Significance
Research on the chemical origin of life comprises one of the most exciting topics in contemporary science. Prebiotic chemistry provided evidence that precursors of both nucleic acids and proteins might be formed in the prebiotic environment. Yet, studies on nonenzymatic replication—a central mechanism driving chemical evolution—focused largely on each class of these molecules separately. This paper reveals a successful attempt to replicate simple nucleopeptide chimeras. Most importantly, different mechanisms control the replication of complementary chimeras, leading to a clear selection of one over the other. We propose that related processes may have led to the emergence of the first functional nucleic-acid–peptide assemblies, which further developed into biological assemblies such as the ribosomes and viruses.
Keywords: chemical evolution, nucleic-acid–peptide conjugates, self-replication, molecular networks
Abstract
Many fundamental cellular and viral functions, including replication and translation, involve complex ensembles hosting synergistic activity between nucleic acids and proteins/peptides. There is ample evidence indicating that the chemical precursors of both nucleic acids and peptides could be efficiently formed in the prebiotic environment. Yet, studies on nonenzymatic replication, a central mechanism driving early chemical evolution, have focused largely on the activity of each class of these molecules separately. We show here that short nucleopeptide chimeras can replicate through autocatalytic and cross-catalytic processes, governed synergistically by the hybridization of the nucleobase motifs and the assembly propensity of the peptide segments. Unequal assembly-dependent replication induces clear selectivity toward the formation of a certain species within small networks of complementary nucleopeptides. The selectivity pattern may be influenced and indeed maximized to the point of almost extinction of the weakest replicator when the system is studied far from equilibrium and manipulated through changes in the physical (flow) and chemical (template and inhibition) conditions. We postulate that similar processes may have led to the emergence of the first functional nucleic-acid–peptide assemblies prior to the origin of life. Furthermore, spontaneous formation of related replicating complexes could potentially mark the initiation point for information transfer and rapid progression in complexity within primitive environments, which would have facilitated the development of a variety of functions found in extant biological assemblies.
The rich, highly efficient, and specific biochemistry in living cells is orchestrated by molecules belonging to a small number of families, primarily nucleic acids, proteins, fatty acids, and sugars. Many fundamental cellular and viral functions, including replication and translation, are facilitated by synergistic activity in complexes of these molecules, very often involving nucleic acids (DNA, RNA, or their constituent nucleotides/nucleobases) and proteins (or peptides/amino acids). Among the most important examples of such complexes are the nucleosome (which comprises DNA packaging units in eukaryotes), the ribosome (which translates RNA sequences into proteins), and amino acid–charged transfer RNA (t-RNA) conjugates (which are exploited during translation) (1–4). In order to harness such synergistic activity in synthetic materials, several groups (including the authors) have recently studied the coassembly of nucleic acids with (often) positively charged peptides or the self-assembly of premade nucleic-acid–peptide (NA–pep) chimeras (5–12). It is expected that such assemblies could produce new materials for various applications, such as autocatalysis, electron transfer, tissue scaffolding, and (drug) delivery (13–18). Intriguingly, the NA–pep assemblies combine “digital” molecular information for the hybridization of nucleic acids with “analog” instructions that affect peptide aggregation and, as such, are expected to show superior behavior in comparison with related nucleic-acid–only or peptide-only assemblies (19–21).
We now propose that alongside the development of NA–pep conjugate assemblies for new materials, an analysis of the formation of chimeras within complex mixtures, and particularly the selection of specific sequences through replication processes, will offer insight into their emergence in the early chemical evolution. Indeed, several studies have indicated that evolution in prebiotic environments, toward the origin of life, must have involved cooperative interactions among diverse classes of molecules (22–25). Other studies, including the seminal works of Eigen (26) and Kauffman (27), have revealed the possible emergence of synergistic activity in prebiotic autocatalytic networks and, as a consequence, phase transitions toward beneficial cooperative and/or selective behavior (28, 29). Importantly, while it has been shown that highly complex functions emerge by wiring together multiple pathways—driving, for example, elaborate feedback loops—our studies, as well as others, have indicated that multiple unique dynamic features (30–36), including chemical computation (37), can be developed in relatively small networks.
Despite strong evidence for prebiotic pathways that yield nucleobases and peptides—suggesting that molecules of both families were indeed present in early chemical evolution—prebiotic chemistry research has focused largely on studying each class of molecule separately (38). This approach has led to incomplete discussions on the “RNA World,” the “Peptide World,” or the “Metabolism-First World,” with minimal overlap between the different domains. In particular, studies on replicating molecules and replication networks have investigated discrete systems affected only by a single class of molecule—whether nucleic acids, peptides, lipid amphiphiles, or small organic molecules (39–41). Herein, we propose to blur the limits between families of replicators by combining nucleic-acid molecular genetic information with peptide-based assembly “phenotypes.” To this end, we sought to reveal the self-organization and selection processes taking place in mixtures containing short complementary nucleopeptide conjugates (designated RAA and RTT, Fig. 1). Experimental and simulation analyses of the template-directed replication processes within these networks clearly demonstrate that product formation is governed both by nucleobase hybridization and by the formation of unique supramolecular architectures by each of the nucleopeptide conjugates. Unequal assembly-dependent replication capacity induces selectivity toward the formation of RAA. By studying the system in a flow reactor, namely, at far-from-equilibrium conditions, we show how this selectivity can be influenced and maximized—through changes in the physical (flow) and chemical (template and inhibition) conditions—to the point of almost complete extinction of RTT, the weaker replicator. We suggest that, prior to the origin of life, processes such as these may have led to the emergence of simple functional NA–pep assemblies (42), which could facilitate further structural development into the current cellular NA–pep assemblies.
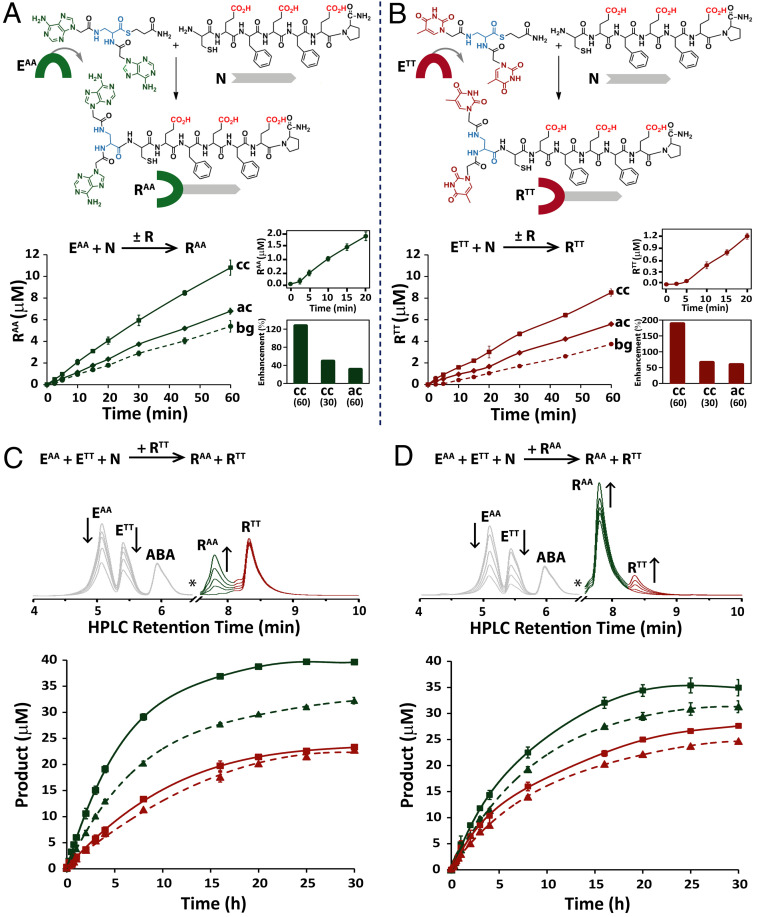
Fig. 1.
Nucleopeptide replication networks. (A and B) NCL reactions forming the RAA and RTT conjugates from their respective electrophile and nucleophile precursors and time-dependent formation of these conjugates in template-free (bg), autocatalytic (ac), and cross-catalytic (cc) reactions. Note that at pH 7.4, the Glu side chain carboxylic acids would be in their deprotonated anionic form. Reactions were carried out with 100 µM EAA or ETT and 100 µM N, either in the absence of a template (bg) or when seeded with the designated amount of template at initiation (ac/cc). Insets show the early stages of the background reactions, highlighting the lag phase typical of product formation through autocatalysis (Top insets) and the rate enhancement (percent) in cross-catalytic reactions seeded with 60 or 30 µM template or in autocatalytic reactions seeded with 60 µM template (Bottom insets). (C and D) A time-dependent analysis of the replicator-assisted product formation of the conjugates RAA (green) and RTT (red) in network reactions initiated with EAA (50 µM), ETT (50 µM), and N (100 µM) and seeded with 20 (dashed lines) or 60 μM (solid lines) RTT (C) or RAA (D). HPLC chromatograms (Top) indicate the increase in RAA and RTT product over time in representative reactions seeded with 60 μM RTT (C) or 60 μM RAA (D); note that RTT and RAA peaks have initial intensity due to seeding (in C and D, respectively), and the * symbols denote minor (≥15%) branched product peaks, removed for clarity (see also SI Appendix, Fig. S20). All reactions were carried out in duplicate, in Hepes buffer (pH 7.4), in the presence of TCEP as a reducing agent (5 mM) and with ABA (30 μM) as the internal standard. Data were acquired by HPLC analysis of aliquots collected at the designated times (SI Appendix, Figs. S17–S20).
Results and Discussion
In nonenzymatic replication, the replicating molecules serve as both templates and catalysts. These replicators either enhance synthesis of their own copies (i.e., self-replication) or participate in reciprocal cross-catalytic cycles with molecules containing complementary structures (39, 40, 43, 44). The most recent studies have focused on harnessing nonlinear replication processes to control information transfer in synthetic networks incorporating multiple reaction pathways and thereby to induce selective product distribution (45–48). Both the replicators studied here, namely, RAA and RTT, are readily produced by native chemical ligation (NCL) of the respective di-nucleobase thioester electrophile, EAA or ETT, with a Cys-containing nucleophilic peptide N (Fig. 1 A and B). These replicators and precursor molecules were synthesized in-house and characterized by NMR, high-performance liquid chromatography (HPLC), and mass spectrometry (SI Appendix, section S2 and Figs. S1–S16).
We started our investigation of RAA and RTT by analyzing their formation in template-free (background) reactions and their replication capacity through either autocatalysis (which is enhanced by seeding at initiation with the respective product as the template) or cross-catalysis (which is enhanced by seeding with the complementary replicator) (Fig. 1 A and B). The background reactions started slowly, but after a lag phase, they progressed at higher rates, revealing a clear signature of autocatalysis (Fig. 1 A and B, Insets). The rates of the template-seeded reactions were higher than the background reactions, and the replication of both RAA and RTT via cross-catalysis pathways was more efficient than that via autocatalysis pathways, potentially indicating the role of complementarity between the two nucleobases for templating the conjugate replication. For RAA formation, the initial rate enhancement via cross-catalysis with RTT (60 μM) was about 125%, which is ∼4-times higher than the rate enhancement observed for its formation via autocatalysis (Fig. 1 A, Inset). Similarly, the rate enhancement of RTT formation via cross-catalysis with RAA (60 μM) was close to 190% (i.e., ∼3-times higher than that for RTT formation via autocatalysis) (Fig. 1 B, Inset). As anticipated for template-assisted reactions, the rate enhancement increased with an increase in the amount of seeded catalyst. Remarkably, the autocatalytic ligation reaction that forms the (potential) replicator containing single adenine and single thymine bases, RAT, was not enhanced versus the respective background reaction (SI Appendix, Fig. S19), empirically signifying the utility of the homodinucleobase motif as a minimal unit for recognition and replication. This finding might explain the lack of sequence specificity recently observed for the replication of conjugates containing a peptide attached to single nucleobase units (49). Since in such a study, the interactions between nucleobases did not play a role in the assembly and replication steps, we now propose that establishing efficient information transfer through base-pairing interactions is an additional requirement to get sequence specificity.
To search for systemic emergent behavior (50), we then studied the binary network formed by RAA and RTT by monitoring their simultaneous production in template-free and template-seeded reaction mixtures containing EAA, ETT, and a stoichiometric amount of N (Fig. 1 C and D and SI Appendix, Fig. S20). In the template-free reaction, the two replicators were formed at similar rates in the early stages, but in the later stages, a moderately larger amount of RAA versus RTT was produced (SI Appendix, Fig. S20F), indicating enrichment of the former via catalytic processes involving the newly formed product/template molecules. Significantly, despite the faster production of both RTT and RAA via cross-catalysis in their separate reaction mixtures (Fig. 1 A and B), seeding with either RTT or RAA resulted in a clear selectivity toward enhanced production of RAA (Fig. 1 C and D). After 4 h of the (60 µM) RTT-seeded reaction, RAA formation was ∼150% higher than RTT formation, whereas after 4 h in the (60 µM) RAA-seeded reaction, formation of RAA was ∼50% higher than that of RTT. The preference for RAA formation in both the autocatalysis and cross-catalysis reaction pathways was found to be concentration dependent, as shown by the higher rate enhancement observed for reactions seeded with 60 μM versus 20 μM of the templates. The enhanced production of RAA reflects asymmetry in the binary network, which could originate from the propensity of RAA and RTT to assemble into distinctly different supramolecular architectures (Fig. 2); this propensity, in turn, confers dissimilar templating capacities toward condensation of the respective building blocks.
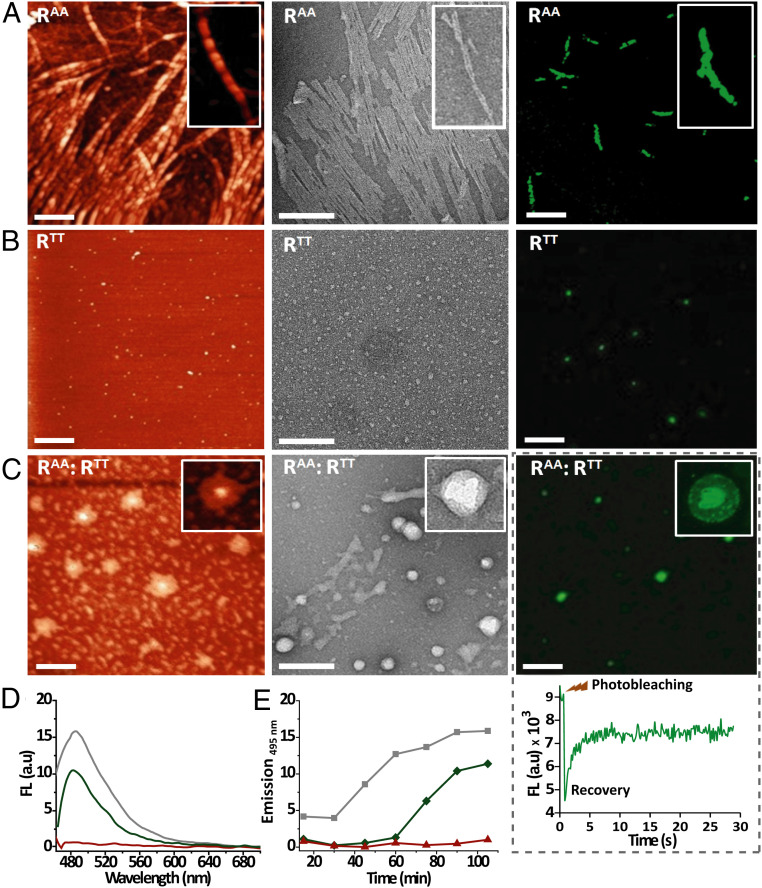
Fig. 2.
RAA and RTT self-assembled nanostructures. (A–C) AFM (Left), TEM (Middle), and fluorescence microscopy (Right) images obtained for assemblies of 60 μM RAA, 60 μM RTT, and equimolar (30 μM each) mixtures of RAA and RTT (RAA:RTT) (Scale bars: AFM, 500 nm; TEM, 200 nm; fluorescence microscopy, 10 µm.) TEM samples were stained with uranyl acetate and fluorescence microscopy samples with ThT. The inset in the right-hand side of C shows the FRAP in the liquid droplets. (D) Emission spectra observed following ThT binding to RAA (green line), RTT (red line), and RAA:RTT (gray line). (E) Kinetics of ThT binding to RAA (green), RTT (red), and RAA:RTT (gray). Experiments were performed in buffer (pH 7.4) at room temperature.
As subtle changes in the sequence or assembly conditions of NA–pep conjugates can lead to the formation of significantly different supramolecular architectures (5, 6), the assembly structures of the replicators RAA and RTT (60 μM)—in separate solutions or when mixed together—were subsequently studied in a set of microscopy and spectroscopy assays (Fig. 2). Atomic force microscopy (AFM) and transmission electron microscopy (TEM) images of RAA revealed (bundled) fiber architectures, 7 to 8 nm wide and up to a few microns long (Fig. 2A). In contrast, images of RTT showed small (diameter ≤ 15 nm) spherical aggregates (Fig. 2B). Staining of the two types of assembly with thioflavin T (ThT) and emission analysis using ensemble fluorescence spectroscopy (Fig. 2 D and E) and confocal microscopy (Fig. 2 A and B, Right) revealed β-sheet–rich domains and stacked nucleobases. Significant characteristic emission was observed for the stained RAA fibers, but only weak emission signals were detected after staining the RTT spherical assemblies, thus further underlining the substantial differences between the molecular arrangements within the two aggregates. Interestingly, the AFM, TEM, and fluorescence microscopy images obtained for equimolar mixtures of RAA and RTT (RAA:RTT, 30 μM each) showed yet another morphology, in which more condensed matter at the center of the assembly was contained within an environment of lower density aggregates, presumably liquid droplets with diameters ranging roughly from 35 to 75 nm (Fig. 2C and SI Appendix, Fig. S22). We then performed fluorescence recovery after photobleaching (FRAP) experiments to determine the diffusion kinetics of ThT in and out of these liquid droplets. Individual droplets were photobleached and imaged for fluorescence recovery over 30 s (Fig. 2C), while other (control) droplets were imaged without bleaching (SI Appendix, Fig. S22C). Significant recovery of fluorescence indicated the propensity of the droplet-like assemblies to allow in/out ThT diffusion, confirming their formation through liquid–liquid phase separation processes, similarly to other synthetic and cellular aggregates (51).
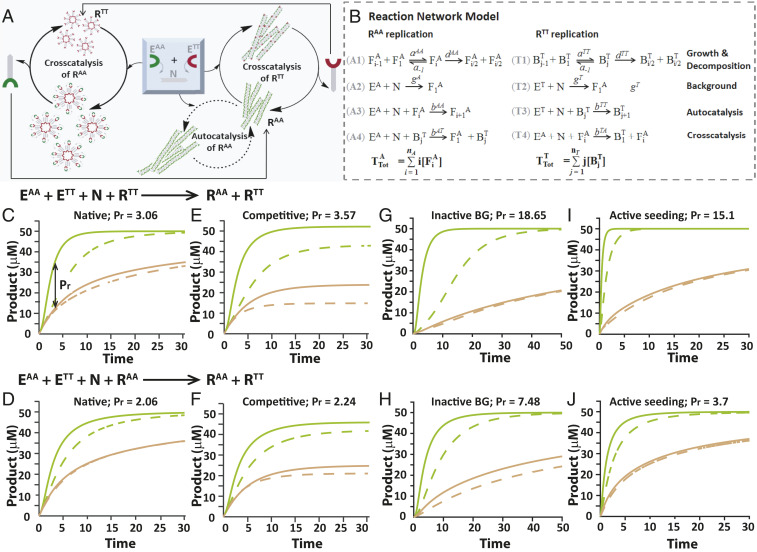
The kinetics of product formation in the individual replication reactions and within the small networks (Fig. 1), taken together with the structural information obtained for RAA and RTT assemblies (Fig. 2), allowed us to draw up the asymmetric network topology that selectively yields higher amounts of RAA versus RTT (Fig. 3A). Based on previous studies with assembly-dependent replication systems (47, 52), we expected the newly formed spheres and fibers to be the dominant catalytic entities. These assemblies can facilitate the association of the precursor molecules at the assembly edges or points of imperfection and consequently enhance their ligation. The network asymmetry emerges because while both RAA and RTT form via cross-catalysis—templated by spheres and fibers, respectively—RAA also replicates in a “selfish” autocatalytic pathway enhanced by the fibers. Moreover, the cross-catalytic pathway leading to the formation of RAA is more efficient than the reciprocal pathway forming RTT (Fig. 1 C and D). Based on this network topology, we constructed a kinetic model representing the multiple active reaction steps: 1) assembly of RAA and RTT into catalytic species and their spontaneous disassembly into smaller aggregates featuring a higher number of catalytic edges (53), 2) background ligations, 3) template-assisted autocatalysis, and 4) template-assisted cross-catalytic reactions (Fig. 3B). This model enabled us to probe the product formation profiles in multiple scenarios simply by manipulating the relevant kinetic constants and/or initial conditions.
Fig. 3.
Simulation analysis of selective product formation within binary nucleopeptide networks. (A) Schematic representation of the network topology and operative catalytic pathways. (B) Reaction model describing the multiple simultaneous processes taking place in the network. F- and B-type species refer to the growing fibers and spheres, respectively, and kinetic constants are described in the SI Appendix, section S7 and Table S1. (C–J) Time-dependent simulation results obtained on the basis of the reaction network model in B and by applying variable initial conditions and/or kinetic constants corresponding to real and putative experimental scenarios (SI Appendix, Table S1). Simulation results in the upper row (C, E, G, and I) and lower row (D, F, H, and J) were obtained for network reactions seeded with RTT and RAA, respectively. In all cases, the production of RAA and RTT is shown in green and brown-red, respectively; product formation in a reaction seeded with 20 or 60 µM of the template is shown in dashed and solid lines, respectively. Native: [EAA] = [ETT] = 50 µM; [N] = 100 µM. Competitive: [EAA] = [ETT] = [N] = 67 µM. Inactive background: same initial conditions as “Native” but with significantly weaker background reactions (g = 0.1). Active seeding: same initial conditions and rate constants as “Native” but with enhanced or reduced seeding (mA or mT set at 5 and/or 25, instead of 10). Pr values, reflecting the RAA to RTT selectivity ratios, are added at the title of each panel; these were calculated for the 60-μM seeded reactions, in which the ratios between RAA and RTT were maximal as indicated, for example, by the double-headed arrow inserted in C.
We ran an elaborate simulation in Matlab (SI Appendix, section S7) (46, 53), starting with a set of initial concentrations that directly matched the experimental conditions. The template-assisted kinetic constants reflected the observed order of catalytic activity by assigning bAT > bAA ∼ bTA > bTT, while other parameters were applied based on experimental data and/or previous simulations of related systems [e.g., fibril-dependent peptide replication (46)] and were grouped together to simplify the analysis when possible (SI Appendix, Table S1). The results of this simple modeling under “native” conditions (Fig. 3 C and D and SI Appendix, Figs. S24 and S25) closely reproduced the replication kinetic profiles observed experimentally for both individual reactions and small networks (Fig. 1). In particular, the selectivity product ratio (Pr = RAA/RTT) in the simulated network (Fig. 3C) nicely matched the experimentally observed ratio. We then applied the simulation analysis to screen for the network growth kinetics that would have been obtained had the network been governed by a different set of interactions. This exercise allowed us to display network behavior that would otherwise have been difficult to predict and thereby guide the design of new experiments in which the selectivity toward one of the replicators would be further enhanced (see below). Consequently, imposing more stringent competition conditions through initiation of the reactions with an understoichiometric amount of N ([EAA] = [ETT] = [N] = 67 μM) led to higher Pr values (Fig. 3 E and F). Studying the network when the template-free background reactions were allegedly slower than in the “native” cases, simply applied by lowering the gA and gT values (Fig. 3B) by one order of magnitude, revealed the higher inherent selectivity of the template-assisted processes toward the production of RAA (Fig. 3 G and H). Finally, simulating the network for cases in which the assembly seed of one of the supramolecular structures—spheres (Fig. 3I and SI Appendix, Fig. S28) or fibers (Fig. 3J and SI Appendix, Fig. S29)—becomes active in a lower aggregation state than the other (achieved by manipulating the minimum seed assembly value m; SI Appendix, Table S1) also dramatically affected the competition, yielding high Pr values toward the selection of RAA (Fig. 3 I and J).
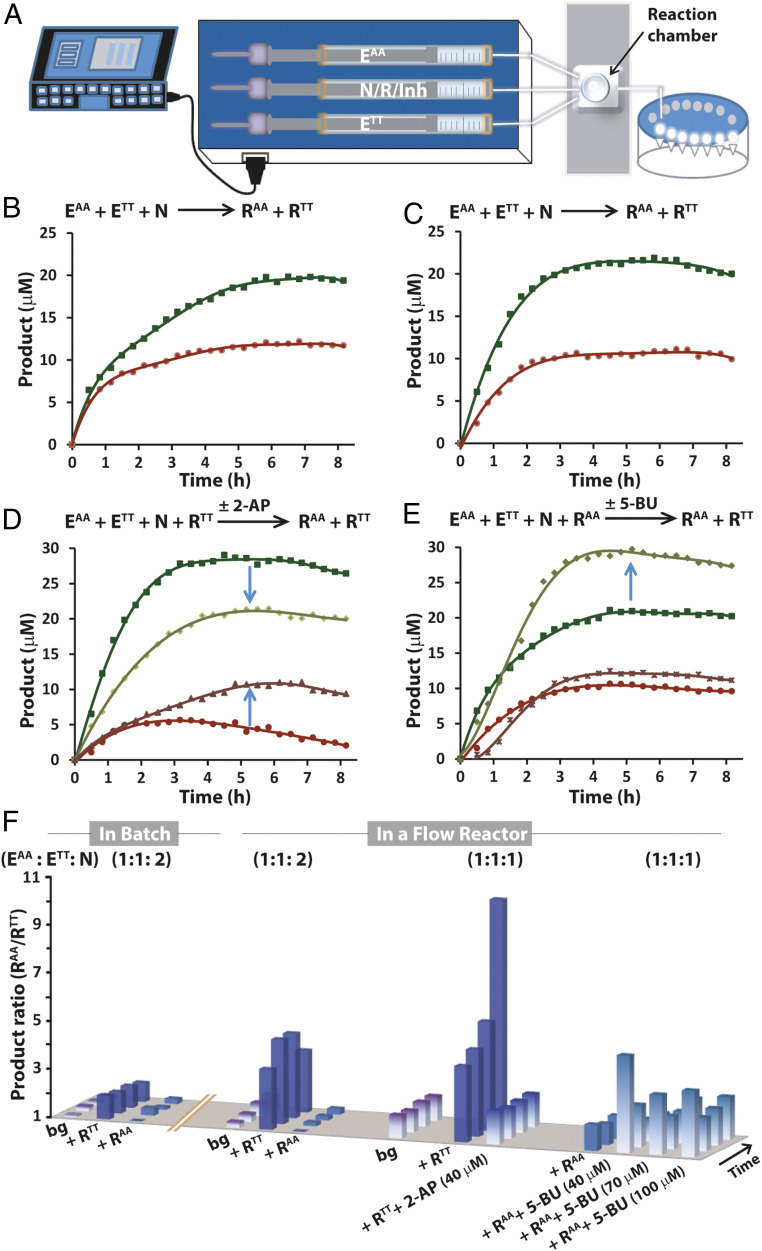
The selection pressure within networks of coexisting replicators can be intensified under conditions of constant flow, in which the catalytic pathways are kept far from equilibrium (Fig. 4) (54). Production of the RAA and RTT replicators in competition experiments was therefore studied over a few hours in a small-volume continuously stirred tank reactor (Fig. 4A). The first sets of experiments were conducted with either stoichiometric (total) amounts of the electrophiles EAA and ETT versus the nucleophile N (1:1:2), similar to the batch reaction experiments (Fig. 1 C and D), or with equal molar amounts of EAA, ETT, and N (1:1:1), thereby imposing stringent competition and potentially enhanced selectivity (Fig. 3 E and F). For each experiment, we studied the background reaction and 20-μM RTT-seeded or 20-μM RAA-seeded reactions. The background experiments revealed more pronounced selectivity toward the formation of RAA in comparison with the selectivity observed in the batch reactions, and the selectivity ratio was indeed further enhanced under the conditions of stringent competition (Fig. 4 B, C, and F). Most dramatically, the seeded reactions (in either the 1:1:2 or 1:1:1 initial concentration ratios) yielded much higher formation of RAA than RTT (SI Appendix, Figs. S30 and S31 and Fig. 4 D–F), revealing a selectivity higher by more than one order of magnitude at steady state for the 1:1:1 case.
Fig. 4.
Selective product formation far from equilibrium. (A) Schematic illustration of the continuously stirred tank reactor. (B, C) Time-dependent product formation in the background reactions, where EAA:ETT:N = 1:1:2 (B) and EAA:ETT:N = 1:1:1 (C). (D) Time-dependent product formation in the RTT-assisted reactions (EAA:ETT:N = 1:1:1) in the absence or presence of 40 µM 2-AP. (E) Time-dependent product formation in the RAA-assisted reactions (EAA:ETT:N = 1:1:1) in the absence or presence of 40 µM 5-BU. (F) Comparison of the product ratio (RAA/RTT) over time for a large set of experiments carried out in batch mode and under flow conditions. All reactions were carried out in Hepes buffer pH 7.4 at room temperature.
To further tune selective product formation by nucleic-acid–related recognition processes, nucleobase analogs, namely 2-aminopurine (2-AP) as an adenine analog and 5-BrUra (5-BU) as a thymine analog, were added to the replicator-seeded reactions. In a series of experimental and computational studies—conducted with free bases or short DNA oligomers—it has been found that 2-AP (and other aminopurines) and 5-BU can specifically base pair with thymine and adenine, respectively (55–61). Each of these aromatic bases would potentially hybridize with the conjugate-containing complementary bases (i.e., 2-AP with RTT and 5-BU with RAA; see the hybridization motifs in SI Appendix, Scheme S1C) and consequently noncovalently inhibit the template-assisted reaction pathways originated by the latter. We could not exclude that, in addition to the direct hybridization with RTT and RAA, 2-AP and 5-BU may undergo nonspecific interactions with the replicators’ supramolecular structures. In any case, as a result of inhibition by 2-AP in the RTT-seeded reaction, we observed lower selectivity due to a decrease in RAA formation, accompanied by an increase in the production of RTT (Fig. 4D). Alternatively, inhibition by 5-BU in the RAA-seeded reaction resulted in higher formation of RAA, leading to pronounced selectivity (Pr ∼5), since RTT formation remained almost unchanged (Fig. 4E). If higher-than-stoichiometric amounts of 5-BU were added to the RAA-seeded reaction, the selectivity toward RAA formation did not change greatly, probably due to the inhibition of additional catalytic pathways in the network.
In summary, we have shown that pronounced selectivity toward the production of one nucleopeptide over another in small, plausibly prebiotic, networks can result from the synergistic activity between a nucleobase domain, delivering interconjugate hybridization, and a peptide domain, which stabilizes different assembly architectures. We hold that this study is significantly relevant to the search for the “origin-of-life molecules”—those potentially prebiotic small molecules and proto-polymers that, through self-organization, could facilitate the transition from nonliving matter to the first primitive life. Recent publications have indicated that peptide and nucleic-acid precursors could be formed in a prebiotic environment through common synthetic routes (62, 63), and thus they support our proposition that synergistic activity developed at a later stage may have led to the selection of functional chimeras. Other recent studies have confirmed synergy in NA–pep systems, affording, for example, peptide formation on short RNA or ribozymes (64–66), enantio-selective RNA precursor synthesis by amino acids (67), peptide nucleic acid–templated replication (68), and more (69). Since our study highlights the effect of synergistic interactions both on the thermodynamics of the hybrid systems, leading to the formation of different assembly structures, and on the kinetics of template-directed replication processes, it may explain the rapid progression in the complexity of systems chemistry in primitive environments, which would have facilitated the development of a large repertoire of functions.
Methods
Synthesis and characterization of the nucleopeptide replicators and their precursor molecules are described in the SI Appendix, section S2.
A time-dependent analysis of the replicator-assisted NCL reactions were carried out with mixtures containing N and the designated electrophile EAA or ETT (100 μM), the applied replicator RAA or RTT (30 or 60 μM), 4-acetamido benzoic acid (ABA, 30 μM), and TCEP (5 mM) at pH 7.4 and room temperature. The reactions were quenched at specific times in 30% acetic acid in water, and the aliquots were analyzed by reversed-phase HPLC (RP-HPLC).
A time-dependent analysis of the replicator-assisted NCL reactions in binary chemical networks (batch mode) were carried out with mixtures containing N (100 μM) and both EAA and ETT (50 μM each), the applied replicator RAA or RTT (20 or 60 μM), ABA (30 μM), and TCEP (5 mM) at pH 7.4 and room temperature. The reactions were quenched at specific times in 30% acetic acid in water, and the aliquots were analyzed by RP-HPLC.
Structural assembly characterization was achieved using the following methods: 1) AFM imaging, performed for samples deposited on freshly cleaved mica surfaces, with topography images acquired by tapping mode and further used for image processing; 2) TEM imaging on copper grids with 200-mesh carbon support covered with the conjugate solutions properly stained with uranyl acetate; and 3) fluorescence microscopy imaging of samples incubated with ThT, deposited on glass slides and covered with coverslip. FRAP experiments were done by instantly initiating a scan with maximal (100%) laser power and then continuing to scan with the optimal laser power for 30 s.
A time-dependent analysis of ThT binding to the self-assembled structures was performed with samples that were allowed 30 min for self-assembly and then incubated for an additional 15 min in presence of 100 µM ThT. Fluorescence spectra were measured at different time intervals, and the typical emission signals of ThT-bound fibers (481 nm) were used to compare the binding efficiency of the different assemblies.
Modeling the mechanisms and kinetics of the network replication reactions supported the network experiments. In order to accurately follow the network behavior, we used the model shown in Fig. 3B. The simulation was applied with rate constants and system parameters that closely correspond to the experimental conditions and initial concentrations used in the actual experiments (SI Appendix, section S7).
A time-dependent analysis of the replicator-assisted reaction networks far from equilibrium were carried out in a flow cell device that contained three inlets fitted with three syringes (Fig. 4A), of which one contained N and R species, the other EAA, and the third ETT and ABA. For experiments that involved inhibition processes, the applied inhibitor was injected from the central inlet. Reactions were quenched by continuously collecting drops from the chamber outlet into 30% acetic acid in water, and the aliquots were analyzed by RP-HPLC.
Supplementary Material
Acknowledgments
We thank Prof. D. G. Lynn (Emory University) for fruitful discussions. The research was supported by the Horizon 2020 Future Emerging Technologies Open (A.d.l.E and G.A.; CLASSY project, Grant Agreement N° 862081), an National Science Foundation (NSF)-US–Israel Binational Science Foundation (BSF) grant (G.A.; BSF-2019745), and the Spanish Ministry of Economy and Competitivity (A.d.l.E; MINECO: CTQ-2014-53673-P, CTQ-2017-89539-P, and EUIN2017-87022). The European Cooperation in Science and Technology Action CM1304 funded a Short-Term Scientific Mission of S.M.-R. to Ben-Gurion University (BGU). A.K.B. received support from the BGU Kreitmann fellowships program.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015285118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Sanbonmatsu K. Y., Large-scale simulations of nucleoprotein complexes: Ribosomes, nucleosomes, chromatin, chromosomes and CRISPR. Curr. Opin. Struct. Biol. 55, 104–113 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yonath A., Polar bears, antibiotics, and the evolving ribosome (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 49, 4341–4354 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Perlmutter J. D., Hagan M. F., Mechanisms of virus assembly. Annu. Rev. Phys. Chem. 66, 217–239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman J. C., Petrov A. S., Frenkel-Pinter M., Penev P. I., Williams L. D., Root of the tree: The significance, evolution, and origins of the ribosome. Chem. Rev. 120, 4848–4878 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Freeman R., et al., Reversible self-assembly of superstructured networks. Science 362, 808–813 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chotera A., Sadihov H., Cohen-Luria R., Monnard P.-A., Ashkenasy G., Functional assemblies emerging in complex mixtures of peptides and nucleic acid-peptide chimeras. Chemistry 24, 10128–10135 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Rha A. K., et al., Electrostatic complementarity drives amyloid/nucleic acid co-assembly. Angew. Chem. Int. Ed. Engl. 59, 358–363 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang T., et al., Structurally ordered nanowire formation from co-assembly of DNA origami and collagen-mimetic peptides. J. Am. Chem. Soc. 139, 14025–14028 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Liu B., et al., Complex molecules that fold like proteins can emerge spontaneously. J. Am. Chem. Soc. 141, 1685–1689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ni R., Chau Y., Tuning the inter-nanofibril interaction to regulate the morphology and function of peptide/DNA co-assembled viral mimics. Angew. Chem. Int. Ed. Engl. 56, 9356–9360 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Buchberger A., Simmons C. R., Fahmi N. E., Freeman R., Stephanopoulos N., Hierarchical assembly of nucleic acid/coiled-coil peptide nanostructures. J. Am. Chem. Soc. 142, 1406–1416 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Humenik M., Scheibel T., Nanomaterial building blocks based on spider silk-oligonucleotide conjugates. ACS Nano 8, 1342–1349 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Raymond D. M., Nilsson B. L., Multicomponent peptide assemblies. Chem. Soc. Rev. 47, 3659–3720 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephanopoulos N., Peptide-oligonucleotide hybrid molecules for bioactive nanomaterials. Bioconjug. Chem. 30, 1915–1922 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Toste F. D., Sigman M. S., Miller S. J., Pursuit of noncovalent interactions for strategic site-selective catalysis. Acc. Chem. Res. 50, 609–615 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korendovych I. V., DeGrado W. F., Catalytic efficiency of designed catalytic proteins. Curr. Opin. Struct. Biol. 27, 113–121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zozulia O., Dolan M. A., Korendovych I. V., Catalytic peptide assemblies. Chem. Soc. Rev. 47, 3621–3639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lerner-Yardeni J., Amit M., Ashkenasy G., Ashkenasy N., Sequence dependent proton conduction in self-assembled peptide nanostructures. Nanoscale 8, 2358–2366 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Goodwin J. T., Mehta A. K., Lynn D. G., Digital and analog chemical evolution. Acc. Chem. Res. 45, 2189–2199 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Bai Y., et al., Achieving biopolymer synergy in systems chemistry. Chem. Soc. Rev. 47, 5444–5456 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Frenkel-Pinter M., Samanta M., Ashkenasy G., Leman L. J., Prebiotic peptides: Molecular hubs in the origin of life. Chem. Rev. 120, 4707–4765 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Gibard C., Bhowmik S., Karki M., Kim E.-K., Krishnamurthy R., Phosphorylation, oligomerization and self-assembly in water under potential prebiotic conditions. Nat. Chem. 10, 212–217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morales-Reina S., et al., Programmed recognition between complementary dinucleolipids to control the self-assembly of lipidic amphiphiles. Chemistry 26, 1082–1090 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Bonfio C., et al., Prebiotic iron-sulfur peptide catalysts generate a pH gradient across model membranes of late protocells. Nat. Catal. 1, 616–623 (2018). [Google Scholar]
- 25.Forsythe J. G., et al., Ester-mediated amide bond formation driven by wet-dry cycles: A possible path to polypeptides on the prebiotic Earth. Angew. Chem. Int. Ed. Engl. 54, 9871–9875 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eigen M., Schuster P., The Hypercycle: A Principle of Natural Self-Organization (Springer, 1979). [DOI] [PubMed] [Google Scholar]
- 27.Kauffman S. A., Autocatalytic sets of proteins. J. Theor. Biol. 119, 1–24 (1986). [DOI] [PubMed] [Google Scholar]
- 28.Matsumura S., et al., Transient compartmentalization of RNA replicators prevents extinction due to parasites. Science 354, 1293–1296 (2016). [DOI] [PubMed] [Google Scholar]
- 29.O’Flaherty D. K., et al., Copying of mixed-sequence RNA templates inside model protocells. J. Am. Chem. Soc. 140, 5171–5178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Semenov S. N., et al., Rational design of functional and tunable oscillating enzymatic networks. Nat. Chem. 7, 160–165 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Maity I., et al., A chemically fueled non-enzymatic bistable network. Nat. Commun. 10, 4636 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semenov S. N., et al., Autocatalytic, bistable, oscillatory networks of biologically relevant organic reactions. Nature 537, 656–660 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Cafferty B. J., et al., Robustness, entrainment, and hybridization in dissipative molecular networks, and the origin of life. J. Am. Chem. Soc. 141, 8289–8295 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Huck J., Kosikova T., Philp D., Compositional persistence in a multicyclic network of synthetic replicators. J. Am. Chem. Soc. 141, 13905–13913 (2019). [DOI] [PubMed] [Google Scholar]
- 35.He M., Lehn J.-M., Time-dependent switching of constitutional dynamic libraries and networks from kinetic to thermodynamic distributions. J. Am. Chem. Soc. 141, 18560–18569 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Zhou Z., Yue L., Wang S., Lehn J.-M., Willner I., DNA-based multiconstituent dynamic networks: Hierarchical adaptive control over the composition and cooperative catalytic functions of the systems. J. Am. Chem. Soc. 140, 12077–12089 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Wagner N., Alasibi S., Peacock-Lopez E., Ashkenasy G., Coupled oscillations and circadian rhythms in molecular replication networks. J. Phys. Chem. Lett. 6, 60–65 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Ruiz-Mirazo K., Briones C., de la Escosura A., Prebiotic systems chemistry: New perspectives for the origins of life. Chem. Rev. 114, 285–366 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Kosikova T., Philp D., Exploring the emergence of complexity using synthetic replicators. Chem. Soc. Rev. 46, 7274–7305 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Bissette A. J., Fletcher S. P., Mechanisms of autocatalysis. Angew. Chem. Int. Ed. Engl. 52, 12800–12826 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Ruiz-Mirazo K., Briones C., de la Escosura A., Chemical roots of biological evolution: The origins of life as a process of development of autonomous functional systems. Open Biol. 7, 170050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang L., Krupkin M., Bashan A., Yonath A., Massa L., Protoribosome by quantum kernel energy method. Proc. Natl. Acad. Sci. U.S.A. 110, 14900–14905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J., Nowak P., Otto S., Dynamic combinatorial libraries: From exploring molecular recognition to systems chemistry. J. Am. Chem. Soc. 135, 9222–9239 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Dadon Z., Wagner N., Ashkenasy G., The road to non-enzymatic molecular networks. Angew. Chem. Int. Ed. Engl. 47, 6128–6136 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Sadownik J. W., Mattia E., Nowak P., Otto S., Diversification of self-replicating molecules. Nat. Chem. 8, 264–269 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Nanda J., et al., Emergence of native peptide sequences in prebiotic replication networks. Nat. Commun. 8, 434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colomer I., Borissov A., Fletcher S. P., Selection from a pool of self-assembling lipid replicators. Nat. Commun. 11, 176 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kosikova T., Philp D., Two synthetic replicators compete to process a dynamic reagent pool. J. Am. Chem. Soc. 141, 3059–3072 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Liu B., et al., Spontaneous emergence of self-replicating molecules containing nucleobases and amino acids. J. Am. Chem. Soc. 142, 4184–4192 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashkenasy G., Hermans T. M., Otto S., Taylor A. F., Systems chemistry. Chem. Soc. Rev. 46, 2543–2554 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Aumiller W. M. Jr, Keating C. D., Phosphorylation-mediated RNA/peptide complex coacervation as a model for intracellular liquid organelles. Nat. Chem. 8, 129–137 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Rubinov B., Wagner N., Rapaport H., Ashkenasy G., Self-replicating amphiphilic beta-sheet peptides. Angew. Chem. Int. Ed. Engl. 48, 6683–6686 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Rubinov B., et al., Transient fibril structures facilitating nonenzymatic self-replication. ACS Nano 6, 7893–7901 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Szathmáry E., Gladkih I., Sub-exponential growth and coexistence of non-enzymatically replicating templates. J. Theor. Biol. 138, 55–58 (1989). [DOI] [PubMed] [Google Scholar]
- 55.Kyogoku Y., Lord R. C., Rich A., The effect of substituents on the hydrogen bonding of adenine and uracil derivatives. Proc. Natl. Acad. Sci. U.S.A. 57, 250–257 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller J. H., Sobell H. M., Infrared demonstration of hydrogen bonding between purine and pyrimidine base analogues in solution. J. Mol. Biol. 24, 345–350 (1967). [DOI] [PubMed] [Google Scholar]
- 57.Sowers L. C., Fazakerley G. V., Eritja R., Kaplan B. E., Goodman M. F., Base pairing and mutagenesis: Observation of a protonated base pair between 2-aminopurine and cytosine in an oligonucleotide by proton NMR. Proc. Natl. Acad. Sci. U.S.A. 83, 5434–5438 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fazakerley G. V., Sowers L. C., Eritja R., Kaplan B. E., Goodman M. F., Structural and dynamic properties of a bromouracil-adenine base pair in DNA studied by proton NMR. J. Biomol. Struct. Dyn. 5, 639–650 (1987). [DOI] [PubMed] [Google Scholar]
- 59.Bull G. D., Thompson K. C., Proton transfer and tautomerism in 2-aminopurine-thymine and pyrrolocytosine-guanine base pairs. Biochemistry 57, 4547–4561 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Brovarets O. O., Hovorun D. M., IR vibrational spectra of H-bonded complexes of adenine, 2-aminopurine and 2-aminopurine+ with cytosine and thymine: Quantum-chemical study. Opt. Spectrosc. 111, 750–757 (2011). [Google Scholar]
- 61.Law S. M., Eritja R., Goodman M. F., Breslauer K. J., Spectroscopic and calorimetric characterizations of DNA duplexes containing 2-aminopurine. Biochemistry 35, 12329–12337 (1996). [DOI] [PubMed] [Google Scholar]
- 62.Patel B. H., Percivalle C., Ritson D. J., Duffy C. D., Sutherland J. D., Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nat. Chem. 7, 301–307 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Islam S., Bučar D.-K., Powner M. W., Prebiotic selection and assembly of proteinogenic amino acids and natural nucleotides from complex mixtures. Nat. Chem. 9, 584–589 (2017). [Google Scholar]
- 64.Turk R. M., Illangasekare M., Yarus M., Catalyzed and spontaneous reactions on ribozyme ribose. J. Am. Chem. Soc. 133, 6044–6050 (2011). [DOI] [PubMed] [Google Scholar]
- 65.Griesser H., Bechthold M., Tremmel P., Kervio E., Richert C., Amino acid-Specific, ribonucleotide-promoted peptide formation in the absence of enzymes. Angew. Chem. Int. Ed. Engl. 56, 1224–1228 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Griesser H., et al., Ribonucleotides and RNA promote peptide chain growth. Angew. Chem. Int. Ed. Engl. 56, 1219–1223 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Hein J. E., Tse E., Blackmond D. G., A route to enantiopure RNA precursors from nearly racemic starting materials. Nat. Chem. 3, 704–706 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Ura Y., Beierle J. M., Leman L. J., Orgel L. E., Ghadiri M. R., Self-assembling sequence-adaptive peptide nucleic acids. Science 325, 73–77 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Frenkel-Pinter M., et al., Mutually stabilizing interactions between proto-peptides and RNA. Nat. Commun. 11, 3137 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.