Abstract
MicroRNAs (miRNAs) have been implicated as regulatory molecules that could play a considerable role in the pathogenesis of different diseases including asthma. This work aims at exploring the role of miR-146a and miR- 106b in the pathogenesis of asthma and their association with asthma severity, IgE, and inflammatory cytokines in asthmatic children. Thirty asthmatic children and twenty age-matched healthy children aged 4-17 years old were enrolled. Expression of plasma miR-146a and miR-106b was measured using quantitative real-time PCR. Plasma levels of interleukin-5 (IL-5) and interleukin-13 (IL-13) were assessed using ELISA. Lung functions were measured by Spirometry. MiR-146a and miR-106b were significantly over-expressed in asthmatic children compared to healthy children. A significant positive correlation between total IgE and both miR-146a and miR-106b was found while no significant correlation could be detected between these miRNAs and asthma severity in asthmatic children. Plasma levels of IL-5 and IL-13 were non-significantly higher in asthmatic children compared to healthy children, and there was no significant correlation between them and both miR-146a and miR-106b expressions in the asthmatic children. The aberrant expression of immune-related miRNAs (miR-146a and miR-106b) and inflammatory cytokines (IL-5 and IL-13) among asthmatic children suggest their probable role in asthma pathogenesis.
Key Words: Asthma, microRNAs, cytokines, children, IL-5, IL-13
Bronchial asthma is considered as one of the most widespread diseases that have a significant impact on patients and their families in both developed and developing countries (1). It involves a reversible airway obstruction associated with difficult breathing and recurrent wheezes (2). Asthma symptoms may be mild, moderate, or severe (3). Insights into the pathophysiology of the inflammation leading to asthma have grown considerably in recent years. Genetic and epigenetic changes play a role in the altered immune responses within the lungs of asthmatic patients (4). Nevertheless, there remain a lot of unexplained aspects of the pathogenesis of asthma, and therefore the molecular mechanisms for the development of asthma and the genetic basis of response to existing therapies remain an area of interest (5).
MicroRNAs (miRNAs) are composed of 19 to 25 nucleotides and control referee posttrans-criptional gene silencing of target genes (6, 7). It is found that abnormalities in functions or expression of some miRNA targets are linked to pulmonary disorders, signifying their participation in these diseases' pathogenesis (8). Expression of miRNAs is not uniform in human airway smooth muscle cells (hASMCs) during an asthmatic attack, also demonstrating dynamic changes in expression in mice’s lungs during allergic airway inflammation (9, 10). In both murine and human allergic asthma, discrete plasma miRNAs are regulated differen-tially, and were related to clinical characters of patients (11), with specific types increased in the former model, namely members of the miR-146 family (miR-146a and miR-146b) (12).
Since the pathophysiology of asthma is complex and involves an interaction between both innate and adaptive immune systems and the respiratory tract lining (13), the role of miRNAs is worth studying. CD4+ T cells participate significantly in asthma, by production of interleukin (IL)-2 as well as IL-4, IL-5, IL-10, and IL-13 and interferon (IFN)-γ. Additionally, the balance between the T-helper subtypes Th1 and Th2 affects immune response; with predominance of the latter, airway inflammation and hyper-responsiveness occur (14). There have been many proven interactions between miRNAs and the immune system (15). T-lymphocyte differentiation is adjusted by miRNAs (16, 17). For example, miR-146a selectively targets regulatory T (Treg) cell–mediated suppression of TH1 responses through targeting signal transducer and activator of transcription 1 (STAT1) (18). The main pathway of innate immunity is the nuclear factor kB (NF-kB) that is targeted by numeral miRNAs, such as miR-146a, which is a NF-kB-dependent miRNA that targets the toll- like receptor 4 (TLR4) signaling pathway (19). Members of the miR-146 family have been demonstrated to down-regulate inflammatory genes expression in multiple cell types, including monocytes (19), fibroblasts (20), endothelial cells (21), airway smooth muscle (22), and epithelial cells (23).
B cell immunoglobulin isotype class switching to IgE is a tightly synchronized step under standard physiological conditions. MiRNAs are shown to have significant roles in regulation of B cell proliferation and function (24). From all the above, it becomes obvious that asthma involves an intricate balance and interaction of pathways, and miRNAs might contribute in the regulation of asthma.
Deciphering roles of miRNAs in pathogenesis of asthma can assist in discovering novel targets for therapy. Our objective was to explore the role of miR-146a and miR-106b in the pathogenesis of asthma in children, and their association with asthma severity, IgE and inflammatory cytokines levels in those patients.
Materials and methods
Study subjects
This case-control study was conducted on fifty children including 30 with asthma and 20 age-matched healthy children. Asthmatic children were recruited from the Pediatric Pulmonary Function Testing Clinic at the Medical Research Centre of Excellency, National Research Center, Egypt. They were diagnosed with asthma, and severity of asthma was classified according to guidelines of the Global Initiative for Asthma (GINA) (25). The patients received control therapy and relievers according to GINA guidelines (25) depending on their grade of asthma. Inclusion criteria of asthmatic children were ages 6-17 years of both sexes. Control group included children without a history of asthma or atopic diseases who presented to the general pediatric clinic with minor non-respiratory complaints. Exclusion criteria of asthmatic children included history of congenital heart diseases, presence of other immunologic diseases, acute upper or lower respiratory tract infections, and mental diseases. The study was in full compliance with the rules and regulations of the Medical Ethical Committee of the National Research Centre, Egypt. Approval number was 16/381.A written informed consent was obtained from all participants' parents and verbal assent was taken from the children according to ICMJE Recommendations for the Protection of Research Participants (26).
Clinical investigations
For anthropometric measurements, height was measured using Harpenden stadiometer to the nearest 0.1 cm (27). Weight was measured by Tanita scale to the nearest 0.1 kg (27). Body mass index (BMI) was calculated according to the equation: BMI= weight (kg)/height (m)2 (27).
For physical examination, measurements of lung function to confirm airflow limitations were performed by flow volume spirometry (JAEGER_MS_IOS, CareFusion, Germany), and the reversibility of bronchial obstruction was determined by a bronchodilator test using inhalation of rapid-acting bronchodilator such as 200-400 µg salbutamol. At least 3 acceptable maneuvers meeting American Thoracic Society (ATS) standards were required, with at least 2 reproducible (forced expiratory volume in one second (FEV1) and forced vial capacity (FVC) within 5% of best) maneuvers required for each test. Outcome measures of lung function included FEV1 and FVC as a percent of predicted (FEV1%
and FVC %) and FEV1 / FVC (28).
RNA extraction and quantitative real - time PCR
MiRNAs were extracted and isolated from plasma of all subjects of the study population using miRNeasy Mini kit (Qiagen, Germany) according to the manufacturer’s instructions (29). For miRNA-specific reverse transcription, miRNA was reverse-transcribed to cDNA using TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems, USA) and using specific primers according to the manufacturer’s instructions. Reverse transcription was performed under the following conditions: 30 min at 16°C, 30 min at 42°C, and followed by 5 min at 85°C. The resulting cDNA was kept at – 80°C until use.
A real-time quantitative PCR (qRT-PCR) was carried out to quantify the expression levels in triplicate of mature miR-146a and miR-106b using TaqMan® MicroRNA Assay kit and TaqMan® Universal Master Mix (Applied Biosystems, USA) using 7500 fast real-time PCR system according to the manufacturer’s instructions. RNU6B was used as endogenous control to normalize the expression levels of target miRNAs. Relative quantification (Rq) of miRNAs expression was calculated using the 2−ΔΔCT threshold cycle method. ΔCt was determined by subtracting the Ct values for RUN6B from the Ct values for the gene of interest. Q RT-PCR was performed under the following conditions: 2 min at 50°C, 10 min at 95°C, followed by 50 cycles at 95°C for 15 s and at 60°C for 1 min (30).
Enzyme linked immunosorbent assay (ELISA)
Plasma IL-5 and IL-13 levels of all study subjects were determined using Human IL-5 and IL-13 ELISA kits (Bioneovan Co., Ltd., Beijing, China) according to the manufacturer’s protocol.
Statistical analysis
Data were statistically analyzed using SPSS version 16.0 software (SPSS Inc., Chicago, Illinois, USA). Independent samples t-test was used to compare gene expression levels of miRNAs between groups. Non-parametric Mann-Whitney U test was used for comparing IL-5 and IL-13 levels between groups. Association between miRNAs expression and age, total IgE, and disease severity of patients was tested using Pearson’s rank correlation while spearman correlation was used to examine the association of miRNAs expression with IL-5 and IL-13 in asthmatic patients. Data were presented as mean ± SEM. A P value of less than 0.05 was considered statistically significant.
Results
Clinical features of asthmatic patients
According to the results of total IgE, the included patients were non-atopic. Among asthmatic children, 9 were intermittent, 10 were mild persistent asthmatic, and 11 were moderate persistent asthmatic. The patients had normal FEV1/FVC due to appropriate treatment protocol as they were well controlled with therapy. The clinical characteristics of asthmatic patients and controls are summarized in Table 1.
Table 1.
The clinical and laboratory characteristics of asthmatic patients and healthy children
| Characteristics | Asthmatic patients | Controls |
|---|---|---|
| Number of cases | 30 | 20 |
| Gender (male/female) | 14/16 | 8/12 |
| Age (years) | 10.63±0.69* | 10.95±0.81* |
| Height (cm) | 137.6±4.5* | 120±5.13* |
| Weight (Kg) | 37.1±3.7* | 48.7±5.7* |
| BMI | 18.7±1* | 32.7±3* |
| pre-FEV1 | 92.5±4.84* | - |
| post-FEV1 | 99.56±4.9* | - |
| pre-FEV1/FVC | 96.2±2.7* | - |
| post-FEV1/FVC | 99.59±1.98* | - |
| TLC, mean±S.E. (10 3 /L) | 7.3±0.46* | - |
| Hb, mean±S.E. (g/dL) | 12.6±0.34* | - |
| Total IgE (IU/mL) | 98.9±27* | - |
| Medications ( corticosteroid s/ beta2- agonist / leukotriene receptor antagonist ), % of patients | 100% | - |
*: Mean±S.E. BMI: Body mass index; FEV1: Forced expiratory volume in one second; FVC: Forced vital capacity; TLC: Total leucocytic count; Hb: Hemoglobin.
The expression pattern of miRNAs and levels of the inflammatory cytokines in plasma of the study population
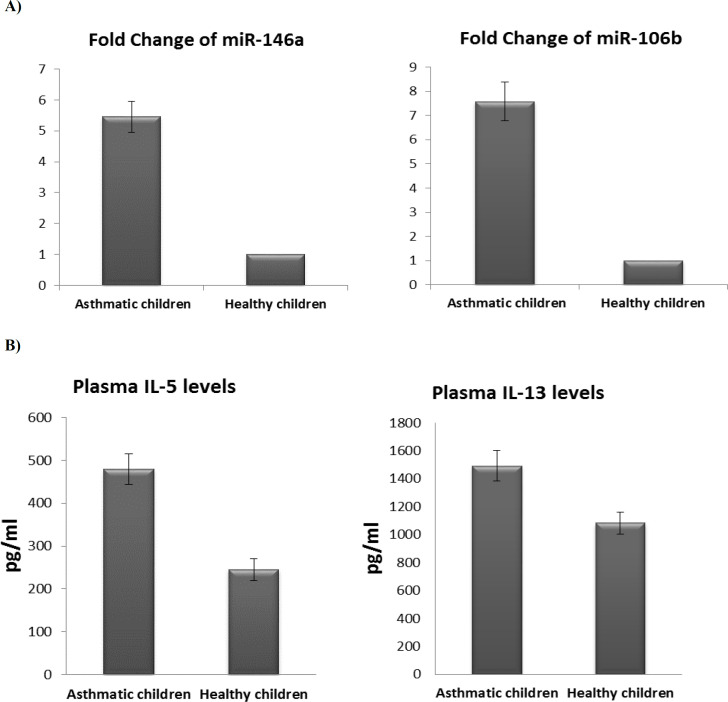
Our findings demonstrated that miR-146a was significantly over-expressed in children with asthma in comparison with healthy controls (P <0.05), with expression levels 5.46-fold higher in asthmatic patients than normal controls (Figure 1A). In addition, our results revealed that the expression pattern of miR-106b was up-regulated in patients with asthma in comparison with healthy control (P <0.05). The fold change of miR-106b expression was 7.58 in asthmatic patients relative to normal controls (Figure 1A).
Fig. 1.
Expression of miRNAs and levels of the inflammatory cytokines in asthmatic and normal children. A) Fold change of miR-146a and miR-106b in asthmatic children relative to healthy controls. Bars show the mean ± SEM fold change. (**: significant, P < 0.05 versus controls, by independent samples t-test); B) Plasma levels of IL-5 and IL-13 in asthmatic and healthy children are shown. Bars show the results as the mean ± SEM. (non-significant, P> 0.05 versus controls, by non-parametric Mann-Whitney U test)
Furthermore, plasma levels of IL-5 and IL-13 were non-significantly higher in asthmatic children compared to healthy children (P > 0.05) (Figure 1B).
Correlation of miRNAs expression with pre-FEV 1 and post-FEV 1 in asthmatic children
A significant positive correlation was noticed between miR-146a and both pre-FEV1 and post-FEV1 in asthmatic children. In addition, miR-106b was highly significantly correlated with pre-FEV1 in asthmatic children (Table 2).
Table 2.
Correlation between miR-146a and miR-106b and pre-FEV1, pre-FEV1/FVC, post-FEV1, and post-FEV1/FVC
| Parameters | Pearson Correlation | P value |
|---|---|---|
| miR-106b expression ~ post-FEV1/FVC | -0.060 | 0.406 |
| miR-146a expression ~ pre-FEV1 | 0.501* | 0.017 |
| miR-146a expression ~ pre-FEV1/FVC | 0.045 | 0.429 |
| miR-146a expression ~ post-FEV1 | 0.412* | 0.045 |
| miR-146a expression ~ post-FEV1/FVC | -0.234 | 0.175 |
| miR-106b expression ~ pre-FEV1 | 0.550* | 0.009 |
| miR-106b expression ~ pre-FEV1/FVC | 0.275 | 0.135 |
| miR-106b expression ~ post-FEV1 | 0.227 | 0.182 |
*Correlation is significant at the 0.05 level (1-tailed).
Correlation of miRNAs expression with the disease activity, total IgE, and age of asthmatic children
As noted in Table 3, no significant correlation was found between miR-146a and miR-106b expression and asthma severity in asthmatic children while a significant positive correlation was observed between total IgE and expressions of both miR-146a and miR-106b in asthmatic children (P <0.05). In addition, a significant negative correlation was revealed between miR-146a expression and age in asthmatic children (P < 0.05) (Table 3).
Table 3.
Correlation of miR-146a and miR-106b expression with asthma severity, total IgE, and age in asthmatic children
| Asthma severity | Total IgE | Age | ||
|---|---|---|---|---|
| miR-146a | Pearson correlation (r) | 0.312 | 0.645* | 0.543*- |
| P value | 0.104 | 0.012 | 0.010 | |
| miR-106b | Pearson correlation (r) | 0.075 | 0.520* | -0.359 |
| P value | 0.384 | 0.041 | 0.072 | |
*Correlation is significant at the 0.05 level (1-tailed).
Correlation of miRNAs expression and the levels of inflammatory cytokines in asthmatic children
There was no significant correlation between miR-146a and miR-106b expression and plasma levels of inflammatory cytokines (IL-5 and IL-13) in asthmatic children (Table 4).
Table 4.
Correlation of miR-146a and miR-106b expression with IL-5 and IL-13 levels in asthmatic children
| IL-5 | IL-13 | ||
|---|---|---|---|
| miR-146a | Spearman correlation (r) | 0.098 | -0.154 |
| P value | 0.354 | 0.277 | |
| miR-106b | Spearman correlation (r) | -0.010 | -0.294 |
| P value | 0.485 | 0.126 | |
*Correlation is significant at the 0.05 level (1-tailed).
Discussion
MiRNAs have critical roles in regulation of genes and multiple gene pathways. Many studies have demonstrated that there may be an association between miRNAs expression and asthma pathogenesis, as well as with lung development and homeostasis (5). Asthma is one of the best examples of complex genetic diseases, and the significance of epigenetic and pharmacogenetic studies is a cornerstone to developing new treatment modalities.
Our study showed that the miR-146a was appreciably over-expressed in asthmatic children compared to healthy controls. This is in line with other studies which found that miR-146a was upregulated in plasma and splenic CD4 T lymphocytes amongst asthmatic patients and murine models of acute asthma, therefore postulating that miR-146a is a proinflammatory factor in asthma (31-33). This is in agreement with a previous study finding that knockout of miR-146a in mice resulted in abnormal T cell activity in inflammation (34). Down-regulation of miR-146a expression that follows treatment with dexamethasone points to the contribution of miR-146a in the process of airway inflammation in asthma (33).
In addition, our results showed that miR-106b was notably upregulated in patients with asthma in comparison with healthy controls. This is in agreement with Specjalski et al. (35) who postulated the possible role of some miRNAs including miR-106 in the pathogenesis of allergic inflammation. Their study explored the up-regulation of miR-106 in blood samples of asthmatic patients, and demonstrated its decrease after completing the buildup phase of Wasp venom immunotherapy. On the other hand, Tang et al. (36) stated that miR-106b expression was much inhibited in OVA-activated murine bone marrow derived dendritic cells (BMDCs). Transfection with a miR-106b inhibitor decreased the production of IL-12 in activated cultured BMDCs and promoted Th2 polarization afterwards (36). Few studies have addressed miR-106b expression regarding airway inflammation, and thus the regulatory role of miR-106b in the inflammatory response remains unclear.
Asthma is a TH2 -driven inflammation resulting in raised TH2 cytokine levels (IL4, IL-5, IL-9, and IL-13) and reduction of anti-inflammatory cytokines (e.g., IL-10) (37). Our study showed that plasma levels of IL-5 and IL-13 were non-significantly higher in asthmatic children in comparison with controls, with no correlation between expression of miR-146a and miR-106b and IL-5 and IL-13. Similarly, Hammad et al. (2018) found no correlation between mir-146a and IL-13 (32). Differentially expressed miRNAs could be essential regulators of Th2 cytokines (38). In contrast, Tang et al. (36) found that miR-106b might play a negative role in Th2 polarization. MiR-146a has been postulated to be necessary for regulatory T (Treg) cells-mediated suppression of TH1, yet not TH2, response. Therefore, miR-146a up regulation as in asthma may potentially boost the Treg cell-mediated suppression of TH1 responses, and results in unopposed TH2 activation (39, 40).
In our study, a significant positive correlation was observed between total IgE and expressions of both miR-146a and miR-106b in asthmatic children. Our findings are consistent with Li et al. (41) who demonstrated that miR-146a increases B cell antibody secretion with IgE class switching via up regulation of 14-3-3σ expression.
We found a lack of significant correlation
between miR-146a and miR-106b expression and severity of asthma. To our knowledge, our research is the first study to find the association among miR-146a and mi-R106b expression and severity of childhood asthma. Similarly, lung biopsy specimens did not indicate different miR-146a expression in patients with mild asthma (42) but showed minor impact on miRNA expression in patients with severe asthma (43). It is likely that the inflammatory changes were too mild, and that changes in expression of miRNA may be more obvious in patients with more severe asthma. Another explanation is that the samples were not taken during an incident of asthma exacerbation.
Our results showed a significant positive correlation between miR-146a in addition to miR-106b and pre-bronchodilator FEV1% and miR-146a only with post- bronchodilator FEV1% in asthmatic children. Also, Hammad et al. found a positive association between miR-146a relative expression and FEV1 (32). A previous study (44) demonstrated a significant positive correlation between miR-106b-5p and FVC%, as well as FEV1/FVC and a lesser number with FEV1% and FVC%, supporting the assumption that miRNAs might affect asthma vulnerability directly through targeting genes and cellular processors which may be proposed in enhancing obstruction of airflow (44).
In conclusion, our study revealed that circulating miR-146a and miR106b were significantly over-expressed in asthmatic children. A significant positive correlation between total IgE and both miR-146a and miR-106b was found while no significant correlations could be detected between these miRNAs and asthma severity in those children. Furthermore, plasma levels of IL-5 and IL-13 were non-significantly higher in asthmatic children compared to healthy children, and there was no significant correlation between them and miR-146a and miR-106b expressions in the asthmatic children. The contribution of miRNAs to the molecular mechanisms in asthma could represent a new therapeutic target in childhood asthma and warrant further study.
Acknowledgment
This study was supported and funded by the National Research Centre, Giza, Egypt.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Hall SC, Agrawal DK. Vitamin D and Bronchial Asthma: An Overview of Data From the Past 5 Years. Clin Ther. 2017;39:917–29. doi: 10.1016/j.clinthera.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18:716–25. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 3.Walter H, Sadeque-Iqbal F, Ulysse R, et al. Effectiveness of school-based family asthma educational programs in quality of life and asthma exacerbations in asthmatic children aged five to 18: a systematic review. JBI Database System Rev Implement Rep. 2016;14:113–38. doi: 10.11124/JBISRIR-2016-003181. [DOI] [PubMed] [Google Scholar]
- 4.Maes T, Cobos FA, Schleich F, et al. Asthma inflammatory phenotypes show differential microRNA expression in sputum. J Allergy Clin Immunol. 2016;137:1433–46. doi: 10.1016/j.jaci.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 5.Midyat L, Gulen F, Karaca E, et al. MicroRNA expression profiling in children with different asthma phenotypes. Pediatr Pulmonol. 2016;51:582–7. doi: 10.1002/ppul.23331. [DOI] [PubMed] [Google Scholar]
- 6.Winter J, Jung S, Keller S, et al. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 7.Christodoulou F, Raible F, Tomer R, et al. Ancient animal microRNAs and the evolution of tissue identity. Nature. 2010;463:1084–8. doi: 10.1038/nature08744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cushing L, Jiang Z, Kuang P, et al. The roles of microRNAs and protein components of the microRNA pathway in lung development and diseases. Am J Respir Cell Mol Biol. 2015;52:397–408. doi: 10.1165/rcmb.2014-0232RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collison A, Mattes J, Plank M, et al. Inhibition of house dust mite-induced allergic airways disease by antagonism of microRNA-145 is comparable to glucocorticoid treatment. J Allergy Clin Immunol. 2011;128:160–7 e4. doi: 10.1016/j.jaci.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Perry MM, Baker JE, Gibeon DS, et al. Airway smooth muscle hyperproliferation is regulated by microRNA- 221 in severe asthma. Am J Respir Cell Mol Biol. 2014;50:7–17. doi: 10.1165/rcmb.2013-0067OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milger K, Gotschke J, Krause L, et al. Identification of a plasma miRNA biomarker signature for allergic asthma: A translational approach. Allergy. 2017;72:1962–71. doi: 10.1111/all.13205. [DOI] [PubMed] [Google Scholar]
- 12.Garbacki N, Di Valentin E, Huynh-Thu VA, et al. MicroRNAs profiling in murine models of acute and chronic asthma: a relationship with mRNAs targets. PLoS One. 2011;6:e16509. doi: 10.1371/journal.pone.0016509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishmael FT. The inflammatory response in the pathogenesis of asthma. J Am Osteopath Assoc. 2011;111:S11–7. [PubMed] [Google Scholar]
- 14.White SR, Loisel DA, Stern R, et al. Human leukocyte antigen-G expression in differentiated human airway epithelial cells: lack of modulation by Th2-associated cytokines. Respir Res. 2013;14 doi: 10.1186/1465-9921-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 16.Rebane A, Akdis CA. MicroRNAs: Essential players in the regulation of inflammation. J Allergy Clin Immunol. 2013;132:15–26. doi: 10.1016/j.jaci.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 17.Kai W, Qian XU, Qun WU. MicroRNAs and Asthma Regulation. Iran J Allergy Asthma Immunol. 2015;14:120–5. [PubMed] [Google Scholar]
- 18.Lu LF, Boldin MP, Chaudhry A, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–29. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taganov KD, Boldin MP, Chang KJ, et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato T, Liu X, Nelson A, et al. Reduced miR-146a increases prostaglandin E(2)in chronic obstructive pulmonary disease fibroblasts. Am J Respir Crit Care Med. 2010;182:1020–9. doi: 10.1164/rccm.201001-0055OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng HS, Sivachandran N, Lau A, et al. MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways. EMBO Mol Med. 2013;5:1017–34. doi: 10.1002/emmm.201202318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larner-Svensson HM, Williams AE, Tsitsiou E, et al. Pharmacological studies of the mechanism and function of interleukin-1beta-induced miRNA-146a expression in primary human airway smooth muscle. Respir Res. 2010;11:68. doi: 10.1186/1465-9921-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perry MM, Moschos SA, Williams AE, et al. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol. 2008;180:5689–98. doi: 10.4049/jimmunol.180.8.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Wan Y, Ji Q, et al. The role of microRNAs in B-cell development and function. Cell Mol Immunol. 2013;10:107–12. doi: 10.1038/cmi.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.(GINA) GIfA. Global Strategy for Asthma Management and Prevention. 2019. Available from: www.ginasthma.org.
- 26.ICMJE. Recommendations for the Protection of Research Participants. 2019. [Google Scholar]
- 27.de Onis M, Onyango AW, Borghi E, et al. Development of a WHO growth reference for school-aged children and adolescents. Bull World Health Organ. 2007;85:660–7. doi: 10.2471/BLT.07.043497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller A. Lung function testing: selection of reference values and interpretative strategies. Am Rev Respir Dis. 1992;146:1368–9. doi: 10.1164/ajrccm/146.5_Pt_1.1368b. [DOI] [PubMed] [Google Scholar]
- 29.Abdel Raouf H, Kholoussi NM, Eissa E, et al. MicroRNAs as Immune Regulators of Inflammation in Children with Epilepsy. Int J Mol Cell Med. 2020;9:188–97. doi: 10.22088/IJMCM.BUMS.9.3.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amr KS, Bayoumi FS, Eissa E, et al. Circulating microRNAs as potential non-invasive biomarkers in pediatric patients with celiac disease. Eur Ann Allergy Clin Immunol. 2019;51:159–64. doi: 10.23822/EurAnnACI.1764-1489.90. [DOI] [PubMed] [Google Scholar]
- 31.Panganiban RP, Wang Y, Howrylak J, et al. Circulating microRNAs as biomarkers in patients with allergic rhinitis and asthma. J Allergy Clin Immunol. 2016;137:1423–32. doi: 10.1016/j.jaci.2016.01.029. [DOI] [PubMed] [Google Scholar]
- 32.Hammad Mahmoud Hammad R, Hamed D, Eldosoky M, et al. Plasma microRNA-21, microRNA-146a and IL-13 expression in asthmatic children. Innate Immun. 2018;24:171–9. doi: 10.1177/1753425918763521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng MJ, Shi F, Qiu C, et al. MicroRNA-181a, -146a and -146b in spleen CD4+ T lymphocytes play proinflammatory roles in a murine model of asthma. Int Immunopharmacol. 2012;13:347–53. doi: 10.1016/j.intimp.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Omran A, Elimam D, Yin F. MicroRNAs: new insights into chronic childhood diseases. Biomed Res Int. 2013;2013:291826. doi: 10.1155/2013/291826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Specjalski K, Maciejewska A, Pawlowski R, et al. Changes in the Expression of MicroRNA in the Buildup Phase of Wasp Venom Immunotherapy: A Pilot Study. Int Arch Allergy Immunol. 2016;170:97–100. doi: 10.1159/000447637. [DOI] [PubMed] [Google Scholar]
- 36.Tang H, Jiang H, Zheng J, et al. MicroRNA-106b regulates pro-allergic properties of dendritic cells and Th2 polarisation by targeting early growth response-2 in vitro. Int Immunopharmacol. 2015;28:866–74. doi: 10.1016/j.intimp.2015.03.043. [DOI] [PubMed] [Google Scholar]
- 37.Greene CM, Gaughan KP. microRNAs in asthma: potential therapeutic targets. Curr Opin Pulm Med. 2013;19:66–72. doi: 10.1097/MCP.0b013e32835a5bc8. [DOI] [PubMed] [Google Scholar]
- 38.Pinkerton M, Chinchilli V, Banta E, et al. Differential expression of microRNAs in exhaled breath condensates of patients with asthma, patients with chronic obstructive pulmonary disease, and healthy adults. J Allergy Clin Immunol. 2013;132:217–9. doi: 10.1016/j.jaci.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 39.Dong X, Zhong N. Pathogenic Roles of MicroRNA in the Development of Asthma. Asthma and Lung Biology: IntechOpen. 2019 [Google Scholar]
- 40.Lu TX, Rothenberg ME. Diagnostic, functional, and therapeutic roles of microRNA in allergic diseases. J Allergy Clin Immunol. 2013;132:3. doi: 10.1016/j.jaci.2013.04.039. 13; quiz 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li F, Huang Y, Huang YY, et al. MicroRNA-146a promotes IgE class switch in B cells via upregulating 14-3-3sigma expression. Mol Immunol. 2017;92:180–9. doi: 10.1016/j.molimm.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 42.Williams AE, Larner-Svensson H, Perry MM, et al. MicroRNA expression profiling in mild asthmatic human airways and effect of corticosteroid therapy. PLoS One. 2009;4:e5889. doi: 10.1371/journal.pone.0005889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suojalehto H, Lindstrom I, Majuri ML, et al. Altered microRNA expression of nasal mucosa in long-term asthma and allergic rhinitis. Int Arch Allergy Immunol. 2014;163:168–78. doi: 10.1159/000358486. [DOI] [PubMed] [Google Scholar]
- 44.Kho AT, Sharma S, Davis JS, et al. Circulating MicroRNAs: Association with Lung Function in Asthma. PLoS One. 2016;11:e0157998. doi: 10.1371/journal.pone.0157998. [DOI] [PMC free article] [PubMed] [Google Scholar]