Abstract
Background
Use of lipoprotein(a) concentrations for identification of individuals at high risk of cardiovascular diseases is hampered by the size polymorphism of apolipoprotein(a), which strongly impacts immunochemical methods, resulting in discordant values. The availability of a reference method with accurate values expressed in SI units is essential for implementing a strategy for assay standardization.
Method
A targeted LC-MS/MS method for the quantification of apolipoprotein(a) was developed based on selected proteotypic peptides quantified by isotope dilution. To achieve accurate measurements, a reference material constituted of a human recombinant apolipoprotein(a) was used for calibration. Its concentration was assigned using an amino acid analysis reference method directly traceable to SI units through an unbroken traceability chain. Digestion time-course, repeatability, intermediate precision, parallelism, and comparability to the designated gold standard method for lipoprotein(a) quantification, a monoclonal antibody-based ELISA, were assessed.
Results
A digestion protocol providing comparable kinetics of digestion was established, robust quantification peptides were selected, and their stability was ascertained. Method intermediate imprecision was below 10% and linearity was validated in the 20–400 nmol/L range. Parallelism of responses and equivalency between the recombinant and endogenous apo(a) were established. Deming regression analysis comparing the results obtained by the LC-MS/MS method and those obtained by the gold standard ELISA yielded y = 0.98*ELISA +3.18 (n = 64).
Conclusions
Our method for the absolute quantification of lipoprotein(a) in plasma has the required attributes to be proposed as a candidate reference method with the potential to be used for the standardization of lipoprotein(a) assays.
Keywords: Lipoprotein(a), apolipoprotein(a), standardization, reference method
Introduction
With multiple studies confirming the causal link between lipoprotein(a) [Lp(a)] concentrations and increased risk of cardiovascular disease (1–4), this past decade has seen a strongly renewed interest in Lp(a) (5–7). Lp(a) is a peculiar lipoprotein composed of apolipoprotein(a) [apo(a)] covalently bound to a low density lipoprotein (LDL)-like particle containing one molecule of apolipoprotein B-100 (8). Apo(a) is a complex protein sharing a high sequence homology with several regions of plasminogen, including the protease domain, and the so-called kringle IV (KIV) and V domains (8). The KIV domain of apo(a) is formed by 10 distinct KIV types numbered from 1 to 10. All KIV types, except KIV type 2 (KIV-2), are present as a single copy, while the KIV-2 repeats vary from 3 to >40 copies, resulting in a large heterogeneity in apo(a) isoform size circulating in plasma (9). Apo(a) concentration is generally inversely correlated with apo(a) size and varies widely between individuals.
The unique structural characteristics and size heterogeneity of apo(a) can have strong influence on the accuracy of immunochemical methods used for Lp(a) measurements (10). Antibodies binding to epitopes in the KIV-2 region will generate multiple antibody–antigen complexes dependent on the size of apo(a). As a consequence, when apo(a) size in the calibrator is smaller or larger than that in the measured sample, Lp(a) concentration will be under or overestimated depending on the method (10).
Improving method comparability requires a strategy for assay standardization based on the development of a reference method calibrated with a high-purity primary reference material, and commutable secondary reference materials with established traceability (11). Efforts in standardization of Lp(a) measurement were initiated 2 decades ago by the International Federation of Clinical Chemistry and Laboratory Medicine (IFCC). A secondary reference material, the WHO/IFCC SRM-2B, was produced in 2000 and its implementation significantly improved between-assay comparability (12).
The concentration of Lp(a) in SRM-2B was assigned by an enzyme-linked immunosorbent assay (ELISA) method (13). This ELISA involves a monoclonal antibody directed to a unique epitope located in KIV type 9, with no cross-reactivity to the KIV-2 region of apo(a), rendering this method insensitive to apo(a) size variability (13). For this reason, it is considered the gold standard for Lp(a) quantification. However, although this ELISA exhibits excellent performance, its accuracy, like that of all methods based on antigen–antibody reactions, may be affected by protein conformational diversity (14).
Recent reviews and guidelines have identified the lack of standardization as a major hindrance to the broader use of Lp(a) for cardiovascular disease risk assessment (5, 6, 15–18). With its ability to accurately quantify proteins in complex matrices, targeted liquid chromatography tandem mass spectrometry (LC-MS/MS) has become a method of choice for the standardization of biomarkers in clinical practice (19–22). Like the ELISA gold standard method that was made independent of apo(a) size polymorphism by selecting a monoclonal antibody not interacting with epitopes in KIV-2, LC-MS/MS can be rendered independent of apo(a) size polymorphism by selecting specific quantification peptides not present in the KIV-2 region. In addition, LC-MS/MS can be traceable to SI units through a calibration strategy such as isotope dilution (22–24), which is a key requirement of a reference method for assay standardization (25).
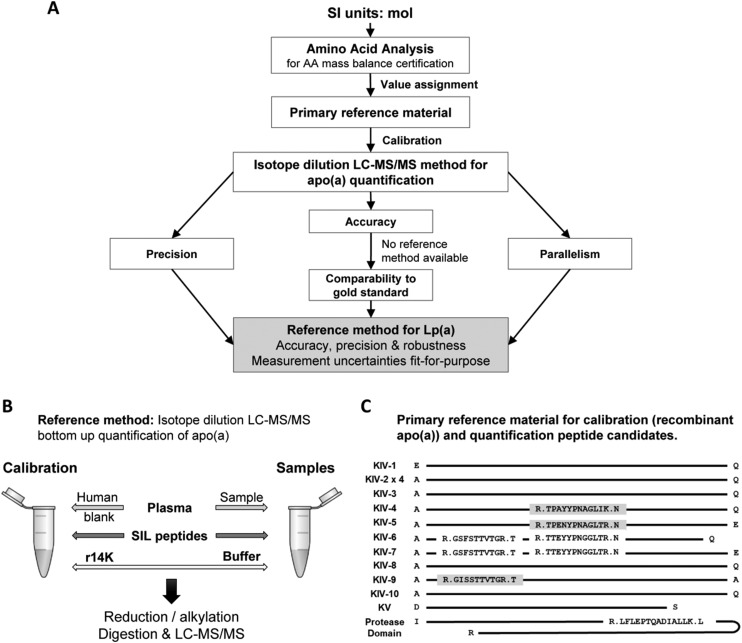
In this study, we developed an accuracy-based method for the absolute quantification of Lp(a) in plasma using a targeted LC-MS/MS approach with an isotope dilution calibration strategy (Fig. 1A) to be proposed as a candidate reference method for the standardization of Lp(a) assays.
Fig. 1.
Development and validation of a reference method for the quantification of apo(a) in plasma. (A) Outline of the method development and validation. (B) Quantification strategy by targeted double isotope dilution LC-MS/MS outlining calibrators and sample preparation. (C) Sequence of the human recombinant apo(a) calibrator. Candidate quantification peptides and their locations are shown. Gray highlight, final quantification peptides; no highlight, candidate peptides not meeting quantification requirements.
Materials and Methods
Complete materials and methods are provided in the Supplemental Materials.
Human Samples
Blood samples from individual donors were collected in lavender top 10 mL K2EDTA tubes. After blood collection, the tubes were inverted several times and let sit on crushed ice for 20–30 min. The blood was then centrifuged at 1500 g for 10 min at 4 °C. Isolated plasma was aliquoted in 2 mL conical polypropylene cryovials and fresh-frozen at -80 °C. A plasma with a low concentration of Lp(a) (4.9 nmol/L by ELISA), further referred to as “blank” plasma, was used as blank matrix. The use of human specimens was approved by the Human Subjects Division at the University of Washington. All donors provided a written informed consent.
Primary Reference Material
A high-purity human recombinant apo(a) with 14 kringles IV (r14K) was selected as primary reference material to calibrate the assay. The full sequence of r14K is provided in Supplemental Fig. 1. The r14K apo(a) was expressed in human embryonic kidney 293 (HEK293) cells, stably transfected with a 14 K-pRK5 expression vector, purified by Lys-Sepharose affinity chromatography, aliquoted and stored at -80°C (26). This expression protocol ensured the proper folding and glycosylation of the recombinant apo(a), which retained the same structural and functional characteristics as the endogenous protein (27–29). The size and purity were ascertained by agarose gel electrophoresis (9), SDS-PAGE electrophoresis, electrospray differential ion mobility analysis and anion-exchange fast protein liquid chromatography.
Determination of the Concentration of the Primary Reference Material
Concentration of the r14K was determined by amino-acid analysis at the National Institute of Standards and Technology (NIST) using a method calibrated with pure higher-order amino-acid (AA) reference standards from the National Metrology Institute of Japan and registered in the Joint Committee for Traceability in Laboratory Medicine (JCTLM) database (30). Gas-phase hydrolysis of the protein was performed using 2 different hydrolysis conditions and 6 AA were quantified by LC-MS/MS. Triplicate independent measurements were performed on 2 different aliquots of the purified r14K. Uncertainties of the 6 AA measurement obtained with the 2 protocols were overlapping and therefore the reported value for r14K concentration was calculated across all 6 AAs and both sets of results.
Method Calibration
Calibrators were prepared using the r14K spiked into “blank” plasma. Double isotope dilution was used for quantification by including the same amount of pure synthetic stable isotope labeled (SIL) peptides in calibrators and in plasma samples. Briefly, r14K was diluted gravimetrically to 6 concentration levels for final calibrator concentrations of 20 to 400 nmol/L and further supplemented with SIL peptides at a final concentration of 100 nmol/L (Fig.1B). All calibrator stocks and intermediates were prepared fresh for each assay. The calibration curve was constructed using linear regression with 1/x weighting without including the blank or the origin.
Digestion Protocol
Samples were prepared by combining 10 µL of plasma, 40 µL of 100 mmol/L ammonium bicarbonate and 20 µL of the SIL peptide working solution (Fig. 1B). After denaturation in 0.5% (w/v) sodium deoxycholate, samples were reduced with dithiothreitol, alkylated with iodoacetamide, and digested with trypsin for 18 h at 37 °C. Digested samples were acidified with formic acid to precipitate sodium deoxycholate and supernatants were analyzed fresh in LC-MS/MS.
Selection of Peptides for Quantification
Candidate peptides for quantification were identified using data independent analysis on trypsin digests of both pure r14K and Lp(a) enriched samples (i.e., reconstituted pellet after apoB-100 precipitation from plasma with 300 nmol/L Lp(a) concentration). From 18 peptides reliably measured in data independent analysis, 6 candidate quantification peptides were selected based on the following criteria: 1) not present in KIV-2 domain of apo(a), 2) absence of AA susceptible to ex-vivo modifications (methionine, cysteine, or terminal glutamic acid or glutamine), 3) absence of homologous peptides in the human proteome, and 4) absence of known human genetic mutations (Fig. 1C).
LC-MS/MS Method
The LC-MS/MS analysis was performed on a Waters Nano-Acquity UPLC system (Waters) coupled to a Thermo ALTIS triple quadrupole mass spectrometer with electrospray ionization (Thermo Fisher). For each peptide and their SIL analogs, measured transitions were summed and a ratio of the chromatographic peak area of endogenous to SIL peptide was calculated in Skyline (31). While Clinical and Laboratory Standards Institute (CLSI) C62-A (32) recommends that clinical LC-MS/MS methods use a single transition for quantification with a second transition as qualifier, we averaged at least 3 transitions for each peptide to increase the robustness of the method. All initial data processing was performed using Skyline, quantification was performed in Excel.
Method Development and Validation
Comparability of the digestion kinetics between r14K and endogenous apo(a) was verified over an 18 h time-course experiment with 3 individual plasma samples with Lp(a) concentrations of 130, 150, and 590 nmol/L and major apo(a) isoform sizes 30, 21, and 12 KIV respectively, and with a 100 nmol/L r14K in “blank” plasma (Supplemental Table 1). For each time-point, samples were digested in triplicate and analyzed in duplicate by LC-MS/MS (n = 6).
Limits of detection (LODs) were calculated as the “blank” plasma response plus 2 standard deviations (SD). The lower limits of quantification (LLOQs) were estimated in several experiments with a maximum allowable bias of 15% and a maximum allowable coefficient of variation (CV) of 20% at LLOQ (CLSI C62-A) (33). Repeatability and intermediate precision were assessed on 5 samples with concentrations (by ELISA) ranging from 20.3 nmol/L to 590.5 nmol/L (Supplemental Table 2), assayed in triplicate on 3 different days, 1 week apart. Intra-day and inter-day CVs were calculated.
Parallelism
Parallelism of responses between r14K and endogenous apo(a) was assessed using a plasma with 150 nmol/L Lp(a) determined by ELISA and a 3-step approach: 1) standard additions of r14K to the plasma sample, 2) serial dilutions of the same plasma sample with “blank” plasma, and 3) preparation of a calibration curve using r14K in “blank” plasma. Parallelism of the 3 regression lines was assessed in 2 independent assays with freshly prepared samples and calibrators by comparing the slopes and their respective 95% confidence intervals (CI).
Assessment of Comparability to the Gold Standard ELISA
In the absence of a reference method, and per JCTLM recommendations, comparability of LC-MS/MS results against the gold standard ELISA was evaluated on a set of 64 individual well-characterized samples. The ELISA was performed as previously reported (13) in duplicate, on 6 different days, and the SD (n = 12) was calculated. For the LC-MS/MS, 2 independent assays were performed as described earlier and samples were digested and injected once per each assay. Methods were compared using weighted-Deming and Pearson least squares regression models. Relative differences to ELISA were evaluated using Bland-Altman difference plot.
Results
The Recombinant Apo(a) as a Primary Calibration Material
A recombinant apo(a) was selected as calibrator because, unlike proteotypic tryptic peptides, it could control variability introduced by sample handling and enzymatic digestion (22, 23, 32). Migration of r14K on agarose gel electrophoresis (9) confirmed that the protein contained 14 KIV repeats (Supplemental Fig. 2). Purity of the preparation was assessed by a combination of orthogonal methods that all indicated purity of 95% or better (Supplemental Fig. 3).
The concentration of r14K was determined at NIST using the amino-acid analysis higher order reference method (Table 1). The 2 hydrolysis conditions yielded indistinguishable results and were combined to calculate r14K concentration. For each AA quantified, CVs across the replicate measurements were below 1.5%, and between-AA CV was 3.8% across all assays. Based on the overall data and using a molecular weight 211,992.4 g/mol, a value of 5.00 ± 0.15 µmol/L was assigned to the r14K as a primary reference material.
Table 1.
Measurement of the concentration of the r14K recombinant apo(a) by amino-acid analysis. Hydrolysis 1, 130 °C for 48 h; hydrolysis 2, 140 °C for 71 h.
| Hydrolysis 1 |
Hydrolysis 2 |
Overall |
||||
|---|---|---|---|---|---|---|
| Amino Acid | Mean (g/L) | % CV | Mean (g/L) | % CV | Mean (g/L) | % CV |
| Phe | 1.10 | 0.72 | 1.11 | 1.20 | 1.11 | 1.00 |
| Leu | 1.11 | 1.80 | 1.06 | 1.70 | 1.09 | 3.20 |
| Ile | 1.14 | 1.90 | 1.06 | 1.50 | 1.10 | 3.90 |
| Val | 1.07 | 0.54 | 1.02 | 0.58 | 1.04 | 2.80 |
| Ala | 0.97 | 0.64 | 0.92 | 0.55 | 0.95 | 2.60 |
| Pro | 1.00 | 0.88 | 0.98 | 0.48 | 0.99 | 1.30 |
| All combined | 1.08 | 5.20 | 1.04 | 5.40 | 1.06 | 5.70 |
| Conc. (µmol/L) | 5.10 | 4.91 | 5.00 | |||
Phe, Phenylalanine; Leu, Leucine; Ile, Isoleucine; Val, Valine; Ala, Alanine; Pro, Proline.
Method Development
In preliminary experiments, 6 candidate quantification peptides were monitored in tryptic digests of r14K and plasma samples, with at least 4 selected reaction monitoring transitions for each endogenous and SIL peptide (Supplemental Table 3). Because the major requirements of the method were accuracy and robustness, stringent criteria were chosen to select the suitable peptides. Three of the 6 peptides, TTEY, GSFS, and LFLE, displayed matrix interferences in all but a single transition for either endogenous or SIL peptide (not meeting CLSI recommendations C62-A (33)) and demonstrated instability under assay conditions (TTEY), or inadequate robustness (GSFS, LFLE) in the intermediate precision study (CV > 10% at LLOQ) and were excluded from consideration. Three peptides, TPEN, TPAY, and GISS, fulfilled CLSI criteria (C62-A) and were selected for further evaluation (Table 2). Data for the excluded peptides are provided in the Supplemental Materials. Moreover, it was suggested that for peptide-based quantification of proteins in LC-MS/MS, combining the results of multiple peptides improves robustness and accuracy of the assay (34, 35). The ultimate measure of Lp(a) concentration was therefore the mean response of the 3 selected peptides.
Table 2.
Characteristics and MS/MS parameters of the 3 quantification peptides selected for the Lp(a) LC-MS/MS method.
| Short- name | Full peptide sequence | Location | Retention time window (min) | Precursor ion (m/z) | Normalized Collision energy (V) | Ion type | Transition (m/z) |
|---|---|---|---|---|---|---|---|
| TPAY | TPAYYPNAGLIK | KIV-4 | 16.5–19.5 | 654.35 | 22.5 | y10 | 1109.6 |
| y9 | 1038.6 | ||||||
| y8 | 875.5 | ||||||
| y7 | 712.4 | ||||||
| TPEN | TPENYPNAGLTR | KIV-5 | 12.0–15.0 | 666.83 | 28.6 | y10 | 1134.6 |
| y8 | 891.5 | ||||||
| y7 | 728.4 | ||||||
| GISS | GISSTTVTGR | KIV-9 | 10.0–13.0 | 489.76 | 17.6 | y8 | 808.4 |
| y7 | 721.4 | ||||||
| y5 | 533.3 | ||||||
| y3 | 333.2 |
To ensure comparable digestion kinetics and to assess the effect of apo(a) isoform size on digestion, time-course experiments were performed. For the 3 peptides, the ratio of endogenous to SIL peptide reached maximum at the first time-point (30 min) and the data did not suggest any trend for overall changes (Supplemental Figs. 4 and 5). Peptide response across the digestion time-course after normalization to t = 18 h overlapped at all time-points independent of apo(a) isoform for all peptides (Supplemental Fig. 6) and no differences were observed in the digestion time-course between samples, thus confirming that the digestion of Lp(a) was robust and that apo(a) isoform size did not affect digestion kinetics.
Parallelism
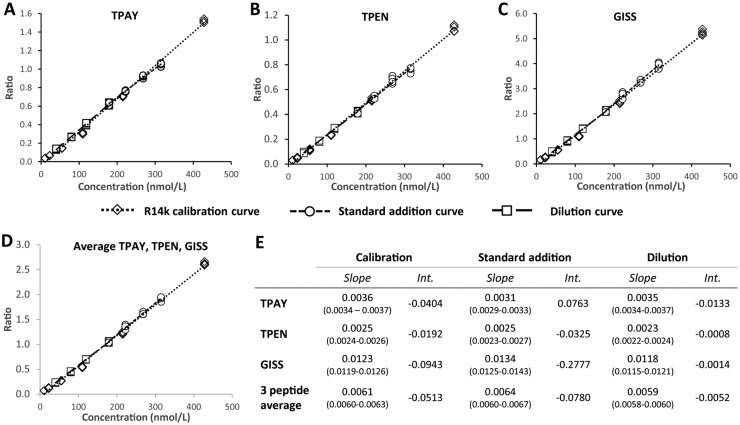
To verify the suitability of r14K as calibrator, parallelism of responses between r14K and endogenous apo(a) was assessed. For TPEN, TPAY, and GISS individually, as well as for their mean, linear regressions of the calibration curve (r14K only), the standard additions curve (r14K added to endogenous apo(a)), and the serial dilutions curve (endogenous apo(a) only) provided similar slopes and intercepts (Fig. 2A–D, Supplemental Fig. 7). The slopes and 95% CI for each individual peptide and their mean were not notably different with overlapping 95%CI (Fig. 2E), confirming the suitability of r14K as calibrator.
Fig. 2.
Parallelism assessment of the recombinant r14K calibrator. (A–D) Plots of the linear regression curves of the ratio of the endogenous to SIL peptide peak areas and the concentration for the 3 final quantification peptides and the 3-peptide mean. Diamonds, the r14K calibration; circles, the standard addition of r14K to a sample; squares, the serial sample dilution.
Method Validation
Linearity, LOD, and LLOQ for the 3 final peptides were determined from a serial dilution of r14K in “blank” plasma in 3 separate assays. Linearity (r2 > 0.994) was determined from 10 nmol/L up to 400 nmol/L; LOD ranged from 4.9 to 10.4 nmol/L, and LLOQ varied between 10 and 20 nmol/L (Supplemental Table 4).
In the repeatability and intermediate precision assessment (Table 3), the Grubbs test for outliers detected 2 outliers for TPEN at the lowest concentration. These outliers were removed from the analysis. Technical replicate imprecision at LLOQ (n = 3 injections) ranged from 0.2% to 12.7% with a mean of 4.1% (SD ± 3.2%) (Table 3). For each measured peptide, intra-day imprecision (n = 9) was below 8% with a mean of 3.9 ± 1.6%, 4.0 ± 1.5% and 3.9 ± 1.5% respectively (Table 3). Inter-day imprecision across concentrations did not exceed 8% for TPEN and TPAY, and 9% for GISS. The results were not found significantly different for the 590.5 nmol/L sample when measured directly or diluted (two-way ANOVA, α = 0.05), with similar CVs for all peptides. For the 3-peptide mean, the intra-day imprecisions matched those of individual peptides (<8%). However, the inter-day imprecision was markedly improved over individual peptides with no sample exceeding 5% (mean 3.2 ± 1.2%). In preliminary experiments, the method was evaluated in another laboratory using a high-throughput normal-flow LC-MS/MS. While intra-day and inter-day CVs were higher, the results were very consistent across platforms with inter-laboratory imprecision <10% and mean bias between the 2 laboratories < 2% (Supplemental Fig. 8, and Supplemental Tables 5 and 6).
Table 3.
Repeatability and intermediate precision of the 3 final quantification peptides and the 3-peptide mean. The mean CV per peptide was additionally calculated as an indicator or each peptide performances.
| Intra-day imprecision %CV (n = 9) |
Inter-day imprecision %CV (n = 27) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample | Day | TPAY | TPEN | GISS | 3-peptide mean | TPAY | TPEN | GISS | 3-peptide mean |
| S01 | 1 | 5.2% | 3.9% | 5.2% | 6.8% | 7.3% | 4.8% | 8.9% | 5.0% |
| 2 | 6.7% | 6.2% | 7.3% | 7.7% | |||||
| 3 | 4.8% | 4.0% | 7.3% | 6.3% | |||||
| S02 | 1 | 4.6% | 5.5% | 3.5% | 5.2% | 7.1% | 6.0% | 5.2% | 4.1% |
| 2 | 6.4% | 7.0% | 4.3% | 7.5% | |||||
| 3 | 7.4% | 5.7% | 4.7% | 7.3% | |||||
| S03 | 1 | 2.6% | 2.7% | 2.8% | 5.5% | 3.8% | 4.1% | 5.0% | 3.1% |
| 2 | 3.2% | 4.8% | 4.0% | 5.9% | |||||
| 3 | 3.8% | 5.0% | 3.4% | 5.5% | |||||
| S05 | 1 | 2.3% | 3.6% | 2.2% | 6.3% | 3.0% | 3.6% | 2.5% | 1.7% |
| 2 | 2.7% | 3.8% | 2.4% | 7.6% | |||||
| 3 | 2.7% | 3.6% | 2.9% | 7.6% | |||||
| S07 | 1 | 2.8% | 2.0% | 2.5% | 4.6% | 3.7% | 1.8% | 3.5% | 2.2% |
| 2 | 3.9% | 1.8% | 3.0% | 5.4% | |||||
| 3 | 3.4% | 1.8% | 2.9% | 5.0% | |||||
| S07-Dil | 1 | 3.0% | 3.5% | 4.2% | 5.3% | 3.5% | 3.9% | 4.8% | 2.9% |
| 2 | 2.4% | 2.3% | 4.0% | 5.4% | |||||
| 3 | 2.4% | 4.1% | 3.5% | 5.3% | |||||
| Mean CV | 3.9% | 4.0% | 3.9% | 6.1% | 4.7% | 4.0% | 5.0% | 3.2% | |
Method Comparison to the ELISA Gold Standard
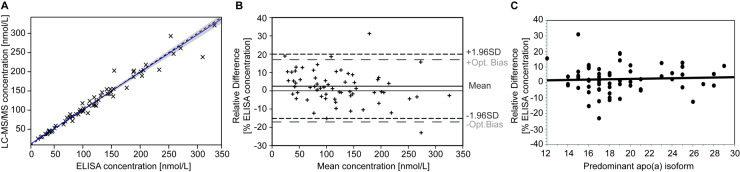
Comparison of results obtained with the LC-MS/MS method and the current gold standard ELISA (13) was performed on a set of 64 well-characterized samples with defined apo(a) isoforms. The secondary reference material WHO/IFCC SRM-2B was additionally evaluated. The value of 104.7 ± 8.4 nmol/L (k = 2 uncertainty) determined by LC-MS/MS was in excellent agreement with the assigned value of 107 nmol/L (Supplemental Methods and Supplemental Table 7). The calibration curves in the 2 assays comparing LC-MS/MS and ELISA, repeated on different days, were indistinguishable (Supplemental Fig. 9). Results of the 64 samples showed a Pearson correlation r2 = 0.958. The weighted-Deming regression analysis demonstrated agreement between the 2 methods with y = 3.18[95%CI: 1.08-5.28] + ELISA*0.98[95%CI: 0.94-1.02] (Fig. 3A). The Bland-Altman difference plot (Fig. 3B) indicated minimal differences of LC-MS/MS values compared to ELISA with a 1.7 nmol/L mean difference (2.5%) [1.96xSD limits of agreement −29.8 to 33.2 nmol/L].
Fig. 3.
Comparison of LC-MS/MS and ELISA methods. (A) Weighted Deming regression: [LC-MS/MS] = 3.18(1.08-5.28) + [ELISA]*0.98(0.94–1.02). Dashed line, regression curve; solid black line, equivalence; gray shaded area, 95% CI. (B) Bland-Altman plot of relative difference between LC-MS/MS and ELISA measurements. Short dashed lines, 95% CI; long dashed lines, the recommended acceptable optimal bias limits. (C) Plot of relative difference between the LC-MS/MS and ELISA measurement vs the size of dominant apo(a) isoform expressed in number of KIV (y = 0.13x−0.4, r2=0.003, N.S., N = 64).
Interestingly, the intercept of the Deming regression was in excellent agreement with the low, but not negligible, concentration of Lp(a) in the “blank” plasma (4.9 nmol/L). However, correction for the “blank” plasma contribution to peak area of endogenous peptides did not influence results of this comparison (Supplemental Table 8). Correlation between the relative bias and the primary apo(a) isoform size expressed in number of KIV (y = 0.13x −0.4, r2 = 0.003, N.S.) indicated that the variation in KIV-2 number accounted for at most 0.3% of the bias variation, confirming that apo(a) size polymorphism did not affect the LC-MS/MS results (Fig. 3C).
Discussion
With the renewed interest in Lp(a) as a causal risk factor for cardiovascular disease, poor method accuracy and lack of method standardization are the major obstacles for the clinical implementation of Lp(a) measurements (5, 10, 16, 36). The complexity of Lp(a) makes it challenging to produce a primary reference material to anchor Lp(a) concentrations to SI units. Because apo(a) is the unique protein component of Lp(a) and its quantification is at the basis of most immunochemical measurements of Lp(a) in plasma, we used this approach to develop an LC-MS/MS-based quantitative method that would meet the requirements of a reference method to be proposed as “candidate reference method” for the standardization of Lp(a) assays.
Requirements of a higher order reference method are defined by the JCTLM (25, 30) and the most important is direct traceability to the SI through an unbroken chain of traceability, established through a suitable calibration strategy. The method should additionally provide equivalence of results between the calibration and the endogenous measurements (i.e., parallelism) and display high levels of accuracy and precision. Double isotope dilution LC-MS/MS is a method of choice for the absolute quantification of proteins using proteolytic peptides as surrogate measurands (22). We selected a recombinant apo(a) as the external primary calibrator that was expressed in human HEK293 cells providing glycosylation and folding patterns similar to endogenous protein, a factor critical for the accuracy of the method (37). We further combined it with SIL peptides as internal standards, an approach that has demonstrated both precision and accuracy (38, 39). Purity and SI-traceable concentration of the recombinant apo(a) calibrator further ensure traceability of the method (24), in contrast to the calibrators of previously published LC-MS/MS methods (40, 41).
Because of its complex, repetitive sequence with a large number of homologous peptides throughout the KIV domains and the variable number of KIV-2 repeats, apo(a) is a major challenge for LC-MS/MS. Previously published methods (40, 41) selected different tryptic peptides, and in particular a peptide LFLEPTQADIALLK found in the protease-like domain of apo(a). However, upon extensive validation on nano- and normal-flow UPLC-MS/MS, this peptide demonstrated lower precision. We therefore selected 3 other peptides with better accuracy, precision, and robustness.
Using these peptides, we demonstrated excellent parallelism between the recombinant apo(a) calibrator and endogenous Lp(a)-associated apo(a), a key parameter for accuracy (42). Our LC-MS/MS method accurately measured the concentration of the WHO/IFCC SRM-2B reference material for Lp(a) that was value-assigned using isolated Lp(a) and amino-acid analysis (43). Similarly, the results obtained by the LC-MS/MS method on 64 clinical samples were in excellent agreement with those obtained by the current gold standard ELISA (13). We additionally demonstrated that, like this ELISA assay, results are not affected by the size polymorphism of apo(a).
Finally, one important requirement defined by the JCTLM for reference methods is the definition of measurement uncertainties that are fit-for-purpose (25). It is commonly accepted that the uncertainties associated to a reference method should be a third of the total allowable error defined for routine assays (24). For Lp(a), defining total allowable error is challenging because of the lack of sound biological variability data to properly define analytical performance specifications (44). Nevertheless, current guidelines from the European Federation of Clinical Chemistry and Laboratory Medicine recommend a desirable 40.3% and an optimum 20.1% total allowable error for Lp(a) routine assays (45). Based on these guidelines, a suitable reference method should have uncertainties below 12%, or optimally below 7%. The 8.0% relative uncertainty estimated for the WHO/IFCC SRM-2B using our LC-MS/MS method is in-line with current recommendations. However, it is critical to note that this uncertainty was calculated using models that do not account for all sources of uncertainty (Supplemental Methods) and that a thorough uncertainty budget calculation is needed.
Altogether, results demonstrate the suitability of recombinant apo(a) as primary reference material for the quantification of Lp(a) in plasma by LC-MS/MS. The method we have developed additionally fulfills quality requirements to be proposed as a candidate reference method for the standardization of Lp(a) assays. However, a clearly defined measurand (i.e., the quantity intended to be measured) is needed to set a new traceability chain (24). The choice of a full-length protein as primary calibrator, while it has notable advantages for accuracy and robustness (22–24), represents a challenge for the definition of the measurand because what constitutes the calibrator and the quantity measured (i.e., proteotypic peptides) differ. We opted for the mean of 3 peptides distributed across apo(a) sequence as a surrogate for the full-length apo(a). To ensure the high quality of the method and to confirm the validity of our choice, we performed an extensive validation and found that the use of the 3-peptide mean was robust across samples and demonstrated comparability to the current designated comparison method.
In summary, our method is directly traceable to SI units, independent of Lp(a) isoform size and demonstrates precision, robustness, and accuracy. These characteristics, coupled with the excellent comparability of results with the current gold standard ELISA, underscore its potential for the standardization of Lp(a) assays. This method will be transferred to a laboratory or agency accredited by the International Organization for Standardization to propose it as candidate reference method to the JCTLM.
Supplemental Material
Supplemental material is available at Clinical Chemistry online.
Supplementary Material
Nonstandard Abbreviations
- AA
amino-acid
- apo(a)
apolipoprotein(a)
- apoB-100
apolipoproteinB-100
- CE
collision energy
- CI
confidence interval
- CLSI
Clinical and Laboratory Standards Institute
- CV
coefficient of variation
- ELISA
enzyme-linked immunosorbent assay
- HEK
human embryonic kidney
- IFCC
International Federation of Clinical Chemistry and Laboratory Medicine
- Ile
isoleucine
- JCTLM
Joint Committee for Traceability in Laboratory Medicine
- KIV-2
Kringle IV type 2
- KIV-9
Kringle IV type 9
- LC-MS/MS
liquid chromatography tandem mass-spectrometry
- LDL
low density lipoprotein
- LOD
limit of detection
- LOQ
limit of quantification
- Lp(a)
Lipoprotein(a)
- MRM
multiple reaction monitoring
- NIST
National Institute of Standards and Technology
- PCSK9
Proprotein convertase subtilisin/kexin type 9
- r14K
human recombinant apo(a) calibrator
- SD
standard deviation
- SI
international system of units
- SIL
stable isotope labelled
- WHO
World Health Organization.
Author Contributions
All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
S.M. Marcovina and T. Vaisar conceived the approach, designed the experiments, and provided financial support; N. Clouet-Foraison and T. Vaisar performed the experiments; M. Lowenthal performed the amino-acid analysis; S.M. Marcovina, M.L. Koschinsky, and M.B. Boffa provided study materials; A.N. Hoofnagle contributed to reviewing the data and commented on the manuscript; N. Clouet-Foraison, S.M. Marcovina, and T. Vaisar wrote the manuscript; all authors were involved in reviewing and commenting on the manuscript.
Authors’ Disclosures or Potential Conflicts of Interest
Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership
M.L. Koschinsky, The University of Western Ontario; M.B. Boffa, The University of Western Ontario; A.N. Hoofnagle, Clinical Chemistry, AACC.
Consultant or Advisory Role
S.M. Marcovina, Roche Diagnostics Germany, Denka Seiken Japan; M.L. Koschinsky, Ayma Therapeutics, Novartis; M.B. Boffa, Ayma Therapeutics.
Stock Ownership
None declared.
Honoraria
M.L. Koschinsky, Novartis, American Heart Association, National Lipid Association.
Research Funding
M.L. Koschinsky, Canadian Institutes of Health Research, Natural Sciences and Engineering Research Council of Canada; A.N. Hoofnagle, funding from NIH and Waters, Inc. to institution; T. Vaisar, National Institutes of Health grant R01-HL-144558.
Expert Testimony
A.N. Hoofnagle, Kilpatrick, Townsend, and Stockton LLP.
Patents
None declared.
Role of Sponsor
The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
Acknowledgments
We acknowledge the assistance from Dr. Michelle A. Emerick and Ms. Jessica O. Becker with the implementation of the assay on the normal-flow LC-MS/MS at the UW Laboratory Medicine.
References
- 1. Willeit P, Ridker PM, Nestel PJ, Simes J, Tonkin AM, Pedersen TR, et al. Baseline and on-statin treatment lipoprotein(a) levels for prediction of cardiovascular events: individual patient-data meta-analysis of statin outcome trials. Lancet 2018;392:1311–20. [DOI] [PubMed] [Google Scholar]
- 2. Arora P, Kalra R, Callas PW, Alexander KS, Zakai NA, Wadley V, et al. Lipoprotein(a) and risk of ischemic stroke in the REGARDS study. Arterioscler Thromb Vasc Biol 2019;39:810–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nordestgaard BG, Langsted A.. Lipoprotein(a) as a cause of cardiovascular disease: insights from epidemiology, genetics, and biology. J Lipid Res 2016;57:1953–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mack S, Coassin S, Rueedi R, Yousri NA, Seppälä I, Gieger C, et al. A genome-wide association meta-analysis on lipoprotein(a) concentrations adjusted for apolipoprotein (a) isoforms. J Lipid Res 2017;58:1834–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tsimikas S. A test in context: lipoprotein(a). Diagnosis, prognosis, controversies, and emerging therapies. J Am Coll Cardiol 2017;69:692–711. [DOI] [PubMed] [Google Scholar]
- 6. Watts GF, Boffa MB.. Lipoprotein(a): lodestar for future clinical trials. Lancet 2018;392:1281–2. [DOI] [PubMed] [Google Scholar]
- 7. Boffa MB, Koschinsky ML.. Lipoprotein(a): truly a direct prothrombotic factor in cardiovascular disease? J Lipid Res 2016;57:745–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koschinsky ML, Marcovina SM.. Structure-function relationships in apolipoprotein(a): insights into lipoprotein(a) assembly and pathogenicity. Curr Opin Lipidol 2004;15:167–74. [DOI] [PubMed] [Google Scholar]
- 9. Marcovina SM, Zhang ZH, Gaur VP, Albers JJ.. Identification of 34 apolipoprotein(a) isoforms: differential expression of apolipoprotein(a) alleles between american blacks and whites. Biochem Biophys Res Commun 1993;191:1192–6. [DOI] [PubMed] [Google Scholar]
- 10. Marcovina SM, Albers JJ.. Lipoprotein(a) measurements for clinical application. J Lipid Res 2016;57:526–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Panteghini M. Traceability, reference systems and result comparability. Clin Biochem Rev 2007;28:97–104. [PMC free article] [PubMed] [Google Scholar]
- 12. Marcovina SM, Albers JJ, Scanu A, Kennedy H, Giaculli F, Berg K, et al. Use of a reference material proposed by the International Federation of Clinical Chemistry and laboratory medicine to evaluate analytical methods for the determination of plasma lipoprotein(a). Clin Chem 2000;46:1956–67. [PubMed] [Google Scholar]
- 13. Marcovina SM, Albers JJ, Gabel BR, Koschinsky ML, Gaur VP.. Effect of the number of apolipoprotein(a) kringle 4 domains on immunochemical measurements of lipoprotein(a). Clin Chem 1995;41:246–55. [PubMed] [Google Scholar]
- 14. Hoofnagle AN, Wener MH.. The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry. J Immunol Methods 2009;347:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kostner KM, Kostner GM, Wierzbicki AS.. Is Lp(a) ready for prime time use in the clinic? A pros-and-cons debate. Atherosclerosis 2018;274:16–22. [DOI] [PubMed] [Google Scholar]
- 16. Tsimikas S, Fazio S, Ferdinand KC, Ginsberg HN, Koschinsky ML, Marcovina SM, et al. NHLBI Working group recommendations to reduce lipoprotein(a)-mediated risk of cardiovascular disease and aortic stenosis. J Am Coll Cardiol 2018;71:177–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. France M, Rees A, Datta D, Thompson G, Capps N, Ferns G, et al. HEART UK statement on the management of homozygous familial hypercholesterolaemia in the United Kingdom. Atherosclerosis 2016;255:128–39. [DOI] [PubMed] [Google Scholar]
- 18. Wilson DP, Jacobson TA, Jones PH, Koschinsky ML, McNeal CJ, Nordestgaard BG, et al. Use of Lipoprotein(a) in clinical practice: A biomarker whose time has come. A scientific statement from the National Lipid Association. J Clin Lipidol 2019;13:374–92. [DOI] [PubMed] [Google Scholar]
- 19. Kaiser P, Akerboom T, Ohlendorf R, Reinauer H.. Liquid chromatography-isotope dilution-mass spectrometry as a new basis for the reference measurement procedure for hemoglobin A1c determination. Clin Chem 2010;56:750–4. [DOI] [PubMed] [Google Scholar]
- 20. van den Broek I, Sobhani K, Van Eyk JE.. Advances in quantifying apolipoproteins using LC-MS/MS technology: implications for the clinic. Expert Rev Proteomics 2017;14:869–80. [DOI] [PubMed] [Google Scholar]
- 21. Smit NPM, Romijn F, van den Broek I, Drijfhout JW, Haex M, van der Laarse A, et al. Metrological traceability in mass spectrometry-based targeted protein quantitation: A proof-of-principle study for serum apolipoproteins A-I and B100. J Proteomics 2014;109:143–61. [DOI] [PubMed] [Google Scholar]
- 22. Villanueva J, Carrascal M, Abian J.. Isotope dilution mass spectrometry for absolute quantification in proteomics: Concepts and strategies. J Proteomics 2014;96:184–99. [DOI] [PubMed] [Google Scholar]
- 23. Brun V, Masselon C, Garin J, Dupuis A.. Isotope dilution strategies for absolute quantitative proteomics. J Proteomics 2009;72:740–9. [DOI] [PubMed] [Google Scholar]
- 24. Josephs RD, Martos G, Li M, Wu L, Melanson JE, Quaglia M, et al. Establishment of measurement traceability for peptide and protein quantification through rigorous purity assessment—a review. Metrologia 2019;56:044006. [Google Scholar]
- 25. Armbruster D, Miller RR.. The Joint Committee for Traceability in Laboratory Medicine (JCTLM): a global approach to promote the standardisation of clinical laboratory test results. Clin Biochem Rev 2007;28:105–13. [PMC free article] [PubMed] [Google Scholar]
- 26. Tam S-P, Zhang X, Koschinsky ML.. Interaction of a recombinant form of apolipoprotein(a) with human fibroblasts and with the human hepatoma cell line HepG2. J Lipid Res 1996;37:518–33. [PubMed] [Google Scholar]
- 27. Koschinsky ML, Tomlinson JE, Zioncheck TF, Schwartz K, Eaton DL, Lawn RM.. Apolipoprotein(a): expression and characterization of a recombinant form of the protein in mammalian cells. Biochemistry 1991;30:5044–51. [DOI] [PubMed] [Google Scholar]
- 28. Koschinsky ML, Marcovina SM, May LF, Gabel BR.. Analysis of the mechanism of lipoprotein(a) assembly. Clin Genet 2008;52:338–46. [DOI] [PubMed] [Google Scholar]
- 29. Gabel BR, Koschinsky ML.. Analysis of the proteolytic activity of a recombinant form of apolipoprotein(a). Biochemistry 1995;34:15777–84. [DOI] [PubMed] [Google Scholar]
- 30.Joint Committee for Traceability in Laboratory Medicine (JCTLM). Database of higher-order reference materials, measurement methods/procedures and services [Internet]. [cited 2020. Jul 9]. Available from: https://www.bipm.org/jctlm/home.do.
- 31. MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010;26:966–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lowenthal MS, Liang Y, Phinney KW, Stein SE.. Quantitative bottom-up proteomics depends on digestion conditions. Anal Chem 2014;86:551–8. [DOI] [PubMed] [Google Scholar]
- 33.Clinical and Laboratory Standards Institute. C62-A: Liquid chromatography - Mass spectrometry Methods; approved guidelines 2014.
- 34. Henderson CM, Lutsey PL, Misialek JR, Laha TJ, Selvin E, Eckfeldt JH, et al. Measurement by a novel LC-MS/MS methodology reveals similar serum concentrations of vitamin D-binding protein in blacks and whites. Clin Chem 2016;62:179–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Toth CA, Kuklenyik Z, Jones JI, Parks BA, Gardner MS, Schieltz DM, et al. On-column trypsin digestion coupled with LC-MS/MS for quantification of apolipoproteins. J Proteomics 2017;150:258–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kronenberg F, Tsimikas S.. The challenges of measuring Lp(a): A fight against Hydra? Atherosclerosis 2019;289:181–3. [DOI] [PubMed] [Google Scholar]
- 37. Cowan KJ, Amaravadi L, Cameron MJ, Fink D, Jani D, Kamat M, et al. Recommendations for selection and characterization of protein biomarker assay calibrator material. AAPS J 2017;19:1550–63. [DOI] [PubMed] [Google Scholar]
- 38. Shuford CM, Walters JJ, Holland PM, Sreenivasan U, Askari N, Ray K, et al. Absolute protein quantification by mass spectrometry: not as simple as advertised. Anal Chem 2017;89:7406–15. [DOI] [PubMed] [Google Scholar]
- 39. Nouri-Nigjeh E, Zhang M, Ji T, Yu H, An B, Duan X, et al. Effects of calibration approaches on the accuracy for LC-MS targeted quantification of therapeutic protein. Anal Chem 2014;86:3575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stefanutti C, Pisciotta L, Favari E, Di Giacomo S, Vacondio F, Zenti MG, et al. Lipoprotein(a) concentration, genetic variants, apo(a) isoform size, and cellular cholesterol efflux in patients with elevated Lp(a) and coronary heart disease submitted or not to lipoprotein apheresis: An Italian case-control multicenter study on Lp(a). J Clin Lipidol 2020;14:487–97.e1. [DOI] [PubMed] [Google Scholar]
- 41. Lassman ME, McLaughlin TM, Zhou H, Pan Y, Marcovina SM, Laterza O, et al. Simultaneous quantitation and size characterization of apolipoprotein(a) by ultra-performance liquid chromatography/mass spectrometry. Rapid Commun Mass Spectrom 2014;28:1101–6. [DOI] [PubMed] [Google Scholar]
- 42. Jenkins R, Duggan JX, Aubry AF, Zeng J, Lee JW, Cojocaru L, et al. Recommendations for validation of LC-MS/MS bioanalytical methods for protein biotherapeutics. AAPS J 2015;17:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dati F, Tate JR, Marcovina SM, Steinmetz A, Laboratory M, et al. First WHO/IFCC International reference reagent for Lipoprotein(a) for immunoassay - Lp(a) SRM 2B. Clin Chem Lab Med 2004;42:670–6. [DOI] [PubMed] [Google Scholar]
- 44. Clouet-Foraison N, Marcovina SM, Guerra E, Aarsand AK, Coşkun A, Díaz-Garzón J, et al. Analytical performance specifications for lipoprotein(a), apolipoprotein B-100, and apolipoprotein A-I using the biological variation model in the EuBIVAS population. Clin Chem 2020;66:727–36. [DOI] [PubMed] [Google Scholar]
- 45. Aarsand AK, Fernandez-Calle P, Webster C, Coskun A, Gonzales-Lao E, Diaz-Garzón J. The EFLM Biological Variation Database. 2018. [cited 2020 Sep 25]. Available from: https://biologicalvariation.eu/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.