Abstract
Due to their pluripotent nature and unlimited cell renewal, stem cells have been proposed as an ideal material for establishing long-term cnidarian cell cultures. However, the lack of unifying principles associated with “stemness” across the phylum complicates stem cells’ identification and isolation. Here, we for the first time report gene expression profiles for cultured coral cells, focusing on regulatory gene networks underlying pluripotency and differentiation. Cultures were initiated from Acropora digitifera tip fragments, the fastest growing tissue in Acropora. Overall, in vitro transcription resembled early larvae, overexpressing orthologs of premetazoan and Hydra stem cell markers, and transcripts with roles in cell division, migration, and differentiation. Our results suggest the presence of pluripotent cell types in cultures and indicate the existence of ancestral genome regulatory modules underlying pluripotency and cell differentiation in cnidaria. Cultured cells appear to be synthesizing protein, differentiating, and proliferating.
Keywords: coral cell culture, RNA-seq, coral stem cells, transcription regulation, cell differentiation
Significance
The inability to establish permanent cnidarian cell lines has directed attention to tissues with a high abundance of stem cells as an ideal material for establishing long-term cultures. Despite this, the lack of unifying principles associated with “stemness” across cnidaria complicates stem cells’ identification and isolation. Here, we report gene expression profiles for cultured coral cells. Our results revealed pluripotent cell types in cultures, identifying coral orthologs of stem cell markers that could be used for further isolation and characterization.
Introduction
Cell–cell interactions are fundamental for body plan establishment and function as they integrate cell type–specific genome regulation during animal development (Peter and Davidson 2011). Coordination and assembly of cell-specific transcription profiles generate interconnected transcriptional networks that in cnidaria generate organismal complexity by deploying morphogenetic borders along an oral–aboral axis (Hayward et al. 2015). In cnidaria, tissue morphogenesis and homeostasis mechanisms are very diverse. The phylum exhibits complex life cycles and diverse developmental mechanisms, spanning planula (P), polyp, and medusa morphologies (Daly et al. 2007). Anthozoans, in particular, display complex polyp morphologies and less regenerative potential, requiring signaling from an organizer region to reconstruct their tissues (Hayward et al. 2015).
At the cellular level, although hydrozoans maintain body structure via the integration of three distinct stem cell lineages established during gastrulation—two epithelial and one interstitial (Hemmrich et al. 2012)—nonhydrozoans cnidarians present two epithelial stem cell lineages lacking the interstitial cell type (i-cells) (review in Gold and Jacobs 2013). Despite this, it is accepted that transdifferentiation of epithelial cells and dedifferentiation of “committed” cell types predates the evolution of cnidarian stem cell systems (Gold and Jacobs 2013). In animals, precursor cells in vivo maintain “stemness” and control differentiation via complex cell–cell interactions within a tissue grade microenvironment known as “stem-cell niche” (Fuchs et al. 2004; Gattazzo et al. 2014). Within the niche, differential deployment of core networks regulates cellular identity by promoting the expression of “pluripotency” genes while repressing developmental signaling pathways (Orkin and Hochedlinger 2011).
In metazoa, stem cells have been reported as early as porifera, with archeocytes and choanocytes identified as the oldest animal stem cell system (Funayama 2010). In cnidaria, undifferentiated stem cells have been reported in a variety of adult (Mydlarz et al. 2008) and larval (Martin and Chia 1982) tissues, with transcriptional profiles of precursor cell types characterized for Hydra (Siebert et al. 2019) and Nematostella (Sebé-Pedrós et al. 2018). Due to the high abundance of stem cells in adult stages, adult tissues have been suggested as an ideal material for the establishment of long-term cnidarian cultures (Rinkevich 2011). However, the lack of unifying principles associated with stemness across the phylum complicates identification and isolation of cnidarian stem cells for culture.
In cnidaria, attempts to establish permanent cnidarian cell lines have failed due to decreasing in vitro viability and proliferation (review in Rinkevich 2011). Despite this, short-term primary cultures have been used to understand the cellular mechanisms underlying fundamental cnidarian processes, such as symbiosis (Barnay-Verdier et al. 2013), calcification (Domart-Coulon et al. 2001; Mass et al. 2012), thermal stress (Nesa and Hidaka 2009), and regeneration (Schmid and Alder 1984). These studies reported inconsistent results regarding survival, proliferation, and viability, reflecting the lack of standardized protocols (reviewed by Rinkevich 2005, 2011). Nonetheless, as primary cell cultures are taken directly from in vivo tissues, they represent a powerful tool to study cellular and physiological processes unattainable using whole organisms.
This paper, reports for the first time gene expression profiles for cultured coral cells, focusing on regulatory gene networks underlying pluripotency and differentiation. We initiated primary cell cultures from Acropora digitifera tip fragments, the fastest growing tissue in Acropora corals (Lirman et al. 2014). A. digitifera has become an emerging model to study coral responses to environmental change (Shinzato et al. 2011) and due to its basal phylogenetic position in cnidaria (Bridge et al. 1992), the species is also an ideal model to study the evolution of animal developmental mechanisms.
Overall, after 4 weeks of culture, we conducted quantitative RNA-seq analysis and compared the in vitro transcriptome with a previously published A. digitifera developmental time series (Reyes-Bermudez et al. 2016). In vitro transcription closely resembled gene expression used in early larvae during the establishment of larval cellular phenotypes. Likewise, we identified upregulation in vitro of orthologs of premetazoan and Hydra stem cell markers (HM) and transcripts with roles in DNA replication, cell division, migration, and differentiation. Our results suggest the existence of 1) pluripotent cell types in cultures and 2) ancestral genome regulatory modules underlying pluripotency and cell differentiation in cnidaria.
Results and Discussion
Cultures Consist of Multicellular Aggregates Actively Dividing and Differentiating
After 4 weeks, cultures consisted of cellular aggregates formed by cells displaying a previously reported unique and nonspecific small round morphology (Reyes-Bermudez and Miller 2009). Cells did not attach to the substrate, and signs of CaCO3 precipitation were not observed. Contrasting with previous results (Lecointe et al. 2013), cultures did not show evidence of decreased proliferation. The fact that one round of subculturing was necessary for 2 weeks after initiation, suggests that cells were actively dividing. Although zooxanthellae were present abundantly at initiation, after 4 weeks, chlorophyll fluorescence was absent. Our results support the idea that the capacity to maintain or reconstruct cell signaling between epitheliums in vitro is critical for the establishment of “long-term” successful primary cultures (reviewed by Rinkevich 2005, 2011).
Moreover, upregulation in vitro of genes with roles in protein synthesis, proliferation, and differentiation indicate that, at least after 4 weeks, a subset of cells was proliferating and differentiating in cultures (table 1). The fact that cultures were initiated from branch tips, the fastest growing tissue in Acropora corals (Lirman et al. 2014), suggest that undifferentiated cell populations in founder tissues might be responsible for in vitro enrichment of corals cells that is a crucial factor for culture viability (reviewed by Rinkevich 2005, 2011). Reduced proliferation in primary cnidarian cultures has been linked to a decrease in the proportion of animal cells as result of culture contamination (Frank et al. 1994).
Table 1.
Gene Ontology (GO) Enrichment Summary in DEGs Only Upregulated in Cells
| GO ID | Node Size | Sample Match | P Adj | Term | Ontology |
|---|---|---|---|---|---|
| GO:0000302 | 990 | 421 | 2.02E−09 | Response to reactive oxygen species | BP |
| GO:0006415 | 152 | 92 | 2.20E−09 | Translational termination | BP |
| GO:0006414 | 369 | 173 | 6.60E−06 | Translational elongation | BP |
| GO:0042755 | 258 | 125 | 0.000134999 | Eating behavior | BP |
| GO:0043462 | 293 | 131 | 0.018485676 | Regulation of ATPase activity | BP |
| GO:0000302 | 990 | 421 | 2.02E−09 | Response to reactive oxygen species | BP |
| GO:0006415 | 152 | 92 | 2.20E−09 | Translational termination | BP |
| GO:0006414 | 369 | 173 | 6.60E−06 | Translational elongation | BP |
| GO:0042755 | 258 | 125 | 0.000134999 | Eating behavior | BP |
| GO:0043462 | 293 | 131 | 0.018485676 | Regulation of ATPase activity | BP |
| GO:0009725 | 2914 | 1,050 | 0.000676117 | Response to hormone stimulus | BP |
| GO:0030865 | 350 | 181 | 4.75E−11 | Cortical cytoskeleton organization | BP |
| GO:0003012 | 826 | 364 | 2.70E−10 | Muscle system process | BP |
| GO:0050905 | 485 | 202 | 0.020936066 | Neuromuscular process | BP |
| GO:0002026 | 161 | 86 | 8.57E−05 | Regulation of the force of heart contraction | BP |
| GO:0060327 | 118 | 69 | 1.68E−05 | Cytoplasmic actin-based contraction involved in cell motility | BP |
| GO:0002027 | 337 | 158 | 3.54E−05 | Regulation of heart rate | BP |
| GO:0000916 | 184 | 95 | 0.000144077 | Actomyosin contractile ring contraction | BP |
| GO:0007109 | 148 | 82 | 1.95E−05 | Cytokinesis, completion of separation | BP |
| GO:0032060 | 158 | 81 | 0.002337013 | Bleb assembly | BP |
| GO:0048739 | 153 | 80 | 0.000935569 | Cardiac muscle fiber development | BP |
| GO:0035277 | 134 | 72 | 0.000968262 | Spiracle morphogenesis, open tracheal system | BP |
| GO:0007427 | 218 | 105 | 0.00265387 | Epithelial cell migration, open tracheal system | BP |
| GO:0046664 | 128 | 70 | 0.000572738 | Dorsal closure, amnioserosa morphology change | BP |
| GO:0007395 | 123 | 69 | 0.000180261 | Dorsal closure, spreading of leading edge cells | BP |
| GO:0007496 | 115 | 66 | 0.000106495 | Anterior midgut development | BP |
| GO:0021549 | 341 | 169 | 4.16E−08 | Cerebellum development | BP |
| GO:0030224 | 166 | 95 | 9.18E−08 | Monocyte differentiation | BP |
| GO:0002119 | 798 | 341 | 2.08E−07 | Nematode larval development | BP |
| GO:0030220 | 127 | 72 | 4.86E−05 | Platelet formation | BP |
| GO:0007527 | 125 | 71 | 5.55E−05 | Adult somatic muscle development | BP |
| GO:0045200 | 141 | 77 | 0.000140176 | Establishment of neuroblast polarity | BP |
| GO:0048147 | 35 | 25 | 0.014716604 | Negative regulation of fibroblast proliferation | BP |
| GO:0001767 | 116 | 66 | 0.000170584 | Establishment of lymphocyte polarity | BP |
| GO:0006930 | 130 | 74 | 2.35E−05 | Substrate-dependent cell migration, cell extension | BP |
| GO:0033057 | 527 | 224 | 0.000940241 | Multicellular organismal reproductive behavior | BP |
| GO:0051015 | 403 | 203 | 1.23E−11 | Actin filament binding | MF |
| GO:0008307 | 210 | 109 | 2.95E−06 | Structural constituent of muscle | MF |
| GO:0019829 | 394 | 179 | 2.23E−05 | Cation-transporting ATPase activity | MF |
| GO:0031762 | 119 | 68 | 2.73E−05 | Follicle-stimulating hormone receptor binding | MF |
| GO:0008013 | 155 | 80 | 0.000637174 | Beta–catenin binding | MF |
| GO:0043531 | 316 | 143 | 0.000915446 | ADP binding | MF |
| GO:0005167 | 47 | 30 | 0.017776487 | Neurotrophin TRK receptor binding | MF |
| GO:0005516 | 616 | 286 | 2.94E−11 | Calmodulin binding | MF |
| GO:0051020 | 651 | 279 | 3.30E−06 | GTPase binding | MF |
| GO:0005083 | 586 | 245 | 0.000425907 | Small GTPase regulator activity | MF |
| GO:0019901 | 1,428 | 583 | 1.59E−10 | Protein kinase binding | MF |
| GO:0004725 | 363 | 157 | 0.007252685 | Protein tyrosine phosphatase activity | MF |
Note.—Node size = total number of GO terms in node. Sample match = number of transcripts with GO terms associated to specific nodes.
Cultured Cells Express More Genes in Common with Early Larvae Than with Adult, P, or Embryonic Stages
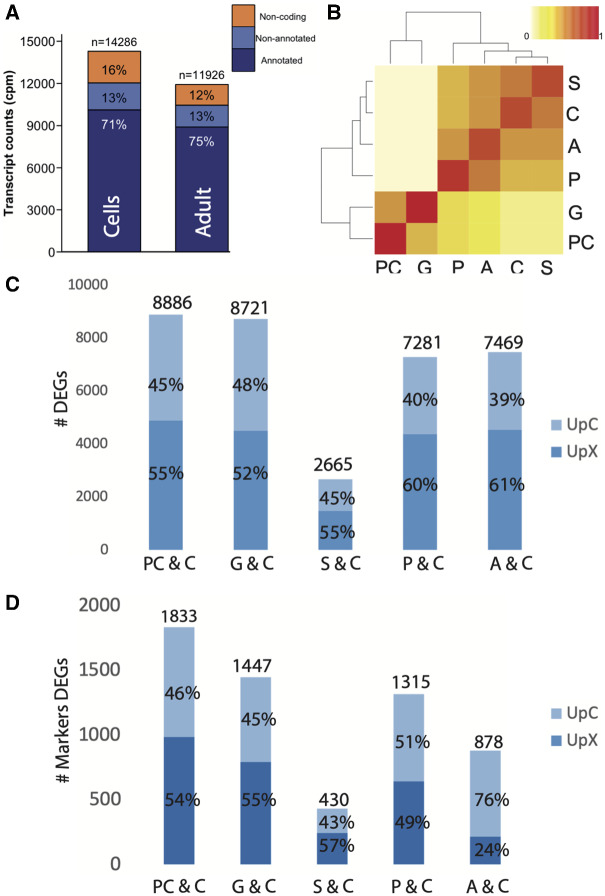
In vitro transcription profile resulted in 14,286 transcripts compared with 11,926 observed in adult tissue (fig. 1A). Transcriptome comparison between cultured cells (C) and data on previously published A. digitifera stages (Reyes-Bermudez et al. 2016), showed that in vitro gene expression is closer to early larvae (sphere[S]) than to adult polyps (adult [A]) (fig. 1B). These results suggest that the abolishment of morphogenetic borders following disassociation of coral polyps resulted in in vitro overexpression of transcriptional networks that resemble those used by S (Reyes-Bermudez et al. 2016). Differential expression analysis supported the observation, showing that C cells expressed more genes in common with S than with other stages (fig. 1C). Clustering of embryonic transcriptomes (blastula—prawnchip-like blastula [PC] and gastrula—G) as a distinct group that differs from the remaining stages, indicates that C cells are most likely cellular lineages originated after gastrulation (fig. 1B).
Fig. 1.
In vitro transcriptome characterization. Cells’ specific transcription profile resulted in 14,286 transcripts compared with 11,926 reported for adult tissue (A). Transcriptome comparison between cultured cells and Acropora digitifera developmental stages showed that although cultures were initiated from adult tissues, their expression profile was closer to early larvae (S) than to any other in vivo stage (B). Differential gene expression analysis revealed a lower number of DEGs in the SvsC comparison (C). Only a fraction of DEGs were identified as orthologs of HM. The lowest percentage was observed in AvsC and the highest in PCvsC (D). Blastula, PC; gastrula, G; early larvae, S; planula, P; adult, A; upregulated in C, UpC; upregulated in vivo, UpX.
Moreover, enrichment in the subset of differentially expressed genes (DEGs) upregulated in vitro with molecules involved in diverse morphogenetic and differentiation processes (table 1) indicates uncoordinated overexpression of genome regulatory programs following the loss of morphogenetic borders (Beloussov 2015). Results revealed a transcriptionally heterogenous C cell population that differed transcriptionally from the in vivo system. Significant transcriptional changes that reflect the emergence of more active and proliferative subgroups have been reported in cell cultures over time (Januszyk et al. 2015).
Cells Expressed Orthologs of Premetazoan and HM
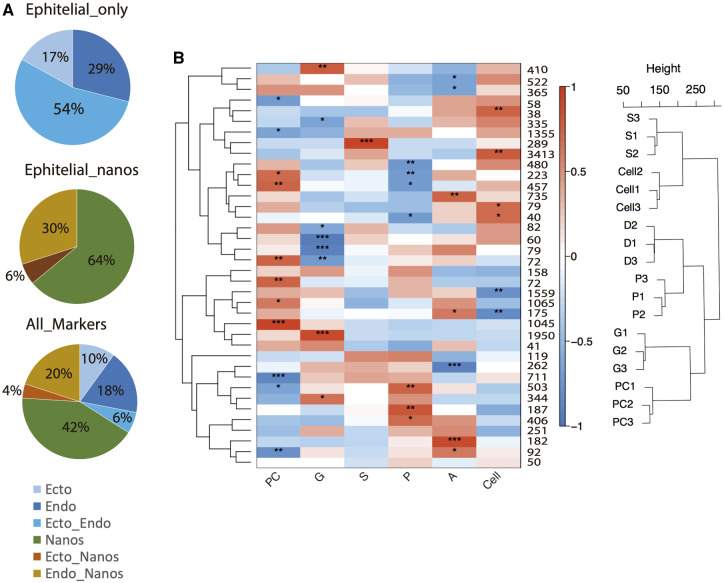
Consistent with 500 Myr of independent cnidarian evolution (Steele et al. 2011), only a small fraction of DEGs were identified as orthologs of HM. We observed over expression of a higher number of HM in Blastula (PC) (fig. 1D), which is consistent with the predominant pluripotent cellular phenotypes present at the stage. Interestingly, Acropora HM orthologs upregulated in vitro were enriched with markers overexpressed by Hydra’s i-cells (nanos) (fig. 2A), which, in a strict sense, is the only true characterized cnidarian stem cell population (Frank et al. 2009). These results may not indicate the presence in coral tissues of stem cell populations homologous to Hydra’s i-cells but most likely indicate the utilization of conserved molecules underlying pluripotency and cell differentiation in Acropora.
Fig. 2.
Orthologs of HM and coexpression networks. The HM fraction expressed in vitro was enriched by both endodermal and i-cell HM (A). Transcripts (18,264) were assigned to 38 different gene modules that ranged from 38 to 3,413 transcripts and grouped in two main coexpression clusters (C1 and C2). Eigengenes were calculated for each module and although we were able to identify discrete gene expression patterns, in most cases. significant module–trait correlations were observed in a stage-specific fashion. *P value ≤0.05, **P value ≤0.01, ***P value ≤0.01. Blastula, PC; gastrula, G; early larvae, S; planula, P; adult, A; cultured cells, C.
Similarly, we observed enrichment in cultures of orthologs of endodermal markers such as Brachyury (Yasuoka et al. 2016) and Hedgehog (Matus et al. 2008) (supplementary material S1–S4, Supplementary Material online). Whether this reflects endodermal enrichment in vitro or utilization of conserved transcriptional networks by different lineages, is not clear. For example, although i-cells are thought to be a “recent” cnidarian innovation, their transcriptome is phylogenetically older than those of Hydra’s epithelial lineages (Hemmrich et al. 2012), suggesting recruitment of ancestral regulatory networks during Hydra’s i-cell evolution. Our results support this idea and suggest that regulatory networks associated with maintenance of pluripotency and differentiation are not fixed entities linked to specific cell types, but dynamic modules recruited and modified multiple times by natural selection during cnidarian diversification.
Likewise, identification of premetazoan and metazoan stem cell markers in the subset of DEGs exclusively upregulated in vitro (table 2) is consistent with the idea that animal stem cell systems were built upon ancestral regulatory gene networks present in the last common metazoan ancestor (Alié et al. 2015). Upregulation in vitro of Acropora orthologs with roles in cell cycle, replication, chromosome maintenance, stress response, DNA repair, as well as diverse transcripts coding RNA-binding proteins, such as a Musashi-1 ortholog (table 2), imply that components of regulatory gene networks associated to stemness are being expressed in cultures.
Table 2.
DEGs with Putative Roles in Stem Cell Homeostasis
| ID | Annotation | KEGG | Marker | Function |
|---|---|---|---|---|
| Ancestral genes | ||||
| adi_v1.04467 | Nuclear factor NF-kappa-B p105 subunit—Metazoa | K02580 | Nanos_3529 | Cell growth and differentiation |
| adi_v1.12580 | Heterogeneous nuclear ribonucleoprotein K—Metazoa | K12886 | Nanos_2219 | Pre-mRNA splicing |
| adi_v1.12078 | ATP-dependent RNA helicase DDX56/DBP9 - Eukariota | K14810 | Nanos_1725 | RNA-Helicases |
| adi_v1.19896 | DNA polymerase alpha subunit A—Eukariota | K02320 | Nanos_3477 | DNA replication |
| adi_v1.00008 | Replication factor C subunit 3/5—Eukariota | K10756 | Nanos_2523 | DNA repair |
| adi_v1.17163 | Flap endonuclease-1—Pre-Eukariota | K04799 | Nanos_Ecto_497 | DNA repair |
| Replication | ||||
| adi_v1.13235 | Replication factor C subunit 2/4 | K10755 | Nanos_2940 | Replication |
| adi_v1.00008 | Replication factor C subunit 3/5 | K10756 | Nanos_2523 | Replication |
| adi_v1.19896 | DNA polymerase alpha subunit A (EC:2.7.7.7) | K02320 | Nanos_3477 | Replication |
| adi_v1.02983 | DNA polymerase sigma (EC:2.7.7.7) | K03514 | Nanos_74 | Replication |
| adi_v1.10030 | DNA polymerase zeta (EC:2.7.7.7) | K02350 | Nanos_4979 | Replication |
| adi_v1.24240 | ATP-binding protein involved in chromosome partitioning | K03593 | Nanos_1409 | Replication |
| adi_v1.13671 | DNA polymerase zeta (EC:2.7.7.7) | K02350 | Endo_Nanos_16 | Replication |
| adi_v1.12265 | DNA topoisomerase VI subunit B (EC:5.99.1.3) | K03167 | Endo_Nanos_1274 | Replication |
| adi_v1.19036 | DNA polymerase sigma (EC:2.7.7.7) | K03514 | Endo_Nanos_861 | Replication |
| Cell cycle | ||||
| adi_v1.05785 | Cell cycle arrest protein BUB3 | K02180 | Nanos_5104 | Cell cycle |
| adi_v1.04546 | Cell cycle checkpoint protein | K06662 | Nanos_2749 | Cell cycle |
| XLOC_000501 | Cell division cycle 20-like protein 1, cofactor of APC complex | K03364 | Nanos_7115 | Cell cycle |
| XLOC_019254 | Cell division cycle 20-like protein 1, cofactor of APC complex | K03364 | Nanos_1396 | Cell cycle |
| adi_v1.03932 | Cell division cycle 20-like protein 1, cofactor of APC complex | K03364 | Nanos_10963 | Cell cycle |
| adi_v1.06900 | G1-/S-specific cyclin PLC1 | K06656 | Nanos_1058 | Cell cycle |
| adi_v1.13930 | Centromere protein B | K11496 | Nanos_2602 | Cell cycle |
| adi_v1.09992 | Cell division protein ZapA | K09888 | Endo_Nanos_2012 | Cell cycle |
| adi_v1.24600 | Signal-induced proliferation-associated gene 1 | K08013 | Endo_Nanos_1253 | Cell cycle |
| adi_v1.24600 | Signal-induced proliferation-associated gene 1 | K08013 | Endo_Nanos_1253 | Cell cycle |
| XLOC_020915 | Cell division cycle 20-like protein 1, cofactor of APC complex | K03364 | Ecto_3004 | Cell cycle |
| Helicases | ||||
| adi_v1.01424 | Chromodomain–helicase–DNA-binding protein 7 (EC:3.6.4.12) | K14437 | Nanos_422 | Chromatin remodeling |
| adi_v1.01424 | Chromodomain–helicase–DNA-binding protein 7 (EC:3.6.4.12) | K14437 | Nanos_422 | Chromatin remodeling |
| adi_v1.13340 | RNAi-mediated heterochromatin assembly 1 (EC:3.6.4.13) | K11701 | Endo_Ecto_43 | Chromatin remodeling |
| adi_v1.23884 | ATP-dependent RNA helicase DHX8/PRP22 (EC:3.6.4.13) | K12818 | Nanos_1508 | Splicing/transcription |
| adi_v1.08670 | ATP-dependent RNA helicase DHX15/PRP43 (EC:3.6.4.13) | K12820 | Nanos_806 | Splicing/transcription |
| adi_v1.23237 | ATP-dependent RNA helicase DDX1 (EC:3.6.4.13) | K13177 | Nanos_1406 | Splicing/transcription |
| adi_v1.12078 | ATP-dependent RNA helicase DDX56/DBP9 (EC:3.6.4.13) | K14810 | Nanos_1725 | Ribosome biogenesis |
| Chromosome maintenance | ||||
| adi_v1.24240 | ATP-binding protein involved in chromosome partitioning | K03593 | Nanos_1409 | Chromosome maintenance |
| adi_v1.17702 | Structural maintenance of chromosome 1 | K06636 | Nanos_Ecto_213 | Chromosome maintenance |
| adi_v1.15153 | Structural maintenance of chromosome 4 | K06675 | Nanos_Ecto_50 | Chromosome maintenance |
| adi_v1.01018 | Structural maintenance of chromosome 1 | K06636 | Nanos_653 | Chromosome maintenance |
| adi_v1.14806 | Structural maintenance of chromosome 4 | K06675 | Nanos_3734 | Chromosome maintenance |
| adi_v1.11026 | Structural maintenance of chromosome 4 | K06675 | Nanos_6620 | Chromosome maintenance |
| adi_v1.04706 | Chromosome segregation protein | K03529 | Endo_Nanos_897 | Chromosome maintenance |
| adi_v1.01300 | Chromosome transmission fidelity protein 1 (EC:3.6.4.13) | K11273 | Ecto_1702 | Chromosome maintenance |
| adi_v1.21696 | Chromosome segregation protein | K03529 | Endo_Ecto_309 | Chromosome maintenance |
| DNA repair/stress response | ||||
| adi_v1.23838 | Three prime repair exonuclease 2 (EC:3.1.11.2) | K10791 | Nanos_3244 | DNA repair |
| adi_v1.03868 | DNA damage-inducible protein 1 | K11885 | Nanos_1310 | DNA repair |
| adi_v1.02191 | DNA excision repair protein ERCC-2 (EC:3.6.4.12) | K10844 | Nanos_1749 | DNA repair |
| adi_v1.22737 | DNA excision repair protein ERCC-3 (EC:3.6.4.12) | K10843 | Nanos_3028 | DNA repair |
| adi_v1.22267 | DNA excision repair protein ERCC-4 (EC:3.1.-.-) | K10848 | Nanos_1886 | DNA repair |
| adi_v1.11724 | DNA excision repair protein ERCC-8 | K10570 | Nanos_5126 | DNA repair |
| adi_v1.03203 | DNA excision repair protein ERCC-8 | K10570 | Nanos_6011 | DNA repair |
| adi_v1.11542 | DNA repair protein RAD16 | K15083 | Nanos_9086 | DNA repair |
| adi_v1.06342 | DNA repair protein RAD50 (EC:3.6.-.-) | K10866 | Nanos_2601 | DNA repair |
| adi_v1.19161 | DnaJ homolog subfamily A member 5 | K09506 | Nanos_1092 | Stress response |
| XLOC_001068 | DnaJ homolog subfamily B member 9 | K09515 | Nanos_4628 | Stress response |
| adi_v1.04788 | Heat shock 70 kDa protein 1/8 | K03283 | Nanos_2410 | Stress response |
| adi_v1.02262 | Heat shock 70 kDa protein 1/8 | K03283 | Nanos_2410 | Stress response |
| adi_v1.04284 | Stress-induced-phosphoprotein 1 | K09553 | Nanos_2665 | Stress response |
| RNA-binding proteins | ||||
| adi_v1.16723 | Multiple RNA-binding domain-containing protein 1 | K14787 | Nanos_527 | RNA-binding |
| adi_v1.13400 | oo18 RNA-binding protein | K02602 | Nanos_1849 | RNA-binding |
| adi_v1.10220 | RNA-binding protein 15 | K13190 | Nanos_1931 | RNA-binding |
| adi_v1.03795 | RNA-binding protein 39 | K13091 | Nanos_357 | RNA-binding |
| adi_v1.05305 | RNA-binding protein Musashi | K14411 | Nanos_2716 | RNA-binding |
| adi_v1.03308 | RNA-binding protein 26 | K13192 | Endo_7052 | RNA-binding |
| adi_v1.15031 | U1 small nuclear ribonucleoprotein A | K11091 | Nanos_334 | RNA-binding |
| adi_v1.00705 | U3 small nucleolar ribonucleoprotein protein IMP4 | K14561 | Nanos_985 | RNA-binding |
| adi_v1.06360 | U3 small nucleolar RNA-associated protein 20 | K14772 | Nanos_283 | RNA-binding |
| adi_v1.19916 | U3 small nucleolar RNA-associated protein 21 | K14554 | Nanos_2230 | RNA-binding |
| adi_v1.13965 | U3 small nucleolar RNA-associated protein 24 | K14566 | Nanos_1277 | RNA-binding |
| adi_v1.12414 | U3 small nucleolar RNA-associated protein 5 | K14546 | Nanos_200 | RNA-binding |
| adi_v1.11136 | U3 small nucleolar RNA-associated protein 6 | K14557 | Nanos_1283 | RNA-binding |
| adi_v1.07136 | U3 small nucleolar RNA-associated protein 6 | K14557 | Nanos_1283 | RNA-binding |
| XLOC_015243 | U3 small nucleolar RNA-associated protein 7 | K14768 | Nanos_1007 | RNA-binding |
| adi_v1.17859 | Heterogeneous nuclear ribonucleoprotein K | K12886 | Nanos_2219 | RNA-binding |
| adi_v1.09619 | Heterogeneous nuclear ribonucleoprotein M | K12887 | Nanos_141 | RNA-binding |
| adi_v1.23861 | U4/U6 small nuclear ribonucleoprotein SNU13 | K12845 | Nanos_Ecto_54 | RNA-binding |
| adi_v1.21303 | RNA-binding protein 15 | K13190 | Endo_Nanos_332 | RNA-binding |
| adi_v1.12580 | RNA-binding protein 5/10 | K13094 | Endo_Nanos_738 | RNA-binding |
| adi_v1.07694 | Heterogeneous nuclear ribonucleoprotein L | K13159 | Endo_Nanos_24 | RNA-binding |
| adi_v1.21839 | Small nuclear ribonucleoprotein B and B' | K11086 | Endo_1503 | RNA-binding |
| Ribosome biogenesis | ||||
| adi_v1.12527 | rRNA biogenesis protein RRP5 | K14792 | Nanos_669 | Ribosome biogenesis |
| adi_v1.10921 | rRNA biogenesis protein RRP5 | K14792 | Nanos_669 | Ribosome biogenesis |
| adi_v1.20333 | Regulator of ribosome biosynthesis | K14852 | Nanos_751 | Ribosome biogenesis |
| adi_v1.09262 | Ribosome assembly protein 4 | K14855 | Nanos_1608 | Ribosome biogenesis |
| adi_v1.05788 | Ribosome biogenesis protein BMS1 | K14569 | Nanos_481 | Ribosome biogenesis |
| adi_v1.01894 | Ribosome biogenesis protein MAK21 | K14832 | Nanos_2059 | Ribosome biogenesis |
| adi_v1.05433 | Ribosome biogenesis protein NSA2 | K14842 | Nanos_425 | Ribosome biogenesis |
| XLOC_014614 | Ribosome production factor 1 | K14846 | Nanos_1610 | Ribosome biogenesis |
| adi_v1.04696 | Ribosome biogenesis GTPase A | K14540 | Endo_Nanos_387 | Ribosome biogenesis |
| adi_v1.02489 | Ribosome biogenesis protein MAK21 | K14832 | Endo_Nanos_4397 | Ribosome biogenesis |
Coexpression Modules Reveal Distinct G-/P-Specific Genome Regulatory Programs
Network analysis assembled DEGs in 38 modules within two main coexpression groups, consisting of distinct and diverse stage-specific coexpression clusters (fig. 2B). Coexpression units usually reflect common functionality and regulation (review in Peter and Davidson 2011). Interestingly, most in vitro upregulated DEG’s were coexpressed in PC, S, and A but were significantly downregulated in G and P stages (fig. 2B). Differential usage of enhancers between G and P stages have been reported for Nematostella (Schwaiger et al. 2014), suggesting the existence of distinct G-/P-specific genome regulatory programs in cnidaria. More research is necessary to test this idea as transcriptional networks underlying early morphogenetic transitions in metazoans are variable and, in some cases, taxa-specific (Erwin and Davidson 2009; Davidson 2010).
Finally, transcriptome comparisons using coexpression networks and transcript composition showed slightly different results. Although the topology built using complete transcriptomes clustered S and C as a sister group to A, leaving P as the most dissimilar stage and PC and G as a separate group, the topology based on coexpression data, resolved P and A as a sister group to PC and G (fig. 1D). In both cases, the similarity between C and S was clear, indicating the usage in the two stages of shared genome regulatory programs based on similar transcript composition. On the other hand, differences between topologies reveal that in vivo complexity is strongly dependent on network interactions and supports the idea that body plan morphogenesis and evolution is a “system-level problem” that cannot be understood by looking at developmental conserved genes in isolation (Peter and Davidson 2011).
Conclusion
Our study demonstrated that primary coral cultures are valuable tools for studying genome regulatory programs and revealed the existence of ancestral genome regulatory modules underlying pluripotency and cell differentiation in cnidaria. Caution must be taken to interpret in vitro experiments as C populations are heterogeneous cell types that drastically differ transcriptionally from the in vivo system.
Materials and Methods
Collection of Samples and Tissue Culture
Tip fragments (∼3 cm) from six different colonies were kept in 50 ml falcon tubes containing 0.2 μm filtered seawater with antibiotics (FSWA) (1% Pen/Strep/L-Glu-solution, Sigma–Aldrich and 0.1% Fungizone, Invitrogen) prior the initiation of cultures. Samples were washed 3× with FSWA and then incubated at 32 °C for 4 h to induce bleaching (Desalvo et al. 2010). Fragments were washed (3×) with FSWA and further incubated (2 h/gentle shaking) in calcium-free FSWA (Marshall and Clode 2004). Following tissue dissociation, naked skeletons were removed, and detached tissue centrifuged (1,500 rpm/10 min) and resuspended in 5 ml of FSWA. Cell pellets were gently washed 3× with 5 ml of cell culture media (30% DMEM Gibco, 10% FBS Gibco, 1% Pen/Strep-solution, 0.1% Fungizone, 1% Glutamax, Gibco, 25 mM HEPES pH 8.0, and 55% 0.2 μm filtered seawater) and resuspended in 5 ml of fresh media. Founding cultures were kept individually in 6-well cultured plates and incubated at 23 °C in the dark for 48 h. After that, 1 ml of the original cultures were used to inoculate 4 ml of fresh media in 6-well cultured plates and returned to incubation conditions. Cultures were monitored daily on a standard inverted light microscope fitted with a color digital camera. Media was changed when cultures reached 60% confluence. After 4 weeks, cells from three wells were harvested for RNA extractions.
Sequencing and Data Analysis
Library preparation and data analysis were conducted as reported in Reyes-Bermudez et al. (2016). To identify Acropora HM, we download the T-CDS data set from Hydra vulgaris strain AEP from http://www.compagen.org/datasets.html. Orthologs were determined using OrthoMCL v.1.4 with a BLASTp E value cut-off of 1e−5, a minimum coverage of 70% and an inflation index of 1.5 (Li et al. 2003).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank all members of the Sesoko Station for their assistance during coral spawning. We are also thankful to members of the Hidaka and Mikheyev laboratories for field assistance and technical support, respectively. Sequencing was performed by the OIST DNA-sequencing section. This work was supported by a postdoctoral fellowship awarded to A.R.B. from the Japanese Society for the Promotion of Science and internal funds from the Okinawa Institute of Science and Technology Graduate University awarded to A.S.M.
Data Availability
Raw data for cell cultures can be found under bio-project ID PRJDB9497: BioSamples: SAMD00210801, SAMD00210803, and SAMD00210805. Experiment: DRX207005–DRX207007.
Literature Cited
- Alié A, et al. 2015. The ancestral gene repertoire of animal stem cells. Proc Natl Acad Sci U S A. 112(51):E7093–E7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnay-Verdier S, et al. 2013. Establishment of primary cell culture from the temperate symbiotic cnidarian, Anemonia viridis. Cytotechnology 65(5):697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloussov LV, editor. 2015. Morphogenesis on the multicellular level: patterns of mechanical stresses and main modes of collective cell behavior. In: Morphomechanics of development. Cham (Switzerland): Springer. p. 75–111. [Google Scholar]
- Bridge D, Cunningham CW, Schierwater B, DeSalle R, Buss LW.. 1992. Class-level relationships in the phylum cnidaria: evidence from mitochondrial genome structure. Proc Natl Acad Sci U S A. 89(18):8750–8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly M, et al. 2007. The phylum cnidaria: a review of phylogenetic patterns and diversity 300 years after Linnaeus. Zootaxa 1668(1):127–182. [Google Scholar]
- Davidson EH. 2010. Emerging properties of animal gene regulatory networks. Nature 468(7326):911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desalvo MK, Sunagawa S, Voolstra CR, Medina M.. 2010. Transcriptomic responses to heat stress and bleaching in the elkhorn coral Acropora palmata. Mar Ecol Prog Ser. 402:97–113. [Google Scholar]
- Domart-Coulon IJ, Elbert DC, Scully EP, Calimlim PS, Ostrander GK.. 2001. Aragonite crystallization in primary cell cultures of multicellular isolates from a hard coral, Pocillopora damicornis. Proc Natl Acad Sci U S A. 98(21):11885–11890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin DH, Davidson EH.. 2009. The evolution of hierarchical gene regulatory networks. Nat Rev Genet. 10(2):141–148. [DOI] [PubMed] [Google Scholar]
- Frank U, Plickert G, Müller WA.. 2009. Cnidarian interstitial cells: the dawn of stem cell research. In: Rinkevich B, Matranga V, editors. Stem cells in marine organisms. Dordrecht, Heidelberg, London, New York: Springer. p. 33–59. [Google Scholar]
- Frank U, Rabinowitz C, Rinkevich B.. 1994. In vitro establishment of continuous cell cultures and cell lines from ten colonial cnidarians. Mar Biol. 120(3):491–499. [Google Scholar]
- Fuchs E, Tumbar T, Guasch G.. 2004. Socializing with the neighbors: stem cells and their niche. Cell 116(6):769–778. [DOI] [PubMed] [Google Scholar]
- Funayama N. 2010. The stem cell system in demosponges: insights into the origin of somatic stem cells. Dev Growth Differ. 52(1):1–14. [DOI] [PubMed] [Google Scholar]
- Gattazzo F, Urciuolo A, Bonaldo P.. 2014. Extracellular matrix: a dynamic microenvironment for stem cell niche. Biochim Biophys Acta. 1840:2506–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold DA, Jacobs DK.. 2013. Stem cell dynamics in cnidaria: are there unifying principles? Dev Genes Evol. 223(1–2):53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward DC, Grasso LC, Saint R, Miller DJ, Ball EE.. 2015. The organizer in evolution-gastrulation and organizer gene expression highlight the importance of Brachyury during development of the coral, Acropora millepora. Dev Biol. 399(2):337–347. [DOI] [PubMed] [Google Scholar]
- Hemmrich G, et al. 2012. Molecular signatures of the three stem cell lineages in hydra and the emergence of stem cell function at the base of multicellularity. Mol Biol Evol. 29(11):3267–3280. [DOI] [PubMed] [Google Scholar]
- Januszyk M, et al. 2015. Evaluating the effect of cell culture on gene expression in primary tissue samples using microfluidic-based single cell transcriptional analysis. Microarrays 4(4):540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecointe A, et al. 2013. Scleractinian coral cell proliferation is reduced in primary culture of suspended multicellular aggregates compared to polyps. Cytotechnology 65(5):705–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13(9):2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lirman D, et al. 2014. Growth dynamics of the threatened Caribbean staghorn coral Acropora cervicornis: influence of host genotype, symbiont identity, colony size, and environmental setting. PLoS One 9(9):e107253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall AT, Clode P.. 2004. Effects of calcium-free and low-calcium artificial seawater on polyps of a scleractinian coral Galaxea fascicularis. Coral Reefs. 23(2):277–280. [Google Scholar]
- Martin VJ, Chia FS.. 1982. Fine structure of a scyphozoan planula, Cassiopeia xamachana. Biol Bull. 163(2):320–328. [Google Scholar]
- Mass T, et al. 2012. Aragonite precipitation by ‘proto-polyps’ in coral cell cultures. PLoS One 7(4):e35049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus DQ, Magie CR, Pang K, Martindale MQ, Thomsen GH.. 2008. The Hedgehog gene family of the cnidarian, Nematostella vectensis, and implications for understanding metazoan Hedgehog pathway evolution. Dev Biol. 13(2):501–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mydlarz LD, Holthouse SF, Peters EC, Harvell CD.. 2008. Cellular responses in sea fan corals: granular amoebocytes react to pathogen and climate stressors. PLoS One 3(3):e1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesa B, Hidaka M.. 2009. High zooxanthella density shortens the survival time of coral cell aggregates under thermal stress. J Exp Mar Biol Ecol. 368(1):81–87. [Google Scholar]
- Orkin SH, Hochedlinger K.. 2011. Chromatin connections to pluripotency and cellular reprogramming. Cell 145(6):835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH.. 2011. Evolution of gene regulatory networks controlling body plan development. Cell 144(6):970–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Bermudez A, Miller DJ.. 2009. In vitro culture of cells derived from larvae of the staghorn coral Acropora millepora. Coral Reefs 28(4):859–864. [Google Scholar]
- Reyes-Bermudez A, Villar-Briones A, Ramirez-Portilla C, Hidaka M, Mikheyev AS.. 2016. Developmental progression in the coral Acropora digitifera is controlled by differential expression of distinct regulatory gene networks. Genome Biol Evol. 8(3):851–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkevich B. 2005. Marine invertebrate cell cultures: new millennium trends. Mar Biotechnol. 7:429–439. [DOI] [PubMed] [Google Scholar]
- Rinkevich B. 2011. Cell cultures from marine invertebrates: new insights for capturing endless stemness. Mar Biotechnol. 13(3):345–354. [DOI] [PubMed] [Google Scholar]
- Schmid V, Alder H.. 1984. Isolated, mononucleated, striated muscle can undergo pluripotent transdifferentiation and form a complex regenerate. Cell 38(3):801–809. [DOI] [PubMed] [Google Scholar]
- Schwaiger M, et al. 2014. Evolutionary conservation of the eumetazoan gene regulatory landscape. Genome Res. 24(4):639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, et al. 2018. Cnidarian cell type diversity and regulation revealed by whole-organism single-cell RNA-Seq. Cell 173(6):1520–1534.e20. [DOI] [PubMed] [Google Scholar]
- Shinzato C, et al. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476(7360):320–323. [DOI] [PubMed] [Google Scholar]
- Siebert S, et al. 2019. Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science 365(6451):eaav9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele RE, David CN, Technau U.. 2011. A genomic view of 500 million years of cnidarian evolution. Trends Genet. 27(1):7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuoka Y, Shinzato C, Satoh N.. 2016. The mesoderm-forming gene brachyury regulates ectoderm-endoderm demarcation in the coral Acropora digitifera. Curr Biol. 26(21):2885–2892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data for cell cultures can be found under bio-project ID PRJDB9497: BioSamples: SAMD00210801, SAMD00210803, and SAMD00210805. Experiment: DRX207005–DRX207007.