Significance
For decades, marine biogeographers have been intrigued by the origins of the Indo-Australian Archipelago (IAA) biodiversity hotspot. Yet one important ecological factor remained unexplored: the trophic status of species across the diversity gradient. Here we show how trophic identity crucially underpins coral reef fish diversity patterns via a disproportional concentration of plankton-feeding species in the IAA. This planktivore hotspot, however, vanishes abruptly away from the IAA. Over the recent geological past, planktivorous reef fishes successfully partitioned constant resources promoted by unique oceanographic conditions in the IAA while likely undergoing disproportional extinctions in peripheral regions. This intriguing case of ecological success intertwined with differential extinctions offers key insights into the origins of biodiversity gradients.
Keywords: species richness, macroecology, trophic groups, Indo-Australian Archipelago, extinction
Abstract
One of the most prominent features of life on Earth is the uneven number of species across large spatial scales. Despite being inherently linked to energetic constraints, these gradients in species richness distribution have rarely been examined from a trophic perspective. Here we dissect the global diversity of over 3,600 coral reef fishes to reveal patterns across major trophic groups. By analyzing multiple nested spatial scales, we show that planktivores contribute disproportionally to the formation of the Indo-Australian Archipelago (IAA) marine biodiversity hotspot. Besides being “hotter” at the hotspot, planktivorous fishes display the steepest decline in species numbers with distance from the IAA when compared to other trophic groups. Surprisingly, we did not detect differences in diversification, transition, and dispersal rates in extant species phylogenies that would explain this remarkable gradient in planktivorous fish richness. Thus, we identify two potential complementary drivers for this pattern. First, exceptional levels of partitioning among planktivorous coral reef fishes were driven by temporally stable oceanographic conditions and abundant planktonic resources in the IAA. Second, extinctions of planktivores outside the IAA have been particularly pronounced during Quaternary climate fluctuations. Overall, our results highlight trophic ecology as an important component of global species richness gradients.
The marine realm hosts one of the most remarkable diversity patterns in the world, with a major global biodiversity hotspot in the Indo-Australian Archipelago (IAA) (1–3). While latitudinal trends in marine species richness parallel those observed in terrestrial taxa (3, 4), the longitudinal accumulation of species across the Indo-Pacific is a unique feature of marine systems (3, 5–7). The superimposition of these latitudinal and longitudinal gradients forms a distinct bull’s-eye pattern of species richness peaking in the IAA. This bull’s-eye pattern of marine biodiversity is particularly pronounced in coastal habitats (3), and explanations for its origin have revolved around it being a center of lineage origination, overlap (or accumulation), and survival (reviewed in 5–9).
These explanations are not mutually exclusive, and geological, paleontological, and molecular evidence all points to a complex and temporally dynamic combination of factors operating throughout the Cenozoic, leading to the formation of the modern IAA hotspot. During the Paleocene–Eocene (66 to 33.9 million years ago [Ma]), the marine biodiversity hotspot was situated in the Tethys Sea (10, 11). At that time, the islands in the IAA were just starting to emerge (12, 13) and to accumulate species (14). With tectonic changes in the Oligocene–Miocene (33.9 to 5.3 Ma), the IAA became progressively more complex (12, 13), with new lineages giving rise to most of the extant diversity in multiple marine taxa (e.g., 10, 14–17). Finally, in the last 3 million years (My) the IAA appears to have acted mainly as a center of survival (14), protecting species against extinctions, especially during Quaternary climatic oscillations (18). This historical sequence of events was largely responsible for the genesis of the bull’s-eye pattern of marine species richness distribution.
While history is the key element underpinning the origin of global marine biodiversity patterns, ecological factors may also play an important role. For example, in coral reef fishes, one of the major contributors to the IAA hotspot (3), the availability of shallow-water habitat area, is an important predictor of species richness (1, 19, 20). This area effect overrides the predicted mid-domain model of species ranges stacking in a bounded domain within the Indo-Pacific (2, 21). More recently, species traits have been considered for their role in structuring assemblages (22), and species maximum body size was also found to be a strong predictor of species richness across multiple spatial scales (23). Locations within the IAA tend to have more reef fish species with smaller body sizes (22, 23). In turn, species body size is correlated with dispersal potential (24), which reinforces the disparities in species numbers between the center and the periphery of the marine biodiversity hotspot (25). Thus, the bull’s-eye pattern of reef fish richness is essentially the combination of historical and ecological processes that resulted in the accumulation and maintenance of numerous small-bodied, low-dispersive species in the IAA.
Although many elements of this story have already been revealed, there is a fundamental component missing from the macroevolutionary narrative: trophic status. The trophic identity of species is inherently linked to the pace of species formation in coral reef fishes, with recently derived guilds sustaining higher diversification rates (26). Yet it remains unclear how marine richness gradients are compartmentalized among species with different trophic ecologies. To address this knowledge gap, we assess the trophic component of diversity distributions in coral reef fishes. More specifically, we first describe the global patterns of reef fish species richness across major trophic groups. Subsequently, we explore the relationship between guild richness and distance to the center of marine diversity (IAA) at both global and regional scales, accounting for species body size. Finally, after finding a disproportional accumulation of planktivorous species within the IAA, we investigate the potential evolutionary mechanisms underpinning this pattern. Our results reveal a trophic link to the bull’s-eye pattern of coral reef fish biodiversity distribution.
Results
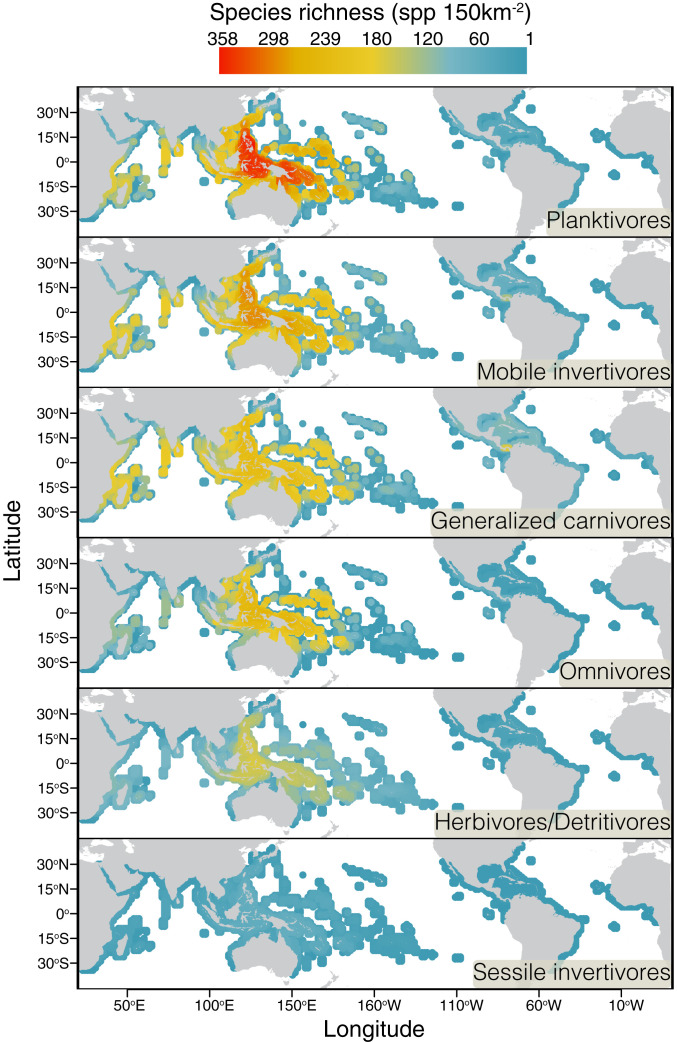
We found remarkable disparities in the distribution of coral reef fish species across trophic groups. In the 13 consensus families examined (i.e., families that occur universally on coral reefs; see Materials and Methods), our global presence–absence dataset revealed species richness to be highest in the IAA in all groups (Fig. 1). However, the absolute number of planktivorous reef fish species exceeds by far those in other trophic groups in grid cells (150 km2) around the IAA (Fig. 1). While over 350 species of planktivores can be found in most IAA grid cells, no other trophic group exceeds 300 species per cell. Indeed, the group with the second-highest number of species per cell (mobile invertivores) has ∼20% less species in the richest cells when compared to planktivores. This accumulation of planktivorous fish species is particularly pronounced around the Philippines, Indonesia, Papua New Guinea, and the Solomon Islands (Fig. 1).
Fig. 1.
Global coral reef fish richness per trophic group. Maps show the absolute number of species per grid cell (n = 2,800) in each classified trophic group. Cell colors correspond to the scale bar and range from low (blue) to intermediate (yellow) and high (red) richness. Grid cell resolution is 150 km2 (see Rabosky et al., 27).
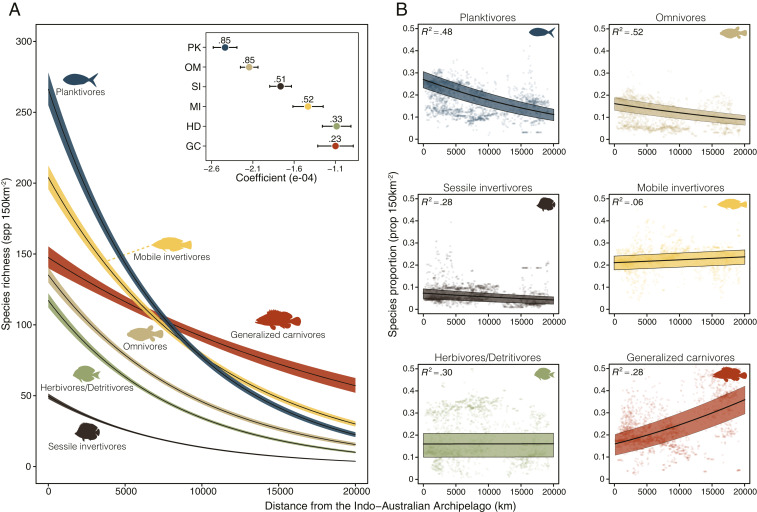
When we modeled species richness per trophic group against the distance to a central point in the IAA (IAAc: latitude = 0°, longitude = 121°E; see Materials and Methods), the disproportional contribution of planktivores to the bull’s-eye pattern became even more evident. As the distance from the IAAc increases, all trophic groups decrease in species richness (Fig. 2A and SI Appendix, Fig. S1). However, planktivores display the steepest decline (Fig. 2A), with a significantly more negative slope than any other trophic group (Fig. 2 A, Inset). Remarkably, this intuitively simple model including only the distance from the IAA and the mean species body size per grid cell was capable of explaining 85% of the global variance in planktivore richness. Besides planktivores, only omnivores had such a high model fit (with a less negative slope), while mobile invertivores, generalized carnivores, herbivores/detritivores, and sessile invertivores had explained variances of 52% or less (Fig. 2A).
Fig. 2.
Global coral reef fish species richness (A) and proportion (B) per guild with distance from the IAA. (A) Number of species per grid cell (mean [black line] ± 95% CI [colored shading]) predicted from a negative binomial model per trophic group. The inset displays the model coefficient (±95% CI) per trophic group along with the R2 value of each model. Lines in the main figure represent the interaction between mean body size and trophic group; therefore, their perceived inclination may not match the size-independent coefficient represented in the Inset. (B) Proportion of species per grid cell in each trophic group (mean [black line] and interquartile range [colored shading]) predicted from beta regression models. Respective pseudo-R2 values are shown in the top left corners. Semitransparent dots represent sampled grid cells (n = 2,800). Predictions from all models were performed with body size fixed in the estimated value for the cells closer to the center of the IAA. Trophic groups: generalized carnivores, red; herbivores/detritivores, green; mobile invertivores, yellow; omnivores, beige; planktivores, blue; sessile invertivores, brown.
Model results were consistent when we used the proportion of species per trophic group as the response variable. Planktivores comprised around 27% (23–30%; interquartile range) of species in IAA cells, a ratio that decreases steeply as one moves away from the IAAc (Fig. 2B). No other trophic group had such a high predicted proportion in cells close to the IAA, and once again, planktivores and omnivores had the highest model fits (48% and 52% of explained variance, respectively). Interestingly, despite the relatively little explained variance, generalized carnivores presented an inverse trend with an increasing proportional richness toward the most peripheral cells, while herbivores/detritivores and mobile invertivores seem to maintain even species proportions globally (Fig. 2B).
It is important to note that the mean species body size per grid cell contributed substantially to model performance. For instance, if we exclude body size as an interactive factor with distance from the IAAc in planktivores, the proportion of explained variance drops from 85 to 34% in our main model. However, this effect is not limited to planktivores. Across all trophic levels, mean species body size tends to be lower in cells closer to the IAAc (SI Appendix, Fig. S2). Therefore, with the exception of herbivores/detritivores, species richness is predicted to be higher in cells that have lower mean species body sizes and are closer to the IAAc (SI Appendix, Fig. S3).
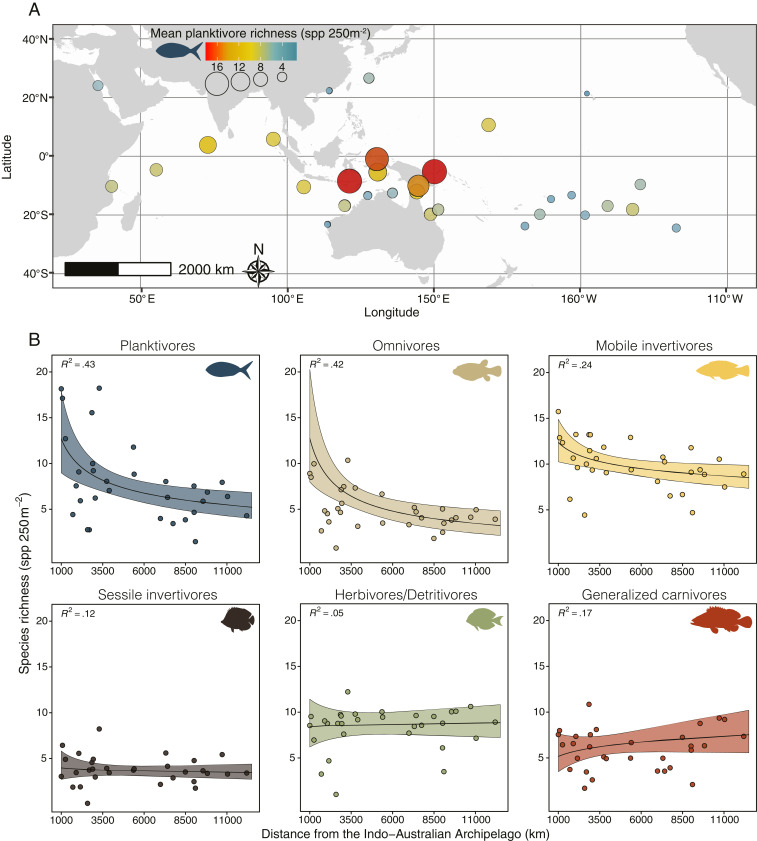
When we analyzed an Indo-Pacific fish survey dataset (848 sites in 31 ecoregions; see Materials and Methods), we detected very similar trends. The mean planktivorous fish richness detected per visual census tended to be substantially higher within the IAA when compared to peripheral sampling locations (Fig. 3A). Furthermore, only visual censuses performed within the IAA (i.e., less than 3,000 km from the IAAc) contained means of 16 to 18 planktivorous species per 250 m2, which is almost double that of most other regions (Fig. 3A). By modeling species richness against distance from the IAAc, we found results that were highly consistent with our global analysis. Besides presenting better model fits, planktivores and omnivores were predicted to have steeper negative slopes with distance from the IAAc than other trophic groups (Fig. 3B). Finally, the results were similar when we analyzed the dataset at the scale of individual sites (SI Appendix, Fig. S4), as opposed to using the sites combined into regions. Alongside our global analysis, these results reveal a robust cross-scale pattern of accumulation of planktivorous species in the IAA.
Fig. 3.
Coral reef fish species richness at the regional scale per trophic group. (A) Mean planktivorous fish species richness per transect in 31 ecoregions (sensu Spalding et al., 28) across the Indo-Pacific. Ecoregions comprise multiple aggregated values from sites (see Materials and Methods), with each site sampled using standardized fish counts covering 250 m2 in area (Edgar and Stuart-Smith, 29). (B) Regional-level mean species richness per transect for each trophic group (from visual surveys, points) at increasing distances from the IAA. Curves show predictions from a generalized linear model (mean [black line] ± 95% CI [colored shading]) with respective R2 values (top left corner). Model predictions were performed with body size fixed in the estimated value for the regions closer to the center of the IAA. Trophic groups: generalized carnivores, red; herbivores/detritivores, green; mobile invertivores, yellow; omnivores, beige; planktivores, blue; sessile invertivores, brown.
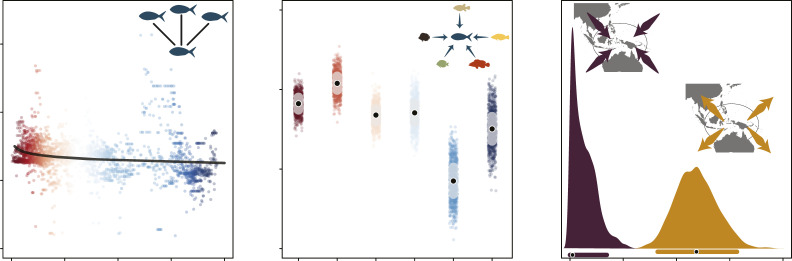
Finally, we assessed the potential evolutionary mechanisms driving the global species richness pattern in planktivores. First, we found that net diversification rates (speciation minus extinction, as calculated from a near-complete reef fish phylogeny; see Materials and Methods) did not present any geographic signal with distance from the IAAc (Fig. 4A and SI Appendix, Fig. S5). Although there seems to be a slight increase in diversification in cells close to the center of the IAA (Fig. 4A), the model fit was very low, suggesting that diversification differences alone would not be sufficient to explain observed richness patterns. Second, the proportion of transitions toward planktivory throughout reef fish evolution was not higher in the IAA when compared to other biogeographic provinces (Fig. 4B). Last, we found substantially higher rates of dispersal in planktivorous lineages going from the IAA toward other regions (Fig. 4C), which is the exact opposite of what would be expected under a scenario of species accumulation due to dispersal.
Fig. 4.
Potential evolutionary mechanisms underpinning the richness gradient in coral reef fish planktivores. (A) Mean net diversification rate per geographic cell (n = 2,800) in planktivorous lineages against distance from the IAA. The black line shows prediction from a generalized linear model with the respective R2 value (top left). (B) Proportion of transitions toward planktivory per biogeographic province. Black points show mean proportions, and shading shows the interquartile range. Semitransparent points represent individual simmap simulations per province (n = 1,000). Provinces: central Pacific (CP), western Indian (WI), tropical eastern Pacific (TEP), eastern Atlantic (EA), and western Atlantic (WA). Colors in A and B depict a gradient of distance from the IAA, with red shades representing locations closest to the IAA and blue shades being farthest from the IAAc. (C) Dispersal rates from (orange) and into (purple) the IAA in planktivorous reef fish lineages. Black points underneath the posterior distributions represent modal values with respective 95% CI (n = 3,600 iterations).
Discussion
Our results suggest that one of the most remarkable gradients in species richness distribution in the world has a strong trophic signal. Although the IAA bull’s-eye pattern of marine species richness has been described for over 50 y (reviewed in 6, 7), an analysis of its trophic characteristics concomitantly with global distribution and survey data has uncovered a hidden pattern in a speciose vertebrate assemblage. Our analyses revealed that while all coral reef fish trophic guilds contain more species in the IAA, planktivores contribute disproportionally to the foundation of this hotspot. This disproportional contribution is consistently recovered across sampling scales and when species proportion per trophic group is considered. Interestingly, although evolutionary processes (speciation, extinction, and dispersal) form the fundamental basis for disparities in species richness globally (30), our phylogenetic analyses did not provide support for any single evolutionary mechanism underlying the planktivore-based hotspot. This suggests that either 1) the drivers of the disproportional richness of planktivorous species in the IAA are not detectable through extant species phylogenies alone (cf. 31, 32) or 2) the pattern has an ecological basis. Below we argue that the explanation may lie in a combination of the two alternatives.
Geological and Oceanographic Drivers.
The Indo-Australian Archipelago is one of the most complex and dynamic geological regions in the tropics (12, 13). Hence, it is widely hypothesized that the exceptional accumulation of marine species within the IAA (3) is ultimately linked to this geological complexity (6). For instance, the large shallow-water habitat area in the IAA has been consistently demonstrated to be a strong predictor of species richness in coral reef fishes (1, 19, 20, 23). However, we show that the effect of the IAA is disproportionally pronounced in planktivorous fishes, which suggests that simple species–area relationships provide only a partial explanation. While the geological complexity of the IAA may, indeed, provide larger coral reef habitat area (20), it also promotes highly dynamic oceanographic conditions (33, 34) which might help explain the high planktivorous fish richness found therein.
The first important element of this oceanographic explanation is associated with the constancy of planktonic resource availability within the IAA. Planktivorous coral reef fishes are heavily reliant on allochthonous food sources that are brought to the reef by complex water movements related to oceanographic currents and tidal regimes (35, 36). Although these particle transportation processes tend to operate mostly on reef-scale topographic features (35), it is likely that the wider-scale oceanographic dynamics in the IAA promotes a constant input of resources for planktivorous fishes. Besides supporting an intense flow of water driven by the strong exchange between Pacific and Indian Ocean waters (33, 34), the IAA is also under the influence of strong upwelling systems and tidal regimes (37). The IAA may thus provide a constant flow of abundant planktonic resources. However, these energetic inputs could only explain the disproportional planktivorous species richness in the IAA if they had been maintained through time (38, 39). Geological evidence provides support for this hypothesis.
Although the initial tectonic history of the IAA dates back to the Eocene (56 to 33.9 Ma), its highest geomorphological complexity was only achieved around 5 Ma (12, 13). Thus, it is reasonable to infer that the major oceanographic processes directly related to the geological features of the archipelago were already in place at that time. This suggests that despite obvious variations related to sea level changes (5), the large-scale oceanography of the IAA has remained relatively constant over the last 5 My. For planktivorous reef fish species, this means a 5 My period with an almost uninterrupted flow of food particles.
Ecological Drivers.
Ecologically, there is clear evidence that planktivorous fishes partition the abundant planktonic food resources in multiple ways. First, reef fish planktivores display clear within-reef spatial distribution patterns (35, 36), with remarkable composition heterogeneity. This heterogeneity is often associated with morphological features that allow some fish species to benefit from the high availability of larger zooplankton in forereef habitats while dealing with increased hydrodynamics or predation pressure (36, 40). Second, planktonic resources are partitioned in time (36). While most planktivorous fish groups are diurnal (e.g., Pomacentridae, Serranidae, Labridae, and Caesioninae), two speciose families are predominantly nocturnal (Apogonidae and Holocentridae). Finally, planktivorous reef fishes may exhibit strong partitioning depending on the resources being targeted. For instance, fairy wrasses (genus Cirrhilabrus) and fusiliers (Caesioninae) appear to target predominantly gelatinous material, in contrast to crustacean zooplankton that is targeted by other planktivorous species (35, 41). Taken together, these lines of evidence suggest that the disproportional amount of planktivorous fish species in the IAA might be the result of successful partitioning of constant and abundant resources.
Evidence from productivity patterns in present-day coral reefs provide further support for the ecological drivers of the IAA planktivorous fish hotspot. In a recent analysis across coral reef fish trophic pathways, Morais and Bellwood (42) revealed that water column-derived productivity may surpass the productivity of any other trophic pathways explored by reef fishes. Although this work was performed on a single coral reef, these pelagic subsidies sustaining high planktivorous fish productivity appear to be widespread, as evidenced by the importance of planktivores to the fish biomass reported by other large-scale studies, particularly in the IAA (43, 44). Alongside the parallel evolution of morphological features permitting water column usage in multiple independent reef fish lineages (45), our results indicate that planktivory can be a successful evolutionary and ecological strategy, provided that a constant supply of planktonic resources is maintained. Hence, the patterns described herein agree with long-standing ecological hypotheses that correlate species diversity and coexistence with resource availability, temporal stability, productivity, and niche partitioning (46, 47).
Evolutionary Drivers.
The resource-related factors explained above appear to provide a compelling case for the differences in species richness between planktivores and other reef fish trophic groups within the IAA. However, explaining the disproportional drop in planktivorous species with distance from the IAA hinges on the understanding that the mechanisms driving such declines are unlikely to be detectable in extant species phylogenies. Past research has shown that the distance from stable coral reef habitats during Quaternary climate fluctuations (last 2.6 My) outweighs present-day environmental factors in explaining global reef fish richness patterns (18). This highlights the potential role of areas that maintained suitable coral reef habitat over geological time as extinction refugia for fishes (18). Our results provide an analogous productivity-based scenario. Given that most historical coral reef refugia lie within the IAA, it seems likely that planktivorous fishes have been disproportionally affected by extinction in areas away from the IAA. In other words, planktivorous species distributions point strongly to differential survival within the IAA vs. peripheral locations during the last 5 My.
The nature of the loss of species in the peripheral locations is hard to establish as it is virtually impossible to detect extinction events and estimate extinction rates without a detailed fossil record (31). The loss of species from peripheral areas may be a result of short-term phenomena, e.g., barriers to establishment, with colonization events being stymied due to unsuitable environmental conditions. Alternatively, they may be longer-term phenomena, with population declines and range contractions leading to local or global extinction. In either case, the loss of species appears to be dependent on the sustained availability of suitable resources. We posit that reef refuges and historically stable oceanographic conditions in the IAA provided constant habitat and food resources for planktivorous fishes through time. This scenario helps to explain 1) why we did not find a geographic signal in the diversification rates of planktivores (Fig. 4A) and 2) our results for transition rates (Fig. 4C). The IAA likely served as a source of surviving planktivorous lineages that recolonized depauperate peripheries after extinction events. It is also interesting to note that one of the very few, and perhaps the sole, documented cases of extinction in a previously abundant reef fish was the loss of the Galapagos planktivorous damselfish Azurina eupalama. Its extinction was apparently driven by changing oceanographic conditions in a geographically peripheral location (48).
Finally, it is important to highlight that our findings apply to fish species that dwell in shallow-water coral reef habitats (less than 30 m deep). Although our knowledge about deeper-water fishes has been increasing at an unprecedented rate, it is still insufficient to draw general conclusions about the large-scale biogeographic patterns of these taxa (49). Mesophotic (30 to 150 m) species distribution data to date suggest that the diversity gradients between the IAA and central Pacific islands are not as steep as in shallow-water taxa (49). However, given that the geological complexity of the IAA extends into deeper waters and that a disproportional number of mesophotic species in the IAA are planktivores (50), we hypothesize that patterns similar to the ones found herein, for shallow-water taxa, will also be found in mesophotic fish species.
In conclusion, our study highlights a unique association between large-scale marine diversity gradients and species trophic identities. Splitting the global coral reef fish species distribution data between trophic groups revealed that planktivores are major contributors to the disparate richness within the IAA biodiversity hotspot. This is likely related to the persistent oceanographic conditions promoted by the geological complexity of the IAA over the last 5 My. By providing an abundant and constant flow of food through time, this oceanographic setting fostered high levels of resource partitioning among planktivorous reef fishes within the IAA. Peripheral regions, on the other hand, almost certainly experienced periods of intense extinction of planktivorous fish lineages associated with habitat loss and oceanographic changes. Despite having been recolonized by some surviving lineages from the IAA, these peripheral regions appear to carry the imprint of past extinctions within planktivorous coral reef fishes.
Materials and Methods
Species Distribution and Survey Data.
We used two independent datasets of coral reef fish distributions: a global presence–absence record of species in 150 km2 resolution grids (27, 51) and a fish community survey dataset (29). The presence–absence dataset was downloaded from a publicly available repository (51) and was built using the AquaMaps algorithm (52). The authors estimated geographic ranges of marine fishes based on species occurrence records and a set of environmental predictors (27). From this presence–absence dataset, we filtered those species that belong to the consensus coral reef fish families (sensu 53). These 13 families are always found on coral reefs irrespective of their biogeographic location (i.e., Acanthuridae, Apogonidae, Blenniidae, Carangidae, Chaetodontidae, Gobiidae, Holocentridae, Labridae, Lutjanidae, Mullidae, Pomacanthidae, Pomacentridae, and Serranidae). Therefore, our focus here was on fish families that universally occur on coral reefs, rather than “reefs” sensu lato, to avoid potentially confounding effects of habitat type. Following previous molecular phylogenetic analyses, we considered Caesioninae a subfamily of Lutjanidae (54) and Microdesminae and Ptereleotrinae part of Gobiidae (55). Altogether, the families considered here comprise ∼3,600 described species. Based on the geographic ranges of these coral reef fishes within the dataset, we calculated the number of overlapping species per grid cell. Subsequently, we divided the species richness per cell according to the classified trophic groups (see Species Trait Data). Finally, we kept only cells that had at least one species per trophic group in the dataset to avoid distribution extremes where very few species occur. Our final presence–absence dataset consisted of 2,800 geographic cells containing the number of species per trophic group along with the respective latitudinal and longitudinal coordinates of the centroid of these cells.
The community survey dataset was downloaded from the publicly available Reef Life Survey website (https://reeflifesurvey.com). This dataset consists of global fish surveys, systematically collected using standardized methods (29). Each individual survey (transect) involves an underwater visual census of fish communities that covers two blocks of 250 m2 each, totaling 500 m2 per survey (29). We averaged the species counts between these two blocks to get the mean number of species in a final area of 250 m2 per transect. Our goal with this dataset was to assess the richness per transect across the Indo-Pacific; therefore, we downloaded the surveys ranging from the western Indian Ocean and Red Sea to the central Pacific islands (Fig. 3A). After filtering data available in these regions that contained a minimum of four transects per ecoregion, we used the data from 848 sites. To calculate the mean richness per site across trophic groups, we categorized all species recorded in the transects according to our defined guilds (see Species Trait Data). Finally, to be able to explore cross-scale patterns of species distributions, we aggregated individual sites into ecoregions (sensu 28) and calculated the mean species richness per trophic group in each region. This regional dataset comprised 31 ecoregions containing at least three sites each.
Species Trait Data.
We used a previously assembled dataset on reef fish ecological traits (26) to assess species-specific trophic identity and body size. The maximum body length (body size) data for each species within this dataset was originally sourced from FishBase (56). For the trophic identity, species were grouped into six major guilds: generalized carnivores, mobile invertivores, omnivores, planktivores, sessile invertivores, and herbivores/detritivores. These guilds are based on species diets in the adult life stages and were previously defined in the literature (57). The major differences, however, are that the original herbivores/macroalgivores group has been merged with the general herbivores/detritivores guild and that we used a broader categorization for carnivores (26). While Mouillot et al. (57) classify species that feed on larger prey (i.e., fish and cephalopods) as piscivores, we adopt a broader category of generalized carnivores to include species that feed more generally on larger elusive prey (including larger crustaceans). Our trophic categorization was used in combination with the distribution and survey datasets to calculate the number of species in each guild per geographic cell (presence–absence data), ecoregion, and site (survey data). In addition to the richness per trophic group, we also calculated the mean species size per guild in each geographic cell, ecoregion, and site using the body size data (26). This body size dataset was then used in our statistical modeling procedures.
Statistical Analyses.
To assess the relationship between guild richness and distance from the center of marine biodiversity, we first calculated the geographical distance (in kilometers) between each grid cell, region, and site to a central point in the IAA (IAAc: latitude = 0°, longitude = 121°E). This was done using the function distHaversine from the 'geosphere' R package (58). Subsequently, we applied negative binomial models to correlate the species richness per grid cell (presence–absence data) with the distance from the IAAc in each trophic guild. These models were built using the gam function within the 'mgcv' R package (59), and all accounted for spatial autocorrelation between geographic cells. We also calculated the proportional guild richness per geographic cell and modeled it against distance from the IAAc using beta regressions implemented in the 'betareg' R package (60). For the community survey dataset, we fitted generalized linear models per trophic group against distance from the IAAc since the richness in this case represents averaged values of multiple transects (site) and sites (ecoregion). Both the site and the ecoregion models were fitted using a gamma distribution for the response variable (mean species richness) with a logarithmic link.
Species body size was recently demonstrated to be a key predictor of coral reef richness across spatial scales (23). Therefore, all of our models were fitted using the mean body size per sampled area as an interactive factor with distance from the IAAc. Since we were interested in isolating the effect of distance from the IAAc in species richness, we performed our main model predictions using the mean body size fixed in the estimated value for the cells, regions, and sites closer to the IAAc. To calculate this fixed value, we first fitted a LOESS (locally estimated scatterplot smoothing) polynomial regression with an α parameter of 0.7 between mean body size and distance to the IAAc. Then we extracted the first estimated value derived from this relationship and used it in our main model predictions. Finally, to assess the effect of varying body size values in our global model results, we performed predictions using the median and the 2.5, 25, and 75% quantiles of the distribution of mean species body size per geographic cell in each trophic group.
Phylogenetic Comparative Methods.
After finding a disproportional contribution of planktivores to the IAA biodiversity hotspot (see Results), we explored the potential evolutionary mechanisms driving this pattern. First, planktivorous species might have accumulated in the IAA as a result of higher diversification within that area. To test this, we calculated the mean tip diversification rate of planktivorous lineages in each geographic cell. The species-specific diversification values were extracted from the Siqueira et al. (26) dataset and were originally calculated using the software BAMM (61). Then we fitted a generalized linear model of planktivorous tip diversification rate against distance to the IAAc to assess whether origination was higher in cells within the IAA. This model was fitted using a gamma distribution for the response variable (net diversification rate) with a logarithmic link.
Alternatively, the disproportional accumulation of planktivores within the IAA might have been the result of higher transition rates toward that guild since transitions to planktivory have been shown to be prevalent throughout reef fish evolution (26). To test this hypothesis, we calculated the proportion of transitions to planktivory between marine biogeographic provinces using stochastic character mappings (62). First, we categorized all consensus species present in the Siqueira et al. (26) phylogenetic tree according to presence or absence in six biogeographic provinces: IAA, central Pacific, western Indian, tropical eastern Pacific, eastern Atlantic, and western Atlantic. Then we pruned the phylogeny to contain only species that were present in each province. Finally, for each pruned tree, we simulated 1,000 stochastic maps of trophic guilds using a modified version of the make.simmap function (63) that considers rate heterogeneity across the tree (26). From the simmap results, we calculated the proportion of transitions toward planktivory from the total trophic transitions.
Last, the planktivorous fish hotspot might have resulted from an accumulation of lineages via dispersal into the IAA. We assessed this hypothesis by applying the GeoSSE model (64), within the 'diversitree' R package (65). This model allows the estimation of dispersal rates associated with geographical states along a phylogenetic tree. Therefore, we built an unconstrained GeoSSE model to calculate dispersal rates out and into the IAA, considering the presence or absence of species within that area. This model was applied to a phylogenetic tree that was pruned to only contain planktivorous species. We used the resulting model coefficients to implement the Bayesian framework of diversitree and sample the posterior distribution of dispersal parameters. We ran 4,000 iterations of the MCMC (Markov Chain Monte Carlo) with exponential priors from a preliminary run of 100 iterations. Finally, we eliminated the initial 10% of samples as burn-in and assessed convergence through the effective sample sizes.
Supplementary Material
Acknowledgments
We thank S. Floeter, C. Hemingson, V. Huertas, M. Mihalitsis, R. Streit, and S. Tebbett for enlightening discussions; two anonymous reviewers for insightful comments; and Rabosky et al. (27) and the Reef Life Survey team for making their data freely available. Funding was provided by the Australian Research Council (D.R.B. and P.F.C., Centre of Excellence for Coral Reef Studies; D.R.B., A.C.S., and R.A.M., grant number LF190100062; P.F.C., grant number DE170100516) and James Cook University (Postgraduate Research Scholarship to A.C.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019404118/-/DCSupplemental.
Data Availability.
Raw data and R scripts generated during the current study are avilable at the Zenodo repository (http://doi.org/10.5281/zenodo.4475349) (66).
References
- 1.Bellwood D. R., Hughes T. P., Regional-scale assembly rules and biodiversity of coral reefs. Science 292, 1532–1535 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Mora C., Chittaro P. M., Sale P. F., Kritzer J. P., Ludsin S. A., Patterns and processes in reef fish diversity. Nature 421, 933–936 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Tittensor D. P., et al., Global patterns and predictors of marine biodiversity across taxa. Nature 466, 1098–1101 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Willig M. R., Kaufman D. M., Stevens R. D., Latitudinal gradients of biodiversity: Pattern, process, scale, and synthesis. Annu. Rev. Ecol. Evol. Syst. 34, 273–309 (2003). [Google Scholar]
- 5.Hoeksema B. W., “Delineation of the Indo-Malayan centre of maximum marine biodiversity: The Coral Triangle” in Biogeography, Time, and Place: Distributions, Barriers, and Islands, Renema W., Ed. (Springer, Dordrecht, Netherlands, 2007), pp. 117–178. [Google Scholar]
- 6.Bellwood D. R., Renema W., Rosen B. R., “Biodiversity hotspots, evolution and coral reef biogeography” in Biotic Evolution and Environmental Change in Southeast Asia, Gower D. J., et al., Eds. (Cambridge University Press, 2012), pp. 216–245. [Google Scholar]
- 7.Briggs J. C., Coincident biogeographic patterns: Indo-West Pacific Ocean. Evolution 53, 326–335 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Gaither M. R., Rocha L. A., Origins of species richness in the Indo-Malay-Philippine biodiversity hotspot: Evidence for the centre of overlap hypothesis. J. Biogeogr. 40, 1638–1648 (2013). [Google Scholar]
- 9.Briggs J. C., Marine centres of origin as evolutionary engines. J. Biogeogr. 30, 1–18 (2003). [Google Scholar]
- 10.Renema W., et al., Hopping hotspots: Global shifts in marine biodiversity. Science 321, 654–657 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Leprieur F., et al., Plate tectonics drive tropical reef biodiversity dynamics. Nat. Commun. 7, 11461 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall R., Cenozoic geological and plate tectonic evolution of SE Asia and the SW pacific: Computer-based reconstructions, model and animations. J. Asian Earth Sci. 20, 353–431 (2002). [Google Scholar]
- 13.Lohman D. J., et al., Biogeography of the Indo-Australian archipelago. Annu. Rev. Ecol. Evol. Syst. 42, 205–226 (2011). [Google Scholar]
- 14.Cowman P. F., Bellwood D. R., The historical biogeography of coral reef fishes: Global patterns of origination and dispersal. J. Biogeogr. 40, 209–224 (2013). [Google Scholar]
- 15.Williams S. T., Duda T. F. Jr, Did tectonic activity stimulate Oligo-Miocene speciation in the Indo-West Pacific? Evolution 62, 1618–1634 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Bellwood D. R., Goatley C. H. R., Bellwood O., The evolution of fishes and corals on reefs: Form, function and interdependence. Biol. Rev. Camb. Philos. Soc. 92, 878–901 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Miller E. C., Hayashi K. T., Song D., Wiens J. J., Explaining the ocean’s richest biodiversity hotspot and global patterns of fish diversity. Proc. Biol. Sci. 285, 20181314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pellissier L., et al., Quaternary coral reef refugia preserved fish diversity. Science 344, 1016–1019 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Bellwood D. R., Hughes T. P., Connolly S. R., Tanner J., Environmental and geometric constraints on Indo-Pacific coral reef biodiversity. Ecol. Lett. 8, 643–651 (2005). [Google Scholar]
- 20.Parravicini V., et al., Global patterns and predictors of tropical reef fish species richness. Ecography 36, 1254–1262 (2013). [Google Scholar]
- 21.Connolly S. R., Bellwood D. R., Hughes T. P., Indo-pacific biodiversity of coral reefs: Deviations from a mid-domain model. Ecology 84, 2178–2190 (2003). [Google Scholar]
- 22.Parravicini V., et al., Strong ‘functional’ divergence of tropical reef fish assemblages along the global diversity gradient. 10.1101/2020.09.18.303008 (20 September 2020). [DOI] [Google Scholar]
- 23.Barneche D. R., et al., Body size, reef area and temperature predict global reef-fish species richness across spatial scales. Glob. Ecol. Biogeogr. 28, 315–327 (2019). [Google Scholar]
- 24.Luiz O. J., et al., Adult and larval traits as determinants of geographic range size among tropical reef fishes. Proc. Natl. Acad. Sci. U.S.A. 110, 16498–16502 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donati G. F. A., et al., A process‐based model supports an association between dispersal and the prevalence of species traits in tropical reef fish assemblages. Ecography 42, 2095–2106 (2019). [Google Scholar]
- 26.Siqueira A. C., Morais R. A., Bellwood D. R., Cowman P. F., Trophic innovations fuel reef fish diversification. Nat. Commun. 11, 2669 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabosky D. L., et al., An inverse latitudinal gradient in speciation rate for marine fishes. Nature 559, 392–395 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Spalding M. D., et al., Marine ecoregions of the world: A bioregionalization of coastal and shelf areas. Bioscience 57, 573–583 (2007). [Google Scholar]
- 29.Edgar G. J., Stuart-Smith R. D., Systematic global assessment of reef fish communities by the Reef Life Survey program. Sci. Data 1, 140007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jablonski D., Roy K., Valentine J. W., Out of the tropics: Evolutionary dynamics of the latitudinal diversity gradient. Science 314, 102–106 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Quental T. B., Marshall C. R., Diversity dynamics: Molecular phylogenies need the fossil record. Trends Ecol. Evol. 25, 434–441 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Louca S., Pennell M. W., Extant timetrees are consistent with a myriad of diversification histories. Nature 580, 502–505 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Gordon A. L., Fine R. A., Pathways of water between the Pacific and Indian oceans in the Indonesian seas. Nature 379, 146–149 (1996). [Google Scholar]
- 34.Gordon A. L., Oceanography of the Indonesian seas and their throughflow. Oceanography (Wash. D.C.) 18, 15–27 (2005). [Google Scholar]
- 35.Hamner W. M., Jones M. S., Carleton J. H., Hauri I. R., Williams D. M., Zooplankton, planktivorous fish, and water currents on a windward reef face: Great barrier reef, Australia. Bull. Mar. Sci. 42, 459–479 (1988). [Google Scholar]
- 36.Hobson E. S., “Trophic relationships of fishes specialized to feed on zooplankters above coral reefs” in The Ecology of Fishes on Coral Reefs, Sale P. F., Ed. (Academic Press, 1991), pp. 69–95. [Google Scholar]
- 37.Robertson R., Ffield A., M2 baroclinic tides in the Indonesian seas. Oceanography (Wash. D.C.) 18, 62–73 (2005). [Google Scholar]
- 38.Sanders H. L., Marine benthic diversity: A comparative study. Am. Nat. 102, 243–282 (1968). [Google Scholar]
- 39.Fischer A. G., Latitudinal variations in organic diversity. Evolution 14, 64–81 (1960). [Google Scholar]
- 40.Hobson E., Chess J., Trophic relationships among fishes and plankton in the lagoon at Enewetak Atoll, Marshall Islands. Fish Bull. 76, 133–153 (1978). [Google Scholar]
- 41.Huertas V., Bellwood D. R., Trophic separation in planktivorous reef fishes: A new role for mucus? Oecologia 192, 813–822 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Morais R. A., Bellwood D. R., Pelagic subsidies underpin fish productivity on a degraded coral reef. Curr. Biol. 29, 1521–1527.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Heenan A., Williams G. J., Williams I. D., Natural variation in coral reef trophic structure across environmental gradients. Front. Ecol. Environ. 18, 69–75 (2020). [Google Scholar]
- 44.Campbell S. J., et al., Fishing restrictions and remoteness deliver conservation outcomes for Indonesia’s coral reef fisheries. Conserv. Lett. 13, e12698 (2020). [Google Scholar]
- 45.Floeter S. R., Bender M. G., Siqueira A. C., Cowman P. F., Phylogenetic perspectives on reef fish functional traits. Biol. Rev. Camb. Philos. Soc. 93, 131–151 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Connell J. H., Orias E., The ecological regulation of species diversity. Am. Nat. 98, 399–414 (1964). [Google Scholar]
- 47.MacArthur R., Species packing and competitive equilibrium for many species. Theor. Popul. Biol. 1, 1–11 (1970). [DOI] [PubMed] [Google Scholar]
- 48.Hawkins J. P., Roberts C. M., Clark V., The threatened status of restricted-range coral reef fish species. Anim. Conserv. 3, 81–88 (2000). [Google Scholar]
- 49.Pyle R., et al., “Fishes: Biodiversity” in Mesophotic Coral Ecosystems, Loya Y., Puglise K., Bridge T., Eds. (Springer, 2019), pp. 749–777. [Google Scholar]
- 50.Pinheiro H. T., et al., Deep reef fishes in the world’s epicenter of marine biodiversity. Coral Reefs 38, 985–995 (2019). [Google Scholar]
- 51.Rabosky D. L., et al., Data from “An inverse latitudinal gradient in speciation rate for marine fishes.” Dryad . 10.5061/dryad.fc71cp4. Accessed 10 July 2019. [DOI] [PubMed]
- 52.Ready J., et al., Predicting the distributions of marine organisms at the global scale. Ecol. Modell. 221, 467–478 (2010). [Google Scholar]
- 53.Bellwood D. R., The Eocene fishes of Monte Bolca: The earliest coral reef fish assemblage. Coral Reefs 15, 11–19 (1996). [Google Scholar]
- 54.Miller T. L., Cribb T. H., Phylogenetic relationships of some common Indo-Pacific snappers (Perciformes: Lutjanidae) based on mitochondrial DNA sequences, with comments on the taxonomic position of the Caesioninae. Mol. Phylogenet. Evol. 44, 450–460 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Tornabene L., Chen Y., Pezold F., Gobies are deeply divided: Phylogenetic evidence from nuclear DNA (Teleostei: Gobioidei: Gobiidae). Syst. Biodivers. 11, 345–361 (2013). [Google Scholar]
- 56.Froese R., Pauly D., FishBase, Version 04/2019. https://www.fishbase.de/. Accessed 10 July 2019.
- 57.Mouillot D., et al., Functional over-redundancy and high functional vulnerability in global fish faunas on tropical reefs. Proc. Natl. Acad. Sci. U.S.A. 111, 13757–13762 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hijmans R. J., geosphere: Spherical trigonometry (R package Version 1.5-10, 2019).
- 59.Wood S. N., Generalized Additive Models: An Introduction with R (Chapman and Hall/CRC, 2017). [Google Scholar]
- 60.Cribari-Neto F., Zeileis A., Beta regression in R. J. Stat. Softw. 34, 1–24 (2010). [Google Scholar]
- 61.Rabosky D. L., Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One 9, e89543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huelsenbeck J. P., Nielsen R., Bollback J. P., Stochastic mapping of morphological characters. Syst. Biol. 52, 131–158 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Revell L. J., Phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012). [Google Scholar]
- 64.Goldberg E. E., Lancaster L. T., Ree R. H., Phylogenetic inference of reciprocal effects between geographic range evolution and diversification. Syst. Biol. 60, 451–465 (2011). [DOI] [PubMed] [Google Scholar]
- 65.FitzJohn R. G., Diversitree: Comparative phylogenetic analyses of diversification in R. Methods Ecol. Evol. 3, 1084–1092 (2012). [Google Scholar]
- 66.Siqueira A. C., Morais R. A., Bellwood D. R., Cowman P. F., Data from “Planktivores as trophic drivers of global coral reef fish diversity patterns” [data set]. Zenodo . 10.5281/zenodo.4475349. Deposited 28 January 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data and R scripts generated during the current study are avilable at the Zenodo repository (http://doi.org/10.5281/zenodo.4475349) (66).