Abstract
Tumor angiogenesis is a crucial step in the further growth and metastasis of solid tumors. However, its regulatory mechanism remains unclear. Here, we showed that TARBP2, an RNA‐binding protein, played a role in promoting tumor‐induced angiogenesis both in vitro and in vivo through degrading the mRNAs of antiangiogenic factors, including thrombospondin1/2 (THBS1/2), tissue inhibitor of metalloproteinases 1 (TIMP1), and serpin family F member 1 (SERPINF1), by targeting their 3′untranslated regions (3′UTRs). Overexpression of TARBP2 promotes tumor cell–induced angiogenesis, while its knockdown inhibits tumor angiogenesis. Clinical cohort analysis revealed that high expression level of TARBP2 was associated with poor survival of lung cancer and breast cancer patients. Mechanistically, TARBP2 physically interacts with the stem‐loop structure located in the 3′UTR of antiangiogenic transcripts, leading to mRNA destabilization by the dsRNA‐binding domains 1/2 (dsRBDs1/2). Notably, the expression level of TARBP2 in human tumor tissue is negatively correlated with the expression of antiangiogenic factors, including THBS1/2, and brain‐specific angiogenesis inhibitor 1 (BAI1). Moreover, TARBP2 expression is strongly associated with tumor angiogenesis in a group of human lung cancer samples. Collectively, our results highlight that TARBP2 is a novel tumor angiogenesis regulator that could promote tumor angiogenesis by selectively downregulating antiangiogenic gene expression.
Keywords: metastasis, mRNA destabilization, TARBP2, thrombospondin1, tumor angiogenesis
We demonstrated that TARBP2 was able to promote tumor angiogenesis through selectively destabilizing the mRNAs of antiangiogenic factor genes via binding to the stem‐loop structure located in the 3′UTRs. Strikingly, our basic research findings have also been confirmed by analyzing the clinical samples and data from human breast and lung cancers. Thus, TARBP2 is likely to be a useful molecular target for antiangiogenic tumor therapy.

Abbreviations
- 3′UTR
3′untranslated region
- ActD
actinomycin D
- ANGPTL4
angiopoietin‐like protein 4
- ARE
AU‐rich element
- BAI1
brain‐specific angiogenesis inhibitor 1
- DRB
5, 6‐dichlorobenzimidazole riboside
- dsRBDs1/2
dsRNA‐binding domains 1/2
- GRE
GU‐rich element
- HUVECs
human umbilical vein endothelial cell
- SERPINF1
serpin family F member 1
- TARBP2
Trans‐activation response (TAR) RNA‐binding protein 2
- THBS1/2
thrombospondin1/2
- TIMP1
tissue inhibitor of metalloproteinases 1
- VEGF
vascular endothelial growth factor
1. INTRODUCTION
Tumor angiogenesis plays a critical role in the growth and metastasis of solid tumors, although the mechanism remains unclear. 1 Tumor cells secrete and release a series of angiogenesis‐related factors that may require the activation of the endothelial cells to form a vascular network within the tumors. 2 , 3 Further, these new vessels will promote the growth of tumor masses and metastasis to distant organs. 4 Angiogenic factors include proangiogenic factors, such as interleukin 1β (IL1β), interleukin 8 (IL8), matrix metalloproteinases (MMPs), vascular endothelial growth factor (VEGF), 5 and antiangiogenic factors, such as THBS1/2, 6 , 7 TIMP1/3, 8 , 9 BAI1, 10 and SERPINF1. 11 The balance between proangiogenic and antiangiogenic factors determines whether angiogenesis is induced or inhibited. 12 Normally, most tumor angiogenesis is caused either by increasing the expression of proangiogenic factor genes or by decreasing the expression of antiangiogenic factor genes. Although recent advances have broadened our understanding of tumor angiogenesis, the mechanism for controlling the post‐transcriptional regulations of angiogenesis factors, in particular, the role and mechanism of RNA‐binding proteins during this process, remains unclear.
TARBP2, a double‐stranded RNA‐binding protein encoded by the TARBP2 gene, 13 was able to regulate microRNA processing and maturation. 14 , 15 , 16 TARBP2 is an essential molecular partner of Dicer1 and required for the RNA interference pathway. 17 Mice deficient of tarbp2 die at weaning, indicating its important role during development. 18 It is believed that TARBP2 functions in many biological and pathological processes, including virus replication, 19 , 20 stress response, 21 and tumor progression. 22 , 23 Higher cytoplasm TARBP2 expression in triple‐negative breast cancer patients was strongly associated with poor survival. 24 TARBP2 can promote tumor metastasis in melanoma 25 and breast cancer by destabilizing the mRNAs of metastasis‐related genes via binding the hairpin structures in the 3′UTR. 26 TARBP2 is also involved in controlling the cleavage and degradation of cancer‐related pre‐mRNAs to regulate lung cancer growth. 27 It has been recently reported that TARBP2 could trigger chemoresistance in breast cancer 28 and hepatocellular carcinoma 29 and thus might act as a tumor promoter. However, evidence from another report showed that TARBP2 could suppress cell proliferation in osteosarcoma. 30 The role and mechanism of TARBP2 in cancer progression remain uncertain. 31
In this study, we identify that TARBP2 is a potent tumor angiogenesis modulator by selectively suppressing the expression of antiangiogenic genes, including THBS1/2, TIMP1, and SERPINF1. TARBP2 expression is increased in human lung and breast cancer tissues, and its expression is negatively correlated with antiangiogenic factors. Ectopic expression of TARBP2 in human lung and breast tumor cells promoted tumor angiogenesis both in vitro and in vivo, whereas knocking down of TARBP2 with shRNAs suppressed tumor angiogenesis. TARBP2 expression was associated with the poor survival of lung cancer and breast cancer patients. 32 By analyzing a group of human lung cancer samples, we found that higher TARBP2 expression is strongly associated with tumor angiogenesis. The results suggest that TARBP2 is a potential tumor angiogenesis inducer involved in regulating the angiogenic pathway by suppression of antiangiogenic gene expression.
2. MATERIALS AND METHODS
2.1. Cell culture
Human lung cancer cell lines (A549, NCI‐H23), breast cancer cell line (MDA‐MB‐468), and liver cancer cell line (HepG2) cells were obtained from ATCC and cultured in RPMI‐1640 or DMEM with 10% FBS plus 1% Peni/Stro, respectively. Immortalized human umbilical vein endothelial cell (HUVEC), HEK293, and HEK293T cells were obtained from National Infrastructure of Cell Line Resource (Beijing, China). The antibodies and reagents were listed in Doc. S1 Supplementary Materials and Methods.
2.2. Wound‐healing assay
1 × 104 HUVECs were seeded in each well of a 96‐well plate in triplicates and incubated at 37℃ for 12 hours. After the cells formed a monolayer, a scratch was created using a pipette tip on the cell monolayer and incubated for 24 hours. The images of the scratch were obtained using a microscope equipped with a charged couple device camera (Zeiss), the distance of the wound was calculated, and the open area was measured.
2.3. Tube formation assay
The tumor cell culture medium was changed to serum‐free RPMI‐1640 or DMEM medium for 48 hours and then collected, centrifuged, and filtered to obtain tumor conditioned medium (CM). The 96‐well plate was coated with 50 µl prethawed Matrigel (BD Biosciences) and incubated for 1 hour in an incubator at 37°C. HUVECs were cultured in endothelial cell medium (ECM) (ScienCell) with 5% serum for 24 hours prior to the assay, and 1 × 104 HUVECs were seeded on the gel with 200 µL of CM concentrated using an ultrafiltration device (Millipore). The tube formation of HUVECs was observed after incubation of 12 hours using a microscope (Zeiss).
2.4. PCRArray
The QIAGEN Human Angiogenesis PCRArray Kit was used to analyze angiogenesis‐related gene expression. Total RNA was extracted with TRIzol from A549/TARBP2 and A549/GFP cells, reverse‐transcribed to cDNA, and added to each well of the PCRArray plates combined with SYBRGreen qRT‐PCR MasterMix. Data analysis was based on the ΔΔCt method, with normalization to three different housekeeping genes.
2.5. RNA‐sequencing analysis
Total RNA was extracted using the TRIzol method. RNA sequencing was completed by Allwegene Technology Inc in Beijing. The cDNA library was then constructed using PCR amplification. RNA‐seq was performed with the PE150 sequencing strategy by the Illumina second‐generation high‐throughput sequencing platform. RNA‐seq reads with inferior quality or adapters were filtered. Clean reads data were processed using Tophat2 and Cufflinks software to complete the alignment of transcriptomes. Genes not expressed in any sample were excluded from further analysis. Differentially expressed genes and transcripts were then filtered for false discovery rate (FDR)‐adjusted P‐values less than or equal to .05. RNA‐seq data have been deposited (PRJNA637758).
2.6. RNA immunoprecipitation
TARBP2/GFP fusion protein was expressed in A549/TARBP2 cells, and then whole‐cell lysates were precleared with isotype IgG, followed by incubation with anti‐GFP antibody at 4℃ for 4 hours. The protein‐RNA complexes were then pulled down by protein G agarose beads (Santa Cruz Biotechnology) and total RNA extracted with TRIzol, followed by detection of angiogenesis‐related genes with RT‐PCR.
2.7. Luciferase reporter assays
Luciferase assay was performed as described previously. 33 pGL3 luciferase reporter constructs containing full‐length or segment of the 3′UTR of different genes were transfected into HEK293 cells along with TARBP2/GFP and GFP‐control constructs, respectively. All transfections were conducted in triplicate and repeated at least three times. The luciferase activity was measured 36 hours after transfection using a dual‐luciferase reporter assay system (Promega).
2.8. RNA‐EMSA and supershift
Twenty femtomole of the 3′‐end biotin‐labeled WT and other probes (Tsingke) (Table S3) were incubated with 15 μg of total cell lysates containing TARBP2/GFP fusion protein for 30 minutes at room temperature with 20 μl of binding buffer containing 10 mmol/L Tris, 50 mmol/L KCl, and 1 mmol/L dithiothreitol (DTT) (pH 7.5). RNA‐protein complexes were resolved by 6% nondenaturing polyacrylamide gel electrophoresis, and the gel was transferred onto a nylon membrane and UV‐cross linked. The membrane was then subjected to detection by a chemiluminescent EMSA kit (Pierce) following the manufacturer's protocol. The supershift assay was performed in the same manner, except that 2 μg anti‐GFP and anti‐TARBP2 antibodies were added into the reaction.
2.9. shRNA lentivirus
Two lentiviral shRNAs (TRCN0000019339; TRCN0000330578) targeting human TARBP2 mRNA were purchased from Sigma. A scramble control shRNA was used as a control. Lentiviral particles were packaged in HEK293T cells by cotransfecting shRNA‐pLKO.1, pCMV‐dR8.2, and pMD2.G constructs. Virus supernatants were collected and centrifuged to discard cell debris and then added to target cells with 1 µg/mL polybrene for overnight. After two rounds of infection, the target cells were selected with puromycin (1.5 µg/mL) for 2 weeks, followed by further study.
2.10. Statistical analysis
Data in bar graphs represent mean ± SEM of at least three biological repeats. Statistical analysis was performed using Student's t‐test by comparing treatment versus vehicle control or otherwise as indicated. P‐value < .05 was considered to be statistically significant.
3. RESULTS
3.1. TARBP2 promotes tumor angiogenesis in vitro and in vivo
To investigate the role of TARBP2 in tumor‐induced angiogenesis, TARBP2/GFP fusion protein was stably expressed in human lung cancer cells A549, NCI‐H23, breast cancer cells MDA‐MB‐468, and liver cancer cells HepG2, respectively, and confirmed by Western blotting (Figure S1A,B). CMs from TARBP2‐overexpressing tumor cells could significantly promote cell migration (Figure 1A) and tube formation (Figure 1B) of HUVEC compared with the control group. Moreover, CMs from different types of tumor cells did not affect the proliferation of endothelial cells (Figure S1C). To examine the impact of TARBP2 on in vivo angiogenesis, TARBP2‐overexpressing A549/TARBP2 and MDA‐MB‐468/TARBP2 cells and their control A549/GFP and MDA‐MB‐468/GFP cells were subcutaneously injected into the back of nude mice to establish tumor models. The tumors with overexpressed TARBP2 were found to grow faster than their control (Figure 1C,D). Metastatic nodules in the lungs of nude mice bearing TARBP2‐overexpressed tumors were more than those of the control group (Figures 1E,F and S1D), which is not surprising and consistent with previous reports. 21 , 26 To evaluate angiogenesis within the tumors, we performed immunohistochemistry (IHC) staining on tumor tissue sections using anti‐CD31 (a classical vascular marker) antibody. Strikingly, there was significant increase in vascular density in both tumor tissues with overexpressed TARBP2 compared with their control groups, respectively (Figure 1G,H). These findings demonstrated that TARBP2 could promote tumor angiogenesis in vitro and in vivo.
FIGURE 1.
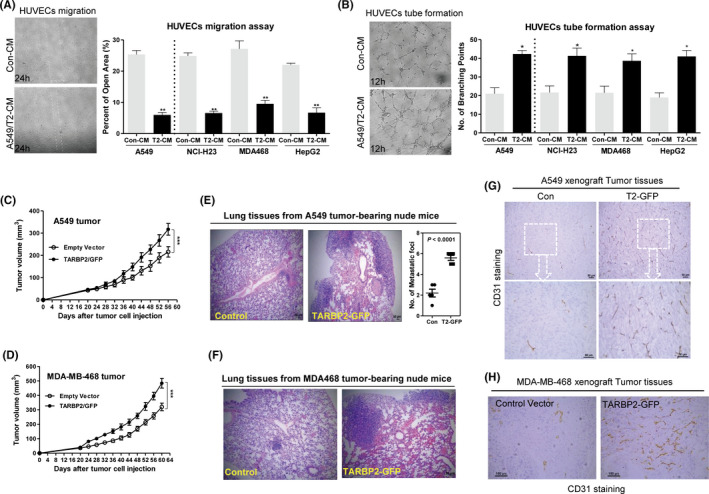
TARBP2 promotes tumor‐induced angiogenesis in vitro and in vivo. A, Wound‐healing assays were performed to detect the cell migration induced by indicated conditioned mediums (CMs) from TARBP2‐overexpressing tumor cells in HUVECs; n = 3. 100×. B, Tube formation assays were also deployed to verify the tube‐formed HUVECs after treatment with CMs from TARBP2‐overexpressing tumor cells; n = 3. 100×. C, D, Tumor growth curves in nude mice received TARBP2‐overexpressing A549/TARBP2 (C), MDA‐MB‐468/TARBP2 (D), and their control cells, respectively; n = 6. E, F, H&E staining of lung tissue sections from nude mice bearing A549 or MDA‐MB‐468 tumors; n = 3. Scale bar: 50 µm. G, H, Representative histological sections from A549 and MDA‐MB‐468 xenografts stained with a specific anti‐CD31 antibody; n = 3. Scale bar: 50 µm. *P < .05, **P < .01, ***P < .0001
3.2. TARBP2 selectively downregulated antiangiogenic factor mRNAs via their 3′UTRs
To clarify how TARBP2 regulates tumor angiogenesis, we performed RNA sequencing using A549/TARBP2 and MDA‐MB‐468/TARBP2, as well as their control A549/GFP and MDA‐MB‐468/GFP cells. The mRNA expression of the antiangiogenic factors, including angiopoietin‐like protein 4 (ANGPTL4), THBS1/2, TIMP1, endostatin (COL18A1), and SERPINF1, was downregulated by TARBP2 (Figure 2A,B) and that of the proangiogenic factors, such as HIF1α, IL8, VEGFA/C, MMP2/14, and IL1β, upregulated (Figure 2C,D, Table S1). We further confirmed their expression by PCR array after extracting the total RNA of A549/TARBP2 and the control cells. Clearly, the antiangiogenic factor mRNAs were downregulated, and proangiogenic factor mRNAs were upregulated in tumor cells (Figure 2E, Table S2). qRT‐PCR was used to verify our RNA‐seq and PCRarray data. Indeed, TARBP2 could downregulate the antiangiogenic factor mRNAs in a time‐dependent manner in both tumor cells (Figures 2F and S2A). However, the upregulated mRNA expression of proangiogenic factors did not show any time dependence (Figures 2G and S2B). At the protein level, the antiangiogenic factor THBS1, which was downregulated by TARBP2, was also downregulated in a time‐dependent manner in two tumor cells (Figure 2H,I). Meanwhile, the protein content of THBS1 in the CMs of two tumor cells with overexpressed TARBP2 was also reduced, as obtained from the ELISA results (Figure S2C,D).
FIGURE 2.
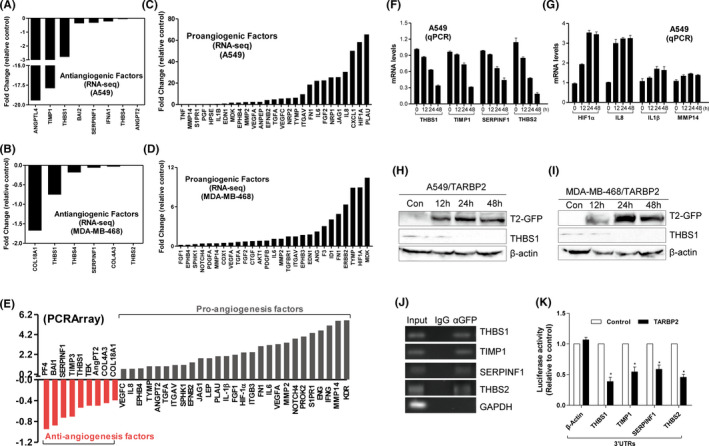
TARBP2 downregulates the expression of antiangiogenic factor mRNAs via targeting 3′UTRs. A, B, RNA‐seq data showing the antiangiogenic factor mRNAs were downregulated in TARBP2‐expressing A549 cells (A) and MDA‐MB‐468 cells (B). C, D, Proangiogenic factor mRNAs were upregulated by TARBP2 in A549 (C) and MDA‐MB‐468 cells (D). E, Human angiogenesis pathway PCR array was performed in A549/TARBP2 cells. F, G, mRNA expressions of the indicated antiangiogenic (F) and proangiogenic (G) genes were measured by qRT‐PCR in TARBP2‐overexpressing A549 cells. H, I, Cell lysates were isolated from the A549/TARBP2 (H) and MDA‐MB‐468/TARBP2 (I) cells followed by Western blot analysis with GFP, THBS1, and β‐actin antibodies. J, RNA‐IP was performed using anti‐GFP antibody or IgG in the extraction of A549/TARBP2 cells. Antiangiogenic factor transcripts were enriched by TARBP2. GAPDH transcript was used as a negative control. K, Luciferase assays were performed by cotransfecting HEK293 cells with reporter constructs containing 3′UTRs of the indicated genes as well as TARBP2 expression vector and empty vector. The ratio of luciferase activity in cells transfected with TARBP2 relative to empty vector was determined, respectively; n = 3. *P < .05
To further investigate how TARBP2 regulates the expression of these mRNAs, RNA pulldown experiment was performed using anti‐GFP antibody and isotype control IgG in A549 cells expressing TARBP2/GFP fusion protein, followed by detecting the bound mRNAs by RT‐PCR. All four antiangiogenic gene mRNAs were amplified by PCR, but not the proangiogenic mRNAs (Figures 2J and S2E). ZNF395 was used as a positive control. We next examined whether TARBP2 was targeting the 3′UTRs of antiangiogenic factor mRNAs. A series of reporter vectors containing the 3′UTRs of antiangiogenic factor genes, including THBS1, TIMP1, SERPINF1, THBS2, were constructed (Figure S2F). Luciferase assay showed that TARBP2 could reduce the luciferase activity of reporters containing different 3′UTRs of antiangiogenic factor genes (Figure 2K). However, TARBP2 could not reduce the activity of the reporter containing 3′TURs of proangiogenic factor genes (Figure S2G). These results suggested that TARBP2 preferentially targeted and bound to the antiangiogenic mRNAs by the 3′UTRs.
3.3. TARBP2 destabilizes the mRNAs of antiangiogenic factors via the dsRBD1/2 domains
Based on the above findings that TARBP2 inhibits the mRNA expression of antiangiogenic genes and the characteristics of TARBP2 destabilizing mRNAs, 26 we hypothesized that TARBP2 might destabilize mRNAs of antiangiogenic genes. To confirm it, TARBP2/GFP was expressed in A549 cells and the de novo mRNA synthesis was blocked with ActD (5 μg/mL) and DRB (5 μg/mL), followed by measuring the remaining mRNAs at different times. 33 The half‐lives of the antiangiogenic mRNAs were shortened about twofold in TARBP2‐overexpressing cells compared with the control cells (Figure 3A). However, TARBP2 overexpression had little impact on the half‐lives of proangiogenic mRNAs (Figure S3A), indicating that TARBP2 specifically reduced the stability of antiangiogenic factor mRNAs rather than that of the proangiogenic factor mRNAs.
FIGURE 3.
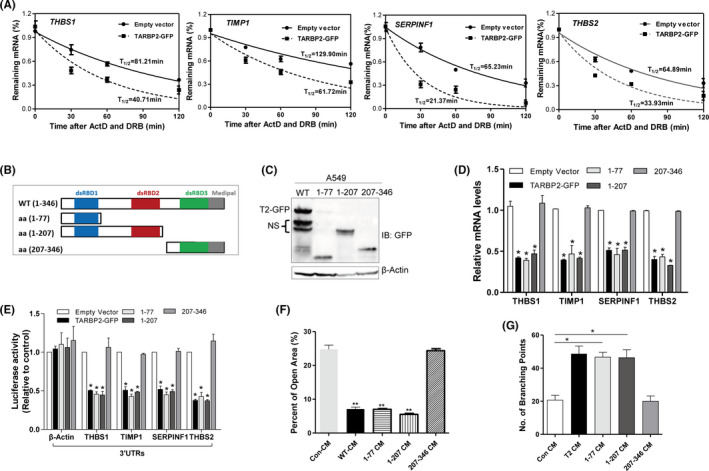
TARBP2 destabilizes the mRNAs of antiangiogenic factors via the dsRBD1/2 domains. A, Half‐lives of indicated antiangiogenic factor genes were measured in TARBP2‐overexpressing A549 cells. B, Schematic representation of the domains in TARBP2 and their truncation mutants. C, Western blot was used to verify the expression of TARBP2 truncation mutants in A549 cells (NS, nonspecific band). D, qRT‐PCR was used to measure the mRNA expression of indicated antiangiogenic genes in A549 cells overexpressing TARBP2 or its mutants; n = 3. E, Luciferase assays were performed by transfecting HEK293 cells with reporter constructs containing 3′UTRs of the indicated genes. The ratio of luciferase activity in cells transfected with TARBP2 and its mutants relative to empty vector was determined, respectively; n = 3. F, Quantification of open area in the wound‐healing assay of HUVECs treated with conditioned mediums (CMs) from A549 cells stably expressing TARBP2 (wt) and its mutants; n = 3. G, Quantification of the number of branching points of HUVECs treated with indicated tumor CMs in the tube formation assay; n = 3. *P < .05, **P < .01
To establish the domains of TARBP2 responsible for the degradation of mRNA, three truncated mutants of TARBP2 were prepared (Figure 3B) and identified by Western blotting (Figure 3C). After transient transfection, wild type (wt), mutant aa 1‐77, and aa 1‐207 suppressed the mRNA expression of antiangiogenic genes (Figure 3D) and the luciferase activities of their 3′UTR reporters (Figure 3E), while aa 207‐346 mutant completely lost the inhibitory function. 34 Besides, the CMs from A549 cells respectively transfected wt, aa 1‐77, and aa 1‐207 mutants could significantly promote the migration (Figures 3F and S3B) and tube formation (Figures 3G and S3C) of HUVEC, but not aa 207‐346 mutant. These results demonstrated that the dsRBD1/2 domains of TARBP2 were essential for the degradation of antiangiogenic mRNAs and the induction of tumor angiogenesis.
3.4. TARBP2 binds to the conserved stem‐loop structure in the 3′UTR of THBS1 for mRNA degradation
TARBP2 is known to destabilize mRNA via the stem‐loop structure. 26 When comparing the 3′UTR sequences of all four antiangiogenic genes among different species, a conserved sequence was identified. Interestingly, a stem‐loop structure could be formed by the conserved sequences when analyzing with the RNAfold web server (Figure S4A‐D). To determine whether these stem‐loop structures are necessary for TARBP2‐mediated degradation of antiangiogenic mRNAs, THBS1 was selected for further analysis. First, we generated a deletion reporter construct by removing the sequence containing the stem‐loop sequence in the 3′UTR of THBS1 (Figure 4A) and then performed reporter assay. TARBP2 suppressed the luciferase activities of reporters with full‐length THBS1 3′UTR and stem‐loop sequence, but did not influence the activity of reporters without the stem‐loop structure, indicating that the stem‐loop sequence is necessary for the decay of mRNA (Figure 4B). To further determine whether the secondary configuration of the stem‐loop structure is necessary for mRNA decay, two 3′UTR mutants of THBS1 were generated. For mutant1, the stem‐loop structure was disrupted by replacing six nucleotides, and mutant2 still retained the stem‐loop structure though four nucleotides were replaced in the stem (Figure 4C). Disruption of the stem‐loop structure in the 3′UTR reduced the inhibitory effect of TARBP2, whereas the mutants with the intact stem‐loop structure were still sensitive to TARBP2 suppression (Figure 4D). RNA‐EMSA was performed to determine whether TARBP2 physically binds to the stem‐loop in the 3′UTR of THBS1. A unique RNA‐protein binding complex formed only by the wt probe but not by the mutant probe of THBS1 (Figure 4E). To confirm TARBP2 binding, anti‐GFP and anti‐TARBP2 antibodies were added to the cytoplasmic proteins and supershift RNA‐EMSA was performed. After adding GFP and TARBP2 antibodies, the binding density was substantially reduced (Figure 4F), suggesting TARBP2 is physically bound to the stem‐loop structure located in the 3′UTR of THBS1. A modified RNA immunoprecipitation–chromatin immunoprecipitation assay (RIP‐ChIP) was performed to confirm that TARBP2 binds to the 3'UTR of THBS1 in vivo. The stem‐loop sequence in the 3′UTR of THBS1 could be amplified in the group pulled down with GFP antibody but not the IgG (Figure 4G), indicating the binding of TARBP2 to the 3′UTR of THBS1 mRNAs inside tumor cells. These data demonstrated that TARBP2 recognized the stem‐loop structure located in the 3′UTRs of antiangiogenic gene transcripts for degradation.
FIGURE 4.
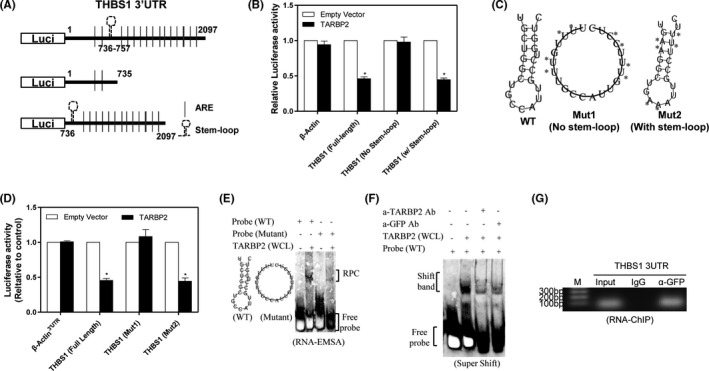
TARBP2 binds to conserved guanine cytosine‐rich stem‐loop structure in THBS1 3′UTR. A, Schematic representation of the luciferase reporter constructs of THBS1 containing truncated 3′UTRs with/without the stem‐loop structure. B, Relative luciferase activities of the indicated reporters were determined by luciferase assay; n = 3. C, The predicted stem‐loop structure of THBS1 (left) in its 3′UTR and mutation strategy (asterisks indicate base substitution). Mutant1 becomes unable to form a stem‐loop structure (middle), while Mutant2 forms a stem‐loop structure (right). D, Relative luciferase activities of the indicated reporters were evaluated by luciferase assay. E, RNA‐EMSA was performed with biotin‐labeled probes corresponding to the THBS1 3′UTR, including WT probe and mutant probe, in the presence of whole‐cell lysates (WCL) extracted from A549/TARBP2 cells. F, Supershift assay was performed using anti‐TARBP2 or anti‐GFP antibodies, followed by performing EMSA assay as described above. G, RNA‐ChIP assay was conducted in A549 cells after TARBP2/GFP fusion protein expression. *P < .05
3.5. Knockdown of TARBP2 increases the stability of antiangiogenic factor transcripts and inhibits tumor angiogenesis
To evaluate the effects of TARBP2 on tumor angiogenesis, we suppressed TARBP2 expression with shRNAs in A549 and MDA‐MB‐468 cells, respectively. The TARBP2 protein was, respectively, reduced by about 65%‐77% by two shRNAs (Figure 5A). Corresponding to the overexpression results, knocking down TARBP2 increased the mRNA expression of antiangiogenic mRNAs within two tumor cells (Figure 5B,C). However, the mRNA expression of the proangiogenic gene did not change after TARBP2 reduced, further suggesting that TARBP2 disrupted the angiogenesis balance in favor of inhibiting antiangiogenic gene expression. As expected, the half‐lives of antiangiogenic mRNAs were increased after knocking down TARBP2 in both tumor cells (Figure 5D,E, S5C‐E). The protein levels of THBS1 were also increased in the CMs of A549/shTARBP2 and MDA‐MB‐468/shTARBP2 cells, as indicated by ELISA data (Figure 5F). The CMs from A549/shTARBP2 and MDA‐MB‐468/shTARBP2 cells could significantly suppress endothelial cell migration (Figure 5G) and tube formation (Figure 5H). Consistent with these in vitro results, TARBP2 knockdown in vivo by injecting shTARBP2‐expressing adenovirus significantly reduced tumor growth (Figure 5I,J, S5F), metastasis (Figure S5G), and the number of CD31‐positive microvessels (Figure 5K) compared with the scrambled control group. Collectively, these results confirmed that the repression of TARBP2 indeed suppressed tumor angiogenesis through stabilizing antiangiogenic transcripts.
FIGURE 5.
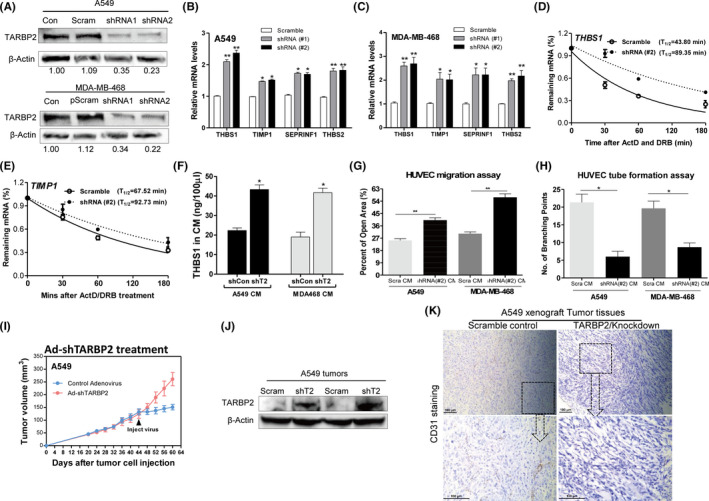
Knocking down TARBP2 increases the stability of antiangiogenic factor transcripts and inhibits tumor angiogenesis. A, Western blot was used to verify the efficiency of TARBP2 knockdown in protein level in A549 cells (upper) and MDA‐MB‐468 cells (lower), respectively. B, C, qRT‐PCR was used to measure the mRNA expression of indicated antiangiogenic genes in A549 cells (B) and MDA‐MB‐468 cells (C) after knockdown of TARBP2; n = 3. D, E, Half‐lives of indicated antiangiogenic factor genes were measured by qRT‐PCR after TARBP2 knockdown in A549 cells. F, ELISA quantification of THBS1 in the serum‐free culture medium of TARBP2 knocked down A549 and MDA‐MB‐468 cells; n = 3. G, Quantification of the open area of HUVECs treated with indicated tumor conditioned mediums (CMs) in the wound‐healing assay; n = 3. H, Quantification of the number of branching points of HUVECs treated with indicated tumor CMs in the tube formation assay; n = 3. *P < .05, **P < .01. I, Tumor growth curves in nude mice after treatment with adenovirus. Black arrows indicate the time point of adenovirus injection. J, Western blot was used to confirm knockdown of TARBP2 in A549 xenografts. K, Representative histological sections from tumors treated with control adenovirus or shTARBP2‐expressing adenovirus; tumor tissues stained with anti‐CD31 antibody. Scale bar, 100 µm
3.6. TARBP2 suppresses antiangiogenic factor gene expression in human tumor tissues and is associated with increased tumor angiogenesis and poor prognosis
To determine the relationship between TARBP2 and antiangiogenic factor gene expression in human tumors, we analyzed the expression of THBS1 in TARBP2‐overexpressing A549 and MDA‐MB‐468 tumors by qRT‐PCR. The mRNA levels of THBS1 were significantly reduced in these two tumor models (Figure S6A). Besides, a total of 17 human lung cancer samples were analyzed by IHC staining using anti‐TARBP2 antibody, of which nine samples showed high TARBP2 expression (TARBP2‐positive) and eight samples showed low TARBP2 expression (TARBP2‐negative). Also, the THBS1 level in TARBP2‐positive tumor tissues was significantly lower than that in TARBP2‐negative tumor tissues (Figure 6A). TARBP2 mRNA expression was increased in human lung cancers (Figure 6B), breast cancers (Figure S6B), and liver cancers (Figure S6D), 35 while THBS1 mRNA levels were reduced (Figures 6C and S6C,D). Moreover, a negative correlation between TARBP2 and THBS1, THBS2, and BAI1 expression was shown in the above human cancers (Figures 6D‐F and S6E‐I). Interestingly, more CD31‐positive microvessels were observed in TARBP2‐positive tumor tissues than in TARBP2‐negative tumor tissues (Figure 6G). Notably, high levels of TARBP2 in tumors from human lung cancer, breast cancer (Figure 6H), and liver cancer (Figure S6J) patients were significantly correlated with poor survival, respectively, while high THBS1 expression in these tumors had better survival (Figure 6I, Figure S6K). Finally, we drew a working model of TARBP2 to show its role in regulating tumor angiogenesis (Figure S6L). TARBP2 directly downregulated antiangiogenic factor mRNAs through destabilizing their mRNAs by dsRBD1/2 domains via binding the stem‐loop structure located in their 3′UTRs, resulting in angiogenesis imbalance in tumor cells.
FIGURE 6.
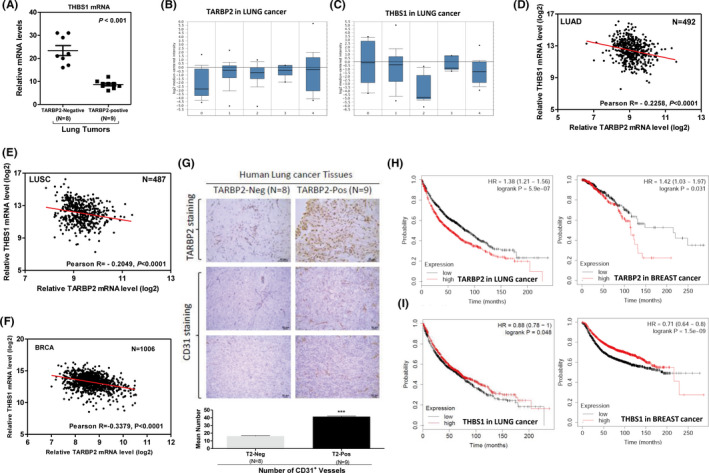
TARBP2 expression was associated with increased tumor angiogenesis and poor prognosis in human cancers. A, qRT‐PCR was used to analyze the THBS1 expression level in TARBP2‐negative (n = 8) or ‐positive (n = 9) human lung tumors. B, C, Comparison of TARBP2 (B) and THBS1 (C) expression between normal tissue (0) (n = 17), lung adenocarcinoma (1) (n = 139), lung carcinoid tumor (2) (n = 20), small cell lung carcinoma (3) (n = 6), and squamous cell lung carcinoma (4) (n = 21). D‐F, Pearson's correlation analysis between TARBP2 and THBS1 expression levels in log2 values in human lung adenocarcinoma (D), lung squamous cell carcinoma (E), and breast cancer patients (F). G, Representative images of IHC staining for TARBP2 (upper) and CD31 (middle) in human lung cancer tissues. Quantification of the number of CD31+ vessels per section from TARBP2‐negative and TARBP2‐positive lung tumor tissues (lower). H, I, Kaplan‐Meier overall survival curves for human lung and breast cancer patients with low and high tumor TARBP2 (H) and THBS1 (I) transcripts, respectively.
4. DISCUSSION
In this study, we identified the previously undiscovered role of TARBP2 in the regulation of tumor angiogenesis, which was confirmed by both in vitro and in vivo experiments using multiple types of tumor cells. Several studies and our results demonstrated TARBP2 promoted the metastatic progression of human lung cancer and breast cancer. 36 To rule out the influence of tumor types, three types of human tumor cells were tested, including human lung cancer, breast cancer, and liver cancer cells. We showed that TARBP2 could significantly promote tumor angiogenesis in several types of human cancers in vitro. In contrast, this could be inhibited by knocking down TARBP2. In vivo studies also confirmed that TARBP2 was able to increase the number of microvessels in human lung tumor and breast tumor xenografts. These findings indicated for the first time that TARBP2 promoted tumor progression through tumor angiogenesis induction.
We used high‐sensitive RNA‐seq together with PCR array to detect angiogenesis‐related gene expression precisely in tumor cells, confirming that TARBP2 specifically downregulates the mRNA expression of antiangiogenic factors. The preferential binding of TARBP2 to antiangiogenic mRNAs but not to proangiogenic mRNAs signified a selective targeting of TARBP2 in regulating tumor angiogenic mRNAs, resulting in an imbalance between antiangiogenic and proangiogenic genes. The upregulation of many proangiogenic mRNAs might be a secondary effect of TARBP2 overexpression because TARBP2 neither enriched the proangiogenic factor transcripts nor targeted their 3′UTRs. Furthermore, the expression of these genes is not affected by TARBP2 knockdown. The broad targets of TARBP2 in the tumor angiogenesis pathway suggested that TARBP2 induces tumor angiogenesis through targeting multiple molecules in tumor cells.
The 3′UTR plays an important role in the RBP‐mediated post‐transcriptional regulation of genes. 37 , 38 , 39 We confirmed that TARBP2 could suppress luciferase activity through the 3′UTRs of antiangiogenic genes and reduced their mRNA stability. It has been reported that the dsRBD1 and dsRBD2 domains are essential for TARBP2‐mediated double‐stranded RNA substrate recognition, binding, and processing. 34 In this study, we demonstrate that the dsRBD1/2 domains are required for the degradation of antiangiogenic genes and tumor angiogenesis induction by truncating the domains of TARBP2. Many elements, such as ARE, GRE, and the stem‐loop structure, are localized in the 3′UTR and responsible for mRNA turnover. 40 , 41 , 42 Our results demonstrated that TARBP2 destabilized THBS1 by binding to the stem‐loop structure in the 3′UTR. There are no consensus stem‐loop sequences among the antiangiogenic genes, which further supports our hypothesis that TARBP2 mainly recognizes the RNA secondary structure in the 3′UTR. The direct binding of TARBP2 to THBS1 and TIMP1 were also confirmed by the high‐throughput sequencing of RNAs isolated by a crosslinking immunoprecipitation experiment. 26
Finally, TARBP2 regulating tumor angiogenesis was further confirmed in human lung tumor samples. Our results demonstrated that TARBP2 is highly expressed in human lung cancer, breast cancer, and liver cancer, which was negatively related to the expression of antiangiogenic mRNAs. The strong association between high TARBP2 expression in tumor samples and poor survival of cancer patients further demonstrated its clinical significance. Collectively, our results identified a new tumor angiogenesis regulator and might lead to improved prognosis using TARBP2 as a target for antiangiogenic cancer therapy.
DISCLOSURE
The authors have no conflict of interest.
Supporting information
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Table S1
Table S2
Table S3
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the CAMS Innovation Fund for Medical Science (No. 2017‐I2M‐1‐016), the PUMC Youth Fund (Grant Number: 3332017105), and the National Natural Science Foundation of China (NSFC) (Grant Number: 81702769).
Zhou M, Lu W, Li B, Liu X, Li A. TARBP2 promotes tumor angiogenesis and metastasis by destabilizing antiangiogenic factor mRNAs. Cancer Sci. 2021;112:1289–1299. 10.1111/cas.14820
Meicen Zhou and Wenbao Lu contributed equally to this work.
REFERENCES
- 1. Kerbel RS. Tumor angiogenesis. N Engl J Med. 2008;358(19):2039‐2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fidler IJ. Critical determinants of metastasis. Semin Cancer Biol. 2002;12(2):89‐96. [DOI] [PubMed] [Google Scholar]
- 3. Husemann Y, Geigl JB, Schubert F, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13(1):58‐68. [DOI] [PubMed] [Google Scholar]
- 4. Viallard C, Larrivee B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis. 2017;20(4):409‐426. [DOI] [PubMed] [Google Scholar]
- 5. Weis SM, Cheresh DA. Tumor angiogenesis: molecular pathways and therapeutic targets. Nat Med. 2011;17(11):1359‐1370. [DOI] [PubMed] [Google Scholar]
- 6. Wang P, Zeng Z, Lin C, et al. Thrombospondin‐1 as a potential therapeutic target: multiple roles in cancers. Curr Pharm Des. 2020;26(18):2116‐2136. [DOI] [PubMed] [Google Scholar]
- 7. Wu XG, Zhou CF, Zhang YM, et al. Cancer‐derived exosomal miR‐221‐3p promotes angiogenesis by targeting THBS2 in cervical squamous cell carcinoma. Angiogenesis. 2019;22(3):397‐410. [DOI] [PubMed] [Google Scholar]
- 8. Zacchigna S, Zentilin L, Morini M, et al. AAV‐mediated gene transfer of tissue inhibitor of metalloproteinases‐1 inhibits vascular tumor growth and angiogenesis in vivo. Cancer Gene Ther. 2004;11(1):73‐80. [DOI] [PubMed] [Google Scholar]
- 9. Zerrouqi A, Pyrzynska B, Febbraio M, Brat DJ, Van Meir EG. P14ARF inhibits human glioblastoma‐induced angiogenesis by upregulating the expression of TIMP3. J Clin Invest. 2012;122(4):1283‐1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu D, Hunter SB, Vertino PM, Van Meir EG. Overexpression of MBD2 in glioblastoma maintains epigenetic silencing and inhibits the antiangiogenic function of the tumor suppressor gene BAI1. Cancer Res. 2011;71(17):5859‐5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beckers J, Herrmann F, Rieger S, et al. Identification and validation of novel ERBB2 (HER2, NEU) targets including genes involved in angiogenesis. Int J Cancer. 2005;114(4):590‐597. [DOI] [PubMed] [Google Scholar]
- 12. Otrock ZK, Mahfouz RA, Makarem JA, Shamseddine AI. Understanding the biology of angiogenesis: review of the most important molecular mechanisms. Blood Cells Mol Dis. 2007;39(2):212‐220. [DOI] [PubMed] [Google Scholar]
- 13. Kozak CA, Gatignol A, Graham K, Jeang KT, McBride OW. Genetic mapping in human and mouse of the locus encoding TRBP, a protein that binds the TAR region of the human immunodeficiency virus (HIV‐1). Genomics. 1995;25(1):66‐72. [DOI] [PubMed] [Google Scholar]
- 14. De Vito C, Riggi N, Cornaz S, et al. A TARBP2‐dependent miRNA expression profile underlies cancer stem cell properties and provides candidate therapeutic reagents in Ewing sarcoma. Cancer Cell. 2012;21(6):807‐821. [DOI] [PubMed] [Google Scholar]
- 15. Sand M, Skrygan M, Georgas D, et al. The miRNA machinery in primary cutaneous malignant melanoma, cutaneous malignant melanoma metastases and benign melanocytic nevi. Cell Tissue Res. 2012;350(1):119‐126. [DOI] [PubMed] [Google Scholar]
- 16. Kim Y, Yeo J, Lee JH, et al. Deletion of human tarbp2 reveals cellular microRNA targets and cell‐cycle function of TRBP. Cell Rep. 2014;9(3):1061‐1074. [DOI] [PubMed] [Google Scholar]
- 17. Daniels SM, Melendez‐Pena CE, Scarborough RJ, et al. Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC Mol Biol. 2009;10:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhong J, Peters AH, Lee K, Braun RE. A double‐stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat Genet. 1999;22(2):171‐174. [DOI] [PubMed] [Google Scholar]
- 19. Ling T, Li SN, Weng GX, et al. TARBP2 negatively regulates IFN‐beta production and innate antiviral response by targeting MAVS. Mol Immunol. 2018;104:1‐10. [DOI] [PubMed] [Google Scholar]
- 20. Ling T, Weng GX, Li J, et al. TARBP2 inhibits IRF7 activation by suppressing TRAF6‐mediated K63‐linked ubiquitination of IRF7. Mol Immunol. 2019;109:116‐125. [DOI] [PubMed] [Google Scholar]
- 21. Daniels SM, Gatignol A. The multiple functions of TRBP, at the hub of cell responses to viruses, stress, and cancer. Microbiol Mol Biol Rev. 2012;76(3):652‐666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Caramuta S, Lee L, Ozata DM, et al. Clinical and functional impact of TARBP2 over‐expression in adrenocortical carcinoma. Endocr Relat Cancer. 2013;20(4):551‐564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bai S, Nunez AL, Wei S, et al. Microsatellite instability and TARBP2 mutation study in upper urinary tract urothelial carcinoma. Am J Clin Pathol. 2013;139(6):765‐770. [DOI] [PubMed] [Google Scholar]
- 24. Lin X, Wu M, Liu P, et al. Up‐regulation and worse prognostic marker of cytoplasmic TARBP2 expression in obstinate breast cancer. Med Oncol. 2014;31(4):868. [DOI] [PubMed] [Google Scholar]
- 25. Sand M, Skrygan M, Georgas D, et al. Expression levels of the microRNA maturing microprocessor complex component DGCR8 and the RNA‐induced silencing complex (RISC) components argonaute‐1, argonaute‐2, PACT, TARBP1, and TARBP2 in epithelial skin cancer. Mol Carcinog. 2012;51(11):916‐922. [DOI] [PubMed] [Google Scholar]
- 26. Goodarzi H, Zhang S, Buss CG, Fish L, Tavazoie S, Tavazoie SF. Metastasis‐suppressor transcript destabilization through TARBP2 binding of mRNA hairpins. Nature. 2014;513(7517):256‐260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fish L, Navickas A, Culbertson B, et al. Nuclear TARBP2 drives oncogenic dysregulation of RNA splicing and decay. Mol Cell. 2019;75(5):967‐81 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang MY, Huang HY, Kuo YL, et al. TARBP2‐enhanced resistance during tamoxifen treatment in breast cancer. Cancers (Basel). 2019;11(2):210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lai HH, Li CW, Hong CC, et al. TARBP2‐mediated destabilization of Nanog overcomes sorafenib resistance in hepatocellular carcinoma. Mol Oncol. 2019;13(4):928‐945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen G, Gu H, Fang T, Zhou K, Xu J, Yin X. Hypoxia‐induced let‐7f‐5p/TARBP2 feedback loop regulates osteosarcoma cell proliferation and invasion by inhibiting the Wnt signaling pathway. Aging (Albany NY). 2020;12(8):6891‐6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu X, Li Z. The role of TARBP2 in the development and progression of cancers. Tumour Biol. 2016;37(1):57‐60. [DOI] [PubMed] [Google Scholar]
- 32. Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725‐731. [DOI] [PubMed] [Google Scholar]
- 33. Lu W, Ning H, Gu L, et al. MCPIP1 selectively destabilizes transcripts associated with an antiapoptotic gene expression program in breast cancer cells that can elicit complete tumor regression. Cancer Res. 2016;76(6):1429‐1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee HY, Zhou K, Smith AM, Noland CL, Doudna JA. Differential roles of human Dicer‐binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013;41(13):6568‐6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data‐mining platform. Neoplasia. 2004;6(1):1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shi Y, Zuo D, Wang X, Han M, Wu Y. shRNAmediated silencing of TARBP2 inhibits NCIH1299 nonsmall cell lung cancer cell invasion and migration via the JNK/STAT3/AKT pathway. Mol Med Rep. 2016;14(4):3725‐3730. [DOI] [PubMed] [Google Scholar]
- 37. Kim DJ, Jang JH, Ham SY, et al. Doxorubicin inhibits PD‐L1 expression by enhancing TTP‐mediated decay of PD‐L1 mRNA in cancer cells. Biochem Biophys Res Commun. 2020;522(2):402‐407. [DOI] [PubMed] [Google Scholar]
- 38. Lipert B, Wilamowski M, Gorecki A, Jura J. MCPIP1, alias Regnase‐1 binds and cleaves mRNA of C/EBPbeta. PLoS One. 2017;12(3):e0174381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Essig K, Kronbeck N, Guimaraes JC, et al. Roquin targets mRNAs in a 3'‐UTR‐specific manner by different modes of regulation. Nat Commun. 2018;9(1):3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vlasova‐St Louis I, Bohjanen PR. Post‐transcriptional regulation of cytokine signaling by AU‐rich and GU‐rich elements. J Interferon Cytokine Res. 2014;34(4):233‐241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hanieh H, Masuda K, Metwally H, et al. Arid5a stabilizes OX40 mRNA in murine CD4(+) T cells by recognizing a stem‐loop structure in its 3'UTR. Eur J Immunol. 2018;48(4):593‐604. [DOI] [PubMed] [Google Scholar]
- 42. Mino T, Murakawa Y, Fukao A, et al. Regnase‐1 and roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell. 2015;161(5):1058‐1073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Table S1
Table S2
Table S3
Supplementary Material


