Abstract
Background
The COVID-19 pandemic has led to an unprecedented demand for testing - for diagnosis and prognosis - as well as for investigation into the impact of the disease on the host metabolism. Sebum sampling has the potential to support both needs by looking at what the virus does to us, rather than looking for the virus itself.
Methods
In this pilot study, sebum samples were collected from 67 hospitalised patients (30 COVID-19 positive and 37 COVID-19 negative) by gauze swab. Lipidomics analysis was carried out using liquid chromatography mass spectrometry, identifying 998 reproducible features. Univariate and multivariate statistical analyses were applied to the resulting feature set.
Findings
Lipid levels were depressed in COVID-19 positive participants, indicative of dyslipidemia; p-values of 0·022 and 0·015 were obtained for triglycerides and ceramides respectively, with effect sizes of 0·44 and 0·57. Partial Least Squares-Discriminant Analysis showed separation of COVID-19 positive and negative participants with sensitivity of 57% and specificity of 68%, improving to 79% and 83% respectively when controlled for confounding comorbidities.
Interpretation
COVID-19 dysregulates many areas of metabolism; in this work we show that the skin lipidome can be added to the list. Given that samples can be provided quickly and painlessly, we conclude that sebum is worthy of future consideration for clinical sampling.
Funding
The authors acknowledge funding from the EPSRC Impact Acceleration Account for sample collection and processing, as well as EPSRC Fellowship Funding EP/R031118/1, the University of Surrey and BBSRC BB/T002212/1. Mass Spectrometry was funded under EP/P001440/1.
Keywords: COVID-19 diagnostics, Sebomics, Multi-variate analysis, Lipidomics, Liquid chromatography-mass spectrometry
Research in context.
Evidence before this study
Previous studies have identified differences in the sebum of individuals with Parkinson's Disease and with Type 1 Diabetes Mellitus that can be detected by mass spectrometry; in the case of Parkinson's Disease changes to metabolism can even be detected by smell. COVID-19 is reported to cause a wide range of metabolic dysregulation, previously detected in blood and breath, suggesting that such disruption of normal metabolism could extend to the skin.
Added value of this study
In this pilot study of 67 participants, dysregulation of the sebum lipidome has been observed in COVID-19 positive participants, so adding skin to the list of organs that are disrupted by the disease. Analysis shows sebum triglycerides in particular as depressed in cases of the disease. This illustrates the additional metabolic information available from this biofluid, as well as suggesting potential for sebum to be investigated as a means of diagnostic testing.
Implications of all the available evidence
The work provides evidence of COVID-19 infection causing dyslipidemia in the stratum corneum. When adjusting for confounding factors, separation between COVID-19 positive and negative participants was also possible, with sensitivity and specificity similar to that seen in other matrices. Given that sebum samples can be provided (and transported) rapidly and painlessly, we conclude that sebum is worthy of future consideration for clinical sampling, both to offer insight into the COVID-19′s impact on skin and barrier function, and also to investigate whether larger cohorts would allow for sufficient stratification to improve sensitivity and selectivity further.
Alt-text: Unlabelled box
1. Introduction
SARS-CoV-2, a novel coronavirus, was identified by the World Health Organization as originating in the Wuhan province of China in late 2019, [1,2] and causes Corona Virus Disease 2019 (COVID-19). Mass testing has been identified by the World Health Organisation as a key weapon in the battle against COVID-19 to contain outbreaks and reduce hospitalisations [3]. Current approaches to testing require the detection of SARS-CoV-2 viral RNA collected from the upper respiratory tract via polymerase chain reaction (PCR). Whilst these types of tests are easily deployable and highly selective for the virus, they suffer from a significant proportion of false negative events; [4] in addition, scarcity of reagents can be an issue for the scale of testing required. Furthermore, PCR approaches are diagnostic rather than prognostic in nature [5].
Approaches that measure the indirect effect of the virus on the host (as opposed to direct measurement of the virus itself) may offer a complementary solution in clinical or mass testing settings; for example, one feasibility study has recently identified derangement of breath biochemistry in COVID-19 patients [6]. As the coronavirus requires lipids for reproduction, COVID-19 can be expected to disrupt the lipidome [7]. Evidence of a dysregulated lipidome has been observed in patients with COVID-19; triglyceride levels have been found to be significantly increased in the serum and plasma of COVID-19 patients, alongside changes in lipoprotein particle size and distribution. This has been characterised as pathogenic in nature and consistent with increased atherosclerotic risk; [8,9] lipidomics therefore offers a promising route to better understanding of - and potentially diagnosis for - COVID-19. Sebum is a biofluid secreted by the sebaceous glands and is rich in lipids. A sample can be collected easily and non-invasively via a gentle swab of skin areas rich in sebum (for example the face, neck or back). Characteristic features have previously been identified from sebum for a limited number of conditions such as Parkinson's Disease and Type 1 Diabetes Mellitus; [10,11,12] sebum changes have also been reported in pregnancy [13]. In addition, whilst the mechanisms for the role of sebum in barrier function are not fully described, sebum lipids provide this function directly and also through commensal bacteria interactions; lipid dysregulation would have implications for skin health [14]. In this work, we explore differences in sebum lipid profiles for patients with and without COVID-19, with a view to exploring sebum's future use as a non-invasive sampling medium, as well as expanding the understanding of sebum as a sampling matrix.
2. Methods
2.1. Background
In May 2020 several UK bodies announced their intention to pool resources and form the COVID-19 International Mass Spectrometry (MS) Coalition [5]. This consortium has the proximal goal of providing molecular level information on SARS-CoV-2 in infected humans, with the distal goal of understanding the impact of the novel coronavirus on metabolic pathways in order to better diagnose and treat cases of COVID-19 infection. This work took place as part of the COVID-19 MS Coalition and all data will be stored and fully accessible on the MS Coalition open repository.
2.2. Participant recruitment and ethics
Ethical approval for this project (IRAS project ID 155921) was obtained via the NHS Health Research Authority (REC reference: 14/LO/1221). The participants included in this study were recruited at NHS Frimley Park NHS Trust, totalling 67 participants, during May and June 2020. Collection of the samples was performed by researchers from the University of Surrey at Frimley Park NHS Foundation Trust hospitals. Participants were identified by clinical staff to ensure that they had the capacity to consent to the study, and were asked to sign an Informed Consent Form; those that did not have this capacity were not sampled. Consenting participants were categorised by the hospital as either “query COVID” (meaning there was clinical suspicion of COVID-19 infection) or “COVID positive” (meaning that a positive COVID test result had been recorded during their admission). All participants were provided with a Patient Information Sheet explaining the goals of the study.
2.3. Sample collection, inactivation and extraction
Patients were sampled immediately upon recruitment to the study. This meant that the range in time between symptom onset and sebum sampling ranged from 1 day to > 1 month, an inevitable consequence of collecting samples in a pandemic situation. Each participant was swabbed on the right side of the upper back, using 15 cm by 7.5 cm gauzes that had each been folded twice to create a four-ply swab. The surface area of sampling was approximately 5 cm × 5 cm, pressure was applied uniformly whilst moving the swab across the upper back for ten seconds. The gauzes were placed into Sterilin polystyrene 30 mL universal containers.
Samples were transferred from the hospital to the University of Surrey by courier within 4 h of collection, whereupon the samples were then quarantined at room temperature for seven days to allow for virus inactivation, avoiding heat treatment given potential for lipid degradation [15]. Finally, the vials were transferred to −80 °C storage until required. Alongside sebum collection, metadata for all participants was also collected covering inter alia sex, age, comorbidities (based on whether the participant was receiving treatment), the results and dates of COVID PCR (polymerase chain reaction) tests, bilateral chest X-Ray changes, smoking status, and whether the participant presented with clinical symptoms of COVID-19. Values for lymphocytes, CRP and eosinophils were also taken - here values within five days of sebum sampling were recorded.
The extraction, storage and reconstitution of the obtained samples followed Sinclair E, Trivedi D, Sarkar D, et al. [16] Samples were analysed over a period of five days. Each day consisted of a run incorporating solvent blank injections (n = 5), pooled QC injections (n = 3), followed by 16 participant samples (triplicate injections of each) with a single pooled QC injection every six injections. Each day's run was completed with pooled QC injections (n = 2) and solvent blanks (n = 3). A triplicate injection of a field blank was also obtained.
2.4. Instrumentation and software
Analysis of samples was carried out using a Dionex Ultimate 3000 HPLC module equipped with a binary solvent manager, column compartment and autosampler, coupled to a Orbitrap Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific, UK) at the University of Surrey‘s Ion Beam Centre. Chromatographic separation was performed on a Waters ACQUITY UPLC BEH C18 column (1·7 µm, 2·1 mm × 100 mm) operated at 55 °C with a flow rate of 0·3 ml min-1.
The mobile phases were as follows: mobile phase A was acetonitrile:water (v/v 60:40) with 0·1% formic acid, whilst mobile phase B was 2-propanol:acetonitrile (v/v, 90:10) with 0·1% formic acid (v/v). An injection volume of 5 µL was used. The initial solvent mixture was 40% B, increasing to 50% B over 1 min, then to 69% B at 3·6 min, with a final ramp to 88% B at 12 min. The gradient was reduced back to 40% B and held for 2 min to allow for column equilibration. Analysis on the Q-Exactive Plus mass spectrometer was performed in split-scan mode with an overall scan range of 150 m/z to 2 000 m/z, and 5 ppm mass accuracy. Split scan was chosen to extend the m/z range from 150 to 2 000 m/z whilst maximising the number of features identified [17,18]. MS/MS validation of features was carried out on Pooled QC samples using data dependent acquisition mode. Operating conditions are summarised in Table S1 (Supplementary Materials).
2.5. Materials and chemicals
The materials and solvents utilised in this study were as follows: gauze swabs (Reliance Medical, UK), 30 mL Sterilin™ tubes (Thermo Scientific, UK), 10 mL syringes (Becton Dickinson, Spain), 2 mL microcentrifuge tubes (Eppendorf, UK), 0.2 µm syringe filters (Corning Incorporated, USA), 200 µL micropipette tips (Starlab, UK) and Qsert™ clear glass insert LC vials (Supelco, UK). Optima™ (LC-MS) grade methanol was used as an extraction solvent, and Optima™ (LC-MS) grade methanol, ethanol, acetonitrile and 2-propanol were used to prepare injection solvents and mobile phases. Formic acid was added to the mobile phase solvents at 0.1% (v/v). Solvents were purchased from Fisher Scientific, UK.
2.6. Data processing
LC-MS outputs (.raw files) were pre-processed for alignment, normalisation and peak identification using Progenesis QI (Non-Linear Dynamics, Waters, Wilmslow, UK), a platform-independent small molecule discovery analysis software for LC-MS data. Peak picking (mass tolerance ±5 ppm), alignment (RT window ±15 s) and area normalisation was carried out with reference to the pooled QC samples. Features identified in MS were initially annotated using accurate mass match with Lipid Blast in Progenesis QI, whilst validation was performed using data dependent MS/MS analysis using LipidSearch (Thermo Fisher Scientific, UK) and Compound Discoverer (Thermo Fisher Scientific, UK), without imputation of missing values. This process yielded an initial peak table with 14,160 features. All those features with a coefficient of variation across all pooled QCs above 20% were removed, as were those that were not present in at least 90% of pooled QC injections. These features were then field blank adjusted: all those features with a signal to noise ratio below 3x were also rejected. The remaining set of 998 features were deemed to be robust, reproducible and suitably distinct from those found in the field blank.
Inclusion criteria were also applied to participant data, requiring both full completion of metadata and also agreement between the result of the PCR COVID-19 test (Y/N) and the clinical diagnosis for COVID-19 (Y/N). Whilst these inclusion criteria reduced the total number of participants from n = 87 to n = 67 (nine participants did not have complete metadata, seven participants presented with clinical symptoms but had a negative RT-PCR result and four participants had a positive RT-PCR result but no symptoms), this was considered worthwhile given the potential for misdiagnosis to confound the development of statistical models.
2.7. Statistical analysis
Data processing and analysis of the pareto-scaled peak:area matrix was conducted through a combination of the R package mixOmics, [19] supplemented by user-written scripts in the statistical programming language R. [20] PLS-DA was used for classification and prediction of data. Separation and classification was based on mahalabonis distance between observations. Leave-one-out cross-validation was used for PLS-DA model validation to test accuracy, sensitivity and specificity; variable importance in projection (VIP) scores were used to assess feature significance.
2.8. Role of funding source
Funding was provided for sample collection by the EPSRC Impact Acceleration Account, as well as EPSRC Fellowship Funding EP/R031118/1. Mass Spectrometry was funded under EP/P001440/1. Sample collection and processing was funded by the University of Surrey and the BBSRC BB/T002212/1. The funding bodies were neither involved in the design of the study nor in the analysis of the data.
3. Results
3.1. Population metadata overview
The study population analysed in this work included 67 participants, comprising 30 participants presenting with COVID-19 clinical symptoms (and an associated positive COVID-19 RT-PCR test) and 37 participants presenting without. A summary of the metadata is shown in Table 1.
Table 1.
Summary of clinical characteristics by participant cohort.
| Parameters | Negative for COVID-19 | Positive for COVID-19 |
|---|---|---|
| n | 37 | 30 |
| Age (mean, standard deviation; years) | 65•0 ± 19•3 | 64•7 ± 19•4 |
| Male / Female (n) | 18 / 19 | 17 / 13 |
| Treated for Hypertension (n) | 17 | 10 |
| Treated for High Cholesterol (n) | 10 | 5 |
| Treated for Type 2 Diabetes Mellitus (n) | 12 | 7 |
| Treated for ischaemic Heart Disease (n) | 7 | 4 |
| Current Smoker (n) | 1 | 0 |
| Ex-Smoker (n) | 10 | 4 |
| Medical Acute Dependency admission (n) | 4 | 11 |
| Intensive Care Unit admission (n) | 0 | 5 |
| Survived Admission (n) | 35 | 27 |
| Time between onset and sebum test (mean, standard deviation; days) | NA | 19 ± 8 |
| Time between positive RT-PCR test and sebum test | NA | 3 ± 7 |
| Lymphocytes (mean, standard deviation; cells / μL) | 1•0 ± 0•5 | 0•6 ± 0•3 |
| C-Reactive Protein (mean, standard deviation; mg / L) | 132•1 ± 95•7 | 181•3 ± 117•2 |
| Eosinophils (mean, standard deviation; 100 / μL) | 0•3 ± 0•4 | 0•2 ± 0•4 |
| Bilateral Chest X-Ray changes (n) | 2 | 21 |
| Continuous Positive Airway Pressure (n) | 2 | 10 |
| O2 required (n) | 10 | 20 |
There were more male participants in the COVID-19 positive group (M:F ratio of 0·57) compared to the participant population overall (M:F ratio of 0·52); given recruitment took place in a hospital environment, this may reflect increased severity amongst males [21]. Age distributions for COVID-19 positive and negative cohorts were almost identical (mean age of 64·7 years and 65·0 years respectively). On average 19 days had elapsed from symptoms onset to sebum swab, consistent with recruitment during hospitalisation. Comorbidities are associated with both hospitalisation and more severe outcomes for COVID-19 infection, but will also alter the metabolome of participants, representing both a causative and confounding factor. The impact on classification accuracy of these comorbidities was tested by stratifying participant data by comorbidity to see if separation improved; this process is described in the following sections. In this pilot study, comorbidities were less well represented in the cohort of COVID-19 positive participants than in the cohort of COVID-19 negative participants.
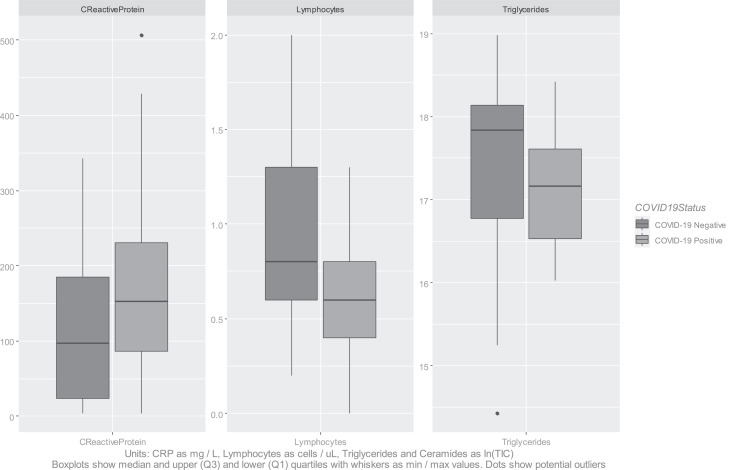
Levels of C-Reactive Protein (CRP) were significantly higher for COVID-19 participants, whilst lymphocyte and eosinophils levels were lower. A two-tailed Mann Whitney U test on the CRP indicator provided a p-value of 0·031, and on the lymphocytes a p-value of 0·004. Effect sizes (calculated by Cohen's D) were 0·56 and 0·85 respectively. COVID-19 positive participants were also more likely to present with bilateral chest X-ray changes (21 out of 30 COVID-19 positive patients, versus 2 out of 37 COVID-19 negative patients). COVID-19 positive participants experienced higher rates of requiring oxygen / CPAP, higher rates of escalation, and lower survival rates (Table 1). These observations were in agreement with literature descriptions of COVID-19 symptoms and progression [22].
3.2. Overview of features identified by liquid chromatography mass spectrometry (LC-MS)
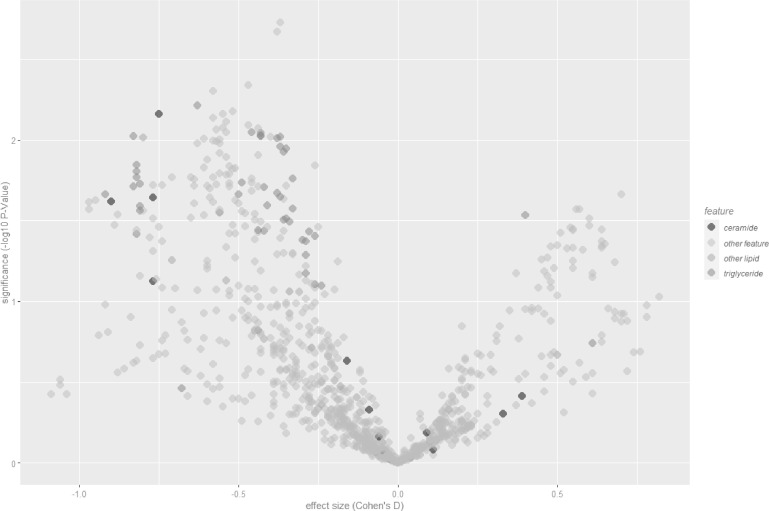
998 features were identified reproducibly by LC-MS (present in greater than 90% of pooled QC LC-MS injections, coefficient of variation below 20% across pooled QCs, signal to noise ratio greater than three) and these formed the basis of the analysis in this work. Differences between COVID-19 positive and negative participants were observed across a range of lipids, with the most consistent difference seen in reduced lipid levels, especially triglycerides (Fig. 1).
Fig. 1.
Volcano plot of features for COVID-19 positive (n = 30) versus negative (n = 37), labelled lipids validated by MS/MS.
Aggregate levels of triglycerides identified by MS/MS were depressed for COVID-19 positive participants, and also for ceramides, albeit fewer lipids of the latter class were identified and validated. The lipids with the most statistically significant differences between the populations are listed in Table S2 (Supplementary Materials) together with their m/z values and p-values, alongside equivalent statistics for other indicators. The distributions of the natural log of aggregated lipid ion counts by class were not characterised as normal by Shapiro-Wilk normality tests [23]. Two-tailed Mann-Whitney U-tests were performed to test the significance of aggregate levels of these lipid classes. These resulted in p-values of 0·022 and 0·015 for triglycerides and ceramides respectively, with effect sizes (calculated by Cohen's D) of 0·44 and 0·57, indicative of medium effect size; the statistical significance of the alteration in levels of triglycerides between positive and negative cohorts is comparable to that for CRP or for lymphocytes (Fig. 2). These results are suggestive of dyslipidemia within the stratum corneum due to COVID-19.
Fig. 2.
Boxplots of diagnostic indicators versus triglyceride levels.
3.3. Population-level clustering analyses
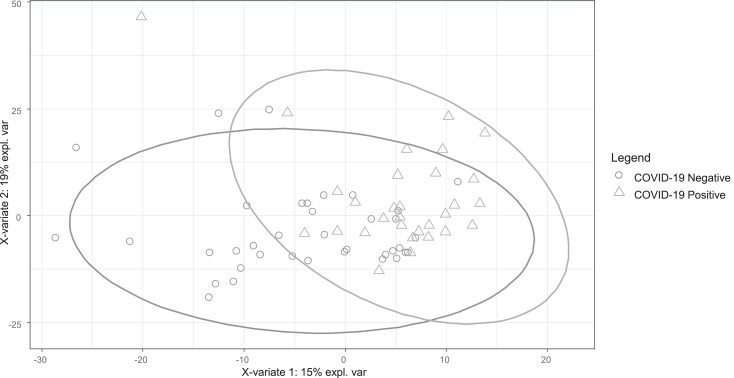
No clustering was identifiable at the total population level by principal component analysis (PCA), i.e. by unsupervised analysis. Partial least squares discriminant analysis (PLS-DA) performed on the same data set revealed limited separation (Fig. 3), with the area under the receiver operating curve (AUROC) over two components of 0.88. AUROC can be inflated when only used on a single training data set, and so a confusion matrix was constructed using a leave-one-out approach. Validating accuracy in this way (Table 2) showed sensitivity of just 57% and specificity of 68%. Given the wide range of comorbidities, this is not unexpected.
Fig. 3.
PLS-DA plot for 67 participants, classified by COVID-19 positive / negative.
Table 2.
Confusion matrix for COVID-19 positive versus negative (all participants).
| All Participants (n = 67) | True COVID-19 Positive (n = 30) | True COVID-19 Negative (n = 37) |
|---|---|---|
| Predicted COVID-19 Positive (%, n) | 57% (17) | 32% (12) |
| Predicted COVID-19 Negative (%, n) | 43% (13) | 68% (25) |
3.4. Investigation of confounding factors
To test the impact of age, diagnostic indicators (CRP, lymphocytes and eosinophils) and time elapsed between onset of symptoms and sebum sampling, these variables were pareto-scaled and included in the matrix for PLS-DA modelling. Variable importance in projection (VIP) scores for lymphocytes, CRP, and eosinophils were 2·47, 1·77 and 0.72 respectively, ranking 1, 90 and 466 out of 1003 total features. As a single feature, depressed lymphocyte levels show high correlation with COVID-19 positive status, consistent with lymphocyte count being both a diagnostic and prognostic biomarker [24]. Age as a vector had a VIP score of just 0·05 (ranking 958 out of 1002 total features), indicating that age is a smaller influencer of stratum corneum lipids than other factors. Days elapsed between onset of symptoms and sebum sampling ranked 102 out of the total features in importance, indicating that although the variation in time between onset and sampling was suboptimal it did not prove a major confounder in this analysis.
Overall, PLS-DA separation improved by the addition of lymphocyte and CRP indicators, with slight model accuracy increases when these two variables were included in the feature matrix (from 62% to 64% accuracy for the overall population, for example). Given that this work focuses on sebum sampling, however, in the analyses that follow only features obtained from sebum are included, i.e. information from other diagnostic indicators is excluded from classification models.
To test whether separation based on sebum alone would improve in smaller / more homogenous groups, separate PLS-DA models were built for each split of the population by comorbidity. If model performance improved (measured by predictive power - Q2Y - and sensitivity and specificity via leave-one-out cross validation) then this could indicate that sebum lipid profiling would perform better if models were constructed based on stratified and matched datasets. Table 3 shows the results for these metrics across the different modelled subsets.
Table 3.
Summary of model parameters for different population subsets.
| Population | Category 1 (n) | Category 2 (n) | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|
| All participants | COVID-19 Positive (30) | COVID-19 Negative (37) | 62% | 57% | 68% |
| Male | COVID-19 Positive (17) | COVID-19 Negative (18) | 66% | 65% | 67% |
| Female | COVID-19 Positive (13) | COVID-19 Negative (19) | 59% | 54% | 63% |
| Type 2 Diabetes Mellitus | COVID-19 Positive (7) | COVID-19 Negative (12) | 73% | 71% | 75% |
| High Cholesterol | COVID-19 Positive (5) | COVID-19 Negative (10) | 87% | 100% | 80% |
| Hypertension | COVID-19 Positive (10) | COVID-19 Negative (17) | 81% | 80% | 82% |
| Ischaemic Heart Disease | COVID-19 Positive (4) | COVID-19 Negative (7) | 68% | 50% | 86% |
| Statins | COVID-19 Positive (11) | COVID-19 Negative (21) | 63% | 55% | 90% |
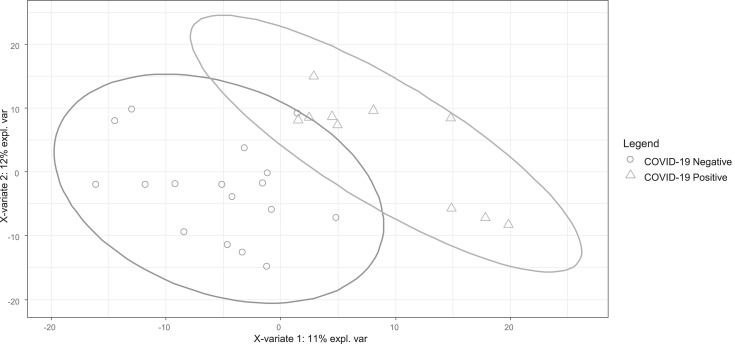
Separation generally improved as the data were grouped more finely and modelled predictive power improved. Based on an average weighted by most significant (in terms of predictive power) comorbidity for these subsets, sensitivity improved to 79% and specificity improved to 83%, with overall accuracy improving to 82%. For example, PLS-DA modelling of the subset of 27 participants under medication for hypertension (Fig. 4) showed both good separation and better sensitivity and specificity (Table 4). These data suggest that comorbidities are confounders in skin lipidomics.
Fig. 4.
PLS-DA plot for 15 participants with hypertension, COVID-19 positive / negative.
Table 4.
Confusion matrix for COVID-19 positive versus negative (participants with hypertension).
| Hypertension (n = 27) | True COVID-19 Positive (n = 10) | True COVID-19 Negative (n = 17) |
|---|---|---|
| Predicted COVID-19 Positive (%, n) | 80% (8) | 18% (3) |
| Predicted COVID-19 Negative (%, n) | 20% (2) | 82% (14) |
Similarly, PLS-DA modelling of the subset of participants under medication for high cholesterol showed good separation (Figure S2 and Table S3, Supplementary Materials), with sensitivity of 100% and specificity of 80%. This subgroup of 15 was treated with lipid-lowering agents, specifically statins. The subgroup of 11 comprising participants undergoing treatment for ischaemic heart disease (IHD) also showed much better separation (Figure S3 and Table S4, Supplementary Materials), with better overall accuracy, with sensitivity and specificity of 50% and 86% respectively. This subgroup received varied medication, but participants presenting with IHD were also being prescribed statins. Finally, the subset of participants under medication for T2DM (Figure S4 and Table S5, Supplementary Materials) also showed both good separation and better sensitivity and specificity (of 71% and 75% respectively). This subgroup of 19 was typically being treated with oral hypoglycaemics, in four cases with insulin alone and in three instances with diet control only.
Model performance (Figure S5 and Table S6, Supplementary Materials) also improved versus the base population for a stratified dataset based on those participants taking statins (sensitivity of 55% and specificity of 90%). Given that statins control cholesterol and lipid levels, this may have provided a more similar “baseline” against which to measure perturbance in the lipidome by COVID-19; patients taking statins which included both participants treated for high cholesterol and also participants with poor diabetic control or history of ischaemic heart disease, where statins are routinely added prophylactically to improve long-term outcomes.
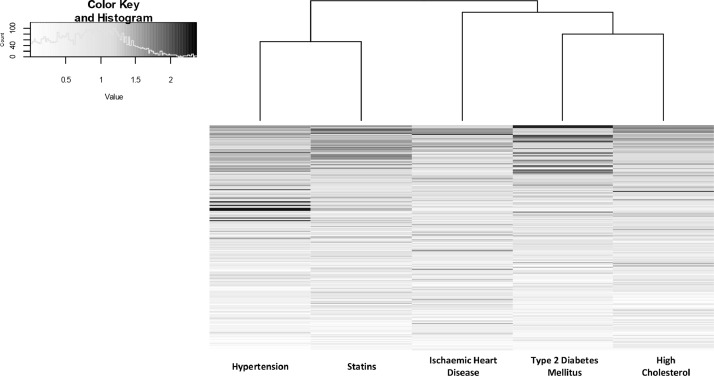
Looking across the models, there was commonality in the features identified as significant in differentiating between COVID-19 positive and negative. Many features featured in all subsets with VIP scores above 2 (dark grey in Fig. 5), but others did not, a possible indicator of overfitting due to the smaller groups when stratified. Where overlap does occur between the features, this may reflect the natural overlap between the subset populations, for example the subset of participants presenting with high cholesterol is largely a subset of the participants receiving treatment by statins.
Fig. 5.
Heat map of VIP scores ranked by commonality to different subgroup PLS-DA models.
Triglycerides made up the majority of lipids with the highest common VIP scores. Triglycerides also dominated the list of lipids validated by MS/MS with the most statistically significant differences between overall COVID-19 positive and negative participants (Table S2, Supplementary Materials). Odd-chain fatty acids were present in eleven of the twenty most statistically significant lipids, unusual within the mammalian lipidome, but consistent with odd-chain fatty acids being more prevalent in sebaceous gland lipids than other tissues [25,26]. MS/MS spectra for selected lipids of high statistical significance with regard to differentiating between COVID-19 positive and negative, including lipids with odd-chain fatty acids, are shown in Figure S1 (Supplementary Materials).
4. Discussion
At the aggregate level, analysis of the metadata for the participants in this study illustrates the challenges involved in constructing a well-designed sample set during a pandemic. Age ranges of participants were large, a wide range of comorbidities were present, and the time between symptom onset and sebum sampling ranged from 1 day to > 1 month, leading to many confounding factors. In addition, participant recruitment of the most severely affected was limited by ethics approval only covering patients who could give informed consent. Definitive separation has not proved possible in this pilot study, given that too few datapoints were available to rigorously stratify by medication or by comorbidity. In addition, the requirement to store the samples for seven days prior to analysis may have altered the lipid profile, albeit – with the exception of squalene – the major lipid classes in sebum including triglycerides have previously been found to be stable at ambient temperatures for up to a month [27]. In future studies storing gauzes in methanol would be preferable to halt enzymatic activity during the storage period [28].
Nonetheless, at the aggregate level, participants with a positive clinical COVID-19 diagnosis present with depressed lipid levels (odd-chain triglycerides in particular), with the possibility of an altered microbiome on the skin surface, reduced barrier function and skin health. Other work has found evidence of dyslipidemia in plasma from COVID-19 positive patients, [29,30,9] although evidence of whether upregulation or downregulation is dominant for these lipid classes is mixed. Plasma triglyceride levels have been found to be elevated in blood plasma for mild cases of COVID-19, but triglyceride levels in plasma may also decline as the severity of COVID-19 increased [31].
It should be remembered, however, that the primary role of skin is barrier function, and lipid expression in the stratum corneum depends on de novo lipogenesis – in fact nonskin sources such as plasma provide only a minor contribution to sebum lipids, [32] which limits the relevance of broader pathway analysis to this biofluid.
Furthermore, these findings suggest that better stratification of participants could yield a clearer separation of positive and negative COVID-19 participants by their lipidomic profile. The overall accuracy in the stratified groups of 82% is comparable to that recently reported using breath biochemistry of 81%, [4] albeit overfitting is a risk in any pilot study with small n. This risk can only be reduced through both a larger training set of data and subsequently testing the models on future independent validation sets, made possible through cohesive efforts such as the work of the MS Coalition.
Another point to note is a possible lack of confounders in the participant population from seasonal respiratory viruses or seasonal environment factors (humidity, temperature). Whilst the COVID-negative patients included patients with respiratory illnesses (e.g. COPD, asthma) and COVID-like symptoms, samples were collected between May and July, when the incidence of respiratory viruses is generally low. Both the common cold and influenza have some symptoms overlap with COVID-19 and may possibly lead to alterations to lipid metabolism that could interfere with the identification of features related to COVID-19 infection. Such viruses within the UK are more prevalent in autumn and winter [33]. Whilst it seems unlikely that seasonal respiratory viruses were a major confounding factor in this work, this is a factor that will need to be taken into account in future studies along with seasonal environmental influences. This may also allow the opportunity to test sebum's selectivity and specificity with regard to other respiratory viruses.
In conclusion, we provide preliminary evidence that COVID-19 infection leads to dyslipidemia in the stratum corneum. We further find that the sebum profiles of COVID positive and negative patients can be separated using the multivariate analysis method PLS-DA, with the separation improving when the patients are segmented in accordance with certain comorbidities. Given that sebum samples can be provided quickly and painlessly, we conclude that sebum is worthy of future consideration for clinical sampling for COVID-19 infection.
Declaration of Competing Interest
Dr Barran and Dr Trivedi report patent PCT/GB2019/052169. The other authors have no interests to declare.
Acknowledgments
Data sharing statement
Participant metadata data with identifiers, alongside mass spectrometry .RAW files will be made available on the Mass Spectrometry Coalition website upon publication of this study. The analytical protocols used as well as sample and participant data will be openly available for all researchers to access. The website URL is https://covid19-msc.org/
Author's contributions
MS was responsible for statistical analysis and authorship of the manuscript. MS and MW extracted the samples, with MW providing additional advice on multivariate analysis. KL, CF and AS collected all samples used in this work; AS and DDW obtained ethical approval. GE and DG facilitated access to participants and collected participant metadata. PB, DT and AP advised on sampling protocol and on mass spectrometry acquisition and protocols. CC and HL assisted with mass spectrometry method development. MB obtained funding for the study, and was responsible for supervision of the research team.
Acknowledgments
The authors acknowledge Samiksha Ghimire from Groningen Medical School for translation of participant information sheets and consent forms into Nepalese. The authors acknowledge Amanda Souza and Ioanna Ntai of Thermo Fisher, as well as Mason Malloy, Patrick Sears and Janella de Jesus of the University of Surrey, for their help with method development. We are grateful to Thanuja Weerasinge (Jay), Manjula Meda, Chris Orchard and Joanne Zamani of Frimley Park NHS Foundation Trust for their help with ethics approvals and access to hospital patients. We also thank the reviewers of this manuscript for their improvements and insights.
Funding
The authors would like to acknowledge funding from the EPSRC Impact Acceleration Account for sample collection, as well as EPSRC Fellowship Funding EP/R031118/1. Mass Spectrometry was funded under EP/P001440/1. Sample collection and processing was funded by the University of Surrey and the BBSRC BB/T002212/1
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.eclinm.2021.100786.
Appendix. Supplementary materials
References
- 1.WHO. WHO advice for international travel and trade in relation to the outbreak of pneumonia caused by a new coronavirus in China. 2019. https://www.who.int/news-room/articles-detail/who-advice-for-international-travel-and-trade-in-relation-to-the-outbreak-of-pneumonia-caused-by-a-new-coronavirus-in-china (accessed July 27, 2020).
- 2.WHO. Novel Coronavirus – China. 2020. https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/(accessed July 27, 2020).
- 3.World Health Organization. Simulation of the effects of COVID-19 testing rates on hospitalizations. https://www.who.int/bulletin/volumes/98/5/20-258186/en/(accessed Sept 29, 2020). [DOI] [PMC free article] [PubMed]
- 4.Surkova E., Nikolayevskyy V., Drobniewski F. False-positive COVID-19 results: hidden problems and costs. Lancet Respir Med. 2020;8:1167–1168. doi: 10.1016/S2213-2600(20)30453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Struwe W., Emmott E., Bailey M. The COVID-19 MS coalition—accelerating diagnostics, prognostics, and treatment. Lancet. 2020;395:1761–1762. doi: 10.1016/S0140-6736(20)31211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruszkiewicz D.M., Sanders D., O'Brien R. Diagnosis of COVID-19 by analysis of breath with gas chromatography-ion mobility spectrometry - a feasibility study. EClinicalMedicine. 2020 doi: 10.1016/j.eclinm.2020.100609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abu-Farha M., Thanaraj T.A., Qaddoumi M.G., Hashem A., Abubaker J., Al-Mulla F. The role of lipid metabolism in COVID-19 virus infection and as a drug target. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21103544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruzzone C., Bizkarguenaga M., Gil-Redondo R. SARS-CoV-2 infection dysregulates the metabolomic and lipidomic profiles of serum. iScience. 2020;23 doi: 10.1016/j.isci.2020.101645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu D., Shu T., Yang X. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl Sci Rev. 2020;7:1157–1168. doi: 10.1093/nsr/nwaa086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinclair E., Trivedi D., Sarkar D., et al. Sebum: a window into dysregulation of mitochondrial metabolism in Parkinson's disease. 2020. DOI:10.26434/chemrxiv.11603613.
- 11.Shetage S.S., Traynor M.J., Brown M.B., Galliford T.M., Chilcott R.P. Application of sebomics for the analysis of residual skin surface components to detect potential biomarkers of type-1 diabetes mellitus. Sci Rep. 2017;7:1–8. doi: 10.1038/s41598-017-09014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barran, Perdita; Sarkar, Depanjan; Trivedi, drupad; Tilo, Kunath; Milne, joy; S.E. Biomarkers and Uses Therof. 2019.
- 13.Chen F., Zhou M., Huang H., Jia Y. Study on the skin status of mid-pregnancy women based on lipidomics. J Cosmet Dermatol. 2020:1–9. doi: 10.1111/jocd.13866. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi T., Voisin B., Kim D.Y. Homeostatic control of sebaceous glands by innate lymphoid cells regulates commensal bacteria equilibrium. Cell. 2019;176:982–997. doi: 10.1016/j.cell.2018.12.031. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loo R.L., Lodge S., Kimhofer T. Quantitative in-vitro diagnostic NMR spectroscopy for lipoprotein and metabolite measurements in plasma and serum: recommendations for analytical artifact minimization with special reference to COVID-19/SARS-CoV-2 samples. J Proteome Res. 2020;19:4428–4441. doi: 10.1021/acs.jproteome.0c00537. [DOI] [PubMed] [Google Scholar]
- 16.Sinclair E., Trivedi D., Sarkar D., et al. Sebum: a Window into dysregulation of mitochondrial metabolism in Parkinson's disease. 2020. DOI:10.26434/chemrxiv.11603613.
- 17.Fall F., Lenuzza N., Lamy E. A split-range acquisition method for the non-targeted metabolomic profiling of human plasma with hydrophilic interaction chromatography - high-resolution mass spectrometry. J Chromatogr B Anal Technol Biomed Life Sci. 2019;1128 doi: 10.1016/j.jchromb.2019.121780. [DOI] [PubMed] [Google Scholar]
- 18.Ranninger C., Schmidt L.E., Rurik M. Improving global feature detectabilities through scan range splitting for untargeted metabolomics by high-performance liquid chromatography-Orbitrap mass spectrometry. Anal Chim Acta. 2016;930:13–22. doi: 10.1016/j.aca.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 19.Rohart F., Gautier B., Singh A. Lê Cao KA. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Core Team. R: a language and environment for statistical computing. 2020.
- 21.Wenham C., Smith J., Morgan R. COVID-19: the gendered impacts of the outbreak. Lancet. 2020;395:846–848. doi: 10.1016/S0140-6736(20)30526-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knight S.R., Ho A., Pius R. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO clinical characterisation protocol: development and validation of the 4C Mortality Score. BMJ. 2020;370:m3339. doi: 10.1136/bmj.m3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yap B.W., Sim C.H. Comparisons of various types of normality tests. J Stat Comput Simul. 2011;81:2141–2155. [Google Scholar]
- 24.Wagner J., DuPont A., Larson S., Cash B., Farooq A. Absolute lymphocyte count is a prognostic marker in Covid-19: a retrospective cohort review. Int J Lab Hematol. 2020 doi: 10.1111/ijlh.13288. n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z., Park H.G., Wang D.H., Kitano R., Kothapalli K.S.D., Brenna J.T. Fatty acid desaturase 2 (FADS2) but not FADS1 desaturates branched chain and odd chain saturated fatty acids. Biochim Biophys Acta - Mol Cell Biol Lipids. 2020;1865 doi: 10.1016/j.bbalip.2019.158572. [DOI] [PubMed] [Google Scholar]
- 26.Kothapalli K., Wang Z., Park H.G., Wang D.H., Kitano R., Brenna J.T. Fatty acid desaturase 2 (FADS2) actions on branched chain and normal odd chain saturated fatty acids. Curr Dev Nutr. 2020;4:1261. doi: 10.1016/j.bbalip.2019.158572. 1261. [DOI] [PubMed] [Google Scholar]
- 27.Stefaniak A.B., Harvey C.J., Wertz P.W. Formulation and stability of a novel artificial sebum under conditions of storage and use. Int J Cosmet Sci. 2010;32:347–355. doi: 10.1111/j.1468-2494.2010.00561.x. [DOI] [PubMed] [Google Scholar]
- 28.Shui G., Cheong W.F., Jappar I.A. Derivatization-independent cholesterol analysis in crude lipid extracts by liquid chromatography/mass spectrometry: applications to a rabbit model for atherosclerosis. J Chromatogr A. 2011;1218:4357—4365. doi: 10.1016/j.chroma.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 29.Song J.W., Lam S.M., Fan X. Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis. Cell Metab. 2020;32:188–202. doi: 10.1016/j.cmet.2020.06.016. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Overmyer K.A., Shishkova E., Miller I.J. Large-scale Multi-omic Analysis of COVID-19 Severity. Cell Syst. 2020:1–18. doi: 10.1016/j.cels.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei X., Zeng W., Su J. Hypolipidemia is associated with the severity of COVID-19. J Clin Lipidol. 2020;14:297–304. doi: 10.1016/j.jacl.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esler W.P., Tesz G.J., Hellerstein M.K. Human sebum requires de novo lipogenesis, which is increased in acne vulgaris and suppressed by acetyl-CoA carboxylase inhibition. Sci Transl Med. 2019;11:1–14. doi: 10.1126/scitranslmed.aau8465. [DOI] [PubMed] [Google Scholar]
- 33.Eyre M.T., Burns R., Kirkby V. Impact of baseline cases of cough and fever on UK COVID-19 diagnostic testing rates: estimates from the Bug Watch community cohort study. medRxiv. 2020 doi: 10.12688/wellcomeopenres.16304.1. 2020.09.03.20187377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.