Abstract
Much like humans, chimpanzees occupy diverse habitats and exhibit extensive behavioural variability. However, chimpanzees are recognized as a discontinuous species, with four subspecies separated by historical geographic barriers. Nevertheless, their range-wide degree of genetic connectivity remains poorly resolved, mainly due to sampling limitations. By analyzing a geographically comprehensive sample set amplified at microsatellite markers that inform recent population history, we found that isolation by distance explains most of the range-wide genetic structure of chimpanzees. Furthermore, we did not identify spatial discontinuities corresponding with the recognized subspecies, suggesting that some of the subspecies-delineating geographic barriers were recently permeable to gene flow. Substantial range-wide genetic connectivity is consistent with the hypothesis that behavioural flexibility is a salient driver of chimpanzee responses to changing environmental conditions. Finally, our observation of strong local differentiation associated with recent anthropogenic pressures portends future loss of critical genetic diversity if habitat fragmentation and population isolation continue unabated.
Subject terms: Structural variation, Evolutionary biology, Classification and taxonomy, Evolutionary ecology, Genotyping and haplotyping
Lester and colleagues use faecal samples and genetic analyses to investigate the genetic connectivity across chimpanzees. Their results indicate that the global pattern of genetic diversity in chimpanzees is largely characterized by a pattern of isolation by distance with several isolated populations exhibiting strong local differentiation.
Introduction
Humans have been characterized as a genetically continuous species1, which is expected to hamper local adaptation2,3. Our species has largely relied on behavioural flexibility4 to become among the most widely distributed species, inhabiting a diverse range of climates and habitats5. Among our closest living relatives, the chimpanzee (Pan troglodytes)6, also exhibits extensive behavioural variation, both at a local and regional scale, and occupies a broad range of habitats and climates, while displaying little associated morphological variation7,8. However, to date, their pattern of genetic diversity is equivocal: some studies provide evidence of connectivity among all populations9,10, while others have concluded that chimpanzees are taxonomically divided into four geographical subspecies11–15. Given the high degree of behavioural variability and flexibility across chimpanzee populations, characterizing range-wide patterns of genetic diversity in chimpanzees is important for understanding how they adapt to changing environmental conditions16. In particular, a signal of genetic connectivity across their range would suggest that, like in humans, local trait fixation is relatively slow in chimpanzees, and behavioural flexibility allows them to quickly and dynamically respond to ecological challenges.
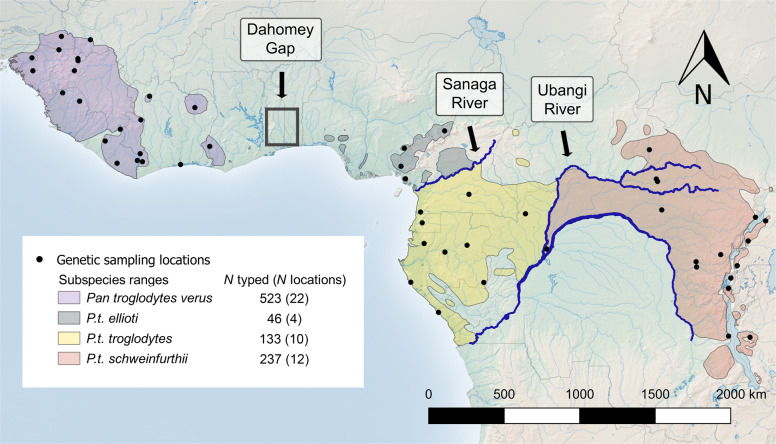
Prior to the recent growth and expansion of human agriculturalist settlements across Africa (ca. 5–2 ka BP)17,18 chimpanzees were nearly continuously distributed across Equatorial Africa7. Rivers appear to be the main barriers to migration7 separating chimpanzees and bonobos (Pan paniscus), as well as three of the four currently recognized chimpanzee subspecies: Nigeria–Cameroon (Pan troglodytes ellioti), central (Pan troglodytes troglodytes) and eastern (Pan troglodytes schweinfurthii) chimpanzees19 (Fig. 1). River systems, however, are dynamic and may become permeable to dispersal during arid periods, or when natural bridges form20. Moreover, the presence of this type of barrier does not preclude the possibility of gene flow occurring around it, which has not been tested on the subspecies-delineating barriers in any previous studies of chimpanzees. Similarly, the Dahomey Gap, an arid, 200-km-wide forest-savannah mosaic that separates western chimpanzees (Pan troglodytes verus) and P. t. ellioti, is also dynamic, as this region hosted rainforest as recently as 4 ka BP (ref. 21; ~160 chimpanzee generations22). Interestingly, discrete geographic clustering of morphological traits along subspecies lines has not been conclusively shown23–25 and, although chimpanzees do show substantial behavioural variation across populations, to date, there are no universal subspecies-specific behaviours, i.e., accumulative stone throwing is unique to P. t. verus, but has only been observed at a few study sites26.
Fig. 1. Distribution map of P. troglodytes and PanAf sampling.
The current approximate chimpanzee subspecies ranges1, sample collection locations and proposed subspecies geographic barriers. Total number of genotyped individuals and number of sampling locations are listed for each subspecies population. Much of the historical population located between P. t. ellioti and P. t. verus populations have been extirpated, creating an extensive sampling gap in the data. Note, samples collected during nationwide studies in Liberia and Equatorial Guinea were included in our spatially explicit analyses and are indicated here as the geographic centre points of the sampling distribution.
In a continuously distributed population, limited dispersal drives a predictable pattern of genetic diversity, known as isolation by distance (IBD), which manifests as a continuous gradient (cline) of declining similarity as geographical distance increases27. Detectable departures from this pattern are evidence of geographic or behavioural barriers that reduce or impede dispersal, thereby leading to increased geographical rates of genetic differentiation. Although chimpanzees are thought to have had a historically continuous geographic distribution across their range7, studies of chimpanzee genetic diversity have tended to show discontinuities consistent with the subspecies classifications11–13, or some degree of population structuring14. To detect geographic clustering of genetic data, these studies relied on several different ‘spatially agnostic’ approaches not accounting for the spatial distribution of samples. These include Bayesian clustering algorithms (e.g., STRUCTURE28), the results of which can be biased when IBD is present in the data29–31 and principal component analyses, a method specifically intended to maximize between-group differences32. However, to date no studies of species-wide chimpanzee population structure have featured spatially explicit methods, which incorporate the geographic location of the genetic samples into the analysis model and assume IBD in the null model9,33. Importantly, when using spatially agnostic methods in the presence of IBD, unbalanced sampling will detect discrete stratification of the data, regardless of the actual pattern9,29,30,33–36, and this cannot be compensated for by analyzing more loci (i.e., a large number of SNPs)37. Mostly due to the logistical challenge of obtaining non-invasive samples from chimpanzees’ wide range, which largely encompasses politically unstable or remote regions, previous studies of chimpanzee population structure have relied on small and clustered datasets, and often made use of zoo and sanctuary samples of unknown provenance, which introduced further uncertainty11–15. Therefore, previous reports of chimpanzee population structure were likely biased by applying spatially agnostic analyses to datasets composed of dispersed and uneven sampling in the presence of IBD9,33, and were possibly further confounded by incorrect knowledge about populations of origin in some samples. In fact, even the most comprehensive studies of chimpanzee population history, which utilized genome-wide sequence data14,15, were based on a relatively small number of samples from captive individuals, and focused more on fitting models of divergence among subspecies than investigating recent genetic connectivity in space.
In this study, we aimed at assessing recent genetic connectivity across the chimpanzees range by analyzing an extensive sample providing an unprecedented level of geographic coverage. As part of the Pan African Programme: The Cultured Chimpanzee (PanAf)38, over an 8-year period, we non-invasively collected and genotyped >5000 wild chimpanzee faecal samples from 55 localities in 18 countries across the entire species range. We scored individual genotypes (allele length polymorphism) at up to 14 microsatellite markers, as these are cost effective and allow for accurate genotyping of non-invasively collected samples39. Previous studies in other species (fruit flies40, fish41,42, birds43, amphibians44, wild boars34, felids45 and beetles46) have demonstrated that 6–14 microsatellite loci are informative enough to even detect subtle population structure. Furthermore, a previous study of how sampling affects detection of chimpanzee population structure found that eleven loci were sufficient to overcome false signals of population structure caused by sampling bias and uncover a signal of IBD9. As microsatellite markers evolve rapidly and are expected to be selectively neutral, they are highly sensitive to patterns of gene flow, especially within shallow evolutionary timescales. For our analyses, we modeled our georeferenced genetic data in space by employing spatially explicit approaches to inform the distribution, thereby avoiding a priori definitions of the population structure or assumptions of homogeneous sampling, thus minimizing biases associated with previous studies.
Results
The PanAf team and collaborators collected 5397 chimpanzee faecal samples from 55 temporary or long-term research sites spanning the geographic range of chimpanzees from 2010 to 2018 (Fig. 1). After DNA extraction and microsatellite amplification, 2497 extracts were successfully typed at up to 14 loci (range: 7–14, mean = 11.3, Supplementary Tables 1.1–1.4) and represent 939 unique individuals from 48 sampling locations (see Supplementary Note 1: ‘Genotype reconstruction’). We found that including individuals typed at seven loci (~8% of all samples) in all analyses did not affect the observed patterns of genetic differentiation compared to limiting the minimum to eight or nine loci, and this inclusion allowed for the addition of several key sampling locations in comparisons of population diversity levels. Reducing the minimum to six loci (n = 69) noticeably reduced differentiation across analyses.
Influence of geographic distance on genetic distance
In an idealized model with uniform population density and dispersal, genetic differentiation among local demes is expected to increase with geographic distance (D). Rousset47 showed that, in a linear one-dimensional (1d) space, FST/(1 – FST) ~ D and FST/(1 – FST) ~ log(D) in an infinite two-dimensional (2d) space, when mutation is negligible. In principle, one may statistically fit these equations to actual data and interpret deviations from fits as signals of non-uniform spatial patterns of density and gene flow.
Actual geographical spaces are neither 1d nor infinite 2d spaces, and populations inhabiting an elongated 2d space are expected to show relationships of FST and D somewhat in-between linear and logarithmic47. In addition, mutation is often not negligible. Microsatellite DNA markers, in particular, have a typically fast mutation rate, and the number of repeats at a given microsatellite marker tends to change according to a stepwise mutation model (SMM)48, which leads to an overestimation of diversity. While a strict SMM lends itself to unbiased estimation of demographic parameters from microsatellite variation (e.g., Goldstein49), it rarely accurately describes the actual mutation process at microsatellite loci. Constraints to the maximum and minimum size number of repeats at a given marker50 are especially important, as they determine an upper limit to the genetic differentiation among pairs of populations. With these limitations in mind, modeling genetic differentiation as a function of geographical distance is still a powerful tool to identify discontinuities in the spatial structure of biological populations.
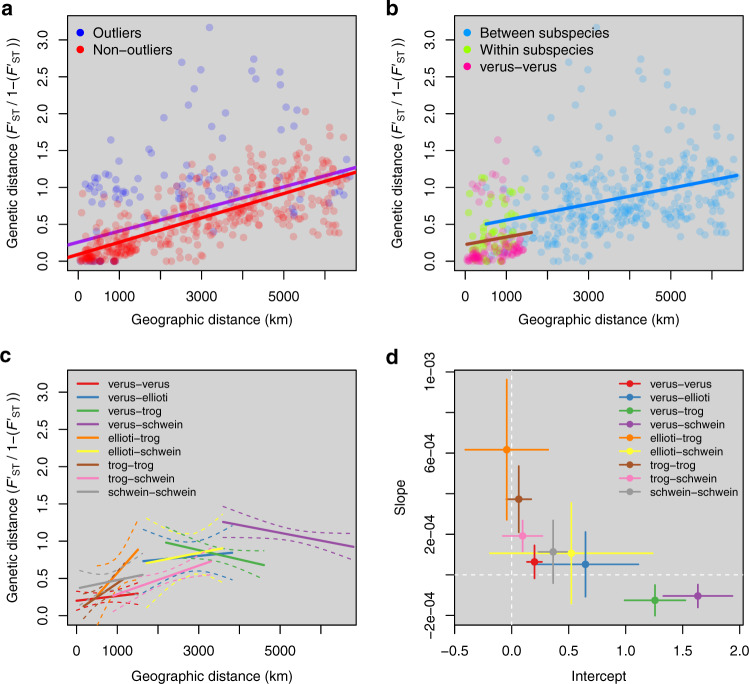
In a global, unstratified test of our data (G ~ D) in which subspecies were not defined, we found that D explained >30% (adjusted r2 = 0.314) of G, and as much as 58% (adjusted r2 = 0.579), when three obvious outlier sites were removed (Mt. Sangbé (Côte d’Ivoire), Gashaka (Nigeria) and Issa (Tanzania)). These sites were consistent outliers in all comparisons and analyses (Supplementary Fig. 3b and Supplementary Fig. 4a, b). A log-transformed model (G ~ log(D)) gave very similar results (adjusted r2 = 0.320 and 0.579 for the global model and for the model excluding outliers, respectively), as it could be expected given that chimpanzee range’s shape is fairly elongated and somewhat intermediate between a 1d and a 2d space.
Full-range data were then stratified according to whether the two sites in each pair belonged to the same or different subspecies (Fig. 2b). However, further analyses in which each subspecies pairs were separately considered (e.g., P. t. verus–P. t. verus, P. t. verus–P. t. troglodytes, etc., Fig. 2c, d) revealed that this pattern was largely driven by the overrepresentation of site pairs within P. t. verus, which is known to have the lowest diversity of all subspecies15,33, resulting in a very low intercept. Fitting G ~ D regressions within each subspecies pair, instead, did not reveal clear evidence of abrupt discontinuities across the chimpanzee range (Fig. 2c, d), but rather highlighted ‘locally’ different patterns. In particular, P. t. verus populations displayed low slope and intercept, which, combined with their low genetic diversity, is consistent with a relatively recent and fast demographic and spatial expansion, while P. t. troglodytes had low intercept but a higher slope (consistent with a more stable pattern of IBD) and there was no clear IBD pattern within P. t. schweinfurthii (where G tends to be high, irrespective of geographic distance, which can be expected given the rugged landscape in the great rift area, where three sampled sites are located). Notably, between-subspecies regressions involving P. t. verus (the most geographically isolated and genetically divergent subspecies15) were essentially flat, suggestive of saturated genetic differentiation at our markers, while the P. t. troglodytes–P. t. schweinfurthii regression was highly similar to within-subspecies regressions, with coefficients in-between P. t. troglodytes and P. t. schweinfurthii in the linear regression and almost identical to within P. t. troglodytes in the log-transformed regression, Fig. 2c, d.
Fig. 2. Linear regressions of genetic distance as a function of geographic distance.
a Linear regressions of genetic distance as a function of geographic distance for all sites with at least six genotyped individuals. Blue dots represent pairwise comparisons involving outlier sites (Mt. Sangbé, Gashaka and Issa, see text). The purple line is fitted to the entire dataset, while the red line is fitted to the dataset excluding the three outlier sampling locations. When excluding the three outlier sites, geographic distance explains 58% of the genetic distance. b Linear regressions of genetic distance as a function of geographic distance for between- and within-subspecies comparisons. Blue dots represent between-subspecies pairwise comparisons with linear regression (blue line). Green and pink dots represent within-subspecies pairwise comparisons with linear regression (brown line) characterized by a noticeably lower y-intercept. Pan troglodytes verus–P. t. verus comparisons (pink dots), which have the lowest genetic diversity of all subspecies, make up 60% of all the within-subspecies comparisons and are largely driving this observed pattern, explaining the source of the stratification in a. c Linear regressions of genetic distance as a function of geographic distance for each subspecies comparison pair. Solid lines represent fitted regressions, and dashed lines enclose 95% confidence intervals. d Estimates (circles) and standard errors (crosses) of intercept and slope for each of the regressions in c.
Historical estimated effective migration rates
To visualize chimpanzee migration patterns and test for spatial genetic structure, we employed the spatially explicit analysis implemented in the Estimated Effective Migration Surfaces (EEMS) software51. This analysis combines spatial and genetic data and relies on MCMC to estimate relative effective migration rates (m; between-deme genetic dissimilarity measured against geographic distance) modeled on a 2d surface, using a mean-centred relative scale. In this model, a ‘flat’ uniform migration surface would indicate perfect IBD across the considered range, and non-uniformity of m in space is interpreted as increased or decreased genetic connectivity. Besides plotting estimated migration surfaces, EEMS results differentiate areas where the probability of m differing from the mean rate is statistically significant (i.e., local m > mean m in >95% of the MCMC samples). Importantly, the EEMS model assumes that microsatellite markers mutate according to a strict SMM and violations of this assumption may lead to an overestimation of effective migration rates, while homoplasy caused by allelic size saturation may lead to an underestimation. Despite having some deviations from strict SMM and the presence of homoplasy that are apparent in our data, we found the EEMS model to be a good fit to our chimpanzee dataset (adjusted r2 = 0.381, Supplementary Fig. 7a). However, the observed genetic dissimilarity tended to be less than predicted by the model for very large distances, i.e., mostly involving P. t. verus vs. P. t. schweinfurthii pairs, and in particular, P. t. verus–Issa comparisons (Supplementary Fig. 7b), similar to our observations in the regression analyses.
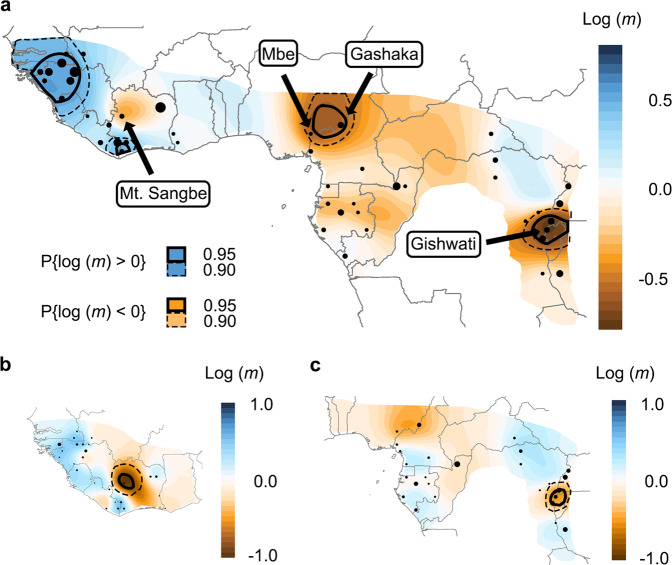
Although we observed spatial variation in m, we did not detect any significant signal of genetic discontinuity, or ‘barriers’ between-subspecies populations (Fig. 3a). Consistent with the divergent results from the G ~ D regression models (Fig. 2a–d), m was generally higher in the P. t. verus range than in the rest of the species range (P. t. ellioti, P. t. troglodytes and P. t. schweinfurthii, hereafter, collectively, ETS) and two patches of significantly high m were identified in the most densely sampled areas of the P. t. verus range. In contrast, significantly low migration was inferred for the region surrounding the Gashaka sampling location within the range of P. t. elliotti and in the mountainous regions of southern Uganda and Rwanda centered around Gishwati in the P. t. schweinfurthii range.
Fig. 3. Estimated effective migration surfaces (EEMS) at different population scales.
a Map of EEMS for the entire species. Estimated effective migration rates (m) are mean centred on a log10 scale. A value of 1 equates to a tenfold rate increase over population average, ranging from areas of low (brown) to high (blue) m. The intensity of the colours represents the relative difference from the population mean rates. Point diameter is proportional to the number of individuals sampled in a given deme (range = 1–72). Solid black lines indicate areas where the posterior probability of m differing from the mean rate is >95 percent and the dashed lines highlight areas that are >90 percent. These can be interpreted as significant effective ‘barriers’ to migration in brown areas and significant effective ‘corridors’ for migration in the blue areas. Two significant effective barriers were present: one corresponding to Gashaka and Mbe (Nigeria) and another originating from Gishwati (Rwanda) and shared with its nearest neighbours. These are localized areas of high differentiation, and when we excluded Mbe and Gishwati from the dataset, these barriers were no longer significant (Extended Data Fig. 6a, c). Notably, historical effective barriers separating the subspecies’ ranges were not detected. Effective migration rates within much of Pan troglodytes verus were significantly higher than average. b Historical EEMS map of P. t. verus. There was a significant barrier associated with Mt. Sangbé in Côte d’Ivoire. When we removed Mt. Sangbé the barrier was no longer significant, but it was still present, suggesting the possibility of reduced historical gene flow across Côte d’Ivoire. c Historical EEMS map of P. t. ellioti, P. t. troglodytes and P. t. schweinfurthii (collectively ETS). As in a, removing Gishwati resulted in the barrier no longer being significant (Extended data Fig. 6c). Rates of m between the panels are relative and not directly comparable.
Since EEMS estimates relative m and the considerable contrast between P. t. verus and ETS may subdue more local patterns, we also performed separated analyses for P. t. verus and for the remaining ETS populations (Fig. 3b, c). The P. t. verus analysis (Fig. 3b) highlighted significantly low m associated with the outlier location of Mt. Sangbé, while the ETS analysis confirmed significantly lower gene flow in the mountainous region in the southeastern portion of the P. t. schweinfurthii range relative to the average ETS migration landscape. To determine whether local effects drove these effective barriers, we removed Mbe, Mt. Sangbé and Gishwati, and re-analyzed the data; all of the barriers were no longer significant (Supplementary Note 2: ‘Spatially explicit analyses (EEMS)’ and Supplementary Fig. 8a–c), indicating that these observed genetic discontinuities stemmed from local effects, and are associated with sites that are highly differentiated from surrounding locations. Importantly, even when outliers were removed, no significant genetic discontinuities were associated with boundaries between-subspecies ranges.
At the population level with ETS excluded, m in P. t. verus were surprisingly uniform (with the exception of Mt. Sangbé), given the high degree of topological, climate and habitat variation in West Africa (Fig. 3b). Meanwhile, ETS displayed a non-uniform effective migration surface, though, apart from the local barrier associated with Gishwati, no areas of significant variation in m were detected (Fig. 3c). A barrier to gene flow was not detected in the eastern extreme of the ETS range across the Albertine Rift, a region consisting of rivers, great lakes, gorges, and steep gradient valleys and mountain ranges—a landscape expected to have increased resistance to migration52. Conversely, latitudinal gene flow in populations east of the rift appeared to be discontinuous, where the mountainous areas of southern Uganda and Rwanda may restrict dispersal.
Diversity rates
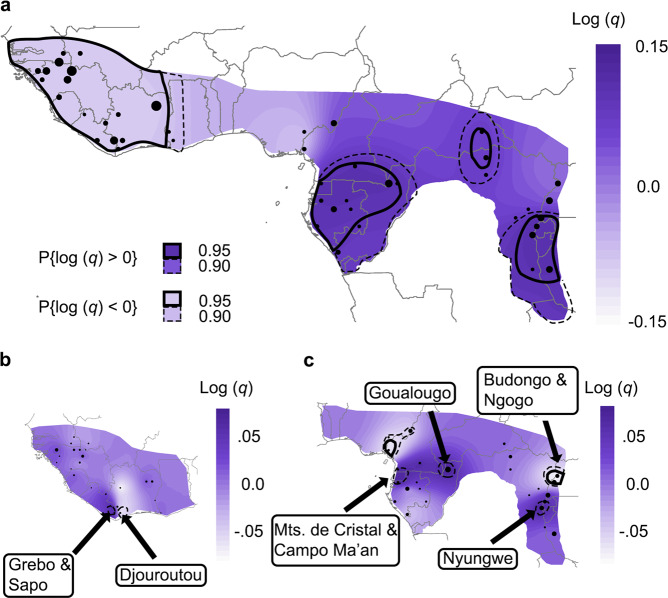
We visualized spatial patterns of chimpanzee genetic diversity by plotting estimated relative diversity rates (q; pairwise within-deme genetic dissimilarity between individuals) from EEMS. At full scale, q in P. t. verus populations were significantly lower than species average (Fig. 4a). We calculated mean q from the EEMS output for both P. t. verus and ETS, and found that ETS was 1.7-fold higher, in agreement with previously published reports15,33. When analyzed alone (Fig. 4b), the overall distribution of diversity in P. t. verus was generally homogeneous, with the exception of very local significant differences in parts of Liberia (Sapo and Grebo) and Côte d’Ivoire (Djouroutou). A focused analysis of ETS (Fig. 4c) revealed that diversity rates tended to be higher in populations closest to the equator, and areas where rates diverged significantly were generally localized between two neighbouring populations. Interestingly, although m was low in the southeastern ETS range, diversity appeared to be relatively high. All of the populations sampled within the P. t. ellioti range had low relative q, with two sites being significantly lower than the average ETS. Two sampling locations in western Uganda (Budongo and Ngogo) in the P. t. schweinfurthii range also displayed significantly lower q.
Fig. 4. EEMS diversity rates in chimpanzees at different population scales.
a Map of diversity rates (q) for the entire species. Diversity rates are mean centred (population average) on a log10 scale, whereby a value of 1 equates to a tenfold rate increase over population mean rate. Point diameter is proportional to the number of individuals sampled in a given deme (range = 1–72). Solid black lines indicate areas where the posterior probability of q differing from the mean rate is >95 percent and the dash lines highlight areas that are >90 percent. Lighter areas indicate populations where q is lower than the mean rate, and darker areas indicate populations where q is higher. Diversity rates were significantly lower in Pan troglodytes verus, while many of the populations in the P. t. troglodytes and P. t. schweinfurthii ranges had significantly higher rates. b Map of q in P. t. verus. Grebo and Sapo (Liberia) had significantly higher diversity rates than the average rate in P. t. verus and Djouroutou (Côte d’Ivoire) had significantly lower rates. Diversity at Mt. Sangbé (Côte d’Ivoire, white area) is also notably low, but not significant. c Map of q in P. t. ellioti, P. t. troglodytes and P. t. schweinfurthii (collectively ETS). Several sites differed significantly, with P. t. ellioti sites in particular displaying lower diversity rates, but overall q were homogeneous. Rates of q between panels are relative and not directly comparable.
Discussion
Our results show an essentially clinal pattern of genetic variation, largely predicted by geographic distance (IBD), with strong local effects driving differentiation in a few highly isolated populations. Importantly, we did not find evidence of major discontinuities conforming to the current taxonomic subdivision. However, although effective barriers were not evident in our data, the P. t. verus population appears to have had a divergent history, as they display much lower genetic diversity in tests of same-subspecies among-site differentiation, while also sharing widespread higher effective migration rates across the subspecies in the whole-species EEMS analysis. Importantly, our observations of high effective migration rates in P. t. verus may simply be a consequence of the overall paucity of diversity in this subspecies. The EEMS model utilizes variation in genetic dissimilarity against geographic distance to estimate effective migration, therefore, high levels of genetic similarity (low diversity) in a geographically clustered subpopulation will appear to have a relatively high migration rate relative to the rest of the sampled populations. Therefore, our observation of relatively high effective migration throughout P. t. verus may indicate higher levels of connectivity in western chimpanzees, but may alternatively or additionally be a signal of a relatively recent (likely northward) population expansion. Indeed, much of the P. t. verus range is savannah mosaic habitat with discontinuous gallery forests53, where chimpanzees tend to have both (1) larger home ranges (over 63 km2)8 than forest-dwelling populations, to seemingly compensate for lower fruit tree density and suitable habitat availability, and (2) a concomitant increase in dispersal distances54,55, thereby possibly driving a signal of increased migration. However, effective migration was uniform across most of the P. t. verus range, as similar rates were also detected among the forest-dwelling populations in southern Liberia and western Côte d’Ivoire, where home ranges have been reported to be between 6 and 37 km2 in size54, suggesting that dynamics other than ecology are involved. This and, especially, the low diversity in P. t. verus, suggest that the high effective migration we observed in this subspecies should be interpreted as evidence of a population bottleneck followed by a recent and fast range expansion. Indeed, this interpretation is corroborated by among-site comparisons of mean standard deviation of allele sizes (SD; large allele-size variation) and Garza Williamson’s M (Supplementary Figs. 5a–d and 6a–d, and see Supplementary Note 2: ‘Detection of a population bottleneck in P. t. verus’ for detailed explanation), whereby the low SD:M ratio observed in P. t. verus suggests a loss of alleles and allele-size ranges, which can be expected for populations that have maintained a relatively small population size for a substantial number of generations (Supplementary Fig. 6a–d)56. A similar pattern was also observed in most P. t. ellioti samples and in the population in the Budongo/Ngogo region of Uganda, while Gashaka has both low SD and moderate M, indicating small long-term population size, possibly, a recent bottleneck. Conversely, populations that maintain a comparatively high SD, but display a low value of M in the central and eastern subspecies (e.g., Gishwati and Issa), are suggested to have experienced a more recent bottleneck (Supplementary Fig. 6a–d).
Our estimates of effective migration rates did not detect significant discontinuities corresponding with the Dahomey Gap, the Sanaga or Ubangi Rivers, the three proposed subspecies-delimiting barriers. Although our sampling scheme endeavoured to collect data from as many localities as was practicable, unavoidable sampling gaps occurred in the P. t. ellioti range, between P. t. ellioti and P. t. troglodytes, and in northwestern DRC between P. t. troglodytes and P. t. schweinfurthii. However, these gaps were of moderate extension and spatial homogeneity in sampling is not an assumption of either the EEMS analysis or our IBD regression, so that it is extremely unlikely that sampling gaps represent a main influence on our results. Our analyses, therefore, indicate that previous observations11–14 of recent genetic discontinuities in chimpanzees likely resulted, at the minimum, from the use of spatially agnostic methods that were biased by geographically clustered genetic samples. Notably, these results do not at all suggest an overall lack of genetic differentiation among chimpanzee populations set apart by geography. Though we found that genetic variation follows a generally clinal pattern across the species, we want to stress that we were specifically focused on patterns occurring on a recent timescale, i.e., within the current interglacial period. Populations are spatially and temporally dynamic, and it is likely that the range of suitable habitat for chimpanzees has fluctuated with the glacial–interglacial cycles, causing repeated population contractions and subsequent expansions57,58. It is important to remark that our analyses utilized microsatellite markers, which, mainly due to both their high mutation rates and polymorphism, can be very effective at revealing subtle genetic structure. However, fast mutation rate, coupled with constraints on maximum and minimum allele size, restrict the information that microsatellites may provide about deeper demographic events, leading to a shallow effective timescale. In particular, a deep divergence between populations, e.g., chimpanzee subspecies, may appear relatively shallow if there are heavy constraints on allele size (which causes differentiation to become asymptotic). However, if previously isolated populations came into contact in the more recent past, demes that are closer to the interface will exchange more migrants, and thus more alleles. The admixed alleles would diffuse along a gradient, since they would become more dispersed as distance from the interface increases. In such cases, recent gene flow and the relative underestimation of deep divergences by microsatellites may, in principle, create a smooth pattern of IBD, whereas other markers might otherwise reveal steeper gradients. This makes microsatellites highly effective for assessing recent patterns of population structure, as signals of ancient contraction–expansion events are flattened by allele size constraints and relatively high mutation rates. Finally, we wish to stress that gradients are not expected to occur by chance alone. Therefore, observations of IBD in our data represent, at least, clear evidence of recent connectivity among populations.
The chimpanzee populations that appear considerably more differentiated than expected by IBD alone, e.g., Mt. Sangbé, Gashaka and Issa, are known to have experienced isolation and decline in recent generations as a consequence of local anthropogenic pressures59–62. Therefore, their observed differentiation most likely stems from loss of alleles due to random drift in extremely small populations.
Our data suggest that, much like humans, chimpanzees likely experienced periods of substantial genetic connectivity across a geographic range encompassing a broad range of habitats, a finding that may point to important evolutionary implications. Environmental heterogeneity and population size act synergistically to influence rates of adaptive evolution. Advantageous mutations are more likely to become fixed in small populations occupying uniform habitats, and are less likely to become fixed in a large population occupying a diverse habitat2. Therefore, as humans and chimpanzees are both widely distributed across a variety of habitat types, evolutionary adaptation is expected to be slow in these species, especially given their long life histories. Hominins have long employed behavioural flexibility to mitigate biological limitations, exploit new resources5, inhabit challenging environments (i.e., use of fire) and migrate out of Africa on several occasions before modern times4,5. Since our results have demonstrated that recent genetic variation in chimpanzees contains little geographic structure on a broad scale, behavioural flexibility may also play an important evolutionary role in their adaptability to environmental heterogeneity63. Chimpanzees have diverse and variable behavioural repertoires64,65 that often vary among nearby communities66, they utilize learned techniques to harvest otherwise unobtainable foods67 and their bevavioural diversity increases with environmental variability68. Indeed, chimpanzees living in arid habitats employ specialized behaviours to regulate the thermal exposure8 and spontaneously innovate novel behaviours in response to increased environmental complexity arising from anthropogenic pressures69–71.
Although our data point to diffuse genetic connectivity in chimpanzees, this signal originates from recent historical patterns in previous generations and does not represent the current potential for gene flow. Indeed, we found that several local chimpanzee populations are very strongly isolated from a genetic perspective, and many more are known to have become fragmented and/or have undergone precipitous declines in recent decades due to anthropogenic factors, such as habitat fragmentation, hunting and disease transmission72–74. P. t. schweinfurthii, P. t. troglodytes and P. t. ellioti are recognized as endangered19, while P. t. verus is recognized as critically endangered73. Though our results indicate a high level of recent genetic connectivity in chimpanzees, we do not suggest that the distinction among the subspecies populations is irrelevant. Each subspecies range hosts populations that contribute unique behaviours to the species as a whole, and our findings suggest that genetic diversity in chimpanzees is distributed throughout the taxon, underscoring the need to maintain regionally driven conservation approaches. However, these results also highlight the need to preserve and restore corridors to facilitate connectivity among remaining populations to avoid inbreeding depression and the accumulation of deleterious alleles that negatively affect fitness. All populations possess unique traits that cumulatively reflect the full genetic and behavioural diversity observed in chimpanzees. Continued isolation and further population decline will irrevocably erode the overall viability of this critical keystone species, ultimately imperiling their long-term fitness. If connectivity between populations is not restored or deteriorates, the geographic distribution we observe in chimpanzees today will shape the genetic structure of future generations75, which may have negative evolutionary consequences2, and ultimately jeopardizes the long-term survival of wild populations.
Methods
Sample collection and preparation
As part of the PanAf, 5397 chimpanzee faecal samples were non-invasively collected at 55 temporary or long-term research sites across the species range (Fig. 1, Supplementary Table 1.3 and Supplementary Fig. 3c), preserved according to the two-step ethanol–silica method76 and stored in the field for up to 2 years. Upon arrival to the lab, samples were stored at −20 °C. DNA extracts were isolated either manually, using the QIAamp Stool Kit (Qiagen), or using an automated process employing the QIAamp 96 PowerFecal QIAcube HT robot (Qiagen), per manufacturer instructions, modified by incorporating a pre-treatment step to improve the DNA quality and yield (Supplementary Note 1: ‘Laboratory methods’). Fourteen unlinked microsatellite loci and one sex-determining locus (amelogenin) were amplified using a two-step multiplex process77 with slight modifications (Supplementary Note 1: ‘Laboratory methods’ and Supplementary Fig. 1). PCR products were analyzed using an ABI Prism 3730 genetic analyzer (Thermo Fisher Scientific) and allele sizes were measured relative to ROX labeled HD400 internal size standard using Genemapper version 5.0 (Thermo Fisher Scientific). Homozygotes were identified by three identical PCR replicates and heterozygotes were confirmed by at least two unambiguous PCR replicates of each allele77 (Supplementary Note 1: ‘Genotype reconstruction’).
Analyses
Cervus 3.0.7 (ref. 78) was used to calculate allele frequencies, determine the minimum number of loci needed to discriminate individuals (PIDsib)79, and to assess the degree of relatedness of individuals within sites (Supplementary Note 1: ‘Genotype reconstruction’ and Supplementary Table 5). Based on the allele frequency data, we determined that we needed eight loci to be 99.9 percent certain that two identical genotypes originated from the same individual rather than full siblings79. In cases where genotypes were unique (not matching to any other genotype), individuals were typed at a minimum of seven loci.
We employed two strategies to assess genetic continuity at the species, subspecies and site levels: (1) we examined the relationship between genetic and geographic distances by applying simple linear regression functions to multiple levels of our data, and (2) we used a spatially explicit analysis that identifies significant spatial deviations from clinal variation (genetic discontinuity). We also performed Mantel, partial Mantel and STRUCTURE analyses on the dataset, as they are standard practice in population structure studies; however, reliable inferences could not be drawn as results were severely confounded by biases in the data. Instead, we provide in-depth details about the modeling and results of these analyses in Supplementary Note 2: ‘Cluster (STRUCTURE) analysis and stratified and partial Mantel tests’ (Supplementary Fig. 2a–c and Supplementary Table 2.1).
We utilized linear regression analyses (G ~ D) to assess the relationship between genetic distance (F′ST/(1 − F′ST))80 and geographic distance (least cost paths; LCPs) at the species, between and within-subspecies and subspecies-comparisons levels. For our analyses, we calculated F′ST as a suitable estimator of FST, which adjusts for differing levels of genetic diversity arising from variation in effective population sizes (Ne) among sampling locations and mutation rate among loci, while not being dependent on within-site diversity80. F′ST/(1 − F′ST) (hereafter G) was modeled as a function of LCP estimators of D, which calculate pairwise distance between sampling localities based on the presence of suitable chimpanzee habitat spanning back 120 ky BP57, and include topological features that could cause resistance to gene flow. We used LCPs specifically to force distance measurements around the presence of large water bodies, such as the Atlantic Ocean, where chimpanzee migration is not possible (Supplementary Fig. 3a, c).
To test for effective barriers to migration and to plot the distribution of chimpanzee genetic diversity across their range, we used the spatially explicit EEMS program51. EEMS estimates historical migration rates that would have given rise to the observed spatial distribution of genetic diversity in the data under idealized conditions, i.e., random mating, neutral selection and constant population size. This method uses georeferenced genetic data to assess diversity in a spatial context; therefore, in contrast to Bayesian clustering algorithms, it does not require sampling homogeneity, ideal for our unbalanced, heterogeneous dataset. Furthermore, the EEMS analysis is not biased by the presence of IBD in the data, rather, decreasing genetic similarity as a function of distance is the null assumption of the model. EEMS generates maps that display mean-centred relative patterns of genetic diversity (q) and relative effective migration (m) rates, therefore, high levels of genetic differentiation appear as divergent from these means.
In a study of both African elephant species Petkova et al.51, demonstrated that a single highly polymorphic locus was able to replicate qualitatively similar effective migration surfaces as when using their entire panel of loci. More importantly, this locus was able to correctly identify the barrier and differentiation between two geographically proximate and closely related species, which is particularly notable as they are known to hybridize where their respective ranges overlap81. In a study of three closely related sympatric beetle species that inhabit forested areas surrounding Kyoto, Japan, the authors used EEMS to examine the spatial distribution of genetic diversity in the presence of habitat discontinuity46. Using a range of nine to ten loci, they detected discrete spatial structuring in two out of the three species, which directly correlated with differing levels of dispersal capability and host species specificity. These results suggest that our use of 14 loci was sufficient for detecting the presence of genetic structure both within subpopulations and between closely related species, especially given the geographic range and depth of sampling in the present study.
We found that the magnitude of differentiation between P. t. verus and ETS populations saturated the signal of regional patterns, so it was necessary to perform analyses of these two populations independently. We found several barriers to m that appeared to be associated with individual populations so we performed additional analyses excluding these (see Supplementary Note 2: ‘Spatially explicit analyses (EEMS)’ and Supplementary Fig. 8a–c). The genetic data analyzed in this study retain the history of hundreds of generations or more, therefore, excluding potential areas of past occupation and migration corridors will bias the results. Since the EEMS model identifies areas where genetic differentiation is higher or lower than expected (IBD), areas where gene flow is restricted or non-existent will be inferred as a significant barrier. Thus, expanding the boundaries of the analysis is necessary and will not lead to erroneous conclusions. The historical range of chimpanzees has likely expanded and contracted over time7, so we accordingly extended the current northern boundaries of the species range by 300 km in our population grids for EEMS. We also included the Dahomey Gap as a functional part of the chimpanzee range, as it has fluctuated in size and featured rainforest as recently as 4 ka BP, a period within the timescale of this study21. For grid density, we performed iterative analyses to ascertain the optimal number of demes that provided the best model fit for analyses at the species (800 demes), P. t. verus (300 demes) and ETS (490 demes) population levels51. For all analyses, to ensure the models reached stability, three independent MCMC chains were run preceded by 500,000 steps of burn-in, at lengths of 6,000,000 repetitions, each beginning with different random seeds.
Statistics and reproducibility
We performed all analyses from a pool of 939 individual genotypes originating from 48 sampling locations with a minimum inter-site distance of 41 km (mean pairwise distance 2843 km; estimated from LCPs). When necessary, we balanced our dataset by imposing a lower limit of 6 and an upper limit of 20 (using a randomized drawing process) on the number of genotypes originating from the same sampling location (see Supplementary Note 2: ‘Isolation by distance (IBD) and distance estimators’). For the EEMS analyses, since balancing was unnecessary, all samples for which geospatial data were recorded (932) were used in the species-wide scale and were divided for focused analyses of P. t. verus (522) and ETS (410). Detailed descriptions of all analyses and p values are provided in the text and Supplementary Note 2: ‘Population genetics analyses’.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We wish to thank the Max Planck Society, Max Planck Society Innovation Fund and the Heinz L. Krekeler Foundation for providing funding to the PanAf project.
We also would like to thank the following authorities who granted permission to conduct research: Agence Nationale des Parcs Nationaux, Gabon, Centre National de la Recherche Scientifique (CENAREST), Gabon, Conservation Society of Mbe Mountains (CAMM), Nigeria, Department of Wildlife and Range Management, Ghana, Direction des Eaux, Forêts et Chasses, Senegal, Eaux et Forets, Mali, Forestry Commission, Ghana, Forestry Development Authority, Liberia, Institut Congolais pour la Conservation de la Nature, DR-Congo, Instituto da Biodiversidade e das Áreas Protegidas (IBAP), Makerere University Biological Field Station (MUBFS), Uganda, Ministere de l’Economie Forestiere, R-Congo, Ministere de la Recherche Scientifique et de l’Innovation, Cameroon, Ministere de la Recherche Scientifique, DR-Congo, Ministère de l’Economie Forestière du gouvernement de la République du Congo, Institut National de Recherche Forestiere, Agence Congolaise de la Faune et des Aires ProtégéesMinistere de l’Agriculture de l’Elevage et des Eaux et Forets, Guinea, Ministere de le Recherche Scientifique et Technologique, R-Congo, Ministere des Eaux et Forets and Ministère de l’Enseignement Supérieur et de la Recherche Scientifique, Cote d’Ivoire, Ministere des Forets et de la Faune, Cameroon, Ministre de l’Environnement et de l’Assainissement et du Developpement Durable du Mali, Ministro da Agricultura e Desenvolvimento Rural, Guinea-Bissau, Ministry of Agriculture, Forestry and Food Security, Sierra Leone, Ministry of Education, Rwanda, National Forestry Authority, Uganda, National Park Service, Nigeria, National Protected area Authority, Sierra Leone, Rwanda Development Board, Rwanda, Société Equatoriale d’Exploitation Forestière (SEEF), Gabon, Tanzania Commission for Science and Technology, Tanzania, Tanzania Wildlife Research Institute, Tanzania, Uganda National Council for Science and Technology (UNCST), Uganda, Uganda Wildlife Authority, Uganda and Ministère de l’Enseignement Supérieur et de la Recherche Scientifique in Côte d’Ivoire.
We thank the following people who were instrumental in providing help with the PanAf project: for analytical guidance, we would like to thank Tomas Marques-Bonet, Aida Andrés Moran, Benjamin Peter, Joshua Schmidt and Lauren C. White. For field site and logistical support, we thank Thierry Aebischer, Alfred Kwabena Assumang, Floris Aubert, Crepin Eyana Ayina, Donatienne Barubiyo, Matthieu Bonnet, Gita Chelluri, Chloe Cipoletta, Katherine Corogenes, Charlotte Coupland, Bryan Curran, Lucy D’Auvergne, Jean Claude Dengui, Theophile Desarmeaux, Karsten Dierks, Emmanuel Dilambaka, Dervla Dowd, Andrew Dunn, Jef Dupain, Henk Eshuis, Marcel K. Eyong, Theo Freeman, John Hart, Martijn Ter Heegde, Veerle Hermans, Inaoyom Imong, Mbangi Kambere, Mohamed Kambi, Deo Kujirakwinja, Vincent Lapeyre, Bradley Larson, Vera Leinert, Joshua M. Linder, Manuel Llana, Eno Nky Manasseh, Giovanna Maretti, Rumen Martín, Michael Masozera, Amelia Meier, Luz C. Miramontes-Sequeiros, Yasmin Moebius, Felix Mulindahabi, Mizuki Murai, Stuart Nixon, Protais Niyigaba, Nadege Wangue Njomen, Emma Normand, Emmanuelle Normand, Nicolas Ntare, Abel Nzeheke, Robinson Orume, Bruno Perodeau, Jodie Preece, Jill Pruetz, Sebastien Regnaut, Alhaji Malikie Siaka, Volker Sommer, Paul Telfer, Emilien Terrade, Alexander Tickle, Richard Tshombe, Hilde Vanleeuwe, Virginie Vergnes, Nouabalé-Ndoki Foundation and Congo Program and Wildlife Conservation Society. For access to the QIAGEN QIAcube HT DNA purification robot and technical support, we thank Dominik Schmidt, Steve Klopfleisch, Nicole Benner, Gabriele Christoffel and Marco Labitzke. We wish to extend a special thanks to Claudia Herf, Christina Kompo, Claudia Feige, Andreas Walther and Rainer Benz for their administrative and IT support, and Anette Nicklisch, Alan Riedel and Katharina Madl for providing genetics laboratory support and assistance.
Author contributions
Study design and project coordination: J.D.L., C.B., H.S.K, P.D. and M.A. coordinated the project. J.D.L, L.V., P.G., M.A., M.S.M. and C.D.B. conceived the study design and wrote the main text with input from all of the co-authors. Field site management, material collection and data generation: A.A., P.A.-V., S.A., E.A.A., E.B., M.B., G.B., R.C., H.C., E.D., T.D., V.E.E., M.E.-N., A.G., A.-C.G., J.H., D.H., R.A.H.-A., K.J.J., S.J., J.J., P.K., M.K., A.K.K., L.K., I.K., K.E.L., J.L., A.L., K.L., S.M., V.M., D.M., G.M., E.N., S.N., C.O., L.J.O., L.P., A.P., M.M.R., A.R., C.S., L.S., A.M.S., F.S., N.T., E.T., J.V.S., M.K.V., E.G.W., J.W., R.M.W., Y.G.Y., K.Y. and K.Z. collected and preserved samples, or provided logistical support with field site management. J.D.L., M.A. and V.S. performed all DNA extraction and PCR amplification. J.D.L and M.A. produced the raw data for reconstruction, and J.D.L and M.S.M reconstructed the genotypes. Population genetic analyses: J.D.L. carried out Cervus, EEMS and Arlequin analyses, with input from L.V., P.G., C.D.B. and M.A. J.D.L did the STRUCTURE analyses. P.G. performed the Garza-Williamson’s M and mean standard deviation of allele sizes investigation and analyses. J.D.L. and P.G. carried out the linear regression analyses. J.D.L. and M.S.M. performed all Mantel tests.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The data from the present study are available from the corresponding authors upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jack D. Lester, Email: jack_lester@eva.mpg.de
Mimi Arandjelovic, Email: arandjel@eva.mpg.de.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-01806-x.
References
- 1.Serre D, Paabo S. Evidence for gradients of human genetic diversity within and among continents. Genome Res. 2004;14:1679–1685. doi: 10.1101/gr.2529604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohta T. Population size and rate of evolution. J. Mol. Evol. 1972;1:305–314. doi: 10.1007/BF01653959. [DOI] [PubMed] [Google Scholar]
- 3.Slatkin, M. Gene flow and selection in a cline. Genetics75, 733–756 (1973). [DOI] [PMC free article] [PubMed]
- 4.Potts R. Variability selection in hominid evolution. Evol. Anthropol. 1998;7:81–96. doi: 10.1002/(SICI)1520-6505(1998)7:3<81::AID-EVAN3>3.0.CO;2-A. [DOI] [Google Scholar]
- 5.Potts R. Hominin evolution in settings of strong environmental variability. Quat. Sci. Rev. 2013;73:1–13. doi: 10.1016/j.quascirev.2013.04.003. [DOI] [Google Scholar]
- 6.Prüfer K, et al. The bonobo genome compared with the chimpanzee and human genomes. Nature. 2012;486:527–531. doi: 10.1038/nature11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill, W. C. O. in The Nomenclature, Taxonomy and Distribution of Chimpanzees, Vol. 1 (ed. Bourne, G. H.) 22–49 (Karger, 1969).
- 8.Pruetz JD, Bertolani P. Chimpanzee (Pan troglodytes verus) behavioral responses to stresses associated with living in a savannah-mosaic environment: implications for hominin adaptations to open habitats. Paleoanthropology. 2009;2009:252–262. doi: 10.4207/PA.2009.ART33. [DOI] [Google Scholar]
- 9.Fünfstück T, et al. The sampling scheme matters: Pan troglodytestroglodytes and P. t. schweinfurthii are characterized by clinal genetic variation rather than a strong subspecies break. Am. J. Phys. Anthropol. 2014;156:181–191. doi: 10.1002/ajpa.22638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer A, Pollack J, Thalmann O, Nickel B, Paabo S. Demographic history and genetic differentiation in apes. Curr. Biol. 2006;16:1133–1138. doi: 10.1016/j.cub.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 11.Gonder MK, et al. A new west African chimpanzee subspecies? Nature. 1997;388:337. doi: 10.1038/41005. [DOI] [PubMed] [Google Scholar]
- 12.Becquet C, Patterson N, Stone AC, Przeworski M, Reich DE. Genetic structure of chimpanzee populations. PLoS Genet. 2007;3:e66. doi: 10.1371/journal.pgen.0030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowden R, et al. Genomic tools for evolution and conservation in the chimpanzee: Pan troglodytesellioti is a genetically distinct population. PLoS Genet. 2012;8:e1002504. doi: 10.1371/journal.pgen.1002504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prado-Martinez J, et al. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Manuel M, et al. Chimpanzee genomic diversity reveals ancient admixture with bonobos. Science. 2016;354:477–481. doi: 10.1126/science.aag2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langergraber KE, et al. Genetic and ‘cultural’ similarity in wild chimpanzees. Proc. R. Soc. B. 2010;278:408–416. doi: 10.1098/rspb.2010.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcin Y, et al. Early anthropogenic impact on Western Central African rainforests 2,600 y ago. Proc. Natl Acad. Sci. USA. 2018;115:3261–3266. doi: 10.1073/pnas.1715336115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vicente M, Schlebusch CM. African population history: an ancient DNA perspective. Curr. Opin. Genet. Dev. 2020;62:8–15. doi: 10.1016/j.gde.2020.05.008. [DOI] [PubMed] [Google Scholar]
- 19.IUCN Red List. IUCN red list of threatened species. http://www.iucnredlist.org (2020).
- 20.Wentworth CK. Natural bridges and glaciation. Am. J. Sci. 1933;26:577–584. doi: 10.2475/ajs.s5-26.156.577. [DOI] [Google Scholar]
- 21.Maley J. The African rainforest: main characteristics of changes in vegetation and climate from the Upper Cretaceous to the Quaternary. Proc. R. Soc. Edinb. 1996;104B:31–73. [Google Scholar]
- 22.Langergraber KE, et al. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc. Natl Acad. Sci. USA. 2012;109:15716–15721. doi: 10.1073/pnas.1211740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shea BT, Coolidge HJ. Craniometric differentiation and systematics in the genus Pan. J. Hum. Evol. 1988;13:671–685. doi: 10.1016/0047-2484(88)90024-3. [DOI] [Google Scholar]
- 24.Albrecht, G. H. & Miller, J. M. A. in Geographic Variation in Primates (eds Kimbel, W. H. & Martin, L. B.) 123–161 (Springer Science & Business Media, 2013).
- 25.Shea, B. T., Leigh, S. R. & Groves, C. P. in Multivariate Craniometric Variation in Chimpanzees (eds Kimbel, W. H. & Martin, L. B.) 265–296 (Springer Science & Business Media, 2013).
- 26.Kühl, H. S. et al. Chimpanzee accumulative stonethrowing. Sci. Rep.6, 22219 (2016). [DOI] [PMC free article] [PubMed]
- 27.Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pritchard, J. K., Wen, X. & Falush, D. Documentation for structure version 2.3 software: version 2.3. 1–39. http://pritchardlab.stanford.edu/structure_software/release_versions/v2.3.4/structure_doc.pdf (2009).
- 30.Meirmans PG. The trouble with isolation by distance. Mol. Ecol. 2012;21:2839–2846. doi: 10.1111/j.1365-294X.2012.05578.x. [DOI] [PubMed] [Google Scholar]
- 31.Perez MF, et al. Assessing population structure in the face of isolation by distance: are we neglecting the problem? Divers. Distrib. 2018;24:1883–1889. doi: 10.1111/ddi.12816. [DOI] [Google Scholar]
- 32.Thalib L, Kitching RL, Bhatti MI. Principal component analysis for grouped data - a case study. Environmetrics. 1999;10:565–574. doi: 10.1002/(SICI)1099-095X(199909/10)10:5<565::AID-ENV360>3.0.CO;2-R. [DOI] [Google Scholar]
- 33.Fischer A, et al. Bonobos fall within the genomic variation of chimpanzees. PLoS ONE. 2011;6:e21605. doi: 10.1371/journal.pone.0021605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frantz AC, Cellina S, Krier A, Schley L, Burke T. Using spatial Bayesian methods to determine the genetic structure of a continuously distributed population: clusters or isolation by distance? J. Appl. Ecol. 2009;46:493–505. doi: 10.1111/j.1365-2664.2008.01606.x. [DOI] [Google Scholar]
- 35.Kalinowski ST. The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human population structure. Heredity. 2010;106:625–632. doi: 10.1038/hdy.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz MK, McKelvey KS. Why sampling scheme matters: the effect of sampling scheme on landscape genetic results. Conserv. Genet. 2008;10:441–452. doi: 10.1007/s10592-008-9622-1. [DOI] [Google Scholar]
- 37.Meirmans PG. Seven common mistakes in population genetics and how to avoid them. Mol. Ecol. 2015;24:3223–3231. doi: 10.1111/mec.13243. [DOI] [PubMed] [Google Scholar]
- 38.Arandjelovic M, et al. Pan African Programme—The cultured chimpanzee. Guidelines for research and data collection. http://panafrican.eva.mpg.de/english/approaches_and_methods.php (2014).
- 39.Arandjelovic M, Vigilant L. Non-invasive genetic censusing and monitoring of primate populations. Am. J. Primatol. 2018;80:e22743. doi: 10.1002/ajp.22743. [DOI] [PubMed] [Google Scholar]
- 40.Arthofer, W., Heussler, C., Krapf, P., Schlick-Steiner, B. C. & Steiner, F. M. Identifying the minimum number of microsatellite loci needed to assess population genetic structure: a case study in fly culturing. Fly12, 13–22 (2018). [DOI] [PMC free article] [PubMed]
- 41.Wenburg JK, Bentzen P, Foote CJ. Microsatellite analysis of genetic population structure in an endangered salmonid: the coastal cutthroat trout (Oncorhyncus clarki clarki) Mol. Ecol. 1998;7:733–749. doi: 10.1046/j.1365-294x.1998.00386.x. [DOI] [PubMed] [Google Scholar]
- 42.Guo, X.-Z. et al. Phylogeography and populationgenetics of Schizothorax o’connori: strong subdivision in the Yarlung Tsangpo River inferred from mtDNA and microsatellite markers. Sci. Rep.6, 29821 (2016). [DOI] [PMC free article] [PubMed]
- 43.Kleinhans C, Willows-Munro S. Low genetic diversity andshallow population structure inthe endangered vulture, Gyps copotheres. Sci. Rep. 2019;9:5536. doi: 10.1038/s41598-019-41755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonato L, et al. Diversity among peripheral populations: genetic and evolutionary differentiation of Salamandra atraat the southern edge of the Alps. J. Zool. Syst. Evol. Res. 2018;56:533–548. doi: 10.1111/jzs.12224. [DOI] [Google Scholar]
- 45.Balkenhol N, et al. A multi-method approach for analyzing hierarchical genetic structures: a case study with cougars Puma concolor. Ecography. 2014;37:552–563. doi: 10.1111/j.1600-0587.2013.00462.x. [DOI] [Google Scholar]
- 46.Kobayashi T, Sota T. Contrasting effects of habitat discontinuity on three closely related fungivorous beetle species with diverging host‐use patterns and dispersal ability. Ecol. Evol. 2019;9:2475–2486. doi: 10.1002/ece3.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rousset, F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics145, 1219–1228 (1997). [DOI] [PMC free article] [PubMed]
- 48.Valdes AM, Slatkin M, Freimer NB. Allele frequencies at microsatellite loci: the stepwise mutation model revisited. Genetics. 1993;133:737–749. doi: 10.1093/genetics/133.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goldstein DB, Ruiz Linares A, Cavalli-Sforza LL, Feldman MW. An evaluation of genetic distances for use with microsatellite loci. Genetics. 1995;139:463–471. doi: 10.1093/genetics/139.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Calabrese PP, Durrett RT, Aquadro CF. Dynamics of microsatellite divergence under stepwise mutation and proportional slippage/point mutation models. Genetics. 2001;159:839–852. doi: 10.1093/genetics/159.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petkova D, Novembre J, Stephens M. Visualizing spatial population structure with estimated effective migration surfaces. Nat. Genet. 2015;48:94–100. doi: 10.1038/ng.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rich, A. M., Wasserman, M. D., Hunt, K. D. & Kaestle, F. A. Chimpanzee (Pan troglodytes schweinfurthii) population spans multiple protected areas in the Albertine Rift. Folia. Primatol. 91, 595–609 (2020). [DOI] [PubMed]
- 53.Baldwin PJ, McGrew WC, Tutin CEG. Wide-ranging chimpanzees at Mt. Assirik, Senegal. Int. J. Primatol. 1982;3:367–385. doi: 10.1007/BF02693739. [DOI] [Google Scholar]
- 54.Lemoine S, et al. Group dominance increases territory size and reduces neighbour pressure in wild chimpanzees. R. Soc. Open Sci. 2020;7:200577. doi: 10.1098/rsos.200577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newton-Fisher NE. The home range of the Sonso community of chimpanzees from the Budongo Forest, Uganda. Afr. J. Ecol. 2003;41:150–156. doi: 10.1046/j.1365-2028.2003.00408.x. [DOI] [Google Scholar]
- 56.Allendorf FW. Genetic drift and the loss of alleles versus heterozygosity. Zoo. Biol. 1986;5:181–190. doi: 10.1002/zoo.1430050212. [DOI] [Google Scholar]
- 57.Barratt, C. D., et al. Late Quaternary habitat suitability models for chimpanzees (Pan troglodytes) since the Last Interglacial (120,000 BP). Preprint at BioRxivhttps://www.biorxiv.org/content/10.1101/2020.05.15.066662v1 (2019).
- 58.Bertola, L. D. et al. Phylogeographic patterns in Africa and high resolution delineation of genetic clades in the lion (Panthera leo). Sci. Rep.6, 30807 (2016). [DOI] [PMC free article] [PubMed]
- 59.Marchesi P, Marchesi N, Fruth B, Boesch C. Census and distribution of chimpanzees in Côte D’Ivoire. Primates. 1995;36:591–607. doi: 10.1007/BF02382880. [DOI] [Google Scholar]
- 60.Sommer V, Adanu J, Faucher I, Fowler A. Nigerian chimpanzees (Pan troglodytes vellerosus) at Gashaka: two years of habituation efforts. Folia Primatol. 2004;75:295–316. doi: 10.1159/000080208. [DOI] [PubMed] [Google Scholar]
- 61.Chancellor RL, Langergraber KE, Ramirez S, Rundus AS, Vigilant L. Genetic sampling of unhabituated chimpanzees (Pan troglodytes schweinfurthii) in Gishwati Forest Reserve, an isolated forest fragment in western Rwanda. Int. J. Primatol. 2012;33:479–488. doi: 10.1007/s10764-012-9591-6. [DOI] [Google Scholar]
- 62.Piel AK, et al. Population status of chimpanzees in the Masito-Ugalla Ecosystem, Tanzania. Am. J. Primatol. 2015;77:1027–1035. doi: 10.1002/ajp.22438. [DOI] [PubMed] [Google Scholar]
- 63.Wessling EG, et al. Seasonal variation in physiology challenges the notion of chimpanzees as a forest-adapted species. Front. Ecol. Evol. 2018;6:1–21. doi: 10.3389/fevo.2018.00060. [DOI] [Google Scholar]
- 64.Whiten A, et al. Cultures in chimpanzees. Nature. 1999;399:682–685. doi: 10.1038/21415. [DOI] [PubMed] [Google Scholar]
- 65.Kühl HS, et al. Human impact erodes chimpanzee behavioral diversity. Science. 2019;363:1453–1455. doi: 10.1126/science.aau4532. [DOI] [PubMed] [Google Scholar]
- 66.Luncz LV, Mundry R, Boesch C. Evidence for cultural differences between neighboring chimpanzee communities. Curr. Biol. 2012;22:922–926. doi: 10.1016/j.cub.2012.03.031. [DOI] [PubMed] [Google Scholar]
- 67.Boesch C, Marchesi P, Marchesi N, Fruth B, Joulian F. Is nut cracking in wild chimpanzees a cultural behaviour? J. Hum. Evol. 1994;26:325–338. doi: 10.1006/jhev.1994.1020. [DOI] [Google Scholar]
- 68.Kalan, A. K. et al. Environmental variability supports chimpanzee behavioural diversity. Nat. Commun. 11, 4451 (2020). [DOI] [PMC free article] [PubMed]
- 69.Hockings, K. J. in Chimpanzees of Bossou and Nimba, 221–219 (Springer Science & Business Media, 2011).
- 70.McCarthy MS, Lester JD, Stanford CB. Chimpanzees (Pan troglodytes) flexibly use introduced species for nesting and bark feeding in a human-dominated habitat. Int. J. Primatol. 2016;38:321–337. doi: 10.1007/s10764-016-9916-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McLennan MR, et al. Surviving at the extreme: chimpanzee ranging is not restricted in a deforested human‐dominated landscape in Uganda. Afr. J. Ecol. 2020;8:e57872. [Google Scholar]
- 72.Junker J, et al. Recent decline in suitable environmental conditions for African great apes. Divers. Distrib. 2012;18:1077–1091. doi: 10.1111/ddi.12005. [DOI] [Google Scholar]
- 73.Kühl HS, et al. The Critically Endangered western chimpanzee declines by 80% Am. J. Primatol. 2017;79:e22681. doi: 10.1002/ajp.22681. [DOI] [PubMed] [Google Scholar]
- 74.Walsh PD, Breuer T, Sanz C, Morgan DB, Doran-Sheehy DM. Potential for Ebola transmission between gorilla and chimpanzee social groups. Am. Nat. 2007;169:684–689. doi: 10.1086/513494. [DOI] [PubMed] [Google Scholar]
- 75.Baden AL, et al. Anthropogenic pressures drive population genetic structuring across a critically endangered lemur species range. Sci. Rep. 2019;9:16276. doi: 10.1038/s41598-019-52689-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nsubuga AM, et al. Factors affecting the amount of genomic DNA extracted from ape faeces and the identification of an improved sample storage method. Mol. Ecol. 2004;13:2089–2094. doi: 10.1111/j.1365-294X.2004.02207.x. [DOI] [PubMed] [Google Scholar]
- 77.Arandjelovic M, et al. Two-step multiplex polymerase chain reaction improves the speed and accuracy of genotyping using DNA from noninvasive and museum samples. Mol. Ecol. Resour. 2009;9:28–36. doi: 10.1111/j.1755-0998.2008.02387.x. [DOI] [PubMed] [Google Scholar]
- 78.Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- 79.Waits LP, Luikart G, Taberlet P. Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Mol. Ecol. 2001;10:249–256. doi: 10.1046/j.1365-294X.2001.01185.x. [DOI] [PubMed] [Google Scholar]
- 80.Meirmans PG, Hedrick PW. Assessing population structure: FST and related measures. Mol. Ecol. Resour. 2010;11:5–18. doi: 10.1111/j.1755-0998.2010.02927.x. [DOI] [PubMed] [Google Scholar]
- 81.Mondol S, et al. New evidence for hybrid zones of forest and savanna elephants in Central and West Africa. Mol. Ecol. 2015;24:6134–6147. doi: 10.1111/mec.13472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data from the present study are available from the corresponding authors upon reasonable request.