Significance
Bacterial noncoding RNAs (ncRNAs) play key roles in many biological processes including gene regulation, RNA processing and modification, and protein synthesis and translocation. OLE RNAs, found in many Gram-positive species, are one of the largest highly structured ncRNA classes whose biochemical functions remain unknown. In Bacillus halodurans, OLE RNAs interact with at least two proteins, OapA and OapB, which are required to assemble a functional OLE ribonucleoprotein (RNP) complex contributing to cellular responses to certain environmental stresses. We established X-ray structural models that reveal the sequence elements and tertiary structural features of OLE RNA that are critical for its specific recognition by OapB, which will aid future exploration of the biological and biochemical functions of the unusual OLE RNP complex.
Keywords: alcohol toxicity, GNRA tetraloop, KOW motif, Mg2+ tolerance, noncoding RNA
Abstract
The OLE (ornate, large, and extremophilic) RNA class is one of the most complex and well-conserved bacterial noncoding RNAs known to exist. This RNA is known to be important for bacterial responses to stress caused by short-chain alcohols, cold, and elevated Mg2+ concentrations. These biological functions have been shown to require the formation of a ribonucleoprotein (RNP) complex including at least two protein partners: OLE-associated protein A (OapA) and OLE-associated protein B (OapB). OapB directly binds OLE RNA with high-affinity and specificity and is believed to assist in assembling the functional OLE RNP complex. To provide the atomic details of OapB–OLE RNA interaction and to potentially reveal previously uncharacterized protein–RNA interfaces, we determined the structure of OapB from Bacillus halodurans alone and in complex with an OLE RNA fragment at resolutions of 1.0 Å and 2.0 Å, respectively. The structure of OapB exhibits a K-shaped overall architecture wherein its conserved KOW motif and additional unique structural elements of OapB form a bipartite RNA-binding surface that docks to the P13 hairpin and P12.2 helix of OLE RNA. These high-resolution structures elucidate the molecular contacts used by OapB to form a stable RNP complex and explain the high conservation of sequences and structural features at the OapB–OLE RNA-binding interface. These findings provide insight into the role of OapB in the assembly and biological function of OLE RNP complex and can guide the exploration of additional possible OLE RNA-binding interactions present in OapB.
Structured noncoding RNAs (ncRNAs) participate in diverse cellular processes across all domains of life (1) and have biological functions distinct from all other known RNA classes. OLE (ornate, large, and extremophilic) RNAs (2, 3) are a unique class of large ncRNAs found in many species of mostly extremophilic Firmicutes that can live in anaerobic environments. OLE RNAs are typically ∼600 nucleotides long and exhibit well-conserved nucleotide sequences and complex structural features (2, 4, 5). These characteristics indicate that OLE RNAs likely perform sophisticated biochemical functions that contribute to one or more important biological processes. However, the precise biochemical functions performed by this unusual ncRNA remain to be experimentally established.
The OLE RNA from the alkaliphilic bacterium Bacillus halodurans is known to partner with at least two proteins, OLE-associated proteins A (OapA) and B (OapB), to form a functional OLE ribonucleoprotein (RNP) complex. This RNP complex is essential for the bacterium to resist certain unfavorable environmental conditions, such as modest levels of ethanol (5%) (4), Mg2+ (as low as 2 mM) (5), or slight cold stress (20 °C) (4). For example, genetic studies in B. halodurans revealed that deletions of the corresponding genes ole, oapA, or both reduced the tolerance of cells to these stresses (4, 6). Mutations to oapB also disrupt the function of the RNP complex (5) as further described below.
OapA, previously also called BH2780 (2) or OAP (4, 7), is a transmembrane protein with no additional experimentally validated functions other than the ability to bind OLE RNA in a 2:1 protein to RNA complex (7). OapA binding to OLE RNA is required for localizing OLE RNA to the cell membrane (7). However, it is likely that this protein partner has one or more additional functions remaining to be discovered that are important for responding to the stresses noted above. Notably, an OapA mutant (called PM1), in which two strictly conserved aspartate residues are mutated to alanine, causes a dominant-negative phenotype wherein cells exhibit even greater sensitivity to the stresses listed above (5, 6). Therefore, we hypothesize that the altered PM1 version of OapA causes a more severe phenotype than its knockout because it impairs proper function of the OLE RNP complex in a manner distinct from its deletion.
This unusual phenotype for PM1 led us to speculate that additional unidentified components of the OLE RNP complex might be present to comprise a sophisticated stress response system located at cell membranes. Indeed, genetic selections under cold stress were used to identify suppressor mutations located in the second protein partner, OapB (5). Mutations that disrupt OapB function alleviate the strong stress phenotypes observed for strains carrying the PM1 version of OapA. These and other results revealed that OapB is a necessary component for assembling a functional OLE RNP complex (5, 8). OapB is known (5) to harbor a putative RNA-binding Kyrpides-Ouzounis-Woese (KOW) motif (9) that is commonly found in ribosomal proteins. Many bacterial species carry oapB genes (also called ybzG) even if they lack the ole and oapA genes. This suggests that OapB has a broader biological function that has not been established previously.
Recent biochemical characterization of OapB from B. halodurans has revealed that the protein directly binds to a specific region of OLE RNA in vitro with very high affinity (8). This binding interaction requires a GNRA (N = A/C/G/U; R = A/G) tetraloop and a single-nucleotide bulge in stem P13 of OLE RNA (8). We sought a high-resolution three-dimensional structural model for the OLE RNA fragment bound to OapB to work toward an in-depth understanding of this portion of the RNP complex. This information can be used to help identify the functions of the highly conserved regions of OLE RNA and the structural organization that is critical for the biochemical and biological functions of the mysterious OLE RNP complex.
Herein, we describe the crystal structures of OapB from B. halodurans at 1.0 Å resolution and OapB in complex with a subdomain of OLE RNA at 2.0 Å resolution. These high-resolution structures reveal the overall architecture of OapB alone and when docked to its OLE RNA–binding target. In addition, the structures reveal the essential binding surfaces formed by OapB that enable its high-affinity association with OLE RNA and explain the importance of a precisely located single-nucleotide bulge in stem P13 of OLE RNA that is required for its specific recognition by OapB. Structural comparisons of OapB with other KOW-motif–containing ribosomal proteins reveals a possible additional OLE RNA-binding site in OapB, which hints at how OapB might assist in the folding of the larger OLE RNP complex.
Results and Discussion
Determining the Structures of OapB and the Complex Formed between OapB and OLE RNA.
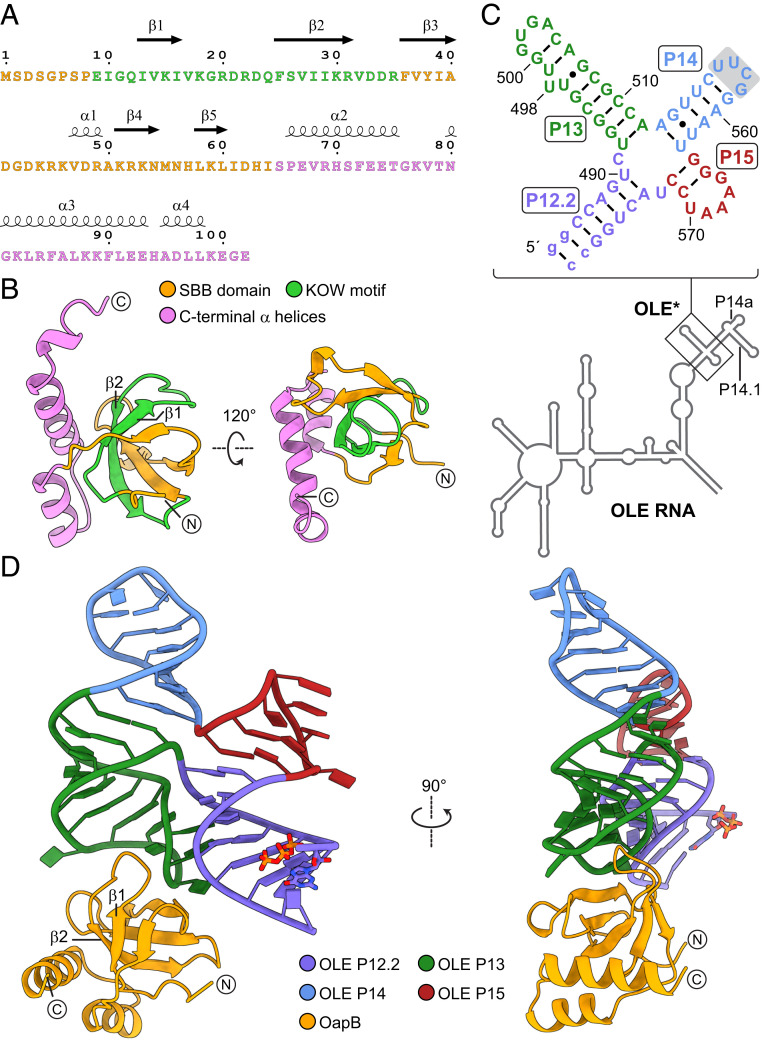
The initial full-length B. halodurans OapB construct carrying an N-terminal hexahistidine (6xHis) affinity tag failed to yield crystals after extensive screening of crystallization conditions. Secondary structure prediction suggests the N-terminal 5 to 10 residues of OapB are unstructured. Therefore, we designed a new construct encoding a near full-length OapB (spanning residue 5 to the C terminus) fused to an N-terminal 6xHis affinity tag. A human rhinovirus (HRV) 3C protease cleavage site was inserted between the affinity tag and the OapB coding region to enable tag removal during protein purification. This preparation yielded a structure of OapB, which was determined up to 1.0 Å resolution by single isomorphous replacement with anomalous scattering (SIRAS), using one native crystal and one derivative containing iodine (SI Appendix, Table S1). The final model contains all the residues in the OapB construct used for crystallization (Fig. 1 A and B).
Fig. 1.
Constructs and structures of OapB and the OapB–OLE* complex. (A) Sequence and secondary structure and (B) crystal structure of B. halodurans OapB. The N-terminal SBB domain is colored orange, the C-terminal helices are colored violet, and the conserved 27-residue KOW motif is colored green. Encircled N and C letters denote the N and C termini of OapB. (C) Sequence and secondary structure model of the OLE* used in crystallization. Stem P14 is capped with a nonnative UUCG tetraloop (shaded box). Stem P12.2 is made more stable with two nonnative G-C base pairs (lowercase letters). (D) Front and side views of the OapB–OLE* complex crystal structure.
Previous biochemical characterization has identified an OLE substructure comprising P12.2, P13, P14, and P15 regions as a minimized RNA element (hereinafter designated “OLE*”) that retains specific and high-affinity binding to OapB (8). To obtain well-diffracting crystals of the OapB–OLE* complex, we screened a series of RNA constructs with varied lengths of the P12.2 stem combined with different sequences of the tetraloop that caps the P14 stem. Each OapB–OLE* complex was reconstituted in vitro using the truncated OapB construct described above and the resulting RNP complexes were purified through size-exclusion chromatography (SI Appendix, Fig. S1).
The complex formed between OapB and a 60-nt OLE* (Fig. 1C) was crystallized to yield two different forms (hereafter referred to as “form I” and “form II”), both with end-to-end stacking of P12.2 helices and diffracting X-rays up to 2.0 Å and 2.1 Å, respectively (SI Appendix, Table S1). The structure was solved by single-wavelength anomalous dispersion (SAD) using crystals soaked with cobalt (III) hexammine. To further improve the electron density map, the experimental phase obtained from the SAD dataset was combined with the phase information from a partial molecular replacement model using the OapB apo structure determined above as a search template. All nucleotides in the OLE* used are observed with well-defined density in the refined maps and are built into the final models of the complex (Fig. 1D and SI Appendix, Fig. S2). Structures from the two crystal forms have the same overall architecture and highly similar OapB–OLE* interfaces (SI Appendix, Fig. S2 D–F). Unless otherwise indicated, the structure descriptions and analyses provided below refer to the OapB–OLE* complex structure determined from crystal form I due to higher data quality and better geometry of the final refined model.
Overview of the OapB Structure Alone and in Complex with an OLE RNA Fragment.
The KOW motif encompasses ∼27 amino acids and is most commonly found in several families of ribosomal proteins (9). The N-terminal portion of OapB, which includes the KOW motif of the protein (amino acids 9 through 35), folds into a small β-barrel (SBB) domain (10), followed by three α helices in the C terminus (Fig. 1B). Specifically, the KOW motif spans β1, β2, and the loop between them (Fig. 1A). Upon binding to OLE RNA, OapB exhibits notable conformational rearrangements in several loop regions, particularly in the β1–β2 loop (SI Appendix, Fig. S3). A series of basic residues at the OLE RNA-binding interface undergo major rotamer changes to better fit the RNA surface (SI Appendix, Fig. S3B). Such an induced fit mechanism is a common theme for many RNA-binding proteins when they form complexes with their cognate RNAs (11, 12).
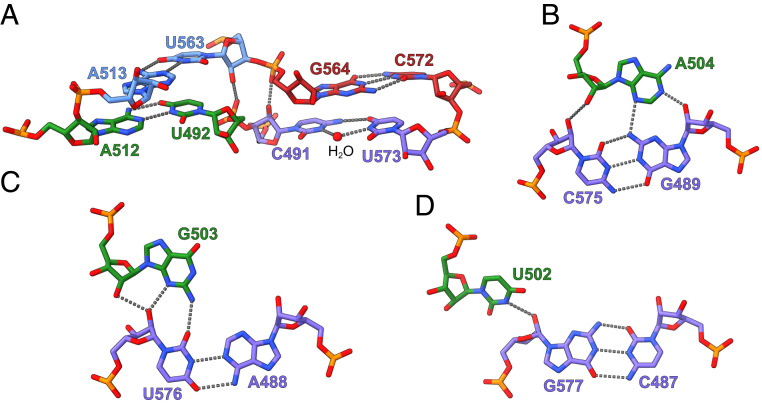
The OLE* contains a near-perfectly base-paired four-way helical (4H) junction, in which four helical segments form two approximately parallel coaxial stacks (Fig. 1D). Stems P13 and P14 stack nearly coaxially, whereas stems P12.2 and P15 form a second coaxial stack. The two stacks are connected by a pair of antiparallel crossovers (Figs. 1D and 2A). Three out of the four internucleotide contacts at the four-way junction are canonical Watson–Crick base pairs. The exception involves two nucleotides at the upper end of stem P12.2, which form a water-mediated noncanonical C-U base pair (Fig. 2A).
Fig. 2.
Tertiary interactions formed by OLE*. (A) Base pairs observed at the 4H junction of OLE*. Nucleotides from stems P12.2, P13, P14, and P15 are colored in purple, green, blue, and red, respectively. The water molecule mediating the noncanonical C491-U573 base pair in stem P12.2 is shown as a red sphere. Hydrogen bonds are shown as gray dashed lines. (B–D) Minor groove base triple interactions formed between P12.2 helix and A504 (B), G503 (C), and U502 (D) in the P13 hairpin tetraloop.
By comparison, a similar 4H junction is also present in the hairpin ribozyme (13), although its two coaxial stacks cross at a ∼60° angle. Single-molecule fluorescence resonance energy transfer (smFRET) analyses of the simple 4H junction derived from the hairpin ribozyme have shown that, without the tertiary contacts in the full-length ribozyme, the 4H junction is highly polymorphic and dynamically samples different stacking conformers (14, 15). In the 4H junction formed by OLE*, the P13-closing GUGA tetraloop, which was previously shown to be the primary OapB-binding site (8), docks into the shallow minor groove of the adjoining P12.2 helix and forms a GNRA tetraloop receptor–like interaction (16) (Fig. 1D). Within the GUGA tetraloop, nucleotide U502 interacts with the sugar edge of G577 (Fig. 2D), nucleotide G503 forms a type II A minor–like interaction (17, 18) with U576 (Fig. 2C), and nucleotide A504 establishes a type I A-minor groove triple motif with the G489-C575 base pair in stem P12.2 (Fig. 2B). These tertiary interactions likely lock the otherwise flexible 4H junction in the specific conformation we observed in the crystal structure.
Our previous mutagenesis analyses suggested that the GCGA tetraloop closing the stem P14.1 of OLE RNA constitutes a weak secondary OapB-binding site (8). Given the pseudosymmetric nature of stems P13 and P14.1 relative to P14 (Fig. 1C and SI Appendix, Fig. S4A), P14.1 likely also coaxially stacks with P14 and thus places the GCGA tetraloop at the distal end of the continuous helix formed by P13, P14, and P14.1, precluding any interactions between the two bound OapB molecules. This is consistent with previous finding that binding of OapB to the two GNRA tetraloops in OLE RNA was not cooperative (8).
It is also interesting to note that additional density is observed in the electron density map generated from crystal form I (SI Appendix, Fig. S5). This density is located at the interface between the P13 hairpin and OapB and is most likely a molecule of HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), the buffer used in the crystallization solution. However, we cannot conclude from our existing data that one of the natural functions of the OLE RNP complex involves the binding of a small molecule metabolite.
The Molecular Basis of OLE RNA Recognition by OapB.
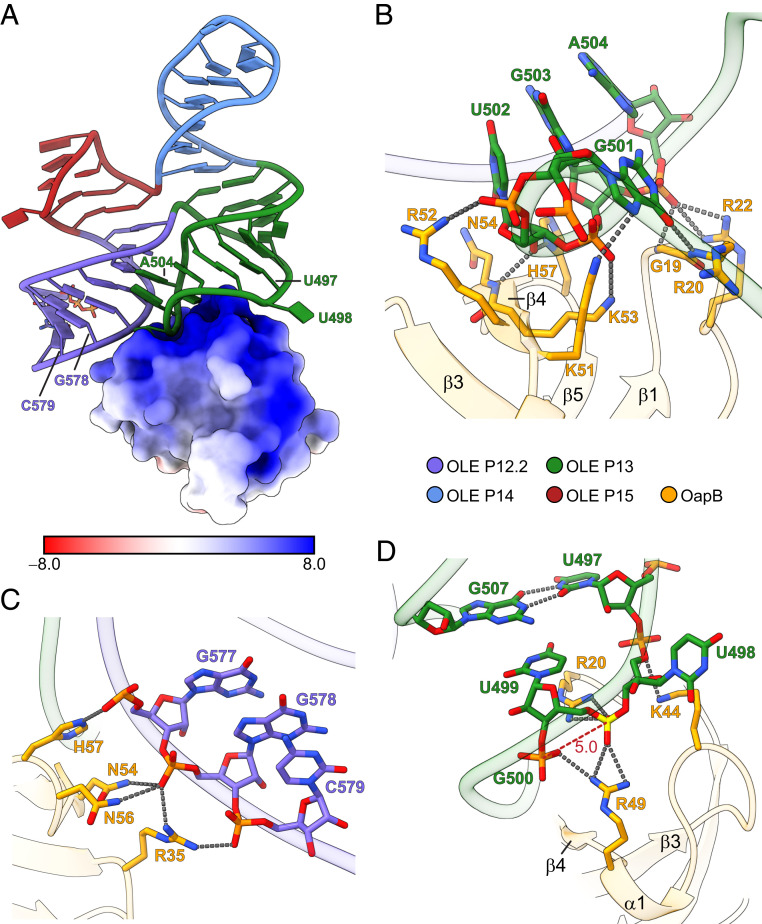
The structure of OapB in complex with the OLE* construct reveals extensive charge and shape complementarity at the protein–RNA binding interface, which buries a total of 680 Å2 of otherwise solvent-accessible surface area upon their association (Fig. 3A). The major groove of hairpin P13 with its end-capped GUGA tetraloop sits on top of a highly basic hill-shaped protrusion composed of β4 and three inter-β-strand loops in OapB (Fig. 3B). The GUGA tetraloop forms an extensive network of hydrogen bonds and salt bridges by contacting various basic amino acid residues mainly through backbone phosphates and 2′-hydroxyl groups (Fig. 3B). The lack of base-specific interactions, except for the two hydrogen bonds mediated by the first G501 in the tetraloop (Fig. 3B), indicates that the specificity of OapB binding to this region is dictated by the tertiary conformation of the GNRA tetraloop, rather than exact sequence within this tetraloop family.
Fig. 3.
Molecular basis of OLE RNA recognition by OapB. (A) Electrostatic surface representation of OapB in complex with OLE*. The unit of electrostatic potential is kT/e−. (B) A close-up view of the OapB–OLE RNA interactions at the P13 GNRA tetraloop region. Hydrogen bonds are shown as gray dashed lines. (C) A close-up view of the interactions between OapB and the helix backbone of P12.2. (D) A close-up view of the OapB–OLE RNA interactions near the bulged U498 nucleotide. The phosphate group connecting U498 and U499 is highlighted in yellow. The phosphate-to-phosphate distance between U499 and G500 is indicated (in Å) and labeled with a red dashed line.
This result is consistent with the fact that the central two nucleotides can mutate without complete loss of protein binding (8). However, replacing the GUGA tetraloop of the P13 hairpin with another common stabilizing tetraloop based on the UNCG (N = A/C/G/U) consensus (19), which is known to adopt a different conformation (20, 21), abolishes OapB binding (8). Thus, the B. halodurans OapB representative places some restrictions on the sequence of the P13 loop sequence and structure. If these constraints are required for OapB binding to OLE RNA in many other bacterial species, this could explain the presence of GNRA tetraloops in the majority of OLE RNAs from other species (5).
Previous genetic selection results revealed that mutations at either of two highly conserved residues in OapB, G19S, and H57Y mitigated the severity of PM1 phenotypes (5). These mutations disrupt the ability of OapB to bind OLE RNA, presumably preventing the proper assembly of the functional OLE RNP complex. Indeed, the OapB–OLE* cocrystal structure provides a basis to understand how these OapB mutations disrupt OapB–OLE RNA binding.
The G19 residue of OapB resides at a strictly conserved position of the SBB domain (9). This amino acid is located in the β1–β2 loop (Figs. 1 A and B and 3B), which projects into the major groove of the OLE RNA P13 hairpin (Fig. 1D). The absence of a side chain at this position of the wild-type protein allows for the close approach of OLE RNA to OapB, and the subsequent formation of two hydrogen bonds between the P13 GUGA tetraloop and the main chain nitrogen atoms of OapB G19 and R20 (Fig. 3B and SI Appendix, Fig. S4B). A G19S mutation likely results in steric clashes that prevent proper contact between OapB and OLE RNA (SI Appendix, Fig. S4B).
As H57 residue carries an imidazole side chain that is inserted between the U502 ribose at the bottom of the P13 GUGA tetraloop and the backbone of the P12.2 helix (SI Appendix, Fig. S4C), the H57Y mutation is expected to cause a loss of two hydrogen bonds otherwise involving the side chain of histidine. In addition, the bulkier side chain of tyrosine is unable to fit within the narrow gap originally occupied by histidine without causing substantial clashes with OLE RNA (SI Appendix, Fig. S4C).
The close proximity of the P12.2 helix with the P13 hairpin tetraloop in the tertiary structure of OLE RNA creates a unified surface for OapB binding to these separate secondary structure features (Fig. 3 A and C). OapB amino acids R35, N54, and N56, in addition to the above-mentioned H57 make multiple contacts with backbone phosphate groups of nucleotides in the P12.2 helix (Fig. 3C). These interactions likely play a role in OLE RNA folding by serving as part of a protein tether that stabilizes the close approach and orientation of the two coaxial stacks (the P12.2–P15 stack and the P13–P14 stack). These contacts appear to work synergistically with the GUGA tetraloop–P12.2 minor groove base triple tertiary interactions to direct the OLE RNA 4H junction to the observed conformation.
The presence of a single-nucleotide bulge in stem P13 (Fig. 1C) is a highly conserved feature of OLE RNA, which was found to be critical for OapB binding (5, 8). In B. halodurans OLE RNA, the base of the bulged nucleotide U498 flips out to extrahelical space, distorting the P13 helix backbone and leading to an unusually short phosphate-to-phosphate distance between U499 and G500 in crystal form I (Fig. 3D) and between U498 and U499 in crystal form II (SI Appendix, Fig. S4D). The crowding of negative charges is neutralized by a pair of arginine residues (R20 and R49) from OapB that clamp the two phosphate groups from both sides (Fig. 3D). Notably, the bulged U498 adopts different conformations in the two crystal forms of OapB–OLE* complex but is not involved in any base-specific interactions with OapB in either form (Fig. 3D and SI Appendix, Fig. S4D). This observation suggests this nucleotide is highly flexible and its identity is unlikely to contribute to the specificity of OLE RNA recognition by OapB.
Previous bioinformatics-derived secondary structure models for OLE RNA assigned the nucleotide equivalent to U497 as the bulged nucleotide (2, 5, 7, 8). Mutagenesis studies with a minimized OLE RNA have shown that substituting U497 with other nucleotides weakens OapB binding by 6- to 10-fold, whereas removing this nucleotide almost completely abolishes binding (8). However, the B. halodurans OLE RNA sequence contains three consecutive uridines at positions 497 through 499, and therefore, it was not initially clear which U nucleotide was bulged in the functional conformation. Specifically, deletion of U497 is equivalent to removing any of the three nucleotides at these positions and, therefore, will be indistinguishable in their effects on OLE RNA structure and function. A deletion of a U nucleotide in this region is expected to eliminate the distorted RNA helical backbone by restoring contiguous base-pairing of P13, thereby displacing the U499 backbone phosphate and breaking its interactions with R20 and R49 of OapB. Mutating U497 to other nucleotides is expected to change its base pairing potential with G507 and consequently shift the positions of the U498 and U499 backbone phosphate groups. However, the suboptimal RNA backbone conformation that results might be partially compensated by the flexibility of the flipped U498, resulting in a less deleterious impact on OapB binding affinity as is observed in biochemical experiments (8). Therefore, it appears that the extrahelical U498 and G507-U497 wobble base pair together place the two backbone phosphate groups for optimal interaction with the OapB arginine dyad.
Comparison of OapB with Ribosomal Proteins Suggests an Additional RNA-Binding Surface Exists.
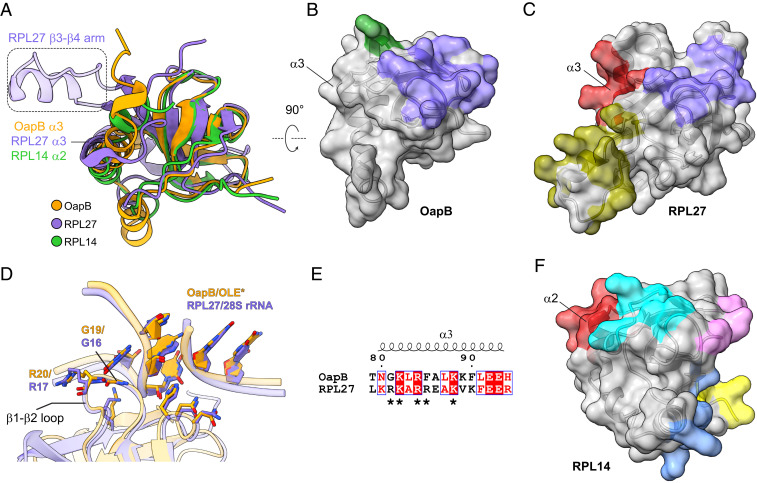
The OLE* construct used for crystallization occupies only a small portion of the total surface area of OapB (Fig. 3A). Extensive surface area with strong positive-charge character remains available to possibly serve as a docking site for an additional RNA structure. We speculate that this area could be utilized when OapB forms a complex with the full-length OLE RNA. To further assess this possibility, we compare the structures and RNA-binding characteristics of OapB with a list of proteins identified to have similar structural motifs by DALI (Distance-matrix ALIgnment) protein structure comparison server (22). Among these proteins, we found that the overall three-dimensional structure of eukaryotic 60S ribosomal protein families L14 and L27 (RPL14 and RPL27) (23) are most similar to that of OapB (Fig. 4A). They all are formed with a KOW motif–containing SBB domain at the N terminus followed by two or three helices at the C terminus.
Fig. 4.
Comparison of OapB with KOW motif–containing ribosomal proteins. (A) Superimposition of OapB, human 60S ribosomal protein L27 (RPL27, PDB 6EK0) and human 60S ribosomal protein L14 (RPL14, PDB 6EK0) structures. (B) Surface representation of OapB. Separate RNA-interacting patches are colored, where purple represents the greatest similarity to the purple region in C. (C) Surface representation of RPL27. Separate RNA-interacting patches are colored. (D) Superimposition of the OapB–OLE* complex with RPL27-28S rRNA complex structures centered on their GNRA tetraloop regions reveals highly similar GNRA tetraloop–recognizing patterns by OapB and RPL27. Nucleotides are shown with filled rings. (E) Structure-based sequence alignment of α3 helices in OapB and RPL27. Amino acid residues in the basic patch (R108, K109, R111, R112, and K115) of RPL27 α3 are indicated with asterisks. The sequence alignment result was rendered using the online server of ESPript 3.0 (41). (F) Surface representations of RPL14.
The residues of OapB that interact with the known OLE RNA–binding site are grouped into two patches on the protein surface. One patch (Fig. 4B, purple) is mainly responsible for interactions with the GUGA tetraloop of P13 and the backbone phosphate groups of the P12.2 helix. Another patch (Fig. 4B, green) participates in specific interactions with the bulged U498 region. In eukaryotic ribosomes, a functionally equivalent region of RPL27 (Fig. 4C, purple) interacts with a GUAA tetraloop and the backbone of an adjacent helix in 28S ribosomal RNA (rRNA) (23) in an almost identical manner as OapB interacts with the GUGA tetraloop and P12.2 helix backbone of OLE RNA (Fig. 4D and SI Appendix, Fig. S6 A and B). Specifically, a Gly–Arg dipeptidyl sequence resides at the tip of the β1–β2 loops of OapB and RPL27. These residues are inserted deeply into the GNRA tetraloop groove (Fig. 4D). Furthermore, the majority of the residues establishing backbone phosphate contacts with a neighboring helix are conserved between OapB and RPL27 (Figs. 3C and 4D), suggesting that there might be a common recognition pattern shared by the GNRA tetraloop-minor groove base triple motifs of OLE RNA and 28S rRNA.
On the opposite sides of both OapB and RPL27, the two proteins carry distinct structural features (Fig. 4 B and C). RPL27 is known to use this face of the protein to form a second RNA-interacting patch (Fig. 4C, olive). Whereas a short helix (α1, V47–R49) within the β3–β4 loop region of OapB makes contact with OLE RNA backbone near the bulged U498 just two nucleotides upstream of the GNRA tetraloop (Figs. 3D and 4B, green), RPL27 establishes interactions with a more distally located 28S rRNA hairpin (SI Appendix, Fig. S6B, olive) using a long arm inserted between β3 and β4 (Fig. 4 A and C, olive). This difference in the RNA-binding characteristics between OapB and RPL27 indicates that the bulged U498 of OLE RNA and a matching structural element in OapB constitute a major specificity determinant of OapB–OLE RNA-binding.
Besides the above two patches, a third site on RPL27 for binding 28S rRNA is provided by helix α3 (Fig. 4C, red and SI Appendix, Fig. S6B), which makes RNA backbone interactions through a basic patch comprising R108, K109, R111, R112, and K115 (Fig. 4E). By contrast, α3 of OapB, which is located in approximately the same spatial position as RPL27 α3 when the two structures are superimposed (Fig. 4A), is currently unoccupied in the OapB–OLE* complex (Fig. 4B). Moreover, structure-guided sequence alignment reveals that the majority of the basic patch residues involved in RPL27 α3–28S rRNA interactions are also present in OapB α3, (Fig. 4E), suggesting it might establish contacts with full-length OLE RNA in a similar fashion as RPL27 α3 binds 28S rRNA. However, the minimized OLE RNA construct used in this study lacks an RNA partner for this putative additional RNA-binding site on OapB. Perhaps this binding site is located elsewhere in the full-sized OLE RNA structure and remains to be discovered.
OapB α3 is an amphipathic helix with multiple basic and polar residues located at the solvent-exposed side and an array of aliphatic and aromatic residues on the opposite side facing a hydrophobic core that holds the N-terminal SBB domain and the two C-terminal α helices together (SI Appendix, Fig. S6C). G42, an invariant amino acid in OapB, sits at the interface between the SBB domain and α3 helix. Genetic selections identified G42V as a suppressor mutation that overcomes the strong phenotypic defect caused by PM1, presumably through the disruption of OapB–OLE RNA interaction and, hence, the integrity of the OLE RNP complex (5). The crystal structure of OapB shows that the space between G42 and α3 is too small to accommodate the side chain of a valine. Therefore, the G42V mutation would alter the position of α3 relative to the SBB domain (SI Appendix, Fig. S6C), which likely needs to be precisely maintained to allow the simultaneous binding of OapB to two discontinuous regions of the full-length OLE RNA molecule.
Like RPL27, RPL14 also employs more surface areas for binding ribosomal RNA, although it uses distinct patterns of molecular recognition contacts compared to OapB (Fig. 4D and SI Appendix, Fig. S6D). One of the five 28S rRNA–contacting patches on the RPL14 surface is formed by residues in the first half of helix α2 (Fig. 4F, red; SI Appendix, Fig. S6D). This region aligns very well with the OapB and RPL27 α3 helices in their structural superimposition (Fig. 4A) and interacts with 28S rRNA using multiple positively charged residues and glutamines at equivalent positions as the residues in the RPL27 basic patch. The structural comparison between OapB and RPL14 again reinforces the hypothesis that OapB α3 helix participates in binding a distal RNA region within the full-length OLE RNA molecule.
Concluding Remarks.
The high-resolution crystal structure of OapB with its cognate OLE RNA substructure reported here provides atomic detail of the architecture and assembly of an intriguing portion of the OLE RNP complex. The OLE RNA fragment bound by OapB includes the stems P12.2, P13, P14, and P15, which together form an almost perfect 4H junction. This region contains RNA tertiary structural motifs that are recurrent in many other large ncRNAs (24), which highlights the importance of RNA tertiary structure to the biological and biochemical functions of OLE RNA.
Three subregions of OapB that are discontinuous in their primary sequences come together to form a bipartite surface for association of the protein with OLE RNA. The binding interface is replete with hydrogen bonding and salt bridge interactions that tightly join the two molecules, which explains the strong binding affinity for the complex (KD∼700 pM) (8). Because most of these protein–RNA contacts are mediated by phosphate and ribose 2′ hydroxyl groups of the RNA phosphodiester backbone, the high specificity of OLE RNA recognition by OapB is primarily dictated by the unique three-dimensional architecture of the OLE* construct, as evident by the remarkable shape complementarity between the two partners at their binding interface (Fig. 3A). Additionally, the OapB–OLE* complex structure explains the disruptive effects of the previously identified mutations in OapB and OLE RNA (5, 8) and provides a basis to understand why each of the three OapB mutations (G19S, G42V, and H57Y) found in genetic selections (5) alleviate the strong dominant-negative phenotype observed with the PM1 strain.
In the OapB–OLE* complex, the amount of OapB surface utilized for RNA binding is markedly lower compared to two structurally similar ribosomal proteins, RPL14 and RPL27. Detailed analyses of their RNA-binding patterns suggest that OapB α3 likely forms an additional RNA-binding site and that this might be important for binding the full-length OLE RNA in the OLE RNP complex. Further analyses involving larger RNA constructs will be needed to evaluate this hypothesis and to characterize the nature of this putative interaction.
Intriguingly, some of the most highly conserved nucleotide positions in this RNA construct are not involved in either binding OapB or in setting up the special 4H junction that is observed in the RNA–protein complex. Most notably, the conserved nucleotides in the loops of P14a and P15, and also those in the junction between P14 and P14a (5, 8), would be located in the natural RNA far from the nucleotides bound by OapB. Therefore, the OLE RNP complex likely exploits these strongly conserved nucleotides to achieve other structural and functional objectives. It is likely that the B. halodurans OLE RNP complex requires OapB binding to assist in forming the local architecture necessary for these other objectives to be met, as well as helping to bring a distal part of OLE RNA near to these conserved nucleotides. Regardless, a combination of bioinformatic, genetic, biochemical, and biophysical approaches will likely be needed to fully reveal the mechanisms and functions of this complex ncRNA and its protein partners.
Materials and Methods
Protein Expression and Purification.
An open reading frame encoding residue 5 to the C terminus of B. halodurans OapB was fused to an N-terminal 6× His affinity tag and HRV 3C protease cleavage sequence and cloned into the pET-11a expression vector (Millipore Sigma) between NdeI and BamHI restrictive enzyme sites. The corresponding OapB protein was overexpressed in Escherichia coli BL21 Star (DE3) cells (Thermo Fisher Scientific) at 17 °C for 18 h.
Pelleted cells expressing recombinant OapB protein were resuspended in buffer A, which is composed of 50 mM Tris HCl (pH 7.5), 500 mM NaCl, 20 mM imidazole (pH 7.5), 5% glycerol, and β-mercaptoethanol (β-ME). The mixture was homogenized using an M110EH Microfluidizer (Microfluidics International Corporation). Cell lysate was clarified by centrifugation using an SS-34 rotor (17,000 rpm for 1 h at 4 °C) and loaded onto a HisTrap HP affinity chromatography column (Cytiva Life Sciences) pre-equilibrated with buffer A. OapB protein was eluted from the HisTrap HP column over 20× column volumes (CV) with a linear gradient from buffer A to buffer B, the latter which is composed of 50 mM Tris HCl (pH 7.5), 500 mM NaCl, 300 mM imidazole (pH 7.5), and 5% glycerol. Eluted protein from multiple fractions were pooled and supplemented with 2 mg of 6× His tagged HRV 3C protease and 1 mM (final concentration) β-ME. Protease cleavage and 6× His tag removal from OapB was performed at 4 °C for 24 h. The protein sample was diluted with equal volume of buffer C, which is composed of 20 mM sodium phosphate (pH 7.0) and 1 mM β-ME, and the mixture was loaded onto a HiTrap Heparin HP column (Cytiva Life Sciences). After washing with 5× CV of buffer C, OapB protein was eluted over 10× CV with a linear gradient from buffer C to buffer D, the latter which is composed of 20 mM sodium phosphate (pH 7.0), 1 M NaCl, and 1 mM β-ME. The eluted OapB protein sample was supplemented with imidazole to a final concentration of 20 mM and passed through a HisTrap HP column again to remove residual 6× His tag, HRV 3C protease, and any uncleaved OapB protein. The flow through was collected, concentrated using an Amicon Ultra centrifugal filter with a 3 kDa cutoff, and further purified using a HiLoad 16/600 Superdex 200 pg size-exclusion chromatography (SEC) column in buffer E, composed of 20 mM Tris⋅HCl (pH 7.5), 500 mM NaCl, and 1 mM Tris(2-carboxyethyl) phosphine hydrochloride (TCEP-HCl).
In Vitro Transcription and OLE* Purification.
The DNA template for in vitro transcription of OLE* was generated by annealing equimolar amounts of two DNA oligonucleotides with complementary sequences (5′-TAA TAC GAC TCA CTA TAG GCC AGT CTG GCG TTT GGT GAC AGC GCC AAG TTC TTC GGA ATT GGG AAA TCC TAC TGG CC-3′ and 5′-[Gm][Gm]C CAG TAG GAT TTC CCA ATT CCG AAG AAC TTG GCG CTG TCA CCA AAC GCC AGA CTG GCC TAT AGT GAG TCG TAT TA-3′). The T7 RNA polymerase (RNAP) promoter sequence in the nontemplate DNA strand is underlined. Two guanosines denoted [Gm] on the 5′ end of template DNA strand are 2'-O-methylated to reduce 3′ end heterogeneity of the RNA transcript (25).
Transcription reactions (3 mL) were assembled using 1.2 nmol annealed DNA template, 300 µg T7 RNAP, 0.3 U inorganic pyrophosphatase (New England Biolabs) and 120 µL 25× RNAsecure RNase inactivation reagent (Thermo Fisher Scientific) in 1× transcription buffer containing 80 mM HEPES-K (pH 7.5), 24 mM MgCl2, 40 mM DTT, 2 mM spermidine, and 4 mM of each ribonucleoside triphosphate. The resulting mixture was incubated at 37 °C for 2.5 h. After removing the pyrophosphate precipitate by centrifugation at 3,000 × g for 10 min, ethylenediaminetetraacetic acid (EDTA) was added to the reaction mix to a final concentration of 50 mM. OLE* was initially purified by extraction three times with acid phenol:chloroform (pH 4.5). The free nucleotides were subsequently removed using a Sephadex G-25 PD-10 desalting column (Cytiva Life Sciences). OLE* was further purified by SEC using a HiLoad 16/600 Superdex 200 pg column in buffer F, containing 20 mM sodium cacodylate (pH 6.5) and 100 mM NaCl.
OapB–OLE* Complex Assembly.
Purified OapB protein and OLE* were mixed in a 2:1 molar ratio and incubated at room temperature for 1 h. OapB–OLE* complex was separated from excessive OapB by SEC as described above using buffer G, containing 20 mM Tris⋅HCl (pH 7.5), 100 mM NaCl, and 1 mM TCEP. Fractions containing OapB–OLE* were pooled and stored at 4 °C for later use.
Crystallization and Data Collection.
Crystallization of OapB was achieved using the sitting-drop vapor-diffusion method at 19 °C in 50 mM Tris⋅HCl (pH 8.5), 100 mM KCl, 10 mM MgCl2, and 30% polyethylene glycol (PEG) 400. Crystals were cryo-protected in buffer containing 50 mM Tris⋅HCl (pH 8.5), 100 mM KCl, 10 mM MgCl2, and 36% PEG 400 before being flash frozen in liquid nitrogen. Iodine-derivative OapB crystals were prepared by soaking the crystals in buffer containing 50 mM Tris⋅HCl (pH 8.5), 100 mM KCl, 10 mM MgCl2, 36% PEG 400, and 500 mM NaI at room temperature for 2 to 5 min. X-ray diffraction data were collected at 100 K at beamlines 24ID-E and 24ID-C of the Advanced Photon Source at Argonne National Laboratory. The datasets for the native and iodine-derivative crystals were initially collected at wavelengths of 0.9792 Å and 1.4586 Å, respectively. Because the diffraction limit of the OapB crystals clearly exceeds the maximum achievable resolutions at these wavelengths, one more dataset was collected using the iodine-derivative crystal at a wavelength of 0.8266 Å to push the resolution limit of the dataset to 1.0 Å.
OapB–OLE* complex preparations were concentrated to about 400 µM and used in crystallization screening. OapB–OLE* complex crystals were grown by the sitting-drop vapor-diffusion method at 19 °C in either 100 mM HEPES (pH 7.0 to 7.5), 200 mM NH4Cl, and 39 to 42.5% 2-Methyl-2,4-pentanediol (MPD) (crystal form I) or 240 mM sodium malonate (pH 6.0 to 7.0) and 20 to 21% PEG 3,350 (crystal form II). Cobalt-derivative OapB–OLE* complex crystals were prepared by soaking crystal form I in buffer containing 100 mM HEPES (pH 7.0), 200 mM NH4Cl, 45% MPD, and 2 mM cobalt (III) hexammine chloride ([Co(NH3)6]Cl3) at 19 °C for 14 h. Data were collected at beamline 24ID-C of the Advanced Photon Source at wavelengths of 0.9791 Å, 1.6058 Å, and 0.9792 Å for native crystal form I, cobalt-derivative crystal form I, and native crystal form II, respectively. All X-ray diffraction datasets were processed with the XDS package (26).
Crystal Structure Determination and Refinement.
Heavy atom sites were identified using SHELXD (27). Eight iodine sites and six cobalt sites were initially found in the iodine-derivative OapB apo and the cobalt-derivative OapB–OLE* datasets, respectively. The OapB apo structure was solved using AutoSol (28) by SIRAS method using one native and one iodine-derivative crystals. Structure of the OapB–OLE* complex from crystal form I was solved using AutoSol by molecular replacement with SAD method (29) using one cobalt-derivative crystal in form I. The structures of native OapB–OLE* complex in crystal form I and II were determined by molecular replacement using the cobalt-derivative crystal form I structure as a search template. The OapB apo structure contains two OapB molecules per asymmetric unit (ASU). The OapB–OLE* complex structure determined from crystal form I contains one OapB–OLE* complex per ASU, whereas the structure determined from crystal form II contains two OapB–OLE* complexs per ASU. The initial models were automatically built using the AutoBuild (30) module of the PHENIX software package (31) and manually rebuilt in Coot (32). ERRASER (33) was used to improve the local RNA geometry in the models of OapB–OLE* complex. Heavy atoms were placed based on refined anomalous difference maps. Hydrogen atoms were automatically added to all the protein residues in the OapB apo structures using phenix.ready_set. The OapB apo structures were refined using phenix.refine (34) with anisotropic individual B factors for all nonhydrogen atoms and automatic optimization of X-ray/stereochemistry and X-ray/ADP weights. The OapB–OLE* complex structures were refined using phenix.refine with automatic optimization of X-ray/stereochemistry and X-ray/ADP weights and Translation-Libration-Screw parameters turned on. Custom geometry restraints for the phosphodiester linkage formed between nonnative (GTP) at position 484 and G at position 485 was manually added in the Selection Editor of phenix.refine. No other geometry or secondary structure restraints were imposed on OLE* during refinement. The final refined models were validated using MolProbity (35). Data collection and refinement statistics are summarized in SI Appendix, Table S1. Molecular representations were prepared using UCSF ChimeraX (36) and PyMOL (Schrödinger, LLC).
Supplementary Material
Acknowledgments
We thank members of the Breaker laboratory for helpful discussions and the staff at beamlines 24-ID-C and 24-ID-E of the Advanced Photon Source for technical assistance with X-ray crystallography data collection. This work was supported by NIH Grant T32GM007223 (to D.L.W.) and NIH Grant P01GM022778 (to R.R.B.). RNA research in the Breaker laboratory is also supported by the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020393118/-/DCSupplemental.
Data Availability
Atomic coordinates and associated structure factors have been deposited in the Protein Data Bank with accession codes 7K9B (native OapB apo structure) (37), 7K9C (iodine-derivative OapB apo structure), 7KKV (native OapB–OLE* complex structure, crystal form I) (38), 7K9D (cobalt-derivative OapB–OLE* complex structure, crystal form I) (39), and 7K9E (native OapB–OLE* complex structure, crystal form II) (40).
References
- 1.Cech T. R., Steitz J. A., The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157, 77–94 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Puerta-Fernandez E., Barrick J. E., Roth A., Breaker R. R., Identification of a large noncoding RNA in extremophilic eubacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 19490–19495 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris K. A., Breaker R. R., Large noncoding RNAs in bacteria. Microbiol. Spectr. 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallace J. G., Zhou Z., Breaker R. R., OLE RNA protects extremophilic bacteria from alcohol toxicity. Nucleic Acids Res. 40, 6898–6907 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris K. A., Zhou Z., Peters M. L., Wilkins S. G., Breaker R. R., A second RNA-binding protein is essential for ethanol tolerance provided by the bacterial OLE ribonucleoprotein complex. Proc. Natl. Acad. Sci. U.S.A. 115, E6319–E6328 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris K. A., Odzer N. B., Breaker R. R., Disruption of the OLE ribonucleoprotein complex causes magnesium toxicity in Bacillus halodurans. Mol. Microbiol. 112, 1552–1563 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Block K. F., Puerta-Fernandez E., Wallace J. G., Breaker R. R., Association of OLE RNA with bacterial membranes via an RNA-protein interaction. Mol. Microbiol. 79, 21–34 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Widner D. L., Harris K. A., Corey L., Breaker R. R., Bacillus halodurans OapB forms a high-affinity complex with the P13 region of the noncoding RNA OLE. J. Biol. Chem. 295, 9326–9334 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kyrpides N. C., Woese C. R., Ouzounis C. A., KOW: A novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem. Sci. 21, 425–426 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Youkharibache P., et al., The small beta-barrel domain: A survey-based structural analysis. Structure 27, 6–26 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Hainzl T., Huang S., Sauer-Eriksson A. E., Structural insights into SRP RNA: An induced fit mechanism for SRP assembly. RNA 11, 1043–1050 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leulliot N., Varani G., Current topics in RNA-protein recognition: Control of specificity and biological function through induced fit and conformational capture. Biochemistry 40, 7947–7956 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Rupert P. B., Ferré-D’Amaré A. R., Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature 410, 780–786 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Hohng S., et al., Conformational flexibility of four-way junctions in RNA. J. Mol. Biol. 336, 69–79 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Walter F., Murchie A. I., Lilley D. M., Folding of the four-way RNA junction of the hairpin ribozyme. Biochemistry 37, 17629–17636 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Wu L., Chai D., Fraser M. E., Zimmerly S., Structural variation and uniformity among tetraloop-receptor interactions and other loop-helix interactions in RNA crystal structures. PLoS One 7, e49225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doherty E. A., Batey R. T., Masquida B., Doudna J. A., A universal mode of helix packing in RNA. Nat. Struct. Biol. 8, 339–343 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Nissen P., Ippolito J. A., Ban N., Moore P. B., Steitz T. A., RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. U.S.A. 98, 4899–4903 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antao V. P., Lai S. Y., Tinoco I. Jr, A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 19, 5901–5905 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheong C., Varani G., Tinoco I. Jr, Solution structure of an unusually stable RNA hairpin, 5′GGAC(UUCG)GUCC. Nature 346, 680–682 (1990). [DOI] [PubMed] [Google Scholar]
- 21.Heus H. A., Pardi A., Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science 253, 191–194 (1991). [DOI] [PubMed] [Google Scholar]
- 22.Holm L., DALI and the persistence of protein shape. Protein Sci. 29, 128–140 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Natchiar S. K., Myasnikov A. G., Kratzat H., Hazemann I., Klaholz B. P., Visualization of chemical modifications in the human 80S ribosome structure. Nature 551, 472–477 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Butcher S. E., Pyle A. M., The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc. Chem. Res. 44, 1302–1311 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Kao C., Zheng M., Rüdisser S., A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA 5, 1268–1272 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheldrick G. M., A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger T. C., et al., Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panjikar S., Parthasarathy V., Lamzin V. S., Weiss M. S., Tucker P. A., On the combination of molecular replacement and single-wavelength anomalous diffraction phasing for automated structure determination. Acta Crystallogr. D Biol. Crystallogr. 65, 1089–1097 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terwilliger T. C., et al., Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams P. D., et al., PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chou F. C., Sripakdeevong P., Dibrov S. M., Hermann T., Das R., Correcting pervasive errors in RNA crystallography through enumerative structure prediction. Nat. Methods 10, 74–76 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afonine P. V., et al., Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen V. B., et al., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goddard T. D., et al., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y., Breaker R. R., Crystal structure of Bacillus halodurans OapB (native) at 1.2 A. Protein Data Bank. https://www.rcsb.org/structure/7K9B. Deposited 28 September 2020.
- 38.Yang Y., Breaker R. R., Crystal structure of Bacillus halodurans OapB in complex with its OLE RNA target (native, crystal form I). Protein Data Bank. https://www.rcsb.org/structure/7KKV. Deposited 28 October 2020.
- 39.Yang Y., Breaker R. R., Crystal structure of Bacillus halodurans OapB in complex with its OLE RNA target (cobalt-derivative, crystal form I). Protein Data Bank. https://www.rcsb.org/structure/7K9D. Deposited 28 September 2020.
- 40.Yang Y., Breaker R. R., Crystal structure of Bacillus halodurans OapB in complex with its OLE RNA target (crystal form II). Protein Data Bank. https://www.rcsb.org/structure/7K9E. Deposited 28 September 2020.
- 41.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and associated structure factors have been deposited in the Protein Data Bank with accession codes 7K9B (native OapB apo structure) (37), 7K9C (iodine-derivative OapB apo structure), 7KKV (native OapB–OLE* complex structure, crystal form I) (38), 7K9D (cobalt-derivative OapB–OLE* complex structure, crystal form I) (39), and 7K9E (native OapB–OLE* complex structure, crystal form II) (40).