to mediate deubiquitination of the epithelial–mesenchymal transition (EMT) transcriptional factor Twist1 to promote lung cancer cell stemness (Li et al., 2020). Zhou Y. et al. (2019) also demonstrated that inhibiting USP4 expression in GBM cells both inhibited their proliferation and induced apoptosis via antagonizing USP4-mediated stimulation of the ERK signaling pathway. Similarly, knockdown of USP4 significantly decreases cell viability in a p53-dependent manner after treating GMB cells with temozolomide (Qin et al., 2019). A comparable role was observed in melanoma where USP4 expression aggravated cisplatin chemoresistance (Guo et al., 2018). Cyclophilin A (CypA) and TβRI stabilization by USP4 occurs in HCC. Enhanced tumor growth mediated by USP4 overexpression and its correlation with several malignant phenotype characteristics provides the fundamental and clinical evidence that USP4 functions as a tumor promoter in HCC (Heo et al., 2014; Li et al., 2018; Qiu et al., 2018).
On the other hand, quite a few studies discovered that USP4 exerts suppressive functions in particular types of cancer (Xiao et al., 2012; Hou et al., 2013; Liang et al., 2013, 2019; Li Y. et al., 2016; Yao et al., 2017; Zhong et al., 2018; Lai et al., 2020). In esophageal cancer, high expression of USP4 is associated with the improved prognosis of patients presenting with small tumors or diagnosed at an early stage (Yao et al., 2017). An association between USP4 and RIP1 was reported to impair NF-κB activation and exacerbate TNF-α–induced apoptosis in head and neck squamous cell carcinoma (HNSCC) (Hou et al., 2013). Activation of NF-κB is also one of the resistance mechanisms contributing to resistance to chemotherapy and ionizing radiation during cancer treatment (Li and Sethi, 2010). Liang et al. (2013) showed that USP4 could block Dox-induced activation of NF-κB at the early stage of the treatment by preventing TAK1 from conjugating K63-linked polyubiquitin chains.
The reduced expression of USP4 in lung adenocarcinoma and lung squamous cell cancer compared with normal samples is associated with poor survival among lung cancer patients (Zhong et al., 2018). The connection between USP4 and NF-κB is identified in lung cancers where stable knockdown of USP4 augments inflammatory responses, stemness properties, chemoresistance, the expression of programmed death ligand 1, and tumor growth (Lai et al., 2020). This inhibiting effect mediated by USP4 in lung cancer is in accordance with the results from Xiao et al. (2012), while in contrast to another study reported USP4 promotes lung cancer cell stemness (Li et al., 2020). Additionally, two studies indicated that USP4 downregulation might also mediate a tumor-suppressing role in breast cancer (Li Y. et al., 2016; Liang et al., 2019). USP4 was found to interact with and stabilize programmed cell death 4 (PCD4) that is involved in the inhibition of breast cancer cell proliferation (Li Y. et al., 2016). Moreover, knockdown of USP4 facilitated the proliferation, migration, and invasion of breast cancer cells, as well as tamoxifen resistance (Liang et al., 2019). Therefore, abnormal expression of USP4 can either increase or decrease tumorigenesis depending on the cell context, suggesting a multidimensional role for USP4 in cancer.
Viral and Bacterial Infections
Aberrant regulation of USP4 has also been reported in viral and bacterial infections. USP4 is downregulated in cells and mice infected by enterovirus 71 (EV71), a single-stranded positive-sense RNA virus of the Picornaviridae family. Overexpression of USP4 inhibits EV71 replication by stabilizing TRAF6 to activate NF-κB inflammatory signaling (Xu et al., 2018). Similarly, in vesicular stomatitis virus (VSV) infection, removing K48-linked polyubiquitin chains from RIG-I allows USP4 to enhance RIG-I-triggered IFN-β signaling, thereby inducing an antiviral immune response (Wang et al., 2013). Reductions in USP4 levels also occur in macrophages infected with Salmonella typhimurium, but the detailed mechanism has not been established to date (Kummari et al., 2015).
Acute Injury and Inflammation
USP4 is expressed in microglia, but its level is reduced after spinal cord injury. Here the downregulation of USP4 promotes microglial activation and subsequent neuronal inflammation through attenuating the deubiquitination of K63-linked TRAF6 and reversing its suppressor role in the NF-κB signaling pathway (Jiang et al., 2017). In intracerebral hemorrhage, another acute form of neurological damage, USP4 regulates the caspase-3/Bax/Bcl-2 axis to enhance apoptosis in neurons (Liu et al., 2017). In contrast, USP4 exhibits a protective role in hepatic ischemia/reperfusion (I/R) injury. Using a mouse model, Zhou J. et al. (2019) showed that USP4 levels were notably elevated in livers subjected to I/R injury, which functioned to alleviate hepatocyte apoptosis and liver inflammation through USP4-induced deubiquitination of TAK1 and inactivation of TAK1/JNK1/2/p38 signaling pathway.
Chronic Fibrosis-Related Diseases
USP4 is also involved in chronic fibrosis-related diseases. USP4 was reported to have a lower expression in the failing human heart compared with normal heart. Overexpression of USP4 can prevent angiotensin II–induced cardiac hypertrophy and fibrosis in vitro and ameliorate cardiac dysfunction in vivo through downregulation of the TAK1/JNK1/2/p38 signaling pathway (He et al., 2016). Deubiquitinating TβRI and stimulation of TGF-β signaling pathway also associate USP4 with liver fibrosis and cirrhosis where it facilitates activation of hepatic stellate cell and EMT in hepatocytes (Wu et al., 2019; Zhu et al., 2019). USP4 can promote pathological scarring in response to trauma in the skin and mesothelial-to-mesenchymal transition (MMT) during peritoneal dialysis through maintaining TβRI stability (Xiao et al., 2015; Zhang J. et al., 2019; Huang et al., 2020).
Metabolic Disorders
TAK1 plays a central role in hepatic steatosis and insulin resistance (Wang et al., 2016). Zhao et al. (2018) and Zhang Y. et al. (2019) demonstrated that deficiency of USP4 in mouse livers disrupts signaling through the TAK1/JNK-mediated insulin receptor substrate/protein kinase B/glycogen synthase kinase 3β (IRS-AKT-GSK3) axis, giving rise to non-alcoholic fatty liver disease (NAFLD). Thus, USP4 functions as a pivotal suppressor in NAFLD and related metabolic disorders.
Immune Disease
In patients with rheumatic heart disease (RHD), an autoimmune disease, USP4 was found to be significantly elevated in CD4+ T cells. The deubiquitination of RORγt and IRF4 and subsequent development and differentiation of TH17 and TH2 cells mediated by USP4 might provide a possible link between USP4 and RHD (Yang et al., 2015; Guo et al., 2017).
Cellular Regulation of USP4
As USP4 has a multifaceted impact on a series of physiological and pathological contexts, its stability, catalysis activity, and substrates affinity are under tight regulation. It can be organized through transcriptional regulations, PTMs, and protein–protein interactions (Figure 4).
FIGURE 4.
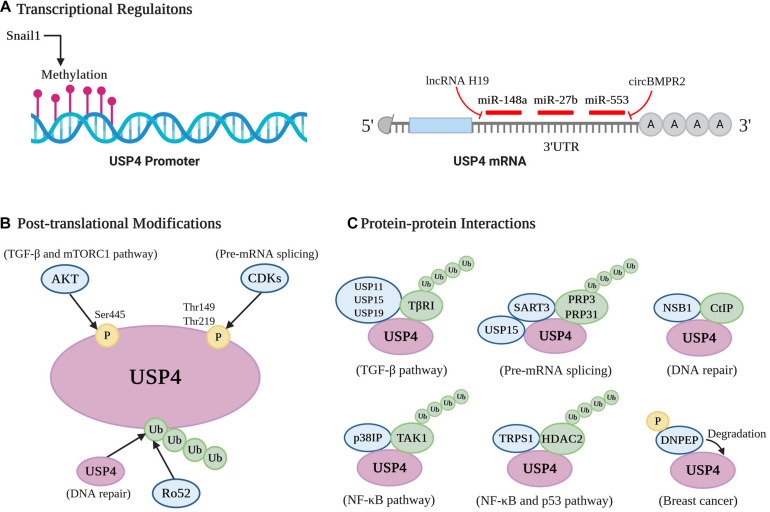
Cellular regulation of USP4. A pictorial representation of the different modes of USP4 regulation [(A) transcriptional regulations; (B) posttranslational modifications; and (C) protein–protein interactions].
Transcriptional Regulations
MiR-148a and miR-27b were reported to destabilize USP4 mRNA by targeting its 3′-UTR in HCC, breast cancer, and liver fibrosis (Heo et al., 2014; Zhang et al., 2017; Wu et al., 2019). Specifically, in liver fibrosis, lower levels of miR-148a and miR-27b lead to the overexpression of USP4 in hepatic stellate cells and hepatocytes. This occurs via upregulation of long non-coding RNA H19, which acts as an miRNA sponge for miR-148a and downregulations of liver X receptor-α and cannabinoid receptor 2, respectively (Wu et al., 2019; Zhu et al., 2019). USP4 is also negatively regulated by miR-553 in breast cancer, and this inhibition could be reversed by circBMPR2, which acts as a sponge to prevent miR-553 associating with USP4 mRNA (Liang et al., 2019). DNA methylation is another transcriptional regulation of USP4. For example, when lung cancer cells interact with macrophages, Snail1 is induced and subsequently silences USP4 by promoter methylation, contributing to the inflammation and stemness associated with macrophage-promoted tumor progression (Lai et al., 2020). Furthermore, Zhong et al. (2018) indicated that hypermethylation and DNA shallow deletion are two possible mechanisms for the decreased USP4 seen in patients with lung adenocarcinoma. Thus, transcriptional mechanisms are important in regulating USP4 and in several pathological situations appear to be involved in silencing its expression.
Posttranslational Modifications
The most prominent PTMs linked with USP4 regulation so far are phosphorylation and ubiquitination (Figure 4B). The enzyme is phosphorylated at Ser445, Thr149, and Thr219 (Zhang et al., 2012; Das et al., 2019; Deng et al., 2019). AKT directly phosphorylates USP4 at Ser445, relocating nuclear USP4 to the cytoplasm and membrane with the help of 14-3-3 protein, as well as enhancing its stability and deubiquitinating activity. Notably, this modification is essential for the USP4-mediated deubiquitination of TβRI and Rheb (Zhang et al., 2012; Deng et al., 2019). Moreover, the stabilization of TβRI and subsequent activation of AKT positively feedback to AKT’s affinity and phosphorylation of USP4. Phosphorylation of USP4 at the alternate residues, Thr149 and Thr219, is mediated by cyclin-dependent kinases and exerts a negative role in pre-mRNA splicing through blocking USP4 interactions with squamous cell carcinoma antigen recognized by T cells 3 (SART3) (Das et al., 2019).
USP4 also possesses various ubiquitination sites, including the zinc-binding motif Cys residues Cys-461, Cys-464, Cys-799, and Cys-802 (Liu et al., 2015; Wijnhoven et al., 2015). USP4 is ubiquitinated by the ubiquitin ligase Ro52, whose deubiquitination, in turn, is facilitated by USP4 (Wada and Kamitani, 2006; Wada et al., 2006). Besides, the ubiquitination level of USP4 is determined by its catalytic activity. Expression studies using an enzyme-dead mutant of USP4 demonstrated that ubiquitination levels were increased compared to wild-type USP4 (Wijnhoven et al., 2015). Of note, USP4 can deubiquitinate itself, and this autodeubiquitination function is crucial for the USP4-CtIP interaction, which occurs in HR (Liu et al., 2015; Wijnhoven et al., 2015). It seems that phosphorylation and ubiquitination are involved in USP4-mediated deubiquitinating processes by acting as a positive or negative regulator. More importantly, the status of these two PTMs may be antagonistic. Zhang et al. (2012) demonstrated that abolishing AKT-mediated phosphorylation increases the level of USP4 conjugation with polyubiquitin chains, consequently impairing enzyme stability and activity.
Protein–Protein Interactions
Several proteins have been identified to mediate the stability and/or activity of USP4 by directly interacting with it (Figure 4C). Foremost, USP4 can bind to itself, whereas other DUBs such as USP11, USP15, and USP19 were observed as binding partners of USP4 that assist in deubiquitinating TβRI (Zhang et al., 2012). During the pre-mRNA splicing process, the U4/U6 recycling protein SART3 functions as a substrate targeting factor of USP4 (Song et al., 2010; Park et al., 2016; Kwon et al., 2017). By binding to the linker between the DUSP and UBL domain of USP4, SART3 can shuttle USP4 to the nucleus from the cytosol through its NLS. The USP4sart3 complex helps to recruit the substrate PRP3, as well as increase the deubiquitinating activity of USP4 (Song et al., 2010; Park et al., 2016). Moreover, as a substrate recruiting factor of USP15 either (Long et al., 2014; Zhang et al., 2016), SART3 can simultaneously bind USP4 and USP15, serving as a platform to deubiquitinate PRP31 and PRP3 (Das et al., 2017). Another protein, NBS1, which is a part of the MRN complex, interacts with the DUSP domain of USP4 and recruits the enzyme to DNA damage sites where it colocalizes with CtIP (Liu et al., 2015).
p38IP was found to be a necessary regulator in USP4-mediated deubiquitination of TAK1. It can specifically bind to the fingers subdomain of USP4 through a cavity-like structure and serves to recruit USP4 to the TAK1 complex (Wang X. D. et al., 2020). This recruitment is dependent on p38IP with a higher affinity to ubiquitinated TAK1 upon TCR, LPS, or TNF-α stimulation (Fan et al., 2011; Wang X. D. et al., 2020). Similarly, trichorhinophalangeal syndrome type I (TRPS1) functions as a scaffold protein involved in the USP4-induced stabilization of HDAC2 through bringing them together to form the TRPS1-USP4-HDAC2 complex (Wang et al., 2018). Finally, USP4 was reported to interact with and be degraded by aspartyl aminopeptidase (DNPEP), a metalloenzyme belonging to the M18 aminopeptidase family, resulting in suppression of its oncogenic role in breast cancer. Moreover, phosphorylation of DNPEP by p21-activated kinase 5 (PAK5) is indispensable in this process (Geng et al., 2020).
Therapeutic Developments for Targeting USP4
Research carried out in the past few decades has uncovered the essential roles of DUBs in disease, especially in cancer, giving sufficient evidence to consider these molecules as potential pharmaceutical targets. USP4 is no exception, but progress in the development of USP4 inhibitors remains slow, with only two compounds reported so far (Okada et al., 2013; Nguyen et al., 2019). Xie et al. (2005) isolated a small compound from Chinese mushroom Thelephora vialis, namely, vialinin A. Vialinin A was initially discovered to exhibit an effective anti-inflammatory activity, but later it was identified as a micromolar inhibitor of USP4 (IC50 = 1.5 μM), USP5/isopeptidase T (IC50 = 5.9 μM), and UCH-L1 (IC50 = 22.3 μM) (Ye et al., 2007; Okada et al., 2013). Treatment with vialinin A not only inhibits colorectal tumor growth but also interferes with USP4-mediated immune responses and pathological scarring (Yang et al., 2015; Guo et al., 2017; Lin et al., 2017; Deng et al., 2019; Zhang J. et al., 2019). NR, a eurhodin dye–containing aminophenazine structure, was recognized as another non-competitive DUB inhibitor with selectivity for USP4 (IC50 = 50 μM). The tumor-promoting role of USP4 in colorectal cancer can also be reversed by NR through decreasing β-catenin stability (Nguyen et al., 2019). These findings suggest that targeting USP4 holds promise as cancer therapy, but highly specific nanomolar-range drugs still need to be discovered and developed.
Conclusion
This review illustrates the current literature on USP4 that sheds light on the prominent and multifaceted role of USP4 in normal and pathological states. USP4 deubiquitinates a wide range of protein substrates under strict and diverse regulatory conditions. Major cancer pathways including p53, Wnt/β-catenin, TGF-β, and NF-κB signaling pathways are modulated by USP4, whereas regulation of TGF-β and NF-κB signaling pathways is also involved in other diseases, including spinal cord injury, liver fibrosis and cirrhosis, and pathological scarring (Xiao et al., 2012; Zhang et al., 2012; Yun et al., 2015; Cao et al., 2016; Hwang et al., 2016; Jiang et al., 2017; Qiu et al., 2018; Qin et al., 2019; Wu et al., 2019; Huang et al., 2020; Lai et al., 2020). Together, these studies provide both insights into USP4 biology as well as its applicability as a potential treatment target for many diseases.
Specifically, in some diseases such as hepatic I/R injury, USP4 is upregulated to exert a protective function (Zhou J. et al., 2019). Moreover, USP4 and its paralog USP15 play both promoting and suppressing roles in cancer (Chou et al., 2017). This highlights the need to carefully evaluate the role of USP4 in individual diseases. Indeed, the present understanding of USP4 is insufficient, and there are some contradictory findings that remain to be explained. For example, USP4 removes K48-linked ubiquitin chains from TRAF6 in cells infected by EV71 (Xu et al., 2018), whereas in TNF-α–induced inflammation, TRAF6 K63-linked polyubiquitinated chains are cleaved (Xiao et al., 2012). Hence, USP4 exerts opposing functions in the activation of the NF-κB signaling pathway. In breast cancer, Wang et al. (2018) reported that USP4 facilitates cell proliferation through promoting the stability of HDAC2, whereas Li Y. et al. (2016) found it inhibits cell growth by deubiquitinating PCD4.
Deciphering the mechanisms determining USP4 specificity toward different types of ubiquitin chains and affinity with different substrates is a far-reaching task that will likely contribute to explaining the multidimensional roles of USP4. As substrates may compete for USP4 binding occurring with USP7, this must also be factored into this analysis. For example, the competition between the tumor suppressor p53 and its E3 ubiquitin ligase HDM2 serves as a switch to fine-tune the cellular level of p53. This is due to a common mode of interaction found in USP7 that all the substrates recognized by the TRAF-like domain share a common P/AxxS motif or ExxS motif (Pozhidaeva and Bezsonova, 2019). Therefore, revealing the interaction modes of USP4 seems to be valuable. USP4 is also subject to diverse regulation within the cell with transcriptional mechanisms being involved in determining USP4 levels, whereas phosphorylation, ubiquitination, and binding partners of USP4 modulate USP4 catalytic activity, subcellular localization, and association with substrate proteins. Different regulations of USP4 may result in the different roles of USP4 in cells or situations. Thus, not only the downstream substrates but also the upstream regulators of USP4 need to be further explored.
Finally, to comment on the slow progress made in developing USP4 as a treatment target. The two inhibitors showing preclinical activity are not USP4-specific and have never been considered for use in clinical trials. Even if suitable inhibitors became available, the use of USP4 as a therapeutic target must be considered carefully as it plays a tumor-suppressing role in some types of cancer. Thus, in addition to identifying more effective USP4 inhibitors targeting USP4, future research should also focus on understanding which cancers would likely benefit from this treatment along with other diseases where USP4 would inhibit progression.
Author Contributions
BH drafted the manuscript. DZ, KZ, and YW drew the pictures. LP and QF revised the manuscript. XM checked and modified the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Mengyuan Huang for the drawings in Figure 2 in this manuscript.
Footnotes
Funding. This work was supported by the 1⋅3⋅5 project for disciplines of excellence–Clinical Research Incubation Project, West China Hospital, Sichuan University.
References
- Al-Aidaroos A. Q., Zeng Q. (2010). PRL-3 phosphatase and cancer metastasis. J. Cell Biochem. 111 1087–1098. 10.1002/jcb.22913 [DOI] [PubMed] [Google Scholar]
- Amerik A. Y., Hochstrasser M. (2004). Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 1695 189–207. 10.1016/j.bbamcr.2004.10.003 [DOI] [PubMed] [Google Scholar]
- Anckar J., Bonni A. (2015). Regulation of neuronal morphogenesis and positioning by ubiquitin-specific proteases in the cerebellum. PLoS One 10:e0117076. 10.1371/journal.pone.0117076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelats C., Wang X.-W., Jermiin L. S., Copeland N. G., Jenkins N. A., Baker R. T. (2003). Isolation and characterization of the mouse ubiquitin-specific protease Usp15. Mammal. Genome 14 31–46. 10.1007/s00335-002-3035-0 [DOI] [PubMed] [Google Scholar]
- Avvakumov G. V., Walker J. R., Xue S., Finerty P. J., Jr., Mackenzie F., et al. (2006). Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8). J. Biol. Chem. 281 38061–38070. 10.1074/jbc.M606704200 [DOI] [PubMed] [Google Scholar]
- Blanchette P., Gilchrist C. A., Baker R. T., Gray D. A. (2001). Association of UNP, a ubiquitin-specific protease, with the pocket proteins pRb, p107 and p130. Oncogene 20 5533–5537. 10.1038/sj.onc.1204823 [DOI] [PubMed] [Google Scholar]
- Cao W. H., Liu X. P., Meng S. L., Gao Y. W., Wang Y., Ma Z. L., et al. (2016). USP4 promotes invasion of breast cancer cells via Relaxin/TGF-β1/Smad2/MMP-9 signal. Eur. Rev. Med. Pharmacol. Sci. 20 1115–1122. [PubMed] [Google Scholar]
- Chou C.-K., Chang Y.-T., Korinek M., Chen Y.-T., Yang Y.-T., Leu S., et al. (2017). The Regulations of Deubiquitinase USP15 and Its Pathophysiological Mechanisms in Diseases. Int. J. Mol. Sci. 18:483. 10.3390/ijms18030483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M., Luna-Vargas M. P. A., Faesen A. C., Sixma T. K. (2014). The DUSP-Ubl domain of USP4 enhances its catalytic efficiency by promoting ubiquitin exchange. Nat. Commun. 5:5399. 10.1038/ncomms6399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H., Nusse R. (2012). Wnt/β-catenin signaling and disease. Cell 149 1192–1205. 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Das T., Kim E. E., Song E. J. (2019). Phosphorylation of USP15 and USP4 regulates localization and spliceosomal deubiquitination. J. Mol. Biol. 431 3900–3912. 10.1016/j.jmb.2019.07.023 [DOI] [PubMed] [Google Scholar]
- Das T., Park J. K., Park J., Kim E., Rape M., Kim E. E., et al. (2017). USP15 regulates dynamic protein-protein interactions of the spliceosome through deubiquitination of PRP31. Nucleic acids res. 45 4866–4880. 10.1093/nar/gkw1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Chen L., Zhao L., Xu Y., Peng X., Wang X., et al. (2019). Ubiquitination of Rheb governs growth factor-induced mTORC1 activation. Cell Res. 29 136–150. 10.1038/s41422-018-0120-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Wang C., Spencer E., Yang L., Braun A., You J., et al. (2000). Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103 351–361. 10.1016/s0092-8674(00)00126-4 [DOI] [PubMed] [Google Scholar]
- DeSalle L. M., Latres E., Lin D., Graner E., Montagnoli A., Baker R. T., et al. (2001). The de-ubiquitinating enzyme Unp interacts with the retinoblastoma protein. Oncogene 20 5538–5542. 10.1038/sj.onc.1204824 [DOI] [PubMed] [Google Scholar]
- Di Fruscio M., Gilchrist C. A., Baker R. T., Gray D. A. (1998). Genomic structure of Unp, a murine gene encoding a ubiquitin-specific protease. Biochimica et Biophys. Acta 1398 9–17. 10.1016/s0167-4781(98)00035-9 [DOI] [PubMed] [Google Scholar]
- Elliott P. R., Liu H., Pastok M. W., Grossmann G. J., Rigden D. J., Clague M. J., et al. (2011). Structural variability of the ubiquitin specific protease DUSP-UBL double domains. FEBS Lett. 585 3385–3390. 10.1016/j.febslet.2011.09.040 [DOI] [PubMed] [Google Scholar]
- Faesen A. C., Dirac A. M., Shanmugham A., Ovaa H., Perrakis A., Sixma T. K. (2011). Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol. Cell 44 147–159. 10.1016/j.molcel.2011.06.034 [DOI] [PubMed] [Google Scholar]
- Fan Y., Yu Y., Shi Y., Sun W., Xie M., Ge N., et al. (2010). Lysine 63-linked polyubiquitination of TAK1 at lysine 158 is required for tumor necrosis factor alpha- and interleukin-1beta-induced IKK/NF-kappaB and JNK/AP-1 activation. J. Biol. Chem. 285 5347–5360. 10.1074/jbc.M109.076976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y. H., Yu Y., Mao R. F., Tan X. J., Xu G. F., Zhang H., et al. (2011). USP4 targets TAK1 to downregulate TNFα-induced NF-κB activation. Cell Death Differ. 18 1547–1560. 10.1038/cdd.2011.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick A., Rolfe M., Chiu M. I. (1998). The human UNP locus at 3p21.31 encodes two tissue-selective, cytoplasmic isoforms with deubiquitinating activity that have reduced expression in small cell lung carcinoma cell lines. Oncogene 16 153–165. 10.1038/sj.onc.1201537 [DOI] [PubMed] [Google Scholar]
- Geng N., Li Y., Zhang W., Wang F., Wang X., Jin Z., et al. (2020). A PAK5-DNPEP-USP4 axis dictates breast cancer growth and metastasis. Int. J. Cancer 146 1139–1151. 10.1002/ijc.32523 [DOI] [PubMed] [Google Scholar]
- Gilchrist C. A., Baker R. T. (2000). Characterization of the ubiquitin-specific protease activity of the mouse/human Unp/Unph oncoprotein. Biochim. et Biophys. Acta 1481 297–309. 10.1016/s0167-4838(00)00134-5 [DOI] [PubMed] [Google Scholar]
- Gilchrist C. A., Gray D. A., Baker R. T. (1997). A ubiquitin-specific protease that efficiently cleaves the ubiquitin-proline bond. J. Biol. Chem. 272 32280–32285. 10.1074/jbc.272.51.32280 [DOI] [PubMed] [Google Scholar]
- Gray D. A., Inazawa J., Gupta K., Wong A., Ueda R., Takahashi T. (1995). Elevated expression of Unph, a proto-oncogene at 3p21.3, in human lung tumors. Oncogene 10 2179–2183. [PubMed] [Google Scholar]
- Guo W., Ma J., Pei T., Zhao T., Guo S., Yi X., et al. (2018). Up-regulated deubiquitinase USP4 plays an oncogenic role in melanoma. J. Cell. Mol. Med. 22 2944–2954. 10.1111/jcmm.13603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Xu P., Ge S., Zhang C., Zheng X., Xu J., et al. (2017). Ubiquitin specific peptidase 4 stabilizes interferon regulatory factor protein and promotes its function to facilitate interleukin-4 expression in T helper type 2 cells. Int. J. Mol. Med. 40 979–986. 10.3892/ijmm.2017.3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K., Chevrette M., Gray D. A. (1994). The Unp proto-oncogene encodes a nuclear protein. Oncogene 9 1729–1731. [PubMed] [Google Scholar]
- Gupta K., Copeland N. G., Gilbert D. J., Jenkins N. A., Gray D. A. (1993). Unp, a mouse gene related to the tre oncogene. Oncogene 8 2307–2310. [PubMed] [Google Scholar]
- Gürtler C., Bowie A. G. (2013). Innate immune detection of microbial nucleic acids. Trends Microbiol. 21 413–420. 10.1016/j.tim.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper S., Besong T. M. D., Emsley J., Scott D. J., Dreveny I. (2011). Structure of the USP15 N-terminal domains: a β-hairpin mediates close association between the DUSP and UBL domains. Biochemistry 50 7995–8004. 10.1021/bi200726e [DOI] [PubMed] [Google Scholar]
- Hayes S. D., Liu H., MacDonald E., Sanderson C. M., Coulson J. M., Clague M. J., et al. (2012). Direct and indirect control of mitogen-activated protein kinase pathway-associated components, BRAP/IMP E3 ubiquitin ligase and CRAF/RAF1 kinase, by the deubiquitylating enzyme USP15. J. Biol. Chem. 287 43007–43018. 10.1074/jbc.M112.386938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Zhao Y.-C., Gao L.-C., Ying X.-Y., Xu L.-W., Su Y.-Y., et al. (2016). Ubiquitin-Specific protease 4 is an endogenous negative regulator of pathological cardiac hypertrophy. Hypertension 67 1237–1248. 10.1161/HYPERTENSIONAHA.116.07392 [DOI] [PubMed] [Google Scholar]
- Heo M. J., Kim Y. M., Koo J. H., Yang Y. M., An J., Lee S.-K., et al. (2014). microRNA-148a dysregulation discriminates poor prognosis of hepatocellular carcinoma in association with USP4 overexpression. Oncotarget 5 2792–2806. 10.18632/oncotarget.1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67 425–479. 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- Hershko A., Heller H., Elias S., Ciechanover A. (1983). Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol. Chem. 258 8206–8214. [PubMed] [Google Scholar]
- Hicke L. (2001). Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2 195–201. 10.1038/35056583 [DOI] [PubMed] [Google Scholar]
- Hou X., Wang L., Zhang L., Pan X., Zhao W. (2013). Ubiquitin-specific protease 4 promotes TNF-α-induced apoptosis by deubiquitination of RIP1 in head and neck squamous cell carcinoma. FEBS Lett. 587 311–316. 10.1016/j.febslet.2012.12.016 [DOI] [PubMed] [Google Scholar]
- Hu M., Li P., Li M., Li W., Yao T., Wu J. W., et al. (2002). Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111 1041–1054. 10.1016/s0092-8674(02)01199-6 [DOI] [PubMed] [Google Scholar]
- Hu M., Li P., Song L., Jeffrey P. D., Chenova T. A., Wilkinson K. D., et al. (2005). Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 24 3747–3756. 10.1038/sj.emboj.7600832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Wang Y., Wang X., Lin L., Wang P., Sun J., et al. (2020). The effects of the transforming growth Factor-β1 (TGF-β1) signaling pathway on cell proliferation and cell migration are mediated by ubiquitin specific protease 4 (USP4) in hypertrophic scar tissue and primary fibroblast cultures. Med. Sci. Monit. 26:e920736. 10.12659/msm.920736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S. J., Lee H. W., Kim H. R., Lee H., Shin C. H., Yun S.-I., et al. (2016). Ubiquitin-specific protease 4 controls metastatic potential through β-catenin stabilization in brain metastatic lung adenocarcinoma. Sci. Rep. 6:21596. 10.1038/srep21596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Yu M., Ou Y., Cao Y., Yao Y., Cai P., et al. (2017). Downregulation of USP4 promotes activation of microglia and subsequent neuronal inflammation in rat spinal cord after injury. Neurochem. Res. 42 3245–3253. 10.1007/s11064-017-2361-2 [DOI] [PubMed] [Google Scholar]
- Jurica M. S., Moore M. J. (2003). Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12 5–14. 10.1016/s1097-2765(03)00270-3 [DOI] [PubMed] [Google Scholar]
- Klinck R., Laberge G., Bisson M., McManus S., Michou L., Brown J. P., et al. (2014). Alternative splicing in osteoclasts and Paget’s disease of bone. BMC Med. Genet. 15:98. 10.1186/s12881-014-0098-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Clague M. J., Urbé S. (2009). Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10 550–563. 10.1038/nrm2731 [DOI] [PubMed] [Google Scholar]
- Komander D., Lord C. J., Scheel H., Swift S., Hofmann K., Ashworth A., et al. (2008). The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol. Cell 29 451–464. 10.1016/j.molcel.2007.12.018 [DOI] [PubMed] [Google Scholar]
- Kulathu Y., Komander D. (2012). Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 13 508–523. 10.1038/nrm3394 [DOI] [PubMed] [Google Scholar]
- Kummari E., Alugubelly N., Hsu C.-Y., Dong B., Nanduri B., Edelmann M. J. (2015). Activity-Based proteomic profiling of deubiquitinating enzymes in Salmonella-infected macrophages leads to identification of putative function of UCH-L5 in inflammasome regulation. PLoS One 10:e0135531. 10.1371/journal.pone.0135531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S.-K., Kim E.-H., Baek K.-H. (2017). RNPS1 is modulated by ubiquitin-specific protease 4. FEBS Lett. 591 369–381. 10.1002/1873-3468.12531 [DOI] [PubMed] [Google Scholar]
- Lai C.-Y., Yeh D.-W., Lu C.-H., Liu Y.-L., Chuang Y.-C., Ruan J.-W., et al. (2020). Epigenetic silencing of ubiquitin specific protease 4 by snail1 contributes to macrophage-dependent inflammation and therapeutic resistance in lung cancer. Cancers 12:E148. 10.3390/cancers12010148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurell C., Velázquez-Fernández D., Lindsten K., Juhlin C., Enberg U., Geli J., et al. (2009). Transcriptional profiling enables molecular classification of adrenocortical tumours. Eur. J. Endocrinol. 161 141–152. 10.1530/EJE-09-0068 [DOI] [PubMed] [Google Scholar]
- Li F., Hu Q., He T., Xu J., Yi Y., Xie S., et al. (2020). The deubiquitinase USP4 stabilizes twist1 protein to promote lung cancer cell stemness. Cancers (Basel) 12:1582. 10.3390/cancers12061582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Sethi G. (2010). Targeting transcription factor NF-kappaB to overcome chemoresistance and radioresistance in cancer therapy. Biochim. Biophys. Acta 1805 167–180. 10.1016/j.bbcan.2010.01.002 [DOI] [PubMed] [Google Scholar]
- Li T., Yan B., Ma Y., Weng J., Yang S., Zhao N., et al. (2018). Ubiquitin-specific protease 4 promotes hepatocellular carcinoma progression via cyclophilin A stabilization and deubiquitination. Cell Death Dis. 9:148. 10.1038/s41419-017-0182-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Jiang D., Zhang Q., Liu X., Cai Z. (2016a). Ubiquitin-specific protease 4 inhibits breast cancer cell growth through the upregulation of PDCD4. Int. J. Mol. Med. 38 803–811. 10.3892/ijmm.2016.2685 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Li Z., Hao Q., Luo J., Xiong J., Zhang S., Wang T., et al. (2016b). USP4 inhibits p53 and NF-κB through deubiquitinating and stabilizing HDAC2. Oncogene 35 2902–2912. 10.1038/onc.2015.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F., Liang J., Wang W. Q., Sun J. P., Udho E., Zhang Z. Y. (2007). PRL3 promotes cell invasion and proliferation by down-regulation of Csk leading to Src activation. J. Biol. Chem. 282 5413–5419. 10.1074/jbc.M608940200 [DOI] [PubMed] [Google Scholar]
- Liang J., Saad Y., Lei T., Wang J., Qi D., Yang Q., et al. (2010). MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J. Exp. Med. 207 2959–2973. 10.1084/jem.20092641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Fan Y., Cheng J., Cheng D., Zhao Y., Cao B., et al. (2013). TAK1 ubiquitination regulates doxorubicin-induced NF-κB activation. Cell. Signal. 25 247–254. 10.1016/j.cellsig.2012.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Song X., Li Y., Ma T., Su P., Guo R., et al. (2019). Targeting the circBMPR2/miR-553/USP4 axis as a potent therapeutic approach for Breast Cancer. Mol. Therapy. Nucleic Acids 17 347–361. 10.1016/j.omtn.2019.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Nie J., Ren J., Liang R., Li D., Wang P., et al. (2017). USP4 interacts and positively regulates IRF8 function via K48-linked deubiquitination in regulatory T cells. FEBS Lett. 591 1677–1686. 10.1002/1873-3468.12668 [DOI] [PubMed] [Google Scholar]
- Liu C., Liu C., Liu H., Gong L., Tao T., Shen Y., et al. (2017). Increased expression of ubiquitin-specific protease 4 participates in neuronal apoptosis after intracerebral hemorrhage in adult rats. Cell. Mol. Neurobiol. 37 427–435. 10.1007/s10571-016-0375-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhang H., Wang X., Tian Q., Hu Z., Peng C., et al. (2015). The Deubiquitylating enzyme USP4 cooperates with CtIP in DNA double-strand break end resection. Cell Rep. 13 93–107. 10.1016/j.celrep.2015.08.056 [DOI] [PubMed] [Google Scholar]
- Long L., Thelen J. P., Furgason M., Haj-Yahya M., Brik A., Cheng D., et al. (2014). The U4/U6 recycling factor SART3 has histone chaperone activity and associates with USP15 to regulate H2B deubiquitination. J. Biol. Chem. 289 8916–8930. 10.1074/jbc.M114.551754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder C., Guthrie C. (2008). Modifications target spliceosome dynamics. Nat. Struct. Mol. Biol. 15 426–428. 10.1038/nsmb0508-426 [DOI] [PubMed] [Google Scholar]
- Mehić M., de Sa V. K., Hebestreit S., Heldin C. H., Heldin P. (2017). The deubiquitinating enzymes USP4 and USP17 target hyaluronan synthase 2 and differentially affect its function. Oncogenesis 6:e348. 10.1038/oncsis.2017.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milojevic T., Reiterer V., Stefan E., Korkhov V. M., Dorostkar M. M., Ducza E., et al. (2006). The ubiquitin-specific protease Usp4 regulates the cell surface level of the A2A receptor. Mol. Pharmacol. 69 1083–1094. 10.1124/mol.105.015818 [DOI] [PubMed] [Google Scholar]
- Murali S. K., Aroankins T. S., Moeller H. B., Fenton R. A. (2019). The deubiquitylase USP4 interacts with the water channel AQP2 to modulate its apical membrane accumulation and cellular abundance. Cells 8:265. 10.3390/cells8030265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. H., Kim T., Nguyen T., Hahn M.-J., Yun S.-I., Kim K. K. (2019). A selective inhibitor of ubiquitin-specific protease 4 suppresses colorectal cancer progression by regulating β-catenin signaling. Cell. Physiol. Biochem. 53 157–171. 10.33594/000000127 [DOI] [PubMed] [Google Scholar]
- Nottrott S., Urlaub H., Lührmann R. (2002). Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins. EMBO J. 21 5527–5538. 10.1093/emboj/cdf544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Ye Y. Q., Taniguchi K., Yoshida A., Akiyama T., Yoshioka Y., et al. (2013). Vialinin A is a ubiquitin-specific peptidase inhibitor. Bioorgan. Med. Chem. Lett. 23 4328–4331. 10.1016/j.bmcl.2013.05.093 [DOI] [PubMed] [Google Scholar]
- Park J. K., Das T., Song E. J., Kim E. E. (2016). Structural basis for recruiting and shuttling of the spliceosomal deubiquitinase USP4 by SART3. Nucleic Acids Res. 44 5424–5437. 10.1093/nar/gkw218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C. M., Fushman D. (2004). Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8 610–616. 10.1016/j.cbpa.2004.09.009 [DOI] [PubMed] [Google Scholar]
- Pozhidaeva A., Bezsonova I. (2019). USP7: structure, substrate specificity, and inhibition. DNA Repair (Amst) 76 30–39. 10.1016/j.dnarep.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin N., Han F., Li L., Ge Y., Lin W., Wang J., et al. (2019). Deubiquitinating enzyme 4 facilitates chemoresistance in glioblastoma by inhibiting P53 activity. Oncol. Lett. 17 958–964. 10.3892/ol.2018.9654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C., Liu Y., Mei Y., Zou M., Zhao Z., Ye M., et al. (2018). Ubiquitin-specific protease 4 promotes metastasis of hepatocellular carcinoma by increasing TGF-β signaling-induced epithelial-mesenchymal transition. Aging 10 2783–2799. 10.18632/aging.101587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V., Díaz-Perales A., Gutiérrez-Fernández A., Garabaya C., Cal S., López-Otín C. (2004). Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases. Biochem. Biophys. Res. Commun. 314 54–62. 10.1016/j.bbrc.2003.12.050 [DOI] [PubMed] [Google Scholar]
- Renatus M., Parrado S. G., D’Arcy A., Eidhoff U., Gerhartz B., Hassiepen U., et al. (2006). Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure 14 1293–1302. 10.1016/j.str.2006.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samara N. L., Datta A. B., Berndsen C. E., Zhang X., Yao T., Cohen R. E., et al. (2010). Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328 1025–1029. 10.1126/science.1190049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboleva T. A., Jans D. A., Johnson-Saliba M., Baker R. T. (2005). Nuclear-cytoplasmic shuttling of the oncogenic mouse UNP/USP4 deubiquitylating enzyme. J. Biol. Chem. 280 745–752. 10.1074/jbc.M401394200 [DOI] [PubMed] [Google Scholar]
- Song E. J., Werner S. L., Neubauer J., Stegmeier F., Aspden J., Rio D., et al. (2010). The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes dev. 24 1434–1447. 10.1101/gad.1925010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storer A. C., Ménard R. (1994). Catalytic mechanism in papain family of cysteine peptidases. Methods Enzymol. 244 486–500. 10.1016/0076-6879(94)44035-2 [DOI] [PubMed] [Google Scholar]
- Tse W. K., Eisenhaber B., Ho S. H., Ng Q., Eisenhaber F., Jiang Y. J. (2009). Genome-wide loss-of-function analysis of deubiquitylating enzymes for zebrafish development. BMC Genom. 10:637. 10.1186/1471-2164-10-637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uras I. Z., List T., Nijman S. M. B. (2012). Ubiquitin-specific protease 4 inhibits mono-ubiquitination of the master growth factor signaling kinase PDK1. PLoS One 7:e31003. 10.1371/journal.pone.0031003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velázquez-Fernández D., Laurell C., Geli J., Höög A., Odeberg J., Kjellman M., et al. (2005). Expression profiling of adrenocortical neoplasms suggests a molecular signature of malignancy. Surgery 138 1087–1094. 10.1016/j.surg.2005.09.031 [DOI] [PubMed] [Google Scholar]
- Vlasschaert C., Xia X., Coulombe J., Gray D. A. (2015). Evolution of the highly networked deubiquitinating enzymes USP4, USP15, and USP11. BMC Evol. Biol. 15:230. 10.1186/s12862-015-0511-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasschaert C., Xia X., Gray D. A. (2016). Selection preserves ubiquitin specific protease 4 alternative exon skipping in therian mammals. Sci. Rep. 6:20039. 10.1038/srep20039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K., Kamitani T. (2006). UnpEL/Usp4 is ubiquitinated by Ro52 and deubiquitinated by itself. Biochem. Biophys. Res. Commun. 342 253–258. 10.1016/j.bbrc.2006.01.144 [DOI] [PubMed] [Google Scholar]
- Wada K., Tanji K., Kamitani T. (2006). Oncogenic protein UnpEL/Usp4 deubiquitinates Ro52 by its isopeptidase activity. Biochem. Biophys. Res. Communications 339 731–736. 10.1016/j.bbrc.2005.11.076 [DOI] [PubMed] [Google Scholar]
- Wang L., Zhao W., Zhang M., Wang P., Zhao K., Zhao X., et al. (2013). USP4 positively regulates RIG-I-mediated antiviral response through deubiquitination and stabilization of RIG-I. J. Virol. 87 4507–4515. 10.1128/JVI.00031-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. X., Zhang X. J., Luo P., Jiang X., Zhang P., Guo J., et al. (2016). Hepatocyte TRAF3 promotes liver steatosis and systemic insulin resistance through targeting TAK1-dependent signalling. Nat. Commun. 7:10592. 10.1038/ncomms10592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. D., Zhao C. S., Wang Q. L., Zeng Q., Feng X. Z., Li L., et al. (2020a). The p38-interacting protein p38IP suppresses TCR and LPS signaling by targeting TAK1. EMBO Rep. 21:e48035. 10.15252/embr.201948035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhou L., Lu J., Jiang B., Liu C., Guo J. (2020b). USP4 function and multifaceted roles in cancer: a possible and potential therapeutic target. Cancer Cell Int. 20:298. 10.1186/s12935-020-01391-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang J., Wu L., Liu W., Wei G., Gong X., et al. (2018). Tricho-rhino-phalangeal syndrome 1 protein functions as a scaffold required for ubiquitin-specific protease 4-directed histone deacetylase 2 de-ubiquitination and tumor growth. Breast Cancer Res. BCR 20:83. 10.1186/s13058-018-1018-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S. J., Gratton H. E., Indrayudha P., Michavila C., Mukhopadhyay R., Maurer S. K., et al. (2018). The structure of the deubiquitinase USP15 reveals a misaligned catalytic triad and an open ubiquitin-binding channel. J. Biol. Chem. 293 17362–17374. 10.1074/jbc.RA118.003857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz I. E., Murray J. M. (2019). Structurally-defined deubiquitinase inhibitors provide opportunities to investigate disease mechanisms. Drug Discov. Today Technol. 31 109–123. 10.1016/j.ddtec.2019.02.003 [DOI] [PubMed] [Google Scholar]
- Wijnhoven P., Konietzny R., Blackford A. N., Travers J., Kessler B. M., Nishi R., et al. (2015). USP4 auto-deubiquitylation promotes homologous recombination. Mol. Cell 60 362–373. 10.1016/j.molcel.2015.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. J., Conze D. B., Li X., Ying S. X., Hanover J. A., Ashwell J. D. (2005). TNF-alpha induced c-IAP1/TRAF2 complex translocation to a Ubc6-containing compartment and TRAF2 ubiquitination. EMBO J. 24 1886–1898. 10.1038/sj.emboj.7600649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Kim T. H., Kim A., Koo J. H., Joo M. S., Kim S. G. (2019). Liver X Receptor α-Induced cannabinoid receptor 2 inhibits ubiquitin-specific peptidase 4 through mir-27b, protecting hepatocytes from TGF-β. Hepatol. Commun. 3 1373–1387. 10.1002/hep4.1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Peng X., Liu F., Tang C., Hu C., Xu X., et al. (2015). AKT regulation of mesothelial-to-mesenchymal transition in peritoneal dialysis is modulated by Smurf2 and deubiquitinating enzyme USP4. BMC Cell Biol. 16:7. 10.1186/s12860-015-0055-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao N., Li H., Luo J., Wang R., Chen H., Chen J., et al. (2012). Ubiquitin-specific protease 4 (USP4) targets TRAF2 and TRAF6 for deubiquitination and inhibits TNFα-induced cancer cell migration. Biochem. J. 441 979–986. 10.1042/BJ20111358 [DOI] [PubMed] [Google Scholar]
- Xie C., Koshino H., Esumi Y., Takahashi S., Yoshikawa K., Abe N. (2005). Vialinin A, a novel 2,2-diphenyl-1-picrylhydrazyl (DPPH) radical scavenger from an edible mushroom in China. Biosci. Biotechnol. Biochem. 69 2326–2332. 10.1271/bbb.69.2326 [DOI] [PubMed] [Google Scholar]
- Xing C., Lu X.-X., Guo P.-D., Shen T., Zhang S., He X.-S., et al. (2016). Ubiquitin-Specific protease 4-Mediated deubiquitination and stabilization of PRL-3 is required for potentiating colorectal oncogenesis. Cancer Res. 76 83–95. 10.1158/0008-5472.CAN-14-3595 [DOI] [PubMed] [Google Scholar]
- Xu C., Peng Y., Zhang Q., Xu X.-P., Kong X.-M., Shi W.-F. (2018). USP4 positively regulates RLR-induced NF-κB activation by targeting TRAF6 for K48-linked deubiquitination and inhibits enterovirus 71 replication. Sci. Rep. 8:13418. 10.1038/s41598-018-31734-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki K., Gohda J., Kanayama A., Miyamoto Y., Sakurai H., Yamamoto M., et al. (2009). Two mechanistically and temporally distinct NF-kappaB activation pathways in IL-1 signaling. Sci. Signal. 2:ra66. 10.1126/scisignal.2000387 [DOI] [PubMed] [Google Scholar]
- Yang J., Xu P., Han L., Guo Z., Wang X., Chen Z., et al. (2015). Cutting edge: ubiquitin-specific protease 4 promotes Th17 cell function under inflammation by deubiquitinating and stabilizing RORγt. J. Immunol. 194 4094–4097. 10.4049/jimmunol.1401451 [DOI] [PubMed] [Google Scholar]
- Yao R., Pu J., Fan R., Zhu W., Ding X., Shen X., et al. (2017). Ubiquitin-specific protease 4 improves the prognosis of the patients in esophageal cancer. Cancer Biomark. Sec. A Dis. Mark. 20 317–323. 10.3233/CBM-170308 [DOI] [PubMed] [Google Scholar]
- Ye Y., Scheel H., Hofmann K., Komander D. (2009). Dissection of USP catalytic domains reveals five common insertion points. Mol. Biosyst. 5 1797–1808. 10.1039/b907669g [DOI] [PubMed] [Google Scholar]
- Ye Y. Q., Koshino H., Onose J., Yoshikawa K., Abe N., Takahashi S. (2007). First total synthesis of vialinin a, a novel and extremely potent inhibitor of TNF-alpha production. Org. Lett. 9 4131–4134. 10.1021/ol701590b [DOI] [PubMed] [Google Scholar]
- Yun S.-I., Kim H. H., Yoon J. H., Park W. S., Hahn M.-J., Kim H. C., et al. (2015). Ubiquitin specific protease 4 positively regulates the WNT/β-catenin signaling in colorectal cancer. Mol. oncol. 9 1834–1851. 10.1016/j.molonc.2015.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S.-I., Kim K. K. (2017). Ubiquitin-specific protease 4 (USP4) suppresses myoblast differentiation by down regulating MyoD activity in a catalytic-independent manner. Cell. Signal. 35 48–60. 10.1016/j.cellsig.2017.03.008 [DOI] [PubMed] [Google Scholar]
- Zhang J., Na S., Pan S., Long S., Xin Y., Jiang Q., et al. (2019a). Inhibition of USP4 attenuates pathological scarring by downregulation of the TGF-β/Smad signaling pathway. Mol. Med. Rep. 20 1429–1435. 10.3892/mmr.2019.10370 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wan J., Xu Z., Hua T., Sun Q. (2019b). Exercise ameliorates insulin resistance via regulating TGFβ-activated kinase 1 (TAK1)-mediated insulin signaling in liver of high-fat diet-induced obese rats. J. Cell. Physiol. 234 7467–7474. 10.1002/jcp.27508 [DOI] [PubMed] [Google Scholar]
- Zhang L., Xing M., Wang X., Cao W., Wang H. (2017). MiR-148a suppresses invasion and induces apoptosis of breast cancer cells by regulating USP4 and BIM expression. Int. J. Clin. Exp. Pathol. 10 8361–8368. [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhou F., Drabsch Y., Gao R., Snaar-Jagalska B. E., Mickanin C., et al. (2012). USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-β type I receptor. Nat. Cell biol. 14 717–726. 10.1038/ncb2522 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Harding R., Hou F., Dong A., Walker J. R., Bteich J., et al. (2016). Structural basis of the recruitment of ubiquitin-specific protease USP15 by Spliceosome Recycling Factor SART3. J. Biol. Chem. 291 17283–17292. 10.1074/jbc.M116.740787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Berger F. G., Yang J., Lu X. (2011). USP4 inhibits p53 through deubiquitinating and stabilizing ARF-BP1. EMBO J. 30 2177–2189. 10.1038/emboj.2011.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Schlesiger C., Masucci M. G., Lindsten K. (2009). The ubiquitin specific protease 4 (USP4) is a new player in the Wnt signalling pathway. J. Cell. Mol. Med. 13 1886–1895. 10.1111/j.1582-4934.2009.00682.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Velasco K., Sompallae R., Pfirrmann T., Masucci M. G., Lindsten K. (2012). The ubiquitin specific protease-4 (USP4) interacts with the S9/Rpn6 subunit of the proteasome. Biochem. Biophys. Res. Commun. 427 490–496. 10.1016/j.bbrc.2012.09.075 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Wang F., Gao L., Xu L., Tong R., Lin N., et al. (2018). Ubiquitin-Specific protease 4 is an endogenous negative regulator of metabolic dysfunctions in nonalcoholic fatty liver disease in mice. Hepatology 68 897–917. 10.1002/hep.29889 [DOI] [PubMed] [Google Scholar]
- Zhong M., Jiang Q., Jin R. (2018). USP4 expression independently predicts favorable survival in lung adenocarcinoma. IUBMB Life 70 670–677. 10.1002/iub.1755 [DOI] [PubMed] [Google Scholar]
- Zhou F., Li F., Fang P., Dai T., Yang B., van Dam H., et al. (2016). Ubiquitin-Specific protease 4 antagonizes osteoblast differentiation through dishevelled. J. Bone Mineral Res. 31 1888–1898. 10.1002/jbmr.2863 [DOI] [PubMed] [Google Scholar]
- Zhou F., Xie F., Jin K., Zhang Z., Clerici M., Gao R., et al. (2017). USP4 inhibits SMAD4 monoubiquitination and promotes activin and BMP signaling. EMBO J. 36 1623–1639. 10.15252/embj.201695372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Zhang X., van Dam H., Ten Dijke P., Huang H., Zhang L. (2012). Ubiquitin-specific protease 4 mitigates Toll-like/interleukin-1 receptor signaling and regulates innate immune activation. J. Biol. Chem. 287 11002–11010. 10.1074/jbc.M111.328187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Qiu T., Wang T., Chen Z., Ma X., Zhang L., et al. (2019a). USP4 deficiency exacerbates hepatic ischaemia/reperfusion injury via TAK1 signalling. Clin. Sci. 133 335–349. 10.1042/CS20180959 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Liang P., Ji W., Yu Z., Chen H., Jiang L. (2019b). Ubiquitin-specific protease 4 promotes glioblastoma multiforme via activating ERK pathway. OncoTargets Therapy 12 1825–1839. 10.2147/OTT.S176582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Luo Z., Pan Y., Zheng W., Li W., Zhang Z., et al. (2019). H19/miR-148a/USP4 axis facilitates liver fibrosis by enhancing TGF-β signaling in both hepatic stellate cells and hepatocytes. J. Cell. Physiol. 234 9698–9710. 10.1002/jcp.27656 [DOI] [PubMed] [Google Scholar]