Diallyl disulfide (DAS) from garlic did not affect the growth dynamics of Pseudomonas aeruginosa PAO1; however, DAS inhibited transcription of most of the quorum sensing (QS) system genes, including lasR, rhlI/rhlR, and pqsABCDE/pqsR; thus, biosynthesis of the signal molecules C4‐HSL (encoded by rhlI) and PQS (encoded by pqsABCDE) was inhibited.DAS suppressed the production of some virulence factors toxic to the host and enhanced the production of some nutrition factors beneficial to the host. These actions of DAS may be due to its thioether group.
Summary
Diallyl sulfide (DAS) and diallyl disulfide (DADS), two constituents of garlic, can inhibit quorum sensing (QS) systems of Pseudomonas aeruginosa. However, the differences in the mechanism of QS inhibition between DAS and DADS, and the functional chemical groups of these sulfides that contribute in QS inhibition have not been elucidated yet. We assumed that the sulfide group might play a key role in QS inhibition. To prove this hypothesis and to clarify these unsolved problems, in this study, we synthesized diallyl ether (DAE), and compared and investigated the effects of DAS and DAE on the growth and production of virulence factors, including Pseudomonas quinolone signal (PQS), elastase and pyocyanin, of P. aeruginosa PAO1. Transcriptome analysis and qRT‐PCR were used to compare and analyse the differentially expressed genes between the different treatment groups (DAS, DAE and control). The results indicated that DAS did not affect the growth dynamics of P. aeruginosa PAO1; however, DAS inhibited transcription of most of the QS system genes, including lasR, rhlI/rhlR and pqsABCDE/pqsR; thus, biosynthesis of the signal molecules C4‐HSL (encoded by rhlI) and PQS (encoded by pqsABCDE) was inhibited. Furthermore, DAS inhibited the transcription of virulence genes regulated by the QS systems, including rhlABC, lasA, lasB, lecA and phzAB, phzDEFG, phzM and phzS that encode for rhamnolipid, exoprotease, elastase, lectin and pyocyanin biosynthesis respectively. DAS also enhanced the expression of the key genes involved in the biosynthesis of three B vitamins: folate, thiamine and riboflavin. In conclusion, DAS suppressed the production of some virulence factors toxic to the host and enhanced the production of some nutrition factors beneficial to the host. These actions of DAS may be due to its thioether group. These findings would be significant for development of an effective drug to control the virulence and pathogenesis of the opportunistic pathogen P. aeruginosa.
Introduction
Pseudomonas aeruginosa is a ubiquitous gram‐negative opportunistic pathogen that causes a variety of infections in human, including chronic infection of the airway in patients with cystic fibrosis (Stover et al., 2000; Martin et al., 2018; Kostylev et al., 2019). P. aeruginosa produces a large number of virulence factors that cause persistent infections (Lee and Zhang, 2015). The bacterium also has many genes that confer strong resistance against antibiotics, and its antibiotic resistance may manifest through mutations and horizontal gene transfer, which pose challenge for effective clinical treatment (Fruci and Poole, 2018; Lopez‐causape et al., 2018; Moura‐Alves et al., 2019). Therefore, it is necessary to develop new treatment strategies for P. aeruginosa infections. It has been reported that at least 3‐6% of the P. aeruginosa genome is regulated by quorum sensing systems based on transcriptomic analysis (Hentzer et al., 2003; Smyth et al., 2010). Therefore, one feasible option is to suppress the virulence and pathogenicity of P. aeruginosa by inhibiting its quorum sensing (QS) systems.
Garlic (Allium sativum) is a common herb, which has been used in folk medicine for thousands of years. Garlic has a wide range of antimicrobial activities, including antibacterial, antifungal and antiviral (Harris et al., 2001). The antifungal activity of garlic is higher than its antibacterial activity (Avato et al., 2000). Garlic distilled oil contains more than 20 organic sulfides with biological properties (Ko et al., 2017), including diallyl sulfide (DAS), diallyl disulfide (DADS), diallyl trisulfide (DATS) and diallyl tetrasulfide (DATTS). The antibacterial activity of the four sulfides against P. aeruginosa increases with increase in the number of disulfide bonds (Tsao and Yin, 2001). Garlic oil also enhances host inflammatory response to improve the ability of clearing P. aeruginosa (Bjarnsholt et al., 2005). A previous study concluded that the allyl group imparts antibacterial property to the four sulfides (Casella et al., 2013). Researchers have also found that garlic extract can inhibit virulence genes regulated by the QS systems of P. aeruginosa (Rasmussen et al., 2005). Later, researchers found that garlic extracts inhibit the production of QS signal molecules and extracellular virulence factors of P. aeruginosa (Harjai et al., 2010), which was verified by in vitro and in vivo experiments (Smyth et al., 2010). Two years later, researchers found that the compound in garlic with QS inhibitory activity against P. aeruginosa was ajoene, a sulfur‐rich compound (Jakobsen et al., 2012). Some other ingredients from garlic were also preliminarily detected to have QS inhibitory activity on luxI/luxR, including DAS, DADS, DATS, DATTS, 2‐vinyl‐4H‐1,3‐dithiine and 3‐vinyl‐3,4‐dihydro‐1,2‐dithiine (Persson et al., 2005; Cady et al., 2012). In our previous study, we verified that DADS in garlic can inhibit QS of P. aeruginosa; DADS inhibits the production of virulence factors in P. aeruginosa by inhibiting the expression of the key genes in the three QS systems, namely las, rhl and pqs (Li et al., 2018, 2019). At the same time, we also detected that DAS from garlic inhibit QS of P. aeruginosa. However, is DAS has the same effect as DADS on QS inhibition of P. aeruginosa? What are the differences in the mechanism of QS inhibition between them? These are problems that have not been clarified. Moreover, the functional chemical groups of these sulfides that acted on the QS inhibition were also unknown. It was assumed that the sulfide group possibly plays the main role in the process of QS inhibition.
To prove this hypothesis and to clarify these unsolved problems, in this study, we synthesized diallyl ether (DAE), and compared and investigated the differences in biological and molecular effects of DAS and DAE on transcriptome of P. aeruginosa PAO1, on cell growth, and on production of its virulence factors. DAS and DAE have the same chemical structure, except for the presence of thioether group in DAS and ether group in DAE. Furthermore, we can analyse the differences in the mechanism of QS inhibition between DAS and DADS and to further identify and verify the chemical groups of these sulfides that are responsible for their QS inhibition activity.
Results
Effects of diallyl sulfide and diallyl ether on the transcriptome of PAO1 strain
Volcano plot analysis
The transcriptome sequencing data of control, DAS‐treated and DAE‐treated groups were compared and analysed in the following manner: DAE vs. DAS, control vs. DAS and control vs. DAE. The details of the differentially expressed genes among the three groups are shown in a volcano plot (Fig. S1). The volcano plot showed that the number of differentially expressed genes between DAE‐treated group and DAS‐treated group was high (Fig. S1A, total 2195, upregulated 1608, downregulated 587). There were many differentially expressed genes in the PAO1 strain between the control group and DAS‐treated group (Fig. S1B, total 2771, upregulated 1905, downregulated 866), while the number of differentially expressed genes between the control group and DAE‐treated group was relatively low (Fig. S1C, total 770, upregulated 349, downregulated 421). The volcano plots indicated that DAS had much more effect on the PAO1 strain than DAE.
Venn diagram analysis
In order to further analyse the relationship between the differentially expressed genes among the different groups, we analysed the differentially expressed genes in the transcriptome by constructing intergroup Venn diagram (Fig. 1B). The Venn diagram shows that 621 genes were differentially expressed after DAS and DAE treatments, but 285 genes among them were differently expressed between DAS and DAE treatments, and 336 genes among them had the same differential expression. More importantly, 1509 differentially expressed genes between control and DAS‐treated group were differentially expressed between DAS and DAE treatment, but not differentially expressed between control and DAE‐treated group. The Venn diagram also showed that the effects of DAS on the transcriptome of the PAO1 strain were much more significant than that of DAE, although some of the changes by both the treatments were same.
Fig. 1.
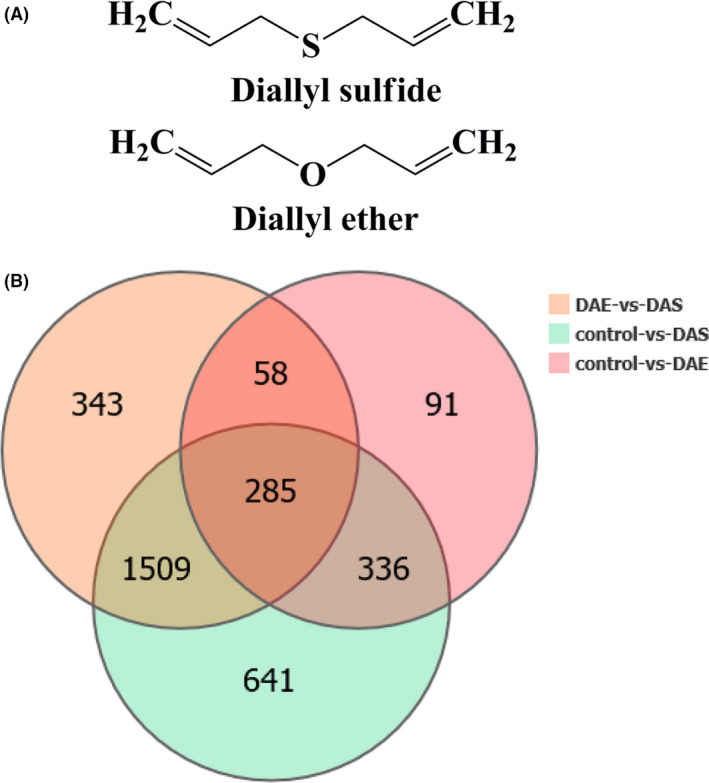
Chemical structural formula of diallyl sulfide (DAS) and diallyl ether (DAE), and Venn diagram of differentially expressed genes (fold change > 2, P value < 0.05) of the PAO1 strain in the control group, DAS‐treated group and DAE‐treated group based on RNA‐seq.
A. Chemical structural formula of DAS and DAE.
B. Venn diagram of differentially expressed genes of the PAO1 strain in the control group, DAS‐treated group, DAE‐treated group and their overlaps based on RNA‐seq.
The Kyoto encyclopedia of Genes and Genomes enrichment analysis
After identifying the differentially expressed genes in the transcriptome of the PAO1 strain after DAS and DAE treatment, we classified and enriched the differentially expressed genes in the three experimental groups in the Kyoto encyclopedia of Genes and Genomes (KEGG) to explore the deep influence and molecular mechanism of DAS on the PAO1 strain. The KEGG enrichment map of the differentially expressed genes in the three experimental groups is shown in Fig. 2. The differentially expressed genes were significantly enriched in 15 KEGG pathways (0.00 < q value < 0.50) (Fig. 2A), when the transcriptome of the DAE‐treated group was compared with that of the DAS‐treated group. The enriched KEGG pathways with significantly downregulated gene transcription were as follows: phenazine biosynthesis, quorum sensing and biosynthesis of siderophore group non‐ribosomal peptides; the enriched KEGG pathways with significantly upregulated gene transcription were as follows: cationic antimicrobial peptide (CAMP) resistance, folate biosynthesis, thiamine metabolism, sulfur relay system, pentose and glucoronate interconversions, beta‐lactam resistance, riboflavin metabolism, ascorbate and aldarate metabolism, lipopolysaccharide biosynthesis, glycerolipid metabolism and sulfur metabolism; many upregulated genes as well as downregulated genes in the KEGG pathway were responsible for biosynthesis of antibiotics. Some of the detailed KEGG pathways are provided in Fig. S2, including phenazine biosynthesis pathway (Fig. S2A), quorum sensing pathway (Fig. S2B), biosynthesis of siderophore group non‐ribosomal peptides (Fig. S2C), folate biosynthesis (Fig. S2D), thiamine metabolism (Fig. S2E) and riboflavin metabolism (Fig. S2F).
Fig. 2.
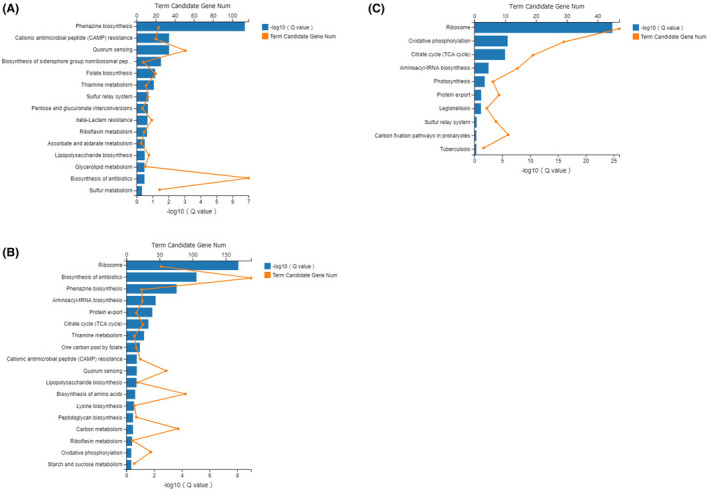
KEGG enrichment column maps of differentially expressed genes of the PAO1 strain in the control group, DAS‐treated group and DAE‐treated group based on RNA‐seq (0.00 < q value < 0.05).
A. DAE‐treated group vs. DAS‐treated group.
B. Control group vs. DAS‐treated group.
C. Control group vs DAE‐treated group.
The differentially expressed genes of the PAO1 strain were significantly enriched in 18 KEGG pathways (0.00 < q value < 0.50) (Fig. 2B), when the transcriptome of control group was compared with that of the DAS‐treated group. Among them, the enriched KEGG pathways with significantly downregulated gene expression were as follows: phenazine biosynthesis and quorum sensing; the enriched KEGG pathways with significantly upregulated gene expression were as follows: ribosome, aminoacyl‐tRNA biosynthesis, protein export, thiamine metabolism, one carbon pool by folate, lipopolysaccharide biosynthesis, biosynthesis of amino acids, lysine biosynthesis and riboflavin metabolism; in other enriched KEGG pathways, there were both upregulated genes as well as downregulated genes. Some of the detailed KEGG pathways are provided in Fig. S3, including phenazine biosynthesis pathway (Fig. S3A), quorum sensing pathway (Fig. S3B), biosynthesis of siderophore group non‐robosomal peptides (Fig. S3C), thiamine metabolism (Fig. S3D), one carbonpool by folate (Fig. S3E) and riboflavin metabolism (Fig. S3F).
The differentially expressed genes of the PAO1 strain were significantly enriched in 10 KEGG pathways (0.00 < q value < 0.50) (Fig. 2C), when the transcriptome of the control group was compared with that of the DAE‐treated group. Among them, no pathway had significantly downregulated gene transcription; the 7 significantly enriched pathways with mainly upregulated genes were as follows: ribosome, oxidative phosphorylation, citrate cycle (TCA cycle), aminoacyl‐tRNA biosynthesis, photosynthesis, protein export and carbon fixation pathways in prokaryotes.
Gene Ontology enrichment analysis
Gene Ontology (GO) enrichment maps of differentially expressed genes in the three experimental groups are provided in Fig. 3. The differentially expressed genes were significantly enriched in 11 GO terms (0.00 < q value < 0.50) (Fig. 3A), when the transcriptome of the DAE‐treated group was compared to that of the DAS‐treated group. The 11 GO terms significantly enriched are as follows: phenazine biosynthetic process, antibiotic biosynthetic process, thiamine metabolic process, Mo‐molybdopterin cofactor biosynthetic process, thiamine biosynthetic process, Mo‐molybdopterin cofactor metabolic process, response to drug, thiamine‐containing compound metabolic process, thiamine‐containing compound biosynthetic process, molybdopterin cofactor metabolic process and prosthetic group metabolic process.
Fig. 3.
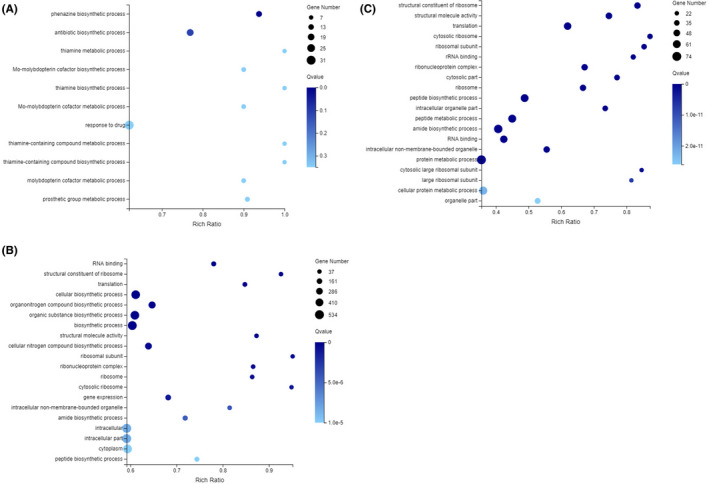
GO enrichment bubble maps of differentially expressed genes in the PAO1 strain in the control group, DAS‐treated group, and DAE‐treated group based on RNA‐seq (0.00 < q value < 0.05).
A. DAE‐treated group vs. DAS‐treated group.
B. Control group vs. DAS‐treated group.
C. Control group vs. DAE‐treated group.
A total of 195 (0.00 < q value < 0.50) of significantly enriched GO terms of the differentially expressed genes were identified when the transcriptome of the control group was compared with that of the DAS‐treated group (Fig. 3B), and 375 (0.00 < q value < 0.50) enriched GO terms of the differentially expressed genes were identified when the control group was compared with that of the DAE‐treated group (Fig. 3C). Most of these significantly enriched GO terms were related to structural constituent of ribosome, RNA binding, translation and cellular biosynthetic process (Fig. 3B and C).
qRT‐PCR validation results
The qRT‐PCR validation map of some differentially expressed key genes of the three QS systems in the PAO1 strain after DAS and DAE treatments are provided in Fig. 4. The map shows that there were significant differences between the transcription levels of all nine genes after DAS treatment based on the independent Student’s t‐test at P < 0.01, as well as significant differences after DAE treatment at P < 0.05. Moreover, the transcription levels of lasR, rhlI, rhlR, pqsA, pqsR, lasA, lasB and phzM were all downregulated with a fold change > 2 after DAS treatment, with the exception of lasI for which a fold change < 2 was observed. However, the transcription levels of lasI, lasR, rhlI, rhlR, pqsA, pqsR and phzM were all downregulated with a fold change < 2 after DAE treatment, but the transcription levels of lasA and lasB were downregulated with a fold change > 2 after DAE treatment. The qRT‐PCR results were consistent with that of the transcriptome data analysis.
Fig. 4.
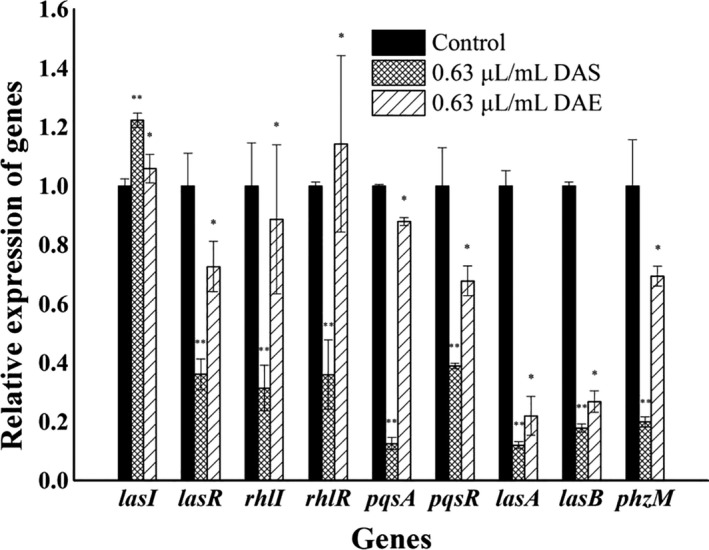
qRT‐PCR validation map of some differentially expressed key genes in quorum sensing systems of the PAO1 strain exposed to 0.63 μl ml−1 of DAS. *, denotes significant difference with P < 0.05 based on the independent Student’s t‐test; **, denotes an extremely significant difference with P < 0.01.
A. PAO1 growth curves of DAS treatment.
B. PAO1 growth curves of DAE treatment.
Growth curves of the PAO1 strain
The growth curves of the PAO1 strain exposed to different concentrations of DAS or DAE are provided in Fig. 5A and B, respectively. The growth curves in Fig. 5A show that the growth curves of the PAO1 strain exposed to 0.16–0.63 μl ml−1 DAS were almost the same as that of the control group, showing a typical growth curve. However, DAS had minor effect on the growth of the PAO1 strain, but it prolonged the lag phase for several hours when the concentration of DAS reached 1.25 μl ml−1 or above. Typical growth curves were also observed when PAO1 was exposed to different concentration (0.16–5 μl ml−1) of DAE (Fig. 5B). Therefore, both DAS and DAE had little effect on the growth of PAO1 under 1.25 μl ml−1 of concentration.
Fig. 5.
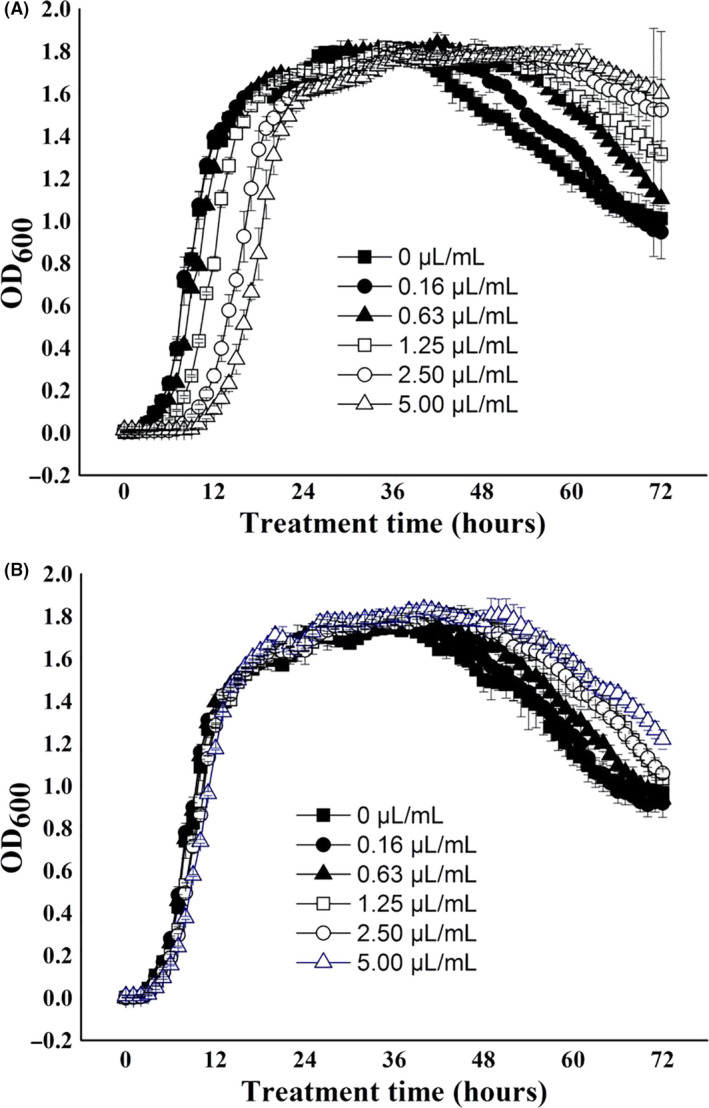
Growth curves of the PAO1 strain exposed to different concentrations of DAS or DAE.
A. PAO1 growth curves of DAS treatment.
B. PAO1 growth curves of DAE treatment.
Determination of the virulence factors of the PAO1 strain. (i) Pseudomonas Quinolone Signal production
The histogram of the relative production of PQS in the PAO1 strain cultures exposed to DAS and DAE for 72 h is provided in Fig. 6A. Figure 6A shows that the PQS concentration in DAS‐treated group was significantly lower than that of the control group, while the PQS concentration in DAE‐treated group was not significantly different than that of the control group. Therefore, DAS inhibited the PQS production of the PAO1 strain and DAE had no effect on the PQS production of the PAO1 strain.
Fig. 6.
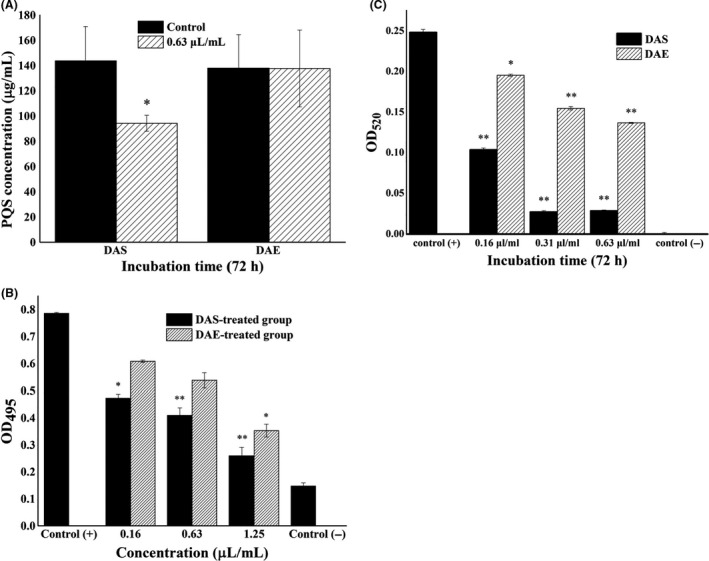
Charts of virulence factors of the PAO1 strain exposed to DAS or DAE.
A. PQS yield determination.
B. Elastase activity determination map.
C. Pyocyanin yield determination. Control (+) group: 0 μl ml−1 DAS‐treated group; Control (−) group: sterilized deionized water; *, denotes a significant difference with P < 0.05 based on the independent Student’s t‐test; **, denotes an extremely significant difference with P < 0.01.
Elastase activity
Elastase activity of the PAO1 strain exposed to different concentrations of DAS or DAE is shown in Fig. 6B. The bar chart shows that the elastase activity of the PAO1 strain was significantly lower than that in the control (+) group (0 μl ml−1 DAS treatment) and significantly higher than that in the control (−)group (sterilized deionized water) both in the three DAS‐treated groups and in the 1.25 μl ml−1 DAE‐treated groups. However, there was no significant difference between the control (+) group and 0.16 or 0.63 μl ml−1 DAE‐treated groups. The elastase activity decreased with the increase of DAS or DAE concentration. In addition, the elastase activity of the DAS‐treated group was lower than that of the DAE‐treated group. Therefore, DAS inhibited the elastase activity, whereas DAE had minor effects on the elastase activity of the PAO1 strain in a concentration dependent manner.
Pyocyanin yield
The dynamic curves of pyocyanin production of the PAO1 strain exposed to different concentrations of DAS or DAE are shown in Fig. 6C. Figure 6C shows that pyocyanin production after incubation for 72 h was the most in the control (+) group (0 μl ml−1 DAS treatment), while the absorbance at 520 nm was nearly zero in the control (−) group (sterilized deionized water). There were significant differences in both DAS‐ and DAE‐treated groups compared with in the control (+) group. In the 0.16–0.63 μl ml−1 DAS‐treated group, pyocyanin production was decreased more than two fold change compared with that of the control (+) group. In contrast, the pyocyanin production was decreased less than two fold change in all DAE‐treated groups after 72 h of incubation. Therefore, both DAS and DAE can inhibit pyocyanin production of the PAO1 strain.
Discussion
The volcano plots show that the transcriptome of the PAO1 strain changed significantly due to DAS treatment, but the change caused by DAE was relatively small; thus, DAS had more effect on the PAO1 strain than DAE. However, Figure 5A and B show that both DAS and DAE had minor effect on the growth dynamics of this bacterium. Effects of DAS on the physiology and metabolism of the PAO1 strain without affecting its growth are analysed below.
There were 336 genes that had the same differential expression after DAS and DAE treatments (Fig. 1B). These changes could have been caused by the carbon skeleton of DAS and DAE. The KEGG enrichment maps in Fig. 2 show these common changes in detail. Five enriched KEGG pathways were same in both of the treatments, including ribosome, oxidative phosphorylation, citrate cycle (TCA cycle), aminoacyl‐tRNA biosynthesis and protein export. The differentially expressed genes in these five pathways were mainly upregulated. These results suggested that the same carbon skeleton structure (diallyl) of DAS and DAE could have stimulated these KEGG pathways and enhanced the expression of the related genes in these pathways. Researchers have reported that diallyl disulfide in garlic has antibacterial activity in vitro, but dipropyl disulfide in leek has no antibacterial activity, so it can be concluded that the allyl group plays a fundamental role in the antibacterial activity of these sulfides (Casella et al., 2013). Our findings also indicated that both DAS and DAE can promote some important physiological functions and metabolic activities of the PAO1 strain, such as ribosome, oxidative phosphorylation, TCA cycle, aminoacyl‐tRNA biosynthesis and protein export. These stimulation and enhancement effects of the DAS and DAE on the PAO1 strain may have resulted from providing nutrition for the strain or they were hormesis phenomena. The concentration of DAS and DAE used in the experiment is very low (0.63 μl ml−1), and LB medium is rich in nutrients. Therefore, a hormesis phenomenon likely occurred. Hormesis refers to the occurrence of negative responses in some organisms to toxic chemicals at high doses (such as inhibition of growth and development) but beneficial responses to toxic chemicals at low doses (Chapman, 2001). Hormesis is a physiological mechanism developed during the long‐term evolution of organisms to adapt to natural selection and improve the survival rate under various low‐level stresses. It is important for rapid recovery of the organism when its self‐stability is destroyed (Calabrese and Baldwin, 2001). Therefore, both 0.63 μl ml−1 DAS and DAE show low toxicity toward the PAO1 strian and simulates its physiological and metabolic functions, including protein synthesis (ribosome, aminoacyl‐tRNA biosynthesis), transportation (protein export), glycolysis (TCA cycle) and respiration (oxidative phosphorylation).
The 1509 differentially expressed genes (Venn diagram, Fig. 1B) between the control and the DAS‐treated group have important analytical significance, because they were differentially expressed between the DAS and DAE treatments but not differentially expressed between the control and DAE‐treated group. The analytical results suggested that these genes were differentially expressed mainly because of the thioether group. The KEGG pathways that were significantly enriched in both treatments (Fig. 2A and in Fig. 2B) reflected the enrichment of these 1509 differentially expressed genes. These genes were enriched in 8 KEGG pathways, including: phenazine biosynthesis, cationic antimicrobial peptide (CAMP) resistance, quorum sensing, folate biosynthesis, thiamine metabolism, riboflavin metabolism, lipopolysaccharide biosynthesis and biosynthesis of antibiotics. Among these pathways, phenazine (Figs S2A and S3A) and quorum sensing (Figs S2B and S3B) pathways were enriched with downregulated genes, while cationic antimicrobial peptide (CAMP) resistance, folate biosynthesis, thiamine metabolism, riboflavin metabolism and lipopolysaccharide biosynthesis were mainly enriched with upregulated genes.
The detailed phenazine pathway (Figs S2A and S3A) shows that majority of the key genes involved in PQS synthesis, pyocyanin synthesis and 1‐hydroxyphenazine synthesis were downregulated in the phenazine pathway, including phzAB, phzD, phzE, phzF, phzG, phzM and phzS. Phenazine compounds are virulence factors of P. aeruginosa, which serve as signal molecules for QS, exert virulence to kill other microbial competitors and influence biofilm formation (Huang et al., 2018). This result was confirmed by PQS (Fig. 6A) and pyocyanin (Fig. 6C) analysis and was consistent with the qRT‐PCR (Fig. 4) analysis. Pyocyanin is a virulence factor of P. aeruginosa, which causes a wide spectrum of cellular damage, including suppression of cell respiration, disruption of calcium homeostasis and inactivation of catalase (Lau et al., 2004; Li et al., 2018). PQS, a signal molecule of QS systems in P. aeruginosa, regulates the expression of some virulence genes (Kostylev et al., 2019). This indicated that DAS inhibited the biosynthesis of phenazine toxins regulated by the QS systems of the PAO1 strain. Therefore, DAS played a positive role in suppressing the virulence and pathogenicity of the bacterium.
The detailed quorum sensing pathway (Figs S1B and S2B) shows that most of the key genes in QS systems were downregulated, including lasR in las system, rhlI/rhlR in rhl system, pqsABCDE/pqsR in pqs system, while the expression level of lasI in las system and pqsH in pqs system was not changed. This result was consistent with the qRT‐PCR analysis (Fig. 4). LasI is the synthetase that synthesizes 3‐Oxododecanoyl Homoserine Lactone (3OC12‐HSL), and LasR is the receptor of this signal molecule (Rampioni et al., 2007; Li et al., 2020). The quorum‐sensing pathway suggested that DAS did not inhibit the synthesis for signal molecule of 3OC12‐HSL but inhibited the receptor of the signal molecule. Similarly, RhlI is the synthetase that synthesizes butyl‐Homoserine Lactone (C4‐HSL), and rhlR is the receptor of this signal molecule (Finch et al., 1998; Li et al., 2020). The quorum‐sensing pathway suggested that DAS inhibited the synthesis of C4‐HSL signal molecule and simultaneously inhibited the receptor of this signal molecule. The pqsABCDE in pqs system are encoding genes for the synthesis of PQS signalling molecules, and pqsR is the encoding gene for the PQS receptor (Kostylev et al., 2019; Li et al., 2020). The quorum‐sensing pathway suggested that DAS inhibited the PQS signal production and suppressed the recognition and transduction of PQS. This result was also confirmed by PQS analysis (Fig. 6A) of the PAO1 strain. The virulence genes regulated by the three QS systems were also downregulated, including rhlABC involved in rhamnolipid biosynthesis, lasA involved in exoprotease synthesis, lasB involved in elastase synthesis and lecA involved in lectin synthesis. Rhamnolipids, a mixture of two or four different homologues, are a type of virulence factors of P. aeruginosa and a kind of biosurfactant with antimicrobial activity to compete with other microorganisms in the environment (Bharali et al., 2013; Varjani and Upasani, 2019). LasB elastase and LasA exoprotease contribute to the pathogenicity of P. aeruginosa, because they have proteolytic activity to hydrolyse bioactive proteins in host tissues (Elston et al., 2007). LecA, a multifunctional virulence factor of P. aeruginosa, is not only an invasion factor, but also an adhesion factor (Zheng et al., 2017). This result was consistent with the qRT‐PCR analysis (Fig. 4) and elastase analysis (Fig. 6B). A previous study reported that four diallyl sulfides (DAS, DADS, DATS, and DATTS) in garlic can inhibit LuxR QS monitoring system (Persson et al., 2005). In our previous study, we found that DADS from garlic inhibited the gene expression of the QS systems and related virulence genes of the PAO1 strain (Li et al., 2019). The difference between DADS and DAS is that DADS inhibited the transcription of all the key genes in the three QS systems, while DAS did not inhibit lasI and pqsH. Therefore, DAS inhibited the biosynthesis of two signal molecules of C4‐HSL and PQS but did not inhibit the biosynthesis of 3OC12‐HSL. Moreover, DAS inhibited the receptors of all the three signal molecules in las, rhl and pqs QS systems. DAS also inhibited the expression of virulence genes and the production of virulence factors regulated by the three QS systems of the PAO1 strain, including rhamnolipid, exoprotease, elastase, lectin and pyocyanin. It can be concluded that DAS inhibited the three QS systems to suppress the related virulence factors of the PAO1 strain and this was mainly due to its thioether group.
Interestingly, the KEGG pathway of biosynthesis of siderophore group non‐ribosomal peptides was significantly enriched in both DAE‐treated group vs DAS‐treated group analysis (0.00 < q value < 0.50) and in control group vs DAS‐treated group analysis (q value = 0.5137168). The detailed pathway showed that six key genes were downregulated in pyochelin biosynthesis from chorismate (Figs S2C and S3C), including four genes (pchDEFG) encoding for pyochelin, while one key gene was upregulated involved in enterochelin biosynthesis from chorismate (Fig. S3F). This result suggested that the pyochelin biosynthesis in the PAO1 strain was inhibited by DAS. Pyochelin, one of the P. aeruginosa iron acquisition systems, binds extracellular trivalent iron ion to transport into the cell with the siderophores (Banin et al., 2005). Pyochelin is also a virulence factor of P. aeruginosa, which is regulated by the QS systems of P. aeruginosa (Kariminik et al., 2017). Pyochelin not only assists bacteria to obtain iron nutrition from the host but also aggravates oxidant injury of the human pulmonary epithelial cells (Britigan et al., 1997; Voss et al., 2000; Lyczak et al., 2002). This indicated that DAS inhibited the virulence factor of pyochelin regulated by the QS systems in the PAO1 strain.
More interestingly, most genes in the folate biosynthesis (Figs S2D and S3D), thiamine metabolism (Figs S2E and S3E) and riboflavin metabolism (Figs S2F and S3F) pathways of the PAO1 strain were upregulated after DAS exposure. Figure S2D shows that there were 26 key genes involved in folate biosynthesis that were significantly upregulated. Figs S2E and S3E show 11 and 14 key genes, respectively, those are involved in thiamine synthesis and were significantly upregulated. Figures S2F and S3F show 10 and 11 key genes, respectively, those are involved in riboflavin biosynthesis and were significantly upregulated. Folate (vitamin B9), thiamine (vitamin B1) and riboflavin (vitamin B2) are essential for mammals, but mammals cannot synthesize them (Xin et al., 2017; Alam et al., 2020). Therefore, folate, thiamine and riboflavin are mainly provided through dietary intake for mammals. The KEGG enrichment showed that DAS enhanced the folate, thiamine and riboflavin biosynthesis and metabolism of the PAO1 strain. P. aeruginosa is a ubiquitous bacterium, which can survive in various environment, including water, soil, air, human skin, respiratory tract and intestines (Pang et al., 2019). The B vitamins of folate, thiamine and riboflavin are generally the main micronutrients both for human and for bacterial metabolism, which primarily serve as precursors of enzymatic cofactors (Obi et al., 2020; Soto‐Martin et al., 2020). DAS promoted the VB1, VB2 and VB9 biosynthesis of the PAO1 strain. This is likely also a hormesis effect, as described above, which is similar to our previous observation of the enhanced swarming motility of P. aeruginosa PAO1 induced by farnesol (Li et al., 2020). Therefore, the promotion of biosynthesis of the three B vitamins is a positive response of P. aeruginosa PAO1 to DAS‐induced stress. This is significant to promote human to obtain essential B vitamins from the metabolites of intestinal microorganisms. Thus, the experimental results suggested that DAS has potential application in the fermentation of these three B vitamins. Can DAS improve the biosynthesis of these three B vitamins by other bacteria? Further research required to confirm this.
In addition, the most differentially expressed genes in the cationic antimicrobial peptide (CAMP) resistance and lipopolysaccharide biosynthesis pathways were upregulated after DAS exposure. These are likely also hormesis effects, which are positive responses in this bacterium to DAS‐induced stress. In the CAMP pathway, the key genes in two two‐component systems of PhoQ‐PhoP and ParS‐ParR were upregulated. The two‐component system of PhoQ‐PhoP is related to antimicrobial peptide and aminoglycoside resistance, and ParS‐ParR system is related to multidrug resistance, QS and phenazine production (Francis et al., 2017). Some genes that were related to amino sugar and nucleotide sugar metabolism and were regulated by the two two‐component systems were also upregulated. In lipopolysaccharides biosynthesis pathway, some genes involved in lipopolysaccharide biosynthesis, and amino sugar and nucleotide sugar metabolism were upregulated. Lipopolysaccharides, the components of the cell envelope, are virulence factors of P. aeruginosa and modulate the immune response of the host (Lam et al., 2011; McCarthy et al., 2017). This suggested that some key genes involved in lipopolysaccharide biosynthesis and amino sugar and nucleotide sugar metabolism of the PAO1 strain were upregulated by DAS, which are significant to modulate host immune response. DAS suppressed the production of some virulence factors harmful to the host and enhanced the production of some nutrition factors beneficial to the host.
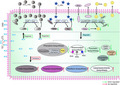
On the basis of the above mentioned analysis, a schematic diagram of the action mechanisms of DAS on the PAO1 strain was proposed in this study (Fig. 7). As shown in Fig. 7, DAS inhibits the expression of some key genes in las, rhl and pqs of the three QS systems of the P. aeruginosa PAO1 strain, including lasR, rhlI, rhlR, pqsABCDE and pqsR. Downregulation of rhlI results in a decrease in the concentration of RhlI (synthetase for C4‐HSL), and this finally leads to the decrease of the concentration of the signal molecules of C4‐HSL. Similarly, the concentration of the PQS molecules decreases due to the downregulated pqsABCDE. Moreover, downregulation of lasR, rhlR and pqsR results in a decrease of the corresponding signal molecule receptors, with decreased LasR for 3OC12‐HSL, RhlR for C4‐HSL and PqsR for PQS. As a result, DAS inhibits the three QS systems of P. aeruginosa PAO1. Furthermore, the biosynthesis pathways of a series of virulence factors regulated by the three QS systems are also inhibited, including phenazine biosynthesis, rhamnolipid biosynthesis, pyochelin biosynthesis, exoprotease of LasA biosynthesis, elastase of LasB biosynthesis and lectin of LecA biosynthesis. On the other hand, DAS enhances the expression of some key genes and corresponding biosynthesis pathways, such as folate biosynthesis, thiamine biosynthesis, riboflavin biosynthesis and lipopolysaccharide biosynthesis. These physiological changes in P. aeruginosa PAO1 caused by DAS, as mentioned in this schematic diagram, are mainly due to its thioether group.
Fig. 7.
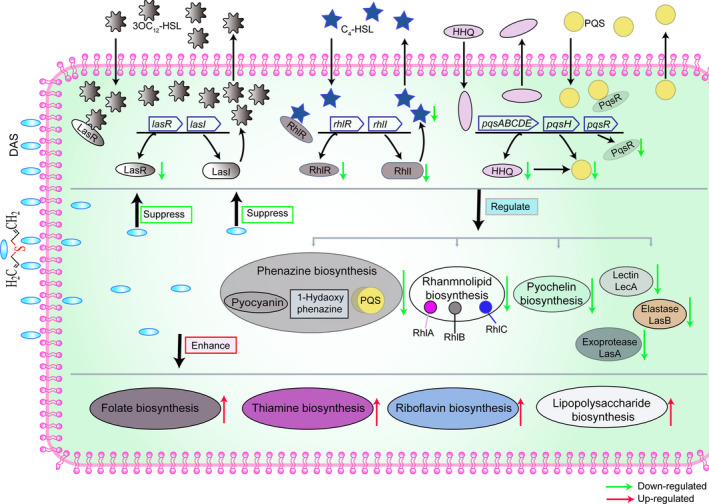
Schematic diagram of the action mechanisms of DAS on the PAO1 strain. DAS induces some various cellular effects in the Pseudomonas aeruginosa PAO1 cells. First, DAS inhibits the transcription level of some key genes in las, rhl, and pqs of the three QS systems of the P. aeruginosa PAO1 strain, including lasR, rhlI, rhlR, pqsABCDE and pqsR. LasI is the synthetase of the 3OC12‐HSL signal molecule, which is encoded by lasI. LasR is the receptor of 3OC12‐HSL and encoded by lasR. Downregulation of the transcription level of lasR leads to a decrease in the LasR receptor of 3OC12‐HSL, preventing 3OC12‐HSL from effectively binding to LasR; this results induced by DAS finally inhibits the subsequent functions of the las QS system. Similarly, RhlI is the synthetase of the C4‐HSL signal molecule, which is encoded by rhlI. RhlR is the receptor of C4‐HSL and encoded by rhlR. Downregulation of the transcription levels of rhlI and rhlR leads to a decrease in the RhlI synthetase and RhlR receptor of C4‐HSL, respectively, which reduces the production of C4‐HSL and RhlR receptor. These results induced by DAS inhibit the functions of the rhl QS system. Similarly, the genes of pqsABCDE encoded the synthetase of the HHQ signal molecule, which is the precursor of the PQS signal molecule. The enzyme encoded by pqsH catalyses HHQ into PQS. PqsR is the receptor of the PQS signal molecule encoded by pqsR. Downregulation of the transcription level of pqsABCDE and pqsR leads to a decrease in HHQ, PQS signal molecules, and PqsR receptors respectively. These results induced by DAS finally inhibit the function of the pqs QS system. Second, the biosynthesis pathways of a series of virulence factors regulated by the three QS systems are also inhibited, including phenazine biosynthesis, rhamnolipid biosynthesis, pyochelin biosynthesis, exoprotease of LasA biosynthesis, elastase of LasB biosynthesis and lectin of LecA biosynthesis. Third, DAS enhances the expression of some key genes and corresponding biosynthesis pathways, such as folate biosynthesis, thiamine biosynthesis, riboflavin biosynthesis and lipopolysaccharide biosynthesis. Abbreviations: DAS, diallyl sulfide; 3OC12‐HSL, N‐(3‐oxododecanoyl)‐l‐homoserine lactone; C4‐HSL, N‐butanoyl homoserine lactone; PQS, 2‐heptyl‐3‐hydroxy‐4‐quinolone; HHQ, 2‐heptyl‐4(1H)‐quinolone.
Similar to DAS, DADS inhibited key genes in the three QS systems of the PAO1 strain (Li et al., 2018, 2019). One difference between DAS and DADS is that DADS inhibits the transcriptions of all key genes in the three QS systems of the PAO1 strain, including lasI and pqsH, whereas DAS does not. The similarities and differences between DADS and DAS in inhibiting P. aeruginosa PAO1 QS systems indicated that DADS has stronger QS inhibitory activity than DAS, as DADS has two thioether groups, whereas DAS has one.
In conclusion, DAS from garlic did not affect the growth dynamics of P. aeruginosa PAO1; however, DAS inhibited transcription of most of the key genes in the QS systems, including lasR of las system, rhlI/rhlR of rhl system and pqsABCDE/pqsR of pqs system. Furthermore, DAS also inhibited the transcription of some virulence genes regulated by the QS systems, including rhlABC, lasA, lasB, lecA, and phzAB, phzD, phzE, phzF, phzG, phzM and phzS. DAS also suppressed the production of the virulence factors. DAS enhanced the expression of the key genes involved in biosynthesis of three B vitamin of folate, thiamine and riboflavin. Inhibition of three QS systems and enhancement of the three B vitamins of P. aeruginosa PAO1 induced by DAS occur mainly because of its thioether group. In addition, DAS simulates and enhances some physiological and metabolic functions in this bacterium, including protein synthesis, transportation, glycolysis and respiration. DAS suppressed some virulence factors harmful to the host but enhanced some nutrition factors beneficial to the host. Therefore, DAS may be a potential oral anti‐inflammatory drug for inhibiting the virulence and pathogenicity of P. aeruginosa by inhibiting QS systems.
Experimental procedures
Strains, media, culturing conditions and reagents
Pseudomonas aeruginosa PAO1 was used for experiments in this study, which was given by the CAS Key Laboratory of Pathogenic Microbiology and Immunology (Beijing, China). Luria‐Bertani (LB) medium and the culturing conditions were the same as described in a previous study (Li et al., 2018). DAS (purity > 98%, SG: 0.89, CAS RN: 592‐88‐1, product encoding: A0235) and DAE (purity > 98%, SG: 0.81, CAS RN: 557‐40‐4, product encoding: A0229) were both purchased from Tokyo Chemical Industry (Tokyo, Japan). The chemical structural formulas of DAS and DAE are provided in Fig. 1A.
High throughput sequencing analysis of transcriptome
Pseudomonas aeruginosa PAO1 cells in log‐phase (1–2 × 108 Colony Forming Unit (CFU) ml−1) were inoculated in 9 equal parts of 100 ml LB medium. Among these cultures, three equal parts were the control group, and the remaining six parts were divided into DAS treatment group and DAE treatment group (three each). The concentration of DAS and DAE was 0.63 μl ml−1 (equivalent to 0.56 mg ml−1 DAS) in the DAS‐treated group and DAE‐treated group, while equal volume of PBS buffer solution was added in the control group. All the experimental cultures were incubated at 37°C and 180 r.p.m. for 5 h. The cultures were centrifuged, and the bacterial precipitates were rapidly frozen at −80°C.
RNA sample preparation, library construction and sequencing were done as described in a previous study (Li et al., 2020). The transcriptome sequencing was performed at BGI Genomics (Shenzhen, China). The sequencing data were filtered with soapnuke v1.5.2 (Li et al., 2008). The clean reads were mapped to the reference genome using hisat2 v2.0.4 (Kim et al., 2015). bowtie2 v2.2.5 (Langmead and Salzberg, 2012) was applied to align the clean reads to the reference coding gene set; then, the transcription level of the gene was calculated by rsem v1.2.12 (Li and Dewey, 2011). Differential expression analysis was performed using the deseq 2 v1.4.5 (Love et al., 2014) with a Q value ≤ 0.05. To evaluate the change in phenotype, GO (http://www.geneontology.org/) and KEGG (http://www.kegg.jp/) enrichment analysis of annotated different expressed gene was performed by phyper (http://en.wikipedia.org/wiki/Hypergeometric_distribution) based on hypergeometric test. The significant levels of terms and pathways were corrected by Q value with a rigorous threshold (Q value ≤ 0.05) by Bonferroni correction (Abdi, 2007). The RNA‐seq data sets are available at the NCBI Gene Expression Omnibus (GEO) database (GSE151292).
qRT‐PCR analysis
For qRT‐PCR, the PAO1 culture condition, control, DAS and DAE treatments were the same as described in Section 2.2. The total RNA extraction, PCR primer design and synthesis, as well as qRT‐PCR reaction were done as described in a previous study (Li et al., 2020). The experiments were performed with three replicates. The independent Student’s t‐test was used to compare the DAS‐ and DAE‐treated groups to the control group.
Growth curves of the PAO1 strain
PAO1 culture condition, control, DAS and DAE treatments were the same as described in section 2.2. Concentration gradients of DAS and DAE used in the growth curve were as follows: 0, 0.16, 0.63, 1.25, 2.50 or 5.00 μl ml−1 (equivalent to 0, 0.14, 0.28, 0.56, 1.11, 2.23 or 4.45 mg ml−1). The inoculation density of PAO1 strain used for the experiment was 1‐2 × 106 CFU/mL, and the incubation time was 72 h. The experiments were performed with three replicates. The independent Student’s t‐test was used to compare the DAS‐ and DAE‐treated groups to the control group.
Determination of the virulence factors of the PAO1 cultures
Pseudomonas Quinolone Signal production
PAO1 culture condition, and control, DAS and DAE treatments were the same as described in section 2.2. PQS from the control group, DAS‐treated group and DAE‐treated group was extracted after 72 h incubation of P. aeruginosa PAO1. The extraction and quantification of PQS were done as described in a previous study (Li et al., 2020).
Elastase activity
PAO1 culture condition, control, DAS and DAE treatments were the same as described in section 2.2. The extraction of elastase and determination of elastase activity were done as described in a previous study (Li et al., 2018). The experiments were performed with three replicates. The independent Student’s t‐test was used to compare the DAS‐ and DAE‐treated groups to the control (+) group.
Pyocyanin production
PAO1 culture condition, control, DAS and DAE treatments were the same as described in section 2.2. After incubation for 72 h, the cultures were sampled and extracted for the pyocyanin. Extraction and quantification of pyocyanin were done as described in a previous study (Li et al., 2018). The experiments were performed with three replicates. The independent Student’s t‐test was used to compare the DAS‐ and DAE‐treated groups to the control (+) group.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Fig. S1. Volcano plots of differentially expressed genes (fold change > 2, P value < 0.05) of the PAO1 strain in the control group, DAS‐treated group, and DAE‐treated group based on RNA‐seq. Each gene is represented by a dot in the map; the up‐regulated genes are represented in red dots and the down‐regulated genes are represented in blue dots. A, DAE‐treated group vs. DAS‐treated group; B, Control group vs. DAS‐treated group; C, Control group vs. DAE‐treated group.
Fig. S2. The significantly enriched KEGG pathways based on transcriptome data of the PAO1 strain (DAE‐treated group vs DAS‐treated group). A, phenazine biosynthesis; B, quorum sensing; C, biosynthesis of siderophore group nonribosomal peptides; D, folate biosynthesis; E, thiamine metabolism; F, riboflavin metabolism.
Fig. S3. The significantly enriched KEGG pathways based on transcriptome data of the PAO1 strain (Control group vs. DAS‐treated group). A, phenazine biosynthesis; B, quorum sensing; C, biosynthesis of siderophore group nonrobosomal peptides; D, one carbon pool by folate; E, thiamine metabolism; F, riboflavin metabolism.
Acknowledgements
This work was funded by Guangdong Basic and Applied Basic Research Foundation (2020A1515010850), Guangzhou Municipal Science and Technology Research Project (201607020020) and GDAS’ Project of Science and Technology Development (2017GDASCX‐0102). The authors thank Pro. Zhang Li‐Xin and Dr. Dai Huan‐Qin for presenting the PAO1 strain. We also thank our team members for their help and support.
Microbial Biotechnology (2021) 14(2), 677–691
Funding informationThis work was funded by Guangdong Basic and Applied Basic Research Foundation (2020A1515010850), Guangzhou Municipal Science and Technology Research Project (201607020020) and GDAS’ Project of Science and Technology Development (2017GDASCX‐0102).
Contributor Information
Xiao‐Bao Xie, Email: xiaobaoxie@126.com.
Qing‐Shan Shi, Email: jigan@gdim.cn.
References
- Abdi, H. (2007) The Bonferonni and Šidák corrections for multiple comparisons. Encycl Meas Stat 1: 1–9. [Google Scholar]
- Alam, C. , Kondo, M. , O'Connor, D.L. , and Bendayan, R. (2020) Clinical implications of folate transport in the central nervous system. Trends Pharmacol Sci 41: 349–361. [DOI] [PubMed] [Google Scholar]
- Avato, P. , Tursi, F. , Vitali, C. , Miccolis, V. , and Candido, V. (2000) Allylsulfide constituents of garlic volatile oil as antimicrobial agents. Phytomedicine 7: 239–243. [DOI] [PubMed] [Google Scholar]
- Banin, E. , Vasil, M.L. , and Greenberg, E.P. (2005) Iron and Pseudomonas aeruginosa biofilm formation. Proc Nat Acad Sci USA 102: 11076–11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharali, P. , Saikia, J.P. , Ray, A. , and Konwar, B.K. (2013) Rhamnolipid (RL) from Pseudomonas aeruginosa OBP1: a novel chemotaxis and antibacterial agent. Colloid Surface B 103: 502–509. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt, T. , Jensen, P.Ø. , Rasmussen, T.B. , Christophersen, L. , Calum, H. , Hentzer, M. , et al. (2005) Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infections. Microbiology 151: 3873–3880. [DOI] [PubMed] [Google Scholar]
- Britigan, B.E. , Rasmussen, G.T. , and Cox, C.D. (1997) Augmentation of oxidant injury to human pulmonary epithelial cells by the Pseudomonas aeruginosa siderophore pyochelin. Infect Immun 65: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady, N.C. , McKean, K.A. , Behnke, J. , Kubec, R. , Mosier, A.P. , Kasper, S.H. , et al. (2012) Inhibition of biofilm formation, quorum sensing and infection in Pseudomonas aeruginosa by natural products‐inspired organosulfur compounds. PLoS One 7: e38492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese, E.J. , and Baldwin, L.A. (2001) U‐shaped responses in biology, toxicology, and public health. Annu Rev Public Health 22: 15–33. [DOI] [PubMed] [Google Scholar]
- Casella, S. , Leonardi, M. , Melai, B. , Fratini, F. , and Pistelli, L. (2013) The role of diallyl sulfides and dipropyl sulfides in the in vitro antimicrobial activity of the essential oil of garlic, Allium sativum L., and Leek, Allium porrum L. Phytother Res 27: 380–383. [DOI] [PubMed] [Google Scholar]
- Chapman, P.M. (2001) The implications of hormesis to ecotoxicology and ecological risk assessment. Hum Exp Toxicol 20: 499–505. [DOI] [PubMed] [Google Scholar]
- Elston, C. , Wallach, J. , and Saulnier, J. (2007) New continuous and specific fluorometric assays for Pseudomonas aeruginosa elastase and LasA protease. Anal Biochem 368: 87–94. [DOI] [PubMed] [Google Scholar]
- Finch, R.G. , Pritchard, D.I. , Bycroft, B.W. , Williams, P. , and Stewart, G.S. (1998) Quorum sensing: a novel target for anti‐infective therapy. J Antimicrob Chemother 42: 569–571. [DOI] [PubMed] [Google Scholar]
- Francis, V.I. , Stevenson, E.C. , and Porter, S.L. (2017) Two‐component systems required for virulence in Pseudomonas aeruginosa . FEMS Microbiol Lett 364: fnx104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruci, M. , and Poole, K. (2018) Aminoglycoside‐inducible expression of the mexAB‐oprM multidrug efflux operon in Pseudomonas aeruginosa: involvement of the envelope stress‐responsive AmgRS two‐component system. PLoS One 13: e0205036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harjai, K. , Kumar, R. , and Singh, S. (2010) Garlic blocks quorum sensing and attenuates the virulence of Pseudomonas aeruginosa . FEMS Immunol Med Microbiol 58: 161–168. [DOI] [PubMed] [Google Scholar]
- Harris, J.C. , Cottrell, S.L. , Plummer, S. , and Lloyd, D. (2001) Antimicrobial properties of Allium sativum (garlic). Appl Microbiol Biotechnol 57: 282–286. [DOI] [PubMed] [Google Scholar]
- Hentzer, M. , Wu, H. , Andersen, J.B. , Riedel, K.B. , Rasmussen, T. , Bagge, N. , et al. (2003) Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J 22: 3803–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. , Zhou, E. , Jiang, C. , Jia, R. , Liu, S. , Xu, D. , et al. (2018) Endogenous phenazine‐1‐carboxamide encoding gene phzH regulated the extracellular electron transfer in biocorrosion of stainless steel by marine Pseudomonas aeruginosa . Electrochem Commun 94: 9–13. [Google Scholar]
- Jakobsen, T.H. , van Gennip, M. , Phipps, R.K. , Shanmugham, M.S. , Christensen, L.D. , Alhede, M. , et al. (2012) Ajoene, a sulfur‐rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob Agents Chemother 56: 2314–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariminik, A. , Baseri‐Salehi, M. , and Kheirkhah, B. (2017) Pseudomonas aeruginosa quorum sensing modulates immune responses: an updated review article. Immun Lett 190: 1–6. [DOI] [PubMed] [Google Scholar]
- Kim, D. , Langmead, B. , and Salzber, S.L. (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, J.‐W. , Park, S.‐H. , Shin, N.‐R. , Shin, J.‐Y. , Kim, J.‐W. , Shin, I.‐S. , et al. (2017) Protective effect and mechanism of action of diallyl disulfide against acetaminophen‐induced acute hepatotoxicity. Food Chem Toxicol 109: 28–37. [DOI] [PubMed] [Google Scholar]
- Kostylev, M. , Kim, D.Y. , Smalley, N.E. , Salukhe, I. , Greenberg, E.P. , and Dandekar, A.A. (2019) Evolution of the Pseudomonas aeruginosa quorum sensing hierarchy. Proc Natl Acad Sci USA 116: 7027–7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, J.S. , Taylor, V.L. , Islam, S.T. , Hao, Y. , and Kocíncová, D. (2011) Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front Microbiol 2: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. , and Salzberg, S.L. (2012) Fast gapped‐read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, G.W. , Hassett, J. , Ran, H. , and Kong, F. (2004) The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med 10: 599–606. [DOI] [PubMed] [Google Scholar]
- Lee, J. , and Zhang, L.H. (2015) The hierarchy quorum sensing network in Pseudomonas aeruginosa . Protein Cell 6: 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. , and Dewey, C.N. RSEM: accurate transcript quantification from RNA‐Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R. , Li, Y. , Kristiansen, K. , and Wang, J. (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24: 713–714. [DOI] [PubMed] [Google Scholar]
- Li, W.R. , Ma, Y.K. , Shi, Q.S. , Xie, X.B. , Sun, T.L. , Peng, H. , et al. (2018) Diallyl disulphide from garlic oil inhibits Pseudomonas aeruginosa virulence factors by inhibiting the transcription of key quorum sensing genes. Appl Microbiol Biotechnol 102: 7555–7564. [DOI] [PubMed] [Google Scholar]
- Li, W.‐R. , Ma, Y.‐K. , Xie, X.‐B. , Shi, Q.‐S. , Wen, X. , Sun, T.‐L. , and Peng, H. (2019) Diallyl disulfide from garlic oil inhibits Pseudomonas aeruginosa quorum sensing systems and corresponding virulence factors. Front Microbiol 9: 3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.R. , Zeng, T.H. , Li, C.L. , Xie, X.B. , and Shi, Q.S. (2020) Inhibition of the pqsABCDE and pqsH in the pqs quorum sensing system and related virulence factors of the Pseudomonas aeruginosa PAO1 strain by farnesol. Int Biodeterior Biodegrad 151: 104956. [Google Scholar]
- Lopez‐Causape, C. , Cabot, G. , del Barrio‐Tofiño, E. , and Oliver, A. (2018) The versatile mutational resistome of Pseudomonas aeruginosa . Front Microbiol 9: 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. & Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2.. Genome Biol, 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak, J.B. , Cannon, C.L. , and Pier, G.B. (2002) Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15: 194–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, L.W. , Robson, C.L. , Watts, A.M. , Gray, A.R. , Wainwright, C.E. , Bell, S.C. , et al. (2018) Expression of Pseudomonas aeruginosa antibiotic resistance genes varies greatly during infections in cystic fibrosis patients. Antimicrob Agents Chem 62: e01379‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, R.R. , Mazon‐Moya, M.J. , Moscoso, J.A. , Hao, Y. , Lam, J.S. , Bordi, C. , et al. (2017) Cyclic‐di‐GMP regulates lipopolysaccharide modification and contributes to Pseudomonas aeruginosa immune evasion. Nat Microbiol 2: 17027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura‐Alves, P. , Puyskens, A. , Stinn, A. , Klemm, M. , Guhlich‐Bornhof, U. , Dorhoi, A. , et al. (2019) Host monitoring of quorum sensing during Pseudomonas aeruginosa infection. Science 366: eaaw1629. [DOI] [PubMed] [Google Scholar]
- Obi, J. , Pastores, S.M. , Ramanathan, L.V. , Yang, J. , and Halpern, N.A. (2020) Treating sepsis with vitamin C, thiamine, and hydrocortisone: exploring the quest for the magic elixir. J Crit Care 57: 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, Z. , Raudonis, R. , Glick, B.R. , Lin, T.J. , and Cheng, Z. (2019) Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv 37: 177–192. [DOI] [PubMed] [Google Scholar]
- Persson, T. , Hansen, T.H. , Rasmussen, T.B. , Skindersø, M.E. , Givskov, M. , and Nielsen, J. (2005) Rational design and synthesis of new quorum‐sensing inhibitors derived from acylated homoserine lactones and natural products from garlic. Org Biomol Chem 3: 253–262. [DOI] [PubMed] [Google Scholar]
- Rampioni, G. , Schuster, M. , Greenberg, E.P. , Bertani, I. , Grasso, M. , Venturi, V. , et al. (2007) RsaL provides quorum sensing homeostasis and functions as a global regulator of gene expression in Pseudomonas aeruginosa . Mol Microbiol 66: 1557–1565. [DOI] [PubMed] [Google Scholar]
- Rasmussen, T.B. , Bjarnsholt, T. , Skindersoe, M.E. , Hentzer, M. , Kristoffersen, P. , Köte, M. , et al. (2005) Screening for quorum‐sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J Bacteriol 187: 1799–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, A.R. , Cifelli, P.M. , Ortori, C.A. , Righetti, K. , Lewis, S. , Erskine, P. , et al. (2010) Quorum sensing in cystic fibrosis‐a pilot randomized controlled trial. Pediatr Pulm 45: 356–362. [DOI] [PubMed] [Google Scholar]
- Soto‐Martin, E.C. , Warnke, I. , Farquharson, F.M. , Christodoulou, M. , Horgan, G. , Derrien, M. , et al. (2020) Vitamin biosynthesis by human gut butyrate‐producing bacteria and cross‐feeding in synthetic microbial communities. MBio 11: e00886‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover, C.K. , Pham, X.Q. , Erwin, A.L. , Mizoguchi, S.D. , Warrener, P. , Hickey, M.J. , et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406: 959–964. [DOI] [PubMed] [Google Scholar]
- Tsao, S.M. , and Yin, M.C. (2001) In vitro activity of garlic oil and four diallyl sulphides against antibiotic‐resistant Pseudomonas aeruginosa and Klebsiella pneumoniae . J Antimicrob Chemother 47: 665–670. [DOI] [PubMed] [Google Scholar]
- Varjani, S. , and Upasani, V.N. (2019) Evaluation of rhamnolipid production by a halotolerant novel strain of Pseudomonas aeruginosa . Bioresource Technol 288: 121577. [DOI] [PubMed] [Google Scholar]
- Voss, J.J.D. , Rutter, K. , Schroeder, B.G. , Su, H. , Zhu, Y.Q. , and Barry, C.E. (2000) The salicylate‐derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc Nat Acad Sci USA 97: 1252–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, Z. , Pu, L. , Gao, W. , Wang, Y. , Wei, J. , Shi, T. , et al. (2017) Riboflavin deficiency induces a significant change in proteomic profiles in HepG2 cells. Sci Rep 7: 45861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, S.S. , Eierhoff, T. , Aigal, S. , Brandel, A. , Thuenauer, R. , Bentzmann, S. , et al. (2017) The Pseudomonas aeruginosa lectin LecA triggers host cell signaling by glycosphingolipid‐dependent phosphorylation of the adaptor protein CrkII. BBA – Mol Cell Res 1864: 1236–1245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Volcano plots of differentially expressed genes (fold change > 2, P value < 0.05) of the PAO1 strain in the control group, DAS‐treated group, and DAE‐treated group based on RNA‐seq. Each gene is represented by a dot in the map; the up‐regulated genes are represented in red dots and the down‐regulated genes are represented in blue dots. A, DAE‐treated group vs. DAS‐treated group; B, Control group vs. DAS‐treated group; C, Control group vs. DAE‐treated group.
Fig. S2. The significantly enriched KEGG pathways based on transcriptome data of the PAO1 strain (DAE‐treated group vs DAS‐treated group). A, phenazine biosynthesis; B, quorum sensing; C, biosynthesis of siderophore group nonribosomal peptides; D, folate biosynthesis; E, thiamine metabolism; F, riboflavin metabolism.
Fig. S3. The significantly enriched KEGG pathways based on transcriptome data of the PAO1 strain (Control group vs. DAS‐treated group). A, phenazine biosynthesis; B, quorum sensing; C, biosynthesis of siderophore group nonrobosomal peptides; D, one carbon pool by folate; E, thiamine metabolism; F, riboflavin metabolism.


