Abstract
Hepatocellular carcinoma (HCC) is the second leading cause of cancer‐related death. High recurrence rates after curative resection and the lack of specific biomarkers for intrahepatic metastases are major clinical problems. Recently, exosomal microRNAs (miRNAs) have been reported to have a role in the formation of the pre‐metastatic niche and as promising biomarkers in patients with malignancy. Here we aimed to clarify the molecular mechanisms of intrahepatic metastasis and to identify a novel biomarker miRNA in patients with HCC. A highly intrahepatic metastatic cell line (HuH‐7M) was established by in vivo selection. HuH‐7M showed increased proliferative ability and suppression of apoptosis and anoikis. HuH‐7M and the parental cell (HuH‐7P) showed the similar expression of epithelial‐mesenchymal transition markers and cancer stem cell markers. In vivo, mice treated with exosomes derived from HuH‐7M showed increased tumorigenesis of liver metastases. Exosomes from HuH‐7M downregulated endothelial cell expression of vascular endothelial‐cadherin (VE‐cadherin) and zonula occludens‐1 (ZO‐1) in non‐cancerous regions of liver and increased the permeability of FITC‐dextran through the monolayer of endothelial cells. The miRNAs (miR‐638, miR‐663a, miR‐3648, and miR‐4258) could attenuate endothelial junction integrity by inhibiting VE‐cadherin and ZO‐1 expression. In patients with HCC, higher serum exosomal miR‐638 expression was associated with tumor recurrence. In conclusion, the miRNAs secreted from a highly metastatic cancer cell can promote vascular permeability via downregulation of endothelial expression of VE‐cadherin and ZO‐1. Serum exosomal miR‐638 expression holds potential for serving as a significant and independent prognostic marker in HCC.
Keywords: exosome, hepatectomy, in vivo selection, liver cancer, microRNA
miR‐638, miR‐663a, miR‐3648, and miR‐4258 downregulate VE‐cadherin and ZO‐1 expression in HUVECs.

Abbreviations
- AFP
alpha‐fetoprotein
- DFS
disease‐free survival
- EpCAM
epithelial cell adhesion molecule
- EthD‐1
ethidium homodimer
- EV
extracellular vesicle
- HCC
hepatocellular carcinoma
- HR
hazard ratio
- miRNA
microRNA
- NC
negative control
- OS
overall survival
- qRT‐PCR
quantitative reverse transcription‐polymerase chain reaction
- VE‐cadherin
vascular endothelial‐cadherin
- ZEB1
Zinc Finger E‐Box Binding Homeobox 1
- ZO‐1
zonula occludens‐1
1. INTRODUCTION
Hepatocellular carcinoma (HCC) is the most frequently occurring primary liver cancer and the second most common cause of cancer‐related death worldwide. 1 Treatment options for HCC include hepatectomy, systemic chemotherapy, transcatheter arterial chemoembolization, and radiofrequency ablation. Hepatectomy is one of the main treatment modalities for HCC, and recent advances in surgical techniques and perioperative management have greatly improved short‐term surgical outcomes. 2 , 3 , 4 , 5 However, the recurrence rate after curative resection is still high, and the most frequent site of tumor relapse is the remnant liver.
Two problems must be solved to improve prognosis. First, the molecular mechanism of intrahepatic metastasis needs to be clarified. Intrahepatic metastasis via portal blood flow, local blood flow, and systemic circulation have been hypothesized. 6 , 7 In this conceptualization, local intrahepatic metastasis arises from the drainage area of the tumor blood flow. Cancer cells passing through the drainage area circulate in the systemic circulation and form intrahepatic metastases. However, the molecular biology of intrahepatic metastasis remains unclear and requires further investigation. Second, serum AFP is widely used as a specific marker in adjunct surveillance, but its disadvantage is low sensitivity. Novel serum biomarkers are urgently required for improving the effectiveness of the surveillance program.
EVs, including exosomes, microvesicles, and apoptotic bodies, are small membrane vesicles secreted from various cell types into the extracellular environment. EVs contain cell‐specific proteins, mRNA, and microRNAs (miRNAs) and are well established mediators of cell‐to‐cell communication, participating in inflammation, immunosuppression, angiogenesis, and drug resistance. 8 , 9 , 10 , 11 In particular, the horizontal transfer of miRNAs has been proposed as a new form of intercellular communication by which donor cells can regulate gene expression of recipient cells. 12 In various carcinomas, miRNAs are reported to be involved in the regulation of the tumor microenvironment, called the “pre‐metastatic niche.” 11 , 13 , 14 , 15 , 16 The contribution of miRNAs to intrahepatic metastasis of HCC is evident, and we and several other groups have reported on the roles of specific miRNAs in intrahepatic metastasis and drug resistance. 17 , 18 , 19 , 20 , 21 , 22 , 23 In addition, exosomal miRNAs have been highlighted as potential biomarkers in patients with malignancy. 24 , 25
In investigating the molecular mechanism of HCC metastasis, comparisons of cell lines of different origins may be complicated to understand because of inter‐line heterogeneity. In vivo selection, with the goal of generating sequential malignant metastatic cell line from the same cell line origin, is an effective method to help clarify the metastatic mechanism. 26 , 27 In the present study, we established a novel, highly intrahepatic metastatic HCC cell line using in vivo selection. Our aim was to clarify the molecular mechanisms of intrahepatic metastasis for the purposes of identifying a novel prognostic marker of HCC.
2. MATERIALS AND METHODS
2.1. Cell culture
We used a luciferase stably expressing human liver cancer cell line: Huh‐7‐Luc (RRID:CVCL_JG51), purchased from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan), and human umbilical vein endothelial cells (HUVECs) purchased from Lonza (Tokyo, Japan). HuH‐7‐Luc was cultured and maintained in DMEM supplemented with 10% fetal bovine serum and 500 µg/mL penicillin streptomycin. HUVECs were cultured and maintained in endothelial cell growth medium 2‐MV.
2.2. Animals
Animal experiments were conducted using male CB17.Cg‐PrkdcscidLystbg‐J/CrlCrlj mice, age 6‐7 wk (Charles River, Japan). All studies were performed in accordance with the ethical guidelines of the Declaration of Helsinki, Japanese Ethical Guidelines for Human Genome/Gene Analysis Research, and Ethical Guidelines for Medical and Health Research Involving Human Subjects at Osaka University. Monitoring of tumor development was conducted by weekly bioluminescence imaging using a Spectrum in vivo imaging system (Caliper Life Sciences, Hopkinton, MA, USA). This study was approved by the Animal Experiments Committee, Osaka University (approval number, 28‐010‐011).
2.3. Establishment of a highly intrahepatic metastatic cell line
A new, highly intrahepatic metastatic cell line was established as follows. A cell suspension containing 1 × 106 HuH‐7‐Luc in a volume of 100 μL was injected into the spleen of mice. Tumor formation in the liver was monitored by in vivo imaging after intraperitoneal luciferin injections. At 3 or 4 wk after injection of cell suspension, liver tumors were harvested and minced, and then incubated with collagenase type I. The medium containing tumor cells was filtered through a 70‐μm cell strainer and centrifuged at 400 g for 5 min at room temperature. Tumor cells were placed in DMEM in a 6‐cm dish. After the cells were grown to confluency, tumor cells were injected into the spleen again. We repeated this procedure for 4 cycles. The cell line of the original HuH‐7‐Luc was designated as HuH‐7P, and the HuH‐7‐Luc after 4 cycles was designated as HuH‐7M (clone A and clone B). The liver tumors were formalin fixed, and paraffin‐embedded tissue sections were examined using hematoxylin and eosin staining.
2.4. Tumorigenesis assay by HuH‐7P and HuH‐7M injection into the spleen
A cell suspension containing 1 × 106 HuH‐7P or HuH‐7M in a volume of 100 μL was injected into the spleen of anesthetized mice, followed by splenectomy. At 28 d from the injection of cell suspension, the liver was harvested.
2.5. In vitro functional analysis
Proliferation was quantified using the Cell Counting Kit‐8 (Dojindo Laboratories). Apoptosis of HuH‐7P and HuH‐7M was detected by annexin V assay as previously described. 28 Cells that were positive for annexin V and propidium iodide were defined as apoptotic cells. Apoptotic cells were counted using the FACS Cant II (BD Biosciences). The anoikis potential of HuH‐7P and HuH‐7M was evaluated using the CytoSelectTM 24‐well Anoikis Assay kit (Cell Biolabs, Inc). Live cells can be detected with calcein AM or MTT, which is a green fluorescent dye. Anoikis‐induced cell death was detected with ethidium homodimer (EthD‐1), which is a red fluorescent dye. Cell viability was assessed using trypan blue (Invitrogen). The cell suspension was mixed with trypan blue at 1:1 and the number of viable cells was measured by an automated cell counter (Cell Counter model R1; Olympus).
2.6. Preparation of exosomes from HuH‐7P and HuH‐7M cell lines
HuH‐7P and HuH‐7M were washed with PBS, and the medium was switched to advanced DMEM. After incubation for 2 d, the medium was centrifuged at 2000 g for 10 min at 4°C. The supernatant was filtered through a 0.22‐mm Stericup (Millipore, Burlington, MA, USA). The conditioned medium was ultracentrifuged at 110 000 g for 70 min at 4°C. The pellets were washed and resuspended in PBS. The fraction containing exosomes was measured for its protein content using the Micro BCA protein assay kit (Thermo Scientific). The size and quality of exosomes were determined using a LM10‐HS NanoSight instrument.
2.7. Tumorigenesis assay by intraperitoneal injection of HuH‐7P‐derived and HuH‐7M‐derived exosomes
A cell suspension containing 1 × 106 HuH‐7P in a volume of 100 μL was injected into the spleen of mice, then splenectomy was performed. In total, 5 μg of exosomes from HuH‐7P or HuH‐7M was intraperitoneally injected 6 times. At 28 d from cell injection, the liver was harvested.
2.8. Immunofluorescence staining
Immunofluorescence staining methods have been previously described. 29 Immunostaining was performed using the following primary antibodies: anti‐VE‐cadherin, anti‐ZO‐1, and anti‐CD31 (Abcam). Sections were stained with the appropriate Alexa Fluor® 488‐conjugated or 647‐conjugated secondary IgGs (Abcam; Cell Signaling).
2.9. PKH67‐labeled exosome transfer
Purified exosomes derived from HuH‐7P and HuH‐7M were labeled with a PKH67 green fluorescent labeling kit (Sigma‐Aldrich) as described previously. 14 HUVECs were incubated for 24 h with PKH67‐labeled exosomes. After counterstaining with DAPI, we analyzed the cells using a fluorescence microscope (BZ‐X700; Keyence).
2.10. HUVEC exosome treatment
HUVECs were incubated for 48 h with HuH‐7P‐ or HuH‐7M‐derived exosomes. Then, we analyzed expression changes in HUVECs by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR), western blotting, and immunocytochemistry.
2.10.1. qRT‐PCR
Total RNA was isolated from exosomes or cell lines using the miRNeasy Mini Kit (Qiagen) and from serum using the miRNeasy Serum/Plasma Advanced Kit (Qiagen). For miRNA, a reverse transcription reaction was performed with the mir‐X miRNA First Strand Synthesis Kit (TaKaRa Bio). For qRT‐PCR, we used the mir‐X miRNA qRT‐PCR TB Green Kit (TaKaRa Bio), and ViiA 7 Software (Thermo Fisher Scientific, Inc). The sequences for the hsa‐miR‐638, miR‐663a, miR‐3648, and miR‐4258 primers were designed based on miRBase (http://www.mirbase.org). Transcript levels were normalized to U6 for cell lines and to cel‐miR‐39 for exosomes and serum. For mRNA, qRT‐PCR was performed as previously described. 30 Complementary DNA was synthesized using the Reverse Transcription System (Promega). Amplification products were quantified using the LightCycler‐DNA master SYBR Green I Kit (Roche Diagnostics), and target gene expression levels were normalized to GAPDH expression levels. The PCR primer sequences were as follows: GAPDH forward primer, 5′‐CAGTATGACTCCACTCACGGC‐3′, and GAPDH reverse primer, 5′‐GAGGGGCCATCCACAGTCTTC‐3′; vascular endothelial‐cadherin (VE‐cadherin) forward primer, 5′‐TTTGCCCAGCCCTACGAACCT‐3′, and VE‐cadherin reverse primer, 5′‐ACCGCCGTCATTGTCTGCCTC‐3′; zonula occludens 1 (ZO‐1) forward primer, 5′‐GCTAAGAGCACAGCAATGGA‐3′, and ZO‐1 reverse primer, 5′‐GCATGTTCAACGTTATCCAT‐3′. Data were analyzed in accordance with the comparative CT method.
2.10.2. Western blotting
Western blot analysis was performed as described previously. 29 Briefly, aliquots of total protein (10 µg) were electrophoresed on sodium dodecyl sulfate polyacrylamide, 10% Tris‐HCl gels (Bio‐Rad Laboratories). The separated proteins were transferred to polyvinylidene difluoride membranes (Bio‐Rad Laboratories) and incubated with primary antibodies overnight at 4°C. Proteins were detected using the following antibodies: anti‐VE‐cadherin (Abcam), anti‐ZO‐1, anti‐E‐cadherin, anti‐Zinc Finger E‐Box Binding Homeobox 1 (ZEB1), anti‐N‐cadherin, anti‐Snail (Cell Signaling Technology, Danvers, MA, USA), anti‐CD13, anti‐CD133, anti‐EpCAM (Abcam) and anti‐β actin (Sigma, Tokyo, Japan).
2.10.3. Immunocytochemical staining
Immunocytochemistry was performed as previously described. 31 Briefly, the cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X‐100, and blocked with 1% bovine serum albumin. Proteins were detected with anti‐VE‐cadherin antibody or anti‐ZO‐1 antibody. After counterstaining with DAPI, the images were captured using a confocal laser scanning microscope (FV 1000‐D (IX); Olympus).
2.10.4. In vitro permeability assay
The permeability of HUVEC monolayers grown on transwell filters (0.4‐μm pore size; BD Biosciences) was assessed by the passage of FITC‐conjugated dextran (FITC‐dextran) (average MW ~70 000; Sigma‐Aldrich). Briefly, HUVECs were seeded into the upper chamber to form a confluent cell monolayer. Then FITC‐dextran was added to the upper chamber. Then medium in the bottom well was collected and the appearance of fluorescence was monitored at 485 nm excitation and 520 nm emission.
2.10.5. miRNA microarray experiments
Microarray analysis of the purified RNAs obtained from HuH‐7P‐ and HuH‐7M‐derived exosomes was performed using the Toray microRNA microarray system. Labeled RNAs were hybridized onto 3D‐Gene Human miRNA Oligo chips (v.21; Toray Industries). Raw and processed data from this analysis were deposited in the Gene Expression Omnibus repository (accession number: GSE151169).
2.10.6. Transfection
Cells were transfected with 10 nmol/L of mirVana mimic (miR‐mimic; miR‐638, miR‐663a, miR‐3648, and miR‐4258) or nontargeting negative control (miR‐NC) oligonucleotides (Ambion) using Lipofectamine RNAiMAX (Invitrogen).
2.10.7. Clinical samples
We analyzed preoperative serum samples from 54 patients with HCC who underwent curative resection (R0) between January 2012 and December 2015 at Osaka University Hospital. The expression levels of miRNAs in serum were measured by qRT‐PCR. Patients with a history of hepatectomy or preoperative therapy were excluded. Clinicopathological features including age, sex, hepatitis B virus surface antigen, anti‐hepatitis C virus antibody, Child‐Pugh classification, AFP levels, des‐γ‐carboxy prothrombin levels, tumor size, number of tumors, histological differentiation, microscopic portal vein invasion, and liver cirrhosis are shown in Table 1. All patients provided written informed consent before inclusion in the study, and the study protocol was approved by the Human Ethics Review Committee of the Graduate School of Medicine, Osaka University (approval number 20109), which adhered to the guidelines of the Declaration of Helsinki.
TABLE 1.
Relationship between clinicopathological factors and serum exosomal miR‐638 expression level
| miR‐638 expression | P value | ||
|---|---|---|---|
| High expression group (n = 27) | Low expression group (n = 27) | ||
| Age | 73 (56‐84) | 70 (56‐81) | .4058 |
| Sex (male/female) | 18/9 | 17/10 | .7756 |
| Hepatitis B virus surface antigen (±) | 4/23 | 4/23 | .9999 |
| Anti‐hepatitis C virus antibody (±) | 10/17 | 13/14 | .4085 |
| Child‐Pugh classification (A/B) | 25/2 | 25/2 | .9999 |
| Alpha‐fetoprotein (ng/ml) | 16 (2‐21 580) | 23 (2‐3100) | .1349 |
| Des‐γ‐carboxy prothrombin (mAU/ml) | 290 (16‐19 101) | 340 (13‐191 285) | .3998 |
| Tumor size (cm) | 4.3 (1.3‐14.5) | 3.0 (1.4‐11.8) | .2400 |
| Number of tumor (multiple/single) | 4/23 | 3/24 | .6849 |
| Histological differentiation (well, moderately/poorly) | 15/12 | 18/9 | .4017 |
| Microscopic portal vein invasion (±) | 8/19 | 7/20 | .7612 |
| Liver cirrhosis (±) | 9/18 | 12/15 | .4017 |
miR, microRNA.
2.10.8. Statistical analysis
Continuous variables are expressed as mean ± standard deviation. Categorical variables were compared using the chi‐square or Fisher exact test, as appropriate, and continuous variables were compared using Student t test. DFS and OS were analyzed using the Kaplan‐Meier method, and differences between survival curves were compared using the log‐rank test. To evaluate the risks associated with the prognostic variables, we performed multivariate analysis using a Cox model, with determination of the hazard ratio and 95% confidence interval. All variables with P < .05 in univariate analysis were considered for inclusion in the multivariate analysis. Statistical analyses were performed using JMP software v.14.0 (SAS Institute Inc).
3. RESULTS
3.1. Establishment of highly intrahepatic metastatic cell line (HuH‐7M) by in vivo selection
Figure 1A shows the schema of in vivo selection. After 4 cycles, a novel, highly intrahepatic metastatic cell line (HuH‐7M) was established. The morphologic appearance of HuH‐7M cell lines was similar to that of the parental line (HuH‐7P). The HuH‐7M formed more metastatic nodules in the liver and showed significant increases in liver metastatic activity compared with HuH‐7P. Figure 1B showed the metastatic nodules after first cycles and fourth cycles of in vivo selection in left and middle panel. The histological analysis of liver tumors revealed that the tumor consisted of moderately differentiated HCC (Figure 1B, right panel).
FIGURE 1.
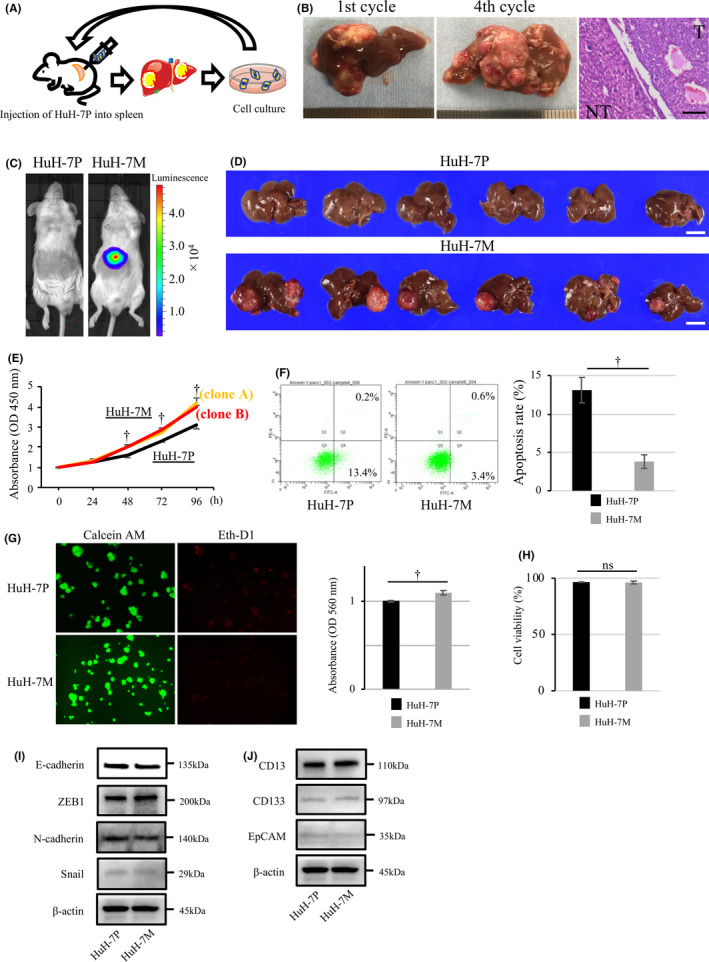
Establishment of a novel highly intrahepatic metastatic cell line (HuH‐7M) by in vivo selection and functional analysis. A, Schema of the method of in vivo selection. B, Metastatic multiple nodules in the liver after first cycle (left panel) and fourth cycle (middle panel) in vivo selection. The pathological examination (right panel). Scale bar, 100 µm. C, Bioluminescence image of mice with injection of HuH‐7P (left image) and HuH‐7M (right image). D, Tumorigenesis assay: the upper panel shows the HuH‐7P injection, and the lower panel shows the HuH‐7M injection. Scale bars, 1.0 cm. E, Proliferation assay: the yellow line indicates the proliferative curve for HuH‐7M clone A and the red line the curve clone B; the black line indicates HuH‐7P. F, Annexin V assay: apoptosis rates of HuH‐7P and HuH‐7M. G, Anoikis assay: cell viability was determined by Calcein AM (green, left panel), whereas anoikis‐like cell death was determined via staining with EthD‐1 (red, left panel). MTT methods also demonstrated viable cells (right panel). OD, optical density. H, Viability assay: cell viability of HuH‐7P and HuH‐7M in cell suspension. I, The expression of epithelial‐mesenchymal transition marker of HuH‐7P or HuH‐7M. J, The expression of cancer stem cell marker of HuH‐7P or HuH‐7M. † P < .01. ns, not significant
3.2. Comparison of tumorigenesis assay
We compared tumorigenesis of HuH‐7P and HuH‐7M in vivo. Both cell lines were simultaneously injected into the spleen, and splenectomy was performed. Using the in vivo imaging system, we observed that increased bioluminescence coincided with liver tumors in HuH‐7M (Figure 1C). The HuH‐7M could form the metastatic nodule of the liver in 67% (6 out of 9) of the mice (Figure 1D, lower panel), whereas the HuH‐7P yielded no liver metastasis (Figure 1D, upper panel).
3.3. Comparison of characteristics of HuH‐7P and HuH‐7M
To investigate the characteristics of HuH‐7P and HuH‐7M, we used proliferation, annexin V, and anoikis assays. In the proliferation assay, 2 clones of HuH‐7M cell lines (clone A and clone B) showed significant increases in proliferative ability compared with the HuH‐7P cell line (Figure 1E). We then used HuH‐7M (clone A) for subsequent experiments. Compared with HuH‐7P, the HuH‐7M showed a more steeply decreased apoptosis rate of 3.4% vs 13.4% (Figure 1F). The HuH‐7M also showed more suppressed anoikis‐like cell death compared with HuH‐7P, based on staining with EthD‐1 (Figure 1G, left panel). In addition, the viability of HuH‐7M was 1.1‐fold greater than that of HuH‐7P, based on MTT colorimetric detection (Figure 1G, right panel). The cell viability of HuH‐7P or HuH‐7M was not significant to be 96.5% and 96.1%, respectively (Figure 1H). The expression change of epithelial‐mesenchymal transition markers of E‐cadherin, ZEB1, N‐cadherin and Snail were not found between HuH‐7P and HuH‐7M cell lines (Figure 1I). The expression of the cancer stem cell markers reported about HCC such as CD13, CD133 and EpCAM was similar between HuH‐7P and HuH‐7M (Figure 1J).
3.4. Exosomes derived from highly intrahepatic metastatic HuH‐7M are linked to increased liver metastasis in vivo
We investigated in vivo the effect on liver metastasis of exosomes derived from the highly intrahepatic metastatic cell line HuH‐7M (Figure 2A). Metastatic nodules in the liver were observed only in mice treated with the HuH‐7M‐derived exosomes (liver metastases: 2 out of 6). No metastatic nodules were found in mice treated with exosomes derived from the original cell line (HuH‐7P; liver metastases: 0 out of 5) (Figure 2B). Furthermore, hepatic metastases were pathologically confirmed as moderately differentiated HCC (Figure 2C). The immunostaining revealed that the expression of VE‐cadherin and ZO‐1 was observed in CD31‐positive endothelial cell in non‐cancerous hepatic regions of mice treated with HuH‐7P‐derived exosomes. Expression of both VE‐cadherin and ZO‐1 was decreased in mice treated with HuH‐7M‐derived exosomes (Figure 2D).
FIGURE 2.
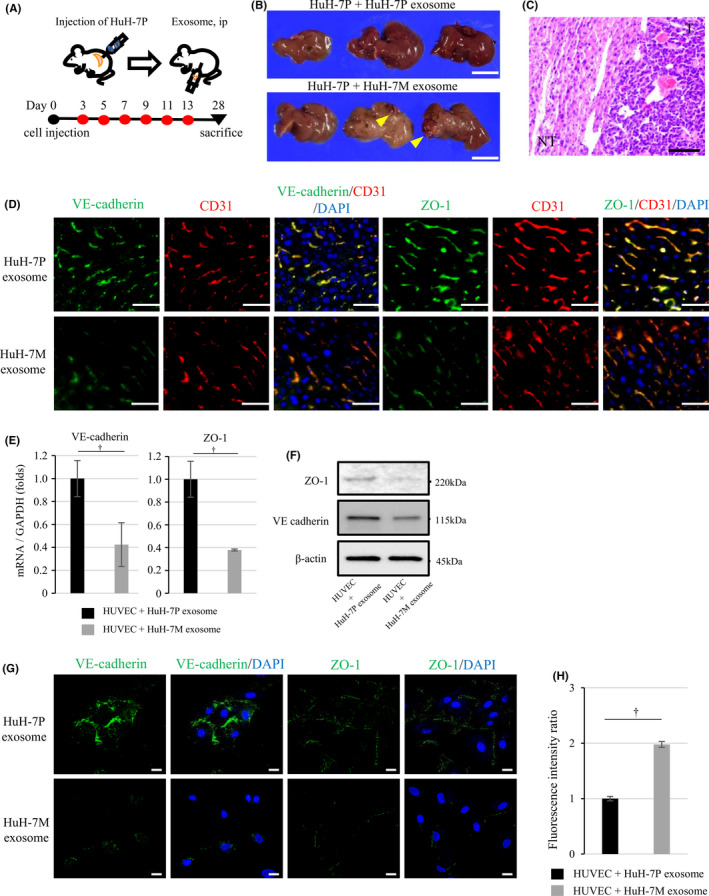
HuH‐7M‐derived exosomes increase intrahepatic metastasis and downregulate VE‐cadherin and ZO‐1 expression of HUVEC. A, Experimental schema and time schedules of HuH‐7P cell line injection and intraperitoneal injection of exosomes. B, Tumorigenesis assay: HuH‐7P plus HuH‐7P‐derived exosomes (upper panel) and HuH‐7P plus HuH‐7M‐derived exosomes (lower panel). Arrowheads indicate hepatic tumors. Scale bars, 1.0 cm. C, The pathological examination of the hepatic tumor. Scale bar, 100 µm. D, Immunostaining of VE‐cadherin and ZO‐1 in CD31‐positive endothelial cell in non‐cancerous hepatic regions of mice treated with HuH‐7P‐derived or HuH‐7M‐derived exosomes. Scale bars, 50 µm. E, VE‐cadherin and ZO‐1 mRNA expression of HUVECs treated with HuH‐7P‐derived or HuH‐7M‐derived exosomes with normalization of GAPDH expression by qRT‐PCR. F, Western blotting of VE‐cadherin and ZO‐1 for HUVECs with HuH‐7P‐derived or HuH‐7M‐derived exosomes. G, Immunocytochemistry of VE‐cadherin and ZO‐1 for HUVECs. Scale bars, 20 µm. H, Permeability of HUVECs treated with HuH‐7P‐derived or HuH‐7M‐derived exosomes. † P < .01
3.5. Exosomes derived from highly intrahepatic metastatic HuH‐7M downregulate endothelial cell VE‐cadherin and ZO‐1 and increase the permeability of endothelial monolayers
To investigate the influence of exosomes on HUVECs, we first confirmed the uptake of PKH67‐labeled exosomes into more than 90% of HUVECs (Figure S1). Next, we explored whether exosomes secreted by highly intrahepatic metastatic HuH‐7M attenuated endothelial cell junction integrity. After 48 h of incubation with HuH‐7P‐ or HuH‐7M‐derived exosomes, HUVEC mRNA expression of VE‐cadherin and ZO‐1 was more steeply reduced with HuH‐7M‐derived exosomes. In HUVECs with HuH‐7M‐derived exosomes, VE‐cadherin expression was downregulated 0.42‐fold more and ZO‐1 expression 0.38‐fold more (Figure 2E). Western blotting analysis showed downregulation of VE‐cadherin and ZO‐1 in HUVECs incubated with HuH‐7M‐derived exosomes (Figure 2F). Immunofluorescence staining revealed that HUVEC monolayers treated with exosomes from HuH‐7M exhibited a marked reduction of VE‐cadherin and ZO‐1 expression in the cell membrane, indicating loss of contact between endothelial cells (Figure 2G). In vitro endothelial permeability assay was performed to measure the traversing of FITC‐dextran as a tracer molecule through the monolayer of HUVECs cultured with HuH‐7P‐derived or HuH‐7M‐derived exosomes. The HUVEC monolayer cultured with HuH‐7M‐derived exosomes allowed the passage of 2 times amounts of FITC‐dextran compared with the HUVEC monolayer cultured with HuH‐7P‐derived exosomes (Figure 2H).
3.6. On microarray analysis, specific miRNAs are elevated in exosomes derived from highly intrahepatic metastatic HuH‐7M cells
Based on the preceding results, we suspected that specific miRNAs of HuH‐7M‐derived exosomes affected endothelial cells and led to changes in VE‐cadherin and ZO‐1 expression. We also hypothesized that these functional changes in endothelial cells facilitated formation of intrahepatic metastatic nodules by cancer cells. To identify candidate exosomal miRNAs that might be involved in intrahepatic metastasis, we performed miRNA microarray studies. We excluded miRNAs that were not detectable, and 22 miRNAs with more than two‐fold expression differences between HuH‐7P‐derived and HuH‐7M‐derived exosomes were identified as candidates (Figure 3A, left panel: heatmap; Figure 3A, right panel: expression data for 6 representative miRNAs: miR‐638, miR‐3648, miR‐663a, miR‐1246, miR‐4258, and miR‐1290). Among these miRNAs, we selected miR‐638, miR‐663a, miR‐3648, and miR‐4258 for subsequent experiments because the expression data for each of them could be validated by qRT‐PCR (Figure 3B).
FIGURE 3.
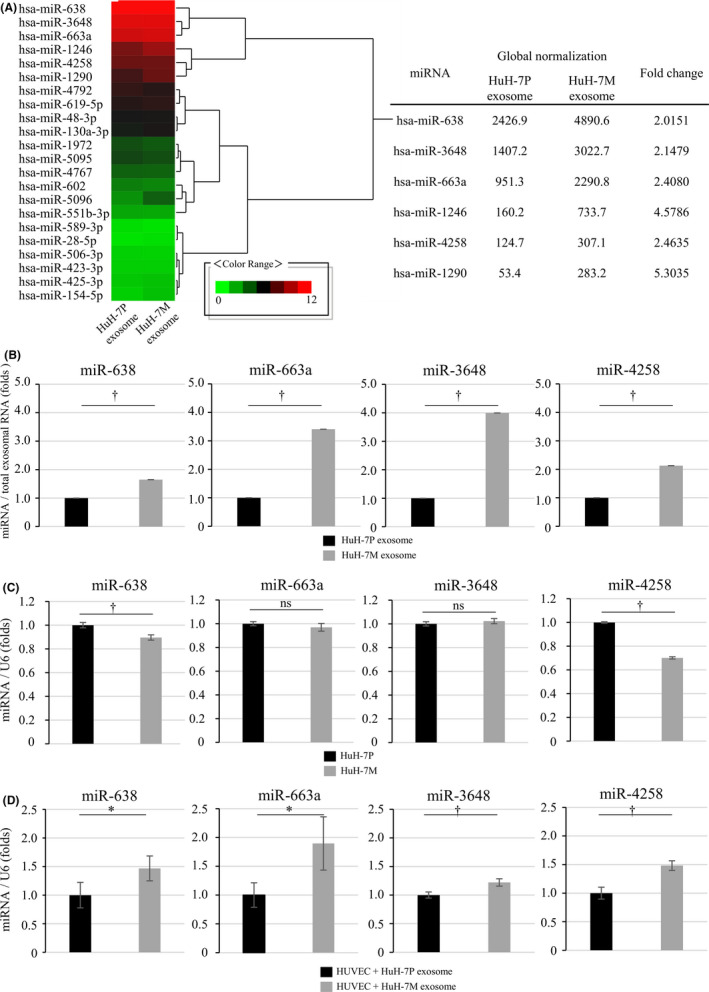
Identification of elevated miRNAs in HuH‐7M‐derived exosomes by microarray analysis and transfer of miRNAs to HUVECs. A, Microarray and heatmap analysis for miRNAs from HuH‐7P and HuH‐7M‐derived exosomes. The expression data and fold changes for the representative 6 miRNAs in accordance with their global normalization are shown in the right panel. B, The expression data for miR‐638, miR‐663a, miR‐3648, and miR‐4258 by qRT‐PCR for validation of the microarray expression data. C, Expression of miR‐638, miR‐663a, miR‐3648, and miR‐4258 in HuH‐7P, and HuH‐7M. D, HUVEC miRNA expression after incubation with HuH‐7P‐derived or HuH‐7M‐derived exosomes. * P < .05; † P < .01; ns, not significant
Expression of these miRNAs was significantly upregulated in HuH‐7M‐derived exosomes, by around 1.65‐ to 4.00‐fold higher compared with exosomes derived from HuH‐7P (Figure 3B). In contrast, expression of miR‐638 and miR‐4258 in the HuH‐7M was significantly decreased compared with expression in the HuH‐7P. Expression of miR‐663a and miR‐3648 did not differ between the 2 cell lines (Figure 3C). After 48 h of incubation of HUVECs with exosomes derived from HuH‐7P or HuH‐7M, the expression of the miR‐638, miR‐663a, miR‐3648, and miR‐4258 was significantly upregulated in HUVECs exposed to exosomes derived from HuH‐7M compared to HUVECs exposed to HuH‐7P‐derived exosomes (Figure 3D).
3.7. MiR‐638, miR‐663a, miR‐3648, and miR‐4258 downregulated VE‐cadherin and ZO‐1 in endothelial cells and increased the permeability of endothelial monolayers
We next explored whether miR‐638, miR‐663a, miR‐3648, or miR‐4258 attenuated endothelial cell junction integrity. Each miRNA was transfected into HUVECs, and the expression of each was confirmed to be elevated in HUVECs. On western blotting, the protein expression of both VE‐cadherin and ZO‐1 was reduced in HUVECs transfected with miR‐mimic (638, 663a, 3648, or 4258) compared with miR‐NC (Figure 4A). Immunofluorescence staining revealed that HUVEC monolayers transfected with miR‐mimic (638, 663a, 3648, or 4258) exhibited a marked reduction in VE‐cadherin and ZO‐1 expression in the cell membrane compared with the negative control (Figure 4B). In vitro endothelial permeability, the HUVEC transfected with miR‐638, miR‐663a, miR‐3648, or miR‐4258 showed the increase passing of FITC‐dextran through the HUVEC monolayer compared with miR‐NC. The amounts of FITC‐dextran after transfection of each miR‐mimic (638, 663a, 3648, or 4258) were 1.3 ~ 6.7 times compared with control (Figure 4C). These experimental results indicated that the 4 miRNAs identified from HuH‐7M‐derived exosomes induced a reduction in endothelial intracellular junction or tight junction proteins in HUVECs. Then, the reduction in intracellular junction or tight junction proteins led to loosen endothelial intercellular adhesions, allowing cancer cells access to form metastatic nodules in the liver.
FIGURE 4.
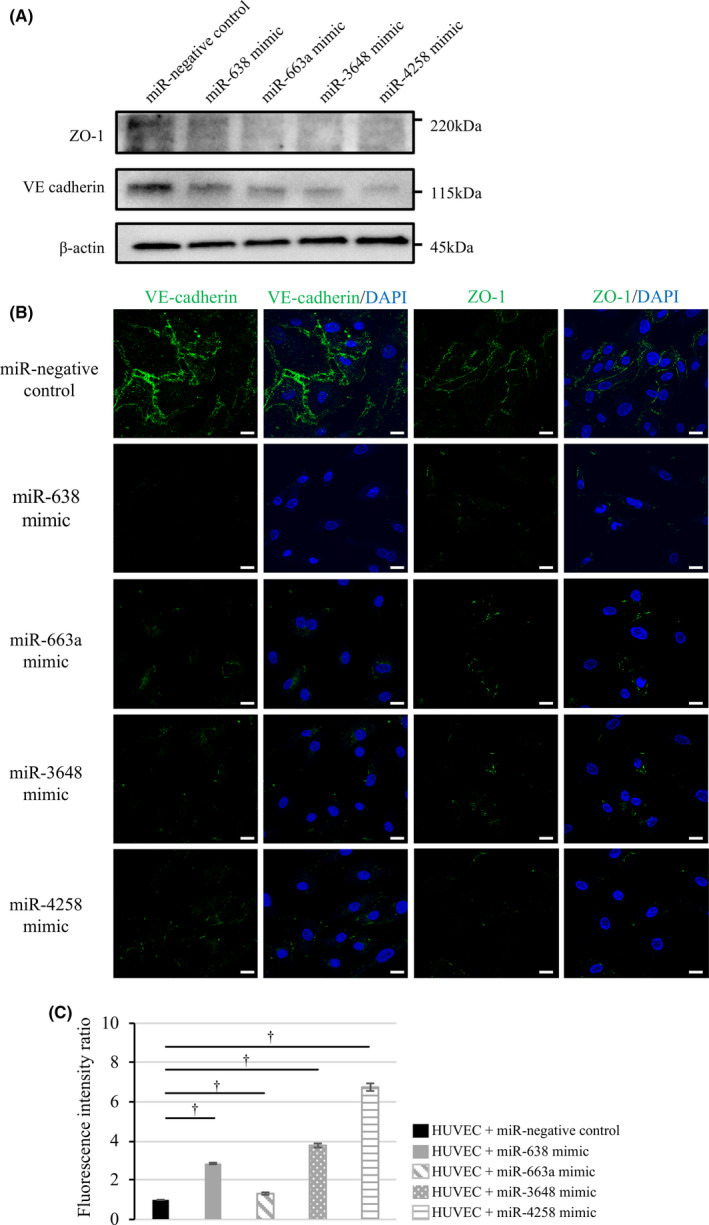
miR‐638, miR‐663a, miR‐3648, and miR‐4258 downregulate VE‐cadherin and ZO‐1 expression in HUVECs and increase the permeability of HUVEC monolayer. A, VE‐cadherin and ZO‐1 expression in HUVECs transfected with miR‐negative control and miR‐mimic (638, 663a, 3648, and 4258). B, Immunocytochemistry of VE‐cadherin and ZO‐1 for transfected HUVECs. Scale bars, 20 µm. C, Permeability of HUVECs transfected with negative control and 4 miR‐mimics. † P < .01
3.8. Clinical significance of serum miR‐638 expression as a novel prognostic marker in HCC
To examine the clinical significance of serum exosomal miR‐638, miR‐663a, miR‐3648, and miR‐4258 expression, we evaluated their expression levels in exosomes of serum that was preoperatively collected from 54 patients with HCC undergoing hepatectomy. The patients were divided into 2 groups in accordance with the median expression value of each miRNA. Kaplan‐Meier survival analysis showed that DFS rates were significantly lower with high vs low expression of miR‐638 (P = .0197). The 2‐y DFS rate was 47.1% in the high expression group and 77.4% in the low expression group (Figure 5A). DFS did not differ by high vs low expression levels of miR‐663a, miR‐3648, or miR‐4258 (Figure 5B‐D). After hepatectomy, 5 patients with distant metastasis and 21 patients with intrahepatic metastasis were observed. The high expression group of miR‐638 demonstrated significantly higher frequency of distant metastasis (11.1%, P = .0064) and intrahepatic metastasis (51.9%, P = .0490) compared with the low expression group (distant metastasis; 7.4%, intrahepatic metastasis; 25.9%). The expression levels of miR‐663a, miR‐3648, and miR‐4258 did not influence the frequency of distant metastasis and intrahepatic metastasis. OS rates were not significantly different with high vs low expression of any of the miRNAs.
FIGURE 5.
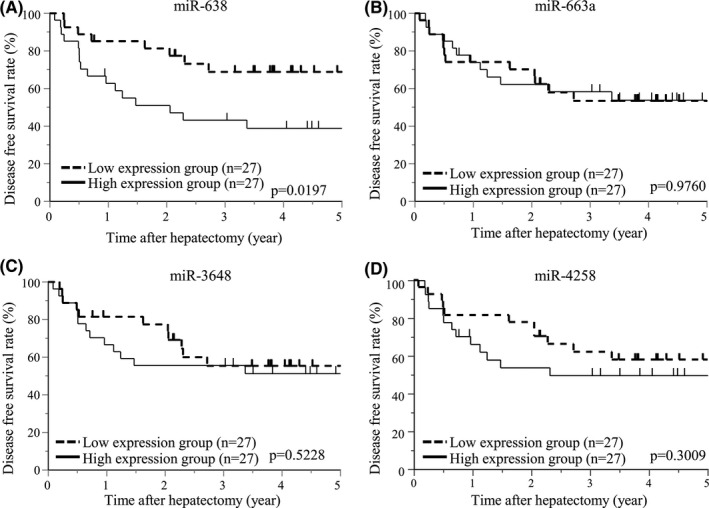
DFS associated with levels of serum exosomal miRNA expression in HCC patients. A, DFS curves for low and high miR‐638 expression by the Kaplan‐Meier method. B, Curves for low and high miR‐663a expression. C, Curves for low and high miR‐3648 expression. D, Curves for low and high miR‐4258 expression
Table 1 summarizes the clinicopathological features of the 2 groups with high vs low miR‐638 expression, which showed no patterns of association. In univariate and multivariate analyses, high miR‐638 expression, being male, having high des‐γ‐carboxy prothrombin values, and having multiple tumors were independent prognostic factors for DFS in HCC (Table 2).
TABLE 2.
Univariate and multivariate analysis of DFS
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age (≥70/<70) | 2.5059 | 0.9972‐6.2974 | .0507 | |||
| Sex (male/female) | 3.0757 | 1.1404‐8.2949 | .0264 | 4.0486 | 1.3768‐11.9048 | .0110 |
| Hepatitis B virus surface antigen (±) | 0.9516 | 0.3264‐2.7742 | .9277 | |||
| Anti‐hepatitis C virus antibody (±) | 1.3645 | 0.6194‐3.0058 | .4405 | |||
| Child‐Pugh classification (A/B) | 1.0035 | 0.2347‐4.2899 | .9962 | |||
| Alpha‐fetoprotein (≥20/<20 ng/mL) | 0.9471 | 0.4290‐2.0908 | .8931 | |||
| Des‐γ‐carboxy prothrombin (≥100/<100 mAU/mL) | 3.7439 | 1.2798‐10.9525 | .0159 | 3.6223 | 1.1524‐11.3859 | .0276 |
| Tumor size (≥5/<5 cm) | 2.3437 | 1.0475‐5.2438 | .0382 | 1.2664 | 0.4991‐3.2135 | .6190 |
| Number of tumors (multiple/single) | 3.1849 | 1.1738‐8.6416 | .0229 | 3.6273 | 1.0927‐12.0402 | .0353 |
| Histological differentiation (poorly/well, moderately) | 2.3048 | 1.0441‐5.0876 | .0387 | 1.7253 | 0.7344‐4.0532 | .2107 |
| Microscopic portal vein invasion (±) | 1.3128 | 0.5650‐3.0502 | .5269 | |||
| Liver cirrhosis (±) | 1.2455 | 0.5650‐2.7457 | .5861 | |||
| miR‐638 expression (high/low) | 2.6354 | 1.1293‐6.1503 | 00250 | 3.2128 | 1.2972‐7.9567 | .0117 |
CI, confidence interval, DFS, disease‐free survival, HR, hazard ratio, miR, microRNA.
4. DISCUSSION
EVs, including exosomes, are derived from various cell types and mediate cell‐to‐cell communication. 8 , 9 Exosomes from malignant cells have various roles, including in tumor growth, cancer progression, angiogenesis, drug resistance, creating an immunosuppressive microenvironment, and metastasis. 10 , 11 , 32 Cancer‐derived exosomes are transferred into normal stromal cells of the target organ and induce an environment suitable for metastasis, which is the formation of pre‐metastatic niches. In the process of initiating the pre‐metastatic niche, cancer‐induced vascular permeability is an important step. Cell‐to‐cell junctions between endothelial cells consist of adherens junctions and tight junctions, which are critical in maintaining vascular integrity. 19 The downregulation or loss of tight junctions and junction‐associated proteins contributes to cancer progression by altering cancer cell migration to the stroma. 13 Loss of these membrane‐associated adhesion molecules is also reported to attenuate endothelial junction integrity, consequently destroying the vascular barrier. 19 miRNAs may play major roles in regulating gene expression in stroma cells of the tumor microenvironment. In breast cancer, for example, exosomal miR‐105, miR‐181c, and miR‐939 destroy vascular endothelial barriers and promote vascular permeability. 13 , 14 , 15 In HCC, exosomal miR‐103 is reported to regulate ZO‐1 and VE‐cadherin. 19 Here, we established a novel, highly metastatic HCC cell line and showed that exosomes facilitate intrahepatic metastasis in vivo. Moreover, miRNA expression analysis identified 4 miRNAs (miR‐638, miR‐663a, miR‐3648, and miR‐4258) that downregulate VE‐cadherin and ZO‐1 in endothelial cells in vitro and in vivo. Among these, miR‐638 is reported to function as a tumor suppressor and inhibits cell migration in HCC. 33 It also influences cancer cell progression through the Wnt/β‐catenin pathway and affects the cell cycle by suppressing cyclin D1 expression. 34 , 35 Another candidate, miR‐663a, also has a tumor suppressor function, showing anti‐proliferative effects and suppression of invasiveness and tumor growth in HCC. 36 Conversely, miR‐3648 is reported to promote cell invasion and migration in bladder cancer. 37 Finally, miR‐4258 is involved in MEN1 parathyroid neoplasia. 38
Our study has demonstrated that these exosomal miRNAs initiate the pre‐metastatic niche by targeting VE‐cadherin and ZO‐1 in endothelial cells. The HuH‐7M‐derived exosomes suppressed the expression VE‐cadherin and ZO‐1 of CD31‐positive endothelial cells in normal liver tissue and HUVEC. Moreover, the HuH‐7M‐derived exosomes increased the passage of labeled dextran through a monolayer of HUVEC. VE‐cadherin forms the endothelial adherens junction. It is an endothelium‐specific member of the cadherin family that binds via its cytoplasmic domain to several protein partners, including p120 and β‐catenin. ZO‐1 is a central component of endothelial tight junctions, binding the transmembrane proteins occludin and the claudins and linking them to cytoskeletal actin. Both VE‐cadherin and ZO‐1 regulate endothelial cell‐cell adhesion and vascular barrier function 39 , 40 and, as we show here, the 4 miRNAs regulated expression of both VE‐cadherin and ZO‐1. Previous reports have shown positive regulation of ZO‐1 and other tight junction proteins by miR‐126, miR‐107, and miR‐21, and negative regulation by miR‐181a, miR‐98, and miR‐150. Regarding VE‐cadherin, miR‐9, miR‐99b, and miR‐181a are reported as regulators. 41 The mechanism regulating endothelial junction proteins is involved with so many miRNAs and a better understanding of it is needed for development of new anti‐angiogenesis therapies.
In HCC, the most common diagnostic marker is AFP. 42 , 43 The American Association Society of Liver Disease also recommends checking serum AFP for surveillance in patients with cirrhosis. 44 However, the false‐negative rate is approximately 40% in early stage HCC, 25 so the need is urgent for non‐invasive biomarkers for HCC surveillance. Several specific miRNAs can act as diagnostic and predictive markers for several malignancies. In our study, we found that expression levels of serum miR‐638 in exosomes could be an independent prognostic marker for patients with HCC. These findings suggest that miR‐638 in serum may be promising for surveillance of HCC recurrence. In our cohort, the 2‐y DFS rate in the group with high miR‐638 expression was 47.1%, compared with 77.4% in the low expression group. In general, the recurrence pattern of HCC can be divided into intrahepatic metastasis and multi‐centric recurrence. The main cause of tumor relapse with HCC within 2 y after curative resection is intrahepatic metastasis. The expression of miR‐638 could distinguish these 2 groups well, likely because of its role in the molecular mechanism of endothelial cell downregulation of VE‐cadherin and ZO‐1.
The methodology of in vivo selection has proved to be useful for studies of cancer because it aims to generate sequential malignant metastases from the same cell line origin. For this reason, it could be a solution to problems with heterogeneity among cell lines. 27 We applied this method to establish a new, highly intrahepatic metastatic HCC cell line from an original HuH‐7‐Luc line. The highly intrahepatic metastatic cells (HuH‐7M) had different characteristics from the original line. HuH‐7M showed slightly increased proliferative ability and suppressed anoikis, a type of programmed cell death induced by the loss of extracellular matrix attachment. We can speculate that HuH‐7M can survive longer within the portal vein and the released exosomes from HuH‐7M downregulate the VE‐cadherin and ZO‐1 of endothelial cells and it results to loosen the cell‐to‐cell junction. Finally, HuH‐7M can easily migrate to the stroma of the target organ, forming a metastatic nodule in the liver.
In conclusion, we found that a novel, highly metastatic cell line is more likely to survive by suppressing anoikis. Specific miRNAs (miR‐638, miR‐663a, miR‐3648, and miR‐4258) in the exosomes from highly metastatic cell line can promote vascular permeability and initiate the pre‐metastatic niche via downregulation of endothelial cell VE‐cadherin and ZO‐1 expression. Among these 4 miRNA candidates, serum exosomal miR‐638 expression shows promise as a significant and independent prognostic marker in patients with HCC.
CONFLICT OF INTEREST
The authors have no conflict of interest.
Supporting information
Fig S1
ACKNOWLEDGMENTS
This work was supported in part by grants from Grant‐in‐Aid for Scientific Research of the Japan Society for the Promotion of Science [(C) 19K09146] and from the SGH foundation. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Yokota Y, Noda T, Okumura Y, et al. Serum exosomal miR‐638 is a prognostic marker of HCC via downregulation of VE‐cadherin and ZO‐1 of endothelial cells. Cancer Sci. 2021;112:1275–1288. 10.1111/cas.14807
REFERENCES
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87‐108. [DOI] [PubMed] [Google Scholar]
- 2. Colvin H, Mizushima T, Eguchi H, Takiguchi S, Doki Y, Mori M. Gastroenterological surgery in Japan: The past, the present and the future. Ann Gastroenterol Surg. 2017;1:5‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Noda T, Eguchi H, Iwagami Y, et al. Minimally invasive liver resection for hepatocellular carcinoma of patients with liver damage B: A propensity score‐based analysis. Hepatol Res. 2018;48:539‐548. [DOI] [PubMed] [Google Scholar]
- 4. Noda T, Eguchi H, Wada H, et al. Short‐term surgical outcomes of minimally invasive repeat hepatectomy for recurrent liver cancer. Surg Endosc. 2018;32:46‐52. [DOI] [PubMed] [Google Scholar]
- 5. Wada H, Eguchi H, Noda T, et al. Selection criteria for hepatic resection in intermediate‐stage (BCLC stage B) multiple hepatocellular carcinoma. Surgery. 2016;160:1227‐1235. [DOI] [PubMed] [Google Scholar]
- 6. Yamanaka N, Okamoto E, Fujihara S, et al. Do the tumor cells of hepatocellular carcinomas dislodge into the portal venous stream during hepatic resection? Cancer. 1992;70:2263‐2267. [DOI] [PubMed] [Google Scholar]
- 7. Sakon M, Nagano H, Nakamori S, et al. Intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. Arch surg. 2002;137:94‐99. [DOI] [PubMed] [Google Scholar]
- 8. Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T. Neutral sphingomyelinase 2 (nSMase2)‐dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. 2013;288:10849‐10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu Y, Gu Y, Cao X. The exosomes in tumor immunity. OncoImmunology. 2015;4:e1027472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mikamori M, Yamada D, Eguchi H, et al. MicroRNA‐155 controls exosome synthesis and promotes gemcitabine resistance in pancreatic ductal adenocarcinoma. Sci Rep. 2017;7:42339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Matsuura Y, Wada H, Eguchi H, et al. Exosomal miR‐155 derived from hepatocellular carcinoma cells under hypoxia promotes angiogenesis in endothelial cells. Dig Dis Sci. 2019;64:792‐802. [DOI] [PubMed] [Google Scholar]
- 12. Tetta C, Ghigo E, Silengo L, Deregibus MC, Camussi G. Extracellular vesicles as an emerging mechanism of cell‐to‐cell communication. Endocrine. 2013;44:11‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou W, Fong MY, Min Y, et al. Cancer‐secreted miR‐105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501‐515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tominaga N, Kosaka N, Ono M, et al. Brain metastatic cancer cells release microRNA‐181c‐containing extracellular vesicles capable of destructing blood‐brain barrier. Nat Commun. 2015;6:6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Di Modica M, Regondi V, Sandri M, et al. Breast cancer‐secreted miR‐939 downregulates VE‐cadherin and destroys the barrier function of endothelial monolayers. Cancer Lett. 2017;384:94‐100. [DOI] [PubMed] [Google Scholar]
- 16. Fang T, Lv H, Lv G, et al. Tumor‐derived exosomal miR‐1247‐3p induces cancer‐associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu H, Chen W, Zhi X, et al. Tumor‐derived exosomes promote tumor self‐seeding in hepatocellular carcinoma by transferring miRNA‐25‐5p to enhance cell motility. Oncogene. 2018;37:4964‐4978. [DOI] [PubMed] [Google Scholar]
- 18. Lin XJ, Fang JH, Yang XJ, et al. Hepatocellular carcinoma cell‐secreted exosomal microrna‐210 promotes angiogenesis in vitro and in vivo. Mol Ther Nucleic Acids. 2018;11:243‐252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fang JH, Zhang ZJ, Shang LR, et al. Hepatoma cell‐secreted exosomal microRNA‐103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology. 2018;68:1459‐1475. [DOI] [PubMed] [Google Scholar]
- 20. Tomimaru Y, Eguchi H, Nagano H, et al. MicroRNA‐21 induces resistance to the anti‐tumour effect of interferon‐alpha/5‐fluorouracil in hepatocellular carcinoma cells. Br J Cancer. 2010;103:1617‐1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tomimaru Y, Eguchi H, Nagano H, et al. Circulating microRNA‐21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. 2012;56:167‐175. [DOI] [PubMed] [Google Scholar]
- 22. Tomokuni A, Eguchi H, Tomimaru Y, et al. miR‐146a suppresses the sensitivity to interferon‐alpha in hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2011;414:675‐680. [DOI] [PubMed] [Google Scholar]
- 23. Koga C, Kobayashi S, Nagano H, et al. Reprogramming using microRNA‐302 improves drug sensitivity in hepatocellular carcinoma cells. Ann Surg Oncol. 2014;21(Suppl 4):S591‐600. [DOI] [PubMed] [Google Scholar]
- 24. Li S, Yao J, Xie M, Liu Y, Zheng M. Exosomal miRNAs in hepatocellular carcinoma development and clinical responses. J Hematol Oncol. 2018;11:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu X, Tao Y, Shan L, et al. The role of MicroRNAs in hepatocellular carcinoma. J Cancer. 2018;9:3557‐3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Clark EA, Golub TR, Lander ES, Hynes RO. Genomic Analysis of Metastasis Reveals an Essential Role for RhoC. Nature. 2000;406:532–535. [DOI] [PubMed] [Google Scholar]
- 27. Kuo CL, Jiang ZY, Wang YW, et al. In vivo selection reveals autophagy promotes adaptation of metastatic ovarian cancer cells to abdominal microenvironment. Cancer Sci. 2019;110:3204‐3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakamura M, Nagano H, Sakon M, et al. Role of the Fas/FasL pathway in combination therapy with interferon‐alpha and fluorouracil against hepatocellular carcinoma in vitro. J Hepatol. 2007;46:77‐88. [DOI] [PubMed] [Google Scholar]
- 29. Noda T, Yamamoto H, Takemasa I, et al. PLOD2 induced under hypoxia is a novel prognostic factor for hepatocellular carcinoma after curative resection. Liver Int. 2012;32:110‐118. [DOI] [PubMed] [Google Scholar]
- 30. Kubo M, Gotoh K, Eguchi H, et al. Impact of CD36 on Chemoresistance in Pancreatic Ductal Adenocarcinoma. Ann Surg Oncol. 2020;27:610‐619. [DOI] [PubMed] [Google Scholar]
- 31. Shinke G, Yamada D, Eguchi H, et al. Role of histone deacetylase 1 in distant metastasis of pancreatic ductal cancer. Cancer Sci. 2018;109:2520‐2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol. 2015;8:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Y, Zhang D, Jiang J, Dong L. Loss of miR‐638 promotes invasion and epithelial‐mesenchymal transition by targeting SOX2 in hepatocellular carcinoma. Oncol Rep. 2017;37:323‐332. [DOI] [PubMed] [Google Scholar]
- 34. Tang KL, Tang HY, Du Y, Tian T, Xiong SJ. MiR‐638 suppresses the progression of oral squamous cell carcinoma through wnt/beta‐catenin pathway by targeting phospholipase D1. Artif Cells Nanomed Biotechnol. 2019;47:3278‐3285. [DOI] [PubMed] [Google Scholar]
- 35. Xue M, Shen J, Cui J, et al. MicroRNA‐638 expression change in osteosarcoma patients via PLD1 and VEGF expression. Exp Ther Med. 2019;17:3899‐3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang C, Chen B, Jiao A, et al. miR‐663a inhibits tumor growth and invasion by regulating TGF‐beta1 in hepatocellular carcinoma. BMC Cancer. 2018;18:1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sun W, Li S, Yu Y, et al. MicroRNA‐3648 is upregulated to suppress TCF21, resulting in promotion of invasion and metastasis of human bladder cancer. Mol Ther Nucleic Acids. 2019;16:519‐530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luzi E, Ciuffi S, Marini F, Mavilia C, Galli G, Brandi ML. Analysis of differentially expressed microRNAs in MEN1 parathyroid adenomas. Am J Transl Res. 2017;9:1743‐1753. [PMC free article] [PubMed] [Google Scholar]
- 39. Dejana E, Orsenigo F, Lampugnani MG. The role of adherens junctions and VE‐cadherin in the control of vascular permeability. J Cell Sci. 2008;121:2115‐2122. [DOI] [PubMed] [Google Scholar]
- 40. Poritz LS, Garver KI, Green C, Fitzpatrick L, Ruggiero F, Koltun WA. Loss of the tight junction protein ZO‐1 in dextran sulfate sodium induced colitis. J Surg Res. 2007;140:12‐19. [DOI] [PubMed] [Google Scholar]
- 41. Zhuang Y, Peng H, Mastej V, Chen W. MicroRNA regulation of endothelial junction proteins and clinical consequence. Mediators Inflamm. 2016;2016:5078627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marubashi S, Nagano H, Wada H, et al. Clinical significance of alpha‐fetoprotein mRNA in peripheral blood in liver resection for hepatocellular carcinoma. Ann Surg Oncol. 2011;18:2200‐2209. [DOI] [PubMed] [Google Scholar]
- 43. Kobayashi S, Tomokuni A, Takahashi H, et al. The clinical significance of alpha‐fetoprotein mRNAs in patients with hepatocellular carcinoma. Gastrointest Tumors. 2017;3:141‐152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Heimbach JK, Kulik LM, Finn RS, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67:358‐380. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1


