Summary
The emergence and rapid spread of multidrug‐resistant bacteria has induced intense research for novel therapeutic approaches. In this study, the Acinetobacter baumannii bacteriophage D2 (vB_AbaP_D2) was isolated, characterized and sequenced. The endolysin of bacteriophage D2, namely Abtn‐4, contains an amphipathic helix and was found to have activity against multidrug‐resistant Gram‐negative strains. By more than 3 log units, A. baumannii were killed by Abtn‐4 (5 µM) in 2 h. In absence of outer membrane permeabilizers, Abtn‐4 exhibited broad antimicrobial activity against several Gram‐positive and Gram‐negative bacteria, such as Staphylococcus aureus, Pseudomonas aeruginosa, Klebsiella pneumonia, Enterococcus and Salmonella. Furthermore, Abtn‐4 had the ability to reduce biofilm formation. Interestingly, Abtn‐4 showed antimicrobial activity against phage‐resistant bacterial mutants. Based on these results, endolysin Abtn‐4 may be a promising candidate therapeutic agent for multidrug‐resistant bacterial infections.
We isolated phage vB_AbaP_D2 from hospital wastewater and purified endolysin from within (referred to as Abtn‐4). Our results suggest that endolysin Abtn‐4 has an intrinsic lytic activity against multidrug resistant A. baumannii. Abtn‐4 also exhibits antimicrobial activity against several Gram‐positive and Gram‐negative bacteria.

Introduction
Acinetobacter species have been recognized as one of the most important nosocomial pathogens (Almasaudi, 2018). Currently, the genus Acinetobacter comprises more than 50 genospecies, of which Acinetobacter baumannii (Acinetobacter genospecies 2), Acinetobacter pittii (Acinetobacter genospecies 3) and Acinetobacter nosocomialis (Acinetobacter genospecies 13TU) are the most clinically significant. A. nosocomialis, A. pittii and A. baumannii have high antibiotic resistance and cause serious nosocomial infections including pneumonia, bacteraemia, meningitis, endocarditis and sepsis (Vijayakumar et al., 2019). Compared to A. nosocomialis and A. pittii, A. baumannii is more resistant to carbapenem and patients with A. baumannii bacteraemia have higher mortality rates (Zhou et al., 2019). In 2017, the World Health Organization (WHO) released a priority list of antibiotic‐resistant pathogens that pose a major threat to global public health (Yuniati et al., 2018). Based on the urgency and the demand for new antimicrobial agents, priority was divided into three tiers: critical, high and medium. The highest ranked Gram‐negative bacteria are carbapenem‐resistant A. baumannii strains, which are also at the top of the critical category. The remarkable abilities of A. baumannii to form biofilms may explain its survival properties, increased virulence and multidrug resistance (Abdi‐Ali et al., 2014; Musafer et al., 2014; Zhao et al., 2015). Based on this, the most urgent need is for new antimicrobial agents capable of overcoming resistant bacteria and biofilms.
Bacteriophages (phages) are the most abundant viruses on the earth, which can specifically infect and lyse their host bacteria (Wittebole et al., 2014). Phages have long been recognized for their ability to combat multidrug‐resistant pathogens and biofilms. However, bacterial cells or biofilm cells may also develop various phage‐resistant mechanisms, seriously limiting the use of phages. Thus, endolysins have been studied as a possible approach to overcome these limitations. Endolysins are phage‐encoded enzymes that degrade the bacterial cell walls at the end of a phage replication cycle (Schmelcher et al., 2012) and have antibacterial activity against Gram‐positive bacteria. In the last decade, endolysins have been investigated as therapeutic agents, applied to a wide range of Gram‐positive bacteria, such as streptococci (Nelson et al., 2001), Enterococcus faecalis (Swift et al., 2016), Bacillus cereus (Schuch et al., 2019), Staphylococcus aureus (Gilmer et al., 2013) and Streptococcus pneumoniae (Jado et al., 2003). Several Gram‐positive phage endolysins have been shown to possess anti‐biofilm abilities including PlyC (Shen et al., 2013), LySMP (Meng et al., 2011) and ClyR (Yang et al., 2016). However, the outer membrane of Gram‐negative bacteria acts as a physical barrier, limiting the antimicrobial activity of endolysins (During et al., 1999; Nikaido, 2003; Briers et al., 2014). Previous studies showed that Gram‐negative bacteria could be killed by the combinations of endolysins and outer membrane permeabilizers (Briers et al., 2011; Oliveira et al., 2014; Wittebole et al., 2014; Guo et al., 2017a,b). Additionally, a few endolysins have an ability to lyse Gram‐negative bacteria by enzymatically active domains or other lytic domains (Morita et al., 2001; Oliveira et al., 2013). Briers and colleagues (Briers et al., 2014; Defraine et al., 2016) developed engineered endolysins (Artilysins) with activity against Gram‐negative bacteria. However, very few studies have reported on the ability of A. baumannii phage endolysins to control biofilms and phage‐resistant bacterial mutants.
In this study, we isolated phage vB_AbaP_D2 from hospital wastewater and purified endolysin from within (referred to as Abtn‐4). Our results suggest that endolysin Abtn‐4 has an intrinsic lytic activity against multidrug‐resistant A. baumannii. Abtn‐4 also exhibits antimicrobial activity against several Gram‐positive and Gram‐negative bacteria, including Staphylococcus aureus, Enterococcus, Salmonella, Pseudomonas aeruginosa and Klebsiella pneumoniae. Moreover, we studied the ability of Abtn‐4 to reduce biofilm formation, testing to see if Abtn‐4 was able to control bacterial biofilms. To the best of our knowledge, there are no other reports on the use of endolysins to combat phage‐resistant mutants. Therefore, the antibacterial efficacy of Abtn‐4 on phage‐resistant mutants was also investigated.
Results
Isolation and characterization of the phage vB_AbaP_D2
The clinical strain of A. baumannii AB9 was used as an indicator to screen phage in sewage samples from four local hospitals. Finally, A. baumannii phage vB_AbaP_D2 was isolated from the sample of the Second Hospital of Dalian Medical University. Phage D2 formed clear zones inside and translucent halos surrounding host strains. Translucent halos became larger after an additional 24 h (Fig. 1A). It is generally suggested that this phenomenon is enzymatic, as many of the lytic phages produce spreading halos around their plaques. Therefore, this phenomenon may indicate the lysis ability of endolysin or depolymerase (Loessner et al., 1995; Wu et al., 2019). We used transmission electron microscopy to classify D2 based on its virion morphology. As seen in Figure 1B, D2 was highly similar to members of the Podoviridae family, with a 47 nm isometric head and a 5 nm short tail. According to the lastest classification by the International Committee on the Taxonomy of Viruses (ICTV), the order of Caudovirales contains five families: Myoviridae, Podoviridae, Siphoviridae, Ackermannviridae and Herelleviridae. So far, the published sequences of Acinetobacter phages belong to three families of Caudovirales: the Myoviridae, Podoviridae and Siphoviridae. About 24% of Acinetobacter phages are podoviruses (Turner et al., 2017; Baginska et al., 2019). Therefore, the identification of a podovirus is relatively rare among the A. baumannii phage.
Fig. 1.

Morphology of phage vB_AbaP_D2.
A. Plaques of vB_AbaP_D2. The phage produced plaques with halos after incubation at 37°C for 12 h with halo size increasing over time.
B. Transmission electron micrograph of vB_AbaP_D2 negatively stained with 2% (w/v) uranyl acetate. The scale bars represent 20 nm.
C. One‐step growth curve of phage vB_AbaP_D2. Values represent the means ± SD (n = 3).
A one‐step growth curve reflects the three most important parameters of phage growth, the latent period, burst size and release period. As illustrated in Figure 1C, the latent and rising period of D2 lasted for 20 and 30 min, respectively, and the average burst size (the number of phage particles released by each infected host cell) was 80 ± 6.
Genome sequencing and ORF identification of phage vB_AbaP_D2
Phage D2 consisted 39 964 nucleotides with 39.23% GC content. Based on Blast‐P, phage D2 had 47 predicted ORFs, of which 24 ORFs were assigned a putative function (Table S1). Putative functions could be categorized into three functional modules: phage structure, host lysis, phage DNA metabolism and modification (Fig. 2).
Fig. 2.
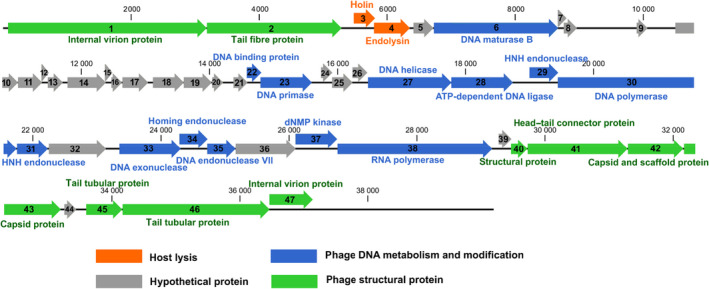
Genome map of the phage vB_AbaP_D2.
Sequence alignment and expression of the Abtn‐4
The putative endolysin protein ORF4 (Abtn‐4) consisted of 185 amino acids (approximately 21 kDa). Conserved domain analysis of CDD and InterProScan showed that Abtn‐4 contained glycoside hydrolase family 19 catalytic domain between residues 76 and 128. Although the sequence similarity of Abtn‐4 and the other three endolysins was high, six of the 18 amino acid mutations were in the conserved region (Fig. 3). The results of TMpred, HMMTOP and TCDB suggested that Abtn‐4 has an amphipathic helix between amino acids 121 and 141 (Fig. 4A). Blast‐P analysis showed that several proteins had sequence similarity with Abtn‐4. According to Phyre2 analysis, Abtn‐4 contained helical architecture, which is the typical structure of lysozyme family (Fig. 4B). Abtn‐4 was expressed and purified (Fig. 4C).
Fig. 3.

Sequence alignment of Abtn‐4 and closely related phage endolysins. The endolysin amino acid sequences used for alignment analysis included Abtn‐4 (GenBank accession no. YP_009624620.1), PlyAB1 (GenBank accession no. YP_008058242.1) (Huang, et al., 2014), LysABP‐01 (GenBank accession no. AHG30899.1) (Thummeepak, et al., 2016) and LysAB3 (GenBank accession no. YP_008060133.1) (Zhang, et al., 2018). Asterisks (*) indicate the mutation residues. Six mutations (green asterisks, amino acids 96, 97, 100, 103, 108 and 111) are found in the conserved domain.
Fig. 4.

Predictive analyses of endolysin Abtn‐4.
A. Hydrophobicity analysis of endolysin Abtn‐4 as predicted by TMpred and HMMTOP; blue and red lines stand for hydropathy and amphipathicity, orange bars mark transmembrane segments.
B. Three‐dimensional structure model of Abtn‐4. The structure model was predicted and built using Phyre2 server.
C. Expression and purification of Abtn‐4. Lane M, molecular weight marker; Lane 1, purified Abtn‐4. Lane 2, unpurified Abtn‐4.
Antibacterial activity of purified Abtn‐4
To test the concentration‐dependent effects of Abtn‐4, A. baumannii strain AB9 was used. Bacterial suspension and different concentrations of Abtn‐4 were incubated in PBS for 120 min at 37°C. As seen in Figure 5A, different concentrations of Abtn‐4, including 2.5, 5 and 7.5 µM, could reduce AB9 titres from about 6 × 108 cfu ml−1 to 2.5 × 107 cfu ml−1 (1 log reduction), 3.6 × 105 cfu ml−1 (3 log reductions) and 5.4 × 104 cfu ml−1 (4 log reduction). Additionally, bacterial counts were reduced by exposure to Abtn‐4 2.5, 5 and 7.5 µM during the first hour. There were no significant reductions in the number of viable cells at the concentration of 0.5 µM. A. baumannii strains AB3, AB7, AB9 were chosen as models to examine the effect of an outer membrane permeabilizer on Abtn‐4 antibacterial activity (Fig. 5B). After being treated with Abtn‐4 (5 µM) in PBS for 2 h at 37°C, at least 99% of the cells of the tested strains were killed (2–3 logs reduction) without outer membrane permeabilizer (EDTA) pretreatment. The Abtn‐4 challenged cells that were treated with EDTA were reduced by about 3–4 log units. However, EDTA alone (0.1 mM) had no significant effect on cell viability, suggesting that EDTA probably increased outer membrane permeability of the cell outer membrane and promoted lytic activity of Abtn‐4.
Fig. 5.

Antimicrobial activity of endolysin Abtn‐4.
A. Dependence of bactericidal activity on Abtn‐4 concentration.
B. Effect of the outer membrane permeabilizer (EDTA) on the lytic ability of Abtn‐4 (5 µM). Values represent the means ± SD (n = 3).
Antibacterial range of phage D2 and endolysin Abtn‐4
A total of 48 bacterial strains (as listed above) were used to analyse the antibacterial range of endolysin Abtn‐4 and phage D2. The results of the antimicrobial range of Abtn‐4 and phage D2 towards these strains are shown in Figure 6 and Table 1. Unexpectedly, Abtn‐4 exhibited broad antimicrobial activity (Fig. 6). The Gram‐negative cells were reduced by 1–3 logs after 2 h of treatment with Abtn‐4 (5 µM). Additionally, the bacterial numbers of 53.8% (7/13) of the tested Gram‐positive strains were significantly lower after Abtn‐4 (5 µM) treatment than after PBS treatment (P < 0.05). However, phage D2 specifically lysed six A. baumannii strains, including AB9, AB10, AB11, AB15, AB16 and DL114 as shown in Table 1. The results of the liquid culture‐based host range assay also showed that phage D2 was able to inhibit the growth of A. baumannii strain AB9, AB10, AB11, AB15, AB16 and DL114 (Fig. S1).
Fig. 6.

The antibacterial range of endolysin Abtn‐4. The bacterial suspension of each strain was treated with Abtn‐4 (5 µM) or PBS for 2 h at 37°C and the viable cell numbers were counted. Values represent the means ± SD (n = 3). Significance was performed using Student’s t‐test. **P < 0.01; *P < 0.05; ns, P > 0.05.
Table 1.
The host range of phage vB_AbaP_D2.
| Organism | Notes | EOP a |
|---|---|---|
| Acinetobacter baumannii | AB1 | – |
| Acinetobacter baumannii | AB3 | – |
| Acinetobacter baumannii | AB5 | – |
| Acinetobacter baumannii | AB7 | – |
| Acinetobacter baumannii | AB9 | + + + |
| Acinetobacter baumannii | AB10 | + + |
| Acinetobacter baumannii | AB11 | + + + |
| Acinetobacter baumannii | AB13 | – |
| Acinetobacter baumannii | AB15 | + + |
| Acinetobacter baumannii | AB16 | + + |
| Acinetobacter baumannii | DL114 | ++ |
| Acinetobacter baumannii | DL112 | – |
| Acinetobacter baumannii | 073 | – |
| Acinetobacter baumannii | ATCC19606 | – |
| Acinetobacter baumannii | ATCC17978 | – |
| Salmonella Enteritidis | SE1 | – |
| Salmonella Typhimurium | CMCC50220 | – |
| Klebsiella pneumoniae | Kp11 | – |
| Klebsiella pneumoniae | Kp12 | – |
| Klebsiella pneumoniae | Kp15 | – |
| Klebsiella pneumoniae | Kp21 | – |
| Klebsiella pneumoniae | Kp22 | – |
| Klebsiella pneumoniae | Kp23 | – |
| Klebsiella pneumoniae | Kp24 | – |
| Klebsiella pneumoniae | Kp25 | – |
| Klebsiella pneumoniae | KP1513 | – |
| Pseudomonas aeruginosa | Pa1 | – |
| Pseudomonas aeruginosa | Pa2 | – |
| Pseudomonas aeruginosa | Pa3 | – |
| Pseudomonas aeruginosa | Pa7 | – |
| Pseudomonas aeruginosa | Pa18 | – |
| Pseudomonas aeruginosa | Pa19 | – |
| Pseudomonas aeruginosa | Pa24 | – |
| Pseudomonas aeruginosa | Pa26 | – |
| Pseudomonas aeruginosa | PA0212 | – |
| Enterococcus faecium | FM1 | – |
| Enterococcus faecium | FM2 | – |
| Enterococcus faecium | FM3 | – |
| Enterococcus faecalis | FS1 | – |
| Enterococcus faecalis | FS2 | – |
| Enterococcus faecalis | FS3 | – |
| Enterococcus faecalis | FS4 | – |
| Staphylococcus aureus | Sau1 | – |
| Staphylococcus aureus | Sau2 | – |
| Staphylococcus aureus | Sau39 | – |
| Staphylococcus aureus | Sau40 | – |
| Staphylococcus aureus | Sau41 | – |
| Staphylococcus aureus | Sau55 | – |
EOP value of 0.5 or more was classified as high efficiency, ‘+++’; 0.1–0.5 as medium efficiency, ‘++’; 0.001–0.1 as ‘low’ efficiency, ‘+’; and 0–0.001 was classified as inefficient, ‘–’.
The biofilm reduction ability of Abtn‐4
In order to investigate whether Abtn‐4 had the ability of reducing biofilm formation, the Abtn‐4 was added before the biofilm matured. Therefore, the Abtn‐4 (5 μM) was added at two growth phases of the biofilms. The two growth phases were the early phase (12 h post‐incubation) and pre‐maturation phase (36 h post‐incubation) respectively. In this experiment, the 96‐well microtitre biofilm assay was used following the crystal violet technique. The biofilm reduction ability of Abtn‐4 was quantified by measuring the OD600. The results showed that Abtn‐4 had a reducing effect on biofilms in the tested strains, and the biofilm reduction rate of the 30 strains was more than 30% (Fig. 7A). According to SEM micrographs, Abtn‐4 could effectively reduce biofilm formation (Fig. 7B). In the Abtn‐4 treatment groups, the biofilm matrixes on coverslips were broken down and only remained a few bacteria.
Fig. 7.

Efficacy of endolysin Abtn‐4 to control biofilms. The Abtn‐4 (5 µM) was added at two timepoints of biofilm growth, namely the early stage (12 h after incubation) and the pre‐mature stage (36 h after incubation). PBS were added instead of Abtn‐4 as control. After 48 h incubation, the biofilm reduction ability of Abtn‐4 was determined using OD600 reduction assay and SEM micrographs.
A. The biofilm reduction ability of Abtn‐4. Values represent the means ± SD (n = 3).
B. SEM micrographs of biofilm dispersion.
The antibacterial effect of Abtn‐4 against phage‐resistant bacterial mutants
During the phage purification process, we found that some colonies appeared on the double‐agar plates. Colonies were picked into LB broth and purified by repeated streaking on LB agar plate. We also found that the colonies still retained phage resistance for more than three passages. As shown in Figure 8A, colonies that occurred on the double‐agar plates were considered as phage‐resistant mutants. We obtained mutant colonies of each phage D2 host strain (AB9, AB10, AB11, AB15, AB16) from the double‐agar plates and named them as AB9R, AB10R, AB11R, AB15R, AB16R respectively. The mutant colonies and their parent strains showed different colony morphology on LB agar plates. Parent strains formed large (diameter 2.5–3 mm), convex, moist and mucoid colonies. The mutant colonies were small (diameter 1–1.5 mm), circular and smooth. The cross streak assay indicated that those colonies were resistant to phage D2 (Fig. 8B). When testing for antibacterial activity, results showed that Abtn‐4 (5 μM) could effectively inhibit the growth of AB9, AB10, AB11, AB15, AB16 and their corresponding phage resistant mutants of AB9R, AB10R, AB11R, AB15R, AB16R (Fig. 9).
Fig. 8.

Isolation of phage‐resistant bacterial mutants.
A. The phage‐resistant colonies on the lawn of phage vB_AbaP_D2 host strain in the double‐layer agar assay.
B. The cross‐streak assay with phage‐resistant mutants growing across phage streak zone.
Fig. 9.

The antibacterial activity of endolysin Abtn‐4 against phage‐resistant bacterial mutants. The bacterial suspensions of AB9, AB10, AB11, AB15, AB16 and their corresponding phage‐resistant mutants AB9R, AB10R, AB11R, AB15R, AB16R treated with Abtn‐4 (5 µM). The control groups were treated with equivalent quantity PBS. The OD600 values were measured and used to evaluate the antibacterial activity of Abtn‐4. Values represent the means ± SD (n = 3).
Discussion
Increasing resistance of pathogenic bacteria to multiple antimicrobial agents has promoted public health sectors across the world to seek alternative antimicrobial agents (Magiorakos et al., 2012). Since endolysins show rapid bactericidal action and low risk of antibacterial resistance, they are promising and novel agents towards the treatment of multidrug‐resistant bacterial infections (Nelson et al., 2012). Generally, peptidoglycan‐degrading enzymes are effective against Gram‐positive bacteria. However, their activity against Gram‐negative bacteria is limited by the outer membrane, which acts as a permeability barrier to restrict access of enzymes to peptidoglycan. Luckily, some Gram‐negative background endolysins have antibacterial activities that use a completely independent enzymatic activity mechanism (Dong et al., 2015; Wang et al., 2017). Several studies have previously described the wide spectrum endolysins from A. baumannii, and growing evidence has indicated that they are capable of much more than just killing parental phage‐specific strains. In such cases, the A. baumannii endolysin LysSi3 was shown to be capable of degrading Gram‐negative ESKAPE pathogens (E. faecium, S. aureus, A. baumannii, P. aeruginosa, K. pneumoniae and other Enterobacteriaceae species) (Antonova et al., 2019). Wu and colleagues (Wu et al., 2018) isolated 40 A. baumannii phages, among which phage PD‐6A3 had a relatively broad host range of 32.4% (179/552) among clinical MDR A. baumannii isolates. The endolysin of A. baumannii phage PD‐6A3 was able to lyse 70.5% (141/200) of the clinical MDRAB isolates. This endolysin also showed lytic activity for the strains of Enterococci, methicillin‐resistant S. aureus, K. pneumonia, P. aeruginosa and E. coli. Huang et al. (2014) also showed that A. baumannii phage endolysin PlyAB1 was able to degrade all 48 clinical pandrug‐resistant A. baumannii isolates within a relatively short time. Moreover, these endolysins have a common feature, they all contain a C‐terminal amphipathic helical region, which plays a key role in lytic activities. Peng et al. (2017) additionally identified that the C‐terminal amphipathic helical region of LysAB2 enhanced the permeability of the outer membrane of A. baumannii. The deletion of C‐terminal amphipathic helical region could reduce the antibacterial activity of these endolysins. Lai et al. (2011) confirmed that the removal of the amphipathic helical domain strongly reduced the bactericidal activity of endolysin LysAB2. After deleting the amphiphilic helical region, the antibacterial effect of endolysin LysAB3 decreased from 95.8% to 33.3% (Zhang et al., 2018a,b). Thandar et al. (2016) showed that the C‐terminal region of the endolysin PlyF307 exhibited higher antibacterial activity against A. baumannii than the intact lysin molecule. Similar to the above endolysins, the endolysin in phage D2 also contained a C‐terminal amphipathic helical region. The similarity between Abtn‐4 and other endolysins suggested that the amphipathic helical region might simultaneously help Abtn‐4 kill Gram‐negative bacteria and broaden their bactericidal spectrum. Furthermore, Abtn‐4 contained a glycoside hydrolase family 19 catalytic domain between residues 76 and 128. It is known that glycoside hydrolase family 19 are able to destroy bacterial cell walls depending upon its activity of hydrolysing β‐1,4‐glycosidic linkage between N‐acetylmuramic acid (NAM) and N‐acetylglucosamine (NAG) (Flach et al. (1992). Therefore, Abtn‐4 might have glycoside hydrolase activity against bacterial peptidoglycan layers.
Our results demonstrated that Abtn‐4 had antibacterial activity against A. baumannii strains in the absence of the outer membrane permeabilizing agent. After being pretreated with EDTA, more than 1 log of cells were killed by Abtn‐4, suggesting that the antibacterial activity of Abtn‐4 could be enhanced by outer membrane permeabilizers. Additionally, Abtn‐4 exhibited concentration‐dependent activities, and at 0.5 μM, Abtn‐4 was not able to inhibit the growth of AB9.
In our results, Abtn‐4 had a wide spectrum of antimicrobial activity. All tested Gram‐negative strains were lysed by Abtn‐4 (5 μM) within 2 h. In addition to strains of A. baumannii, Abtn‐4 could effectively lyse strains of Salmonella, P. aeruginosa and K. pneumoniae. However, the effectiveness of Abtn‐4 against Gram‐positive strains of Enterococcus, especially E. faecium FM1 and FM2, were not obvious. Cell walls of E. faecium contain a thick and rigid peptidoglycan layer (Hancock et al., 2014), which might decrease the lytic abilities of Abtn‐4.
Biofilms are complex surface‐associated communities of bacterial cells and make a significant contribution towards the development of antibiotic resistance across a wide variety of environments (Mah, 2012; He et al., 2015). Previous studies found that the strong biofilm formation abilities of A. baumannii could increase their survivorship in hospital environments (Rodriguez‐Bano et al., 2008). Additionally, it is also very difficult for traditional disinfectants or antibiotics to reduce biofilm formation and dissolve biofilm matrixes (da Costa Luciano et al., 2016). Moreover, biofilms always adhere to indwelling medical devices and cause implant‐associated infections (Hoiby et al., 2015). Therefore, novel approaches are urgently needed to limit biofilm formation and treat biofilm‐associated infections. Phages and phage‐associated enzymes have been extensively studied as strategies for biofilm prevention and eradication. All major Gram‐positive pathogenic endolysins and their efficacy against biofilms have been previously characterized and described. However, few studies have reported on the effectiveness of endolysins from A. baumannii phages against various bacterial biofilms. In our study, the antibacterial activity of Abtn‐4 against Gram‐negative strains suggested that Abtn‐4 might be a promising new tool to control biofilm formation. Our results demonstrated that the biofilms formed by the Gram‐negative strains could be reduced by Abtn‐4. Surprisingly, Abtn‐4 could also reduce biofilm formation of additional Gram‐positive strains, especially those of S. aureus Sau1 and Sau2. These results were further corroborated by SEM micrographs, verifying that Abtn‐4 treated groups showed a clear reduction in biofilm cell densities, with only single cells in representative fields being visible. This indicates efficient reduction of biofilm formation by Abtn‐4. These above results provide evidence towards Abtn‐4 being a highly effective anti‐biofilm agent.
So far, the rapid occurrence of phage‐resistant mutants has become one of the main challenges for the effective treatment by phages (Torres‐Barcelo, 2018). We found that Abtn‐4 was able to inhibit the growth of phage‐resistant bacterial mutants. In our recent studies, phage D2 was mixed with AB9 and AB9R. After coculturing at 37°C for 48 h with shaking, phage D2M was obtained from the mixture. Phage D2M was capable of killing AB9 and AB9R (data not shown here). Interestingly, phage D2M formed a halo around plaques on AB9 and did not form halo on AB9R. Halos are formed due to the degradation of exopolysaccharide by phage depolymerases after phage infection (Oliveira et al., 2017). Therefore, the exopolysaccharide may be the primary receptor for phage D2, and resistant strains can prevent phage D2 adsorption by losing their exopolysaccharide. Without the protection of exopolysaccharide, bacteria may be more likely to be lysed by Abtn‐4.
Altogether, endolysin Abtn‐4 is a promising antimicrobial agent for controlling both Gram‐positive and Gram‐negative bacterial infections. We also believe that endolysin Abtn‐4 is a potentially effective agent to control the formation of biofilms and the growth of phage‐resistant mutants. Our future research will further explore the possibilities that endolysins may be used as a supplement in phage therapies to delay and reduce the appearance of phage‐resistant mutants, thus to extend the durability and effects of phage treatment. Both phages and endolysins should be further studied in order to explore their potential for treating bacterial infections.
Experimental procedures
Bacterial strains and antibiotic susceptibility testing
In total, this study included 48 strains of different bacterial species, including the Gram‐negative A. baumannii (n = 15), Salmonella (n = 2), P. aeruginosa (n = 9) and K. pneumoniae (n = 9), as well as the three Gram‐positive species, Enterococcus faecium (n = 3), E. faecalis (n = 4) and S. aureus (n = 6). Bacterial sources, culture conditions and growth media are listed in Table S2.
The antibiotic susceptibility of clinical isolates was determined by the Clinical Laboratory and Standards Institute (CLSI) broth microdilution method. Briefly, antibiotic stock solutions were twofold serially diluted from 256 μg ml−1 with TSB in a 96‐well plate, and the bacterial strains (105 cfu ml−1) were cultured in the diluted antibiotic solutions. After incubation for 20 h at 37°C with agitation, minimum inhibitory concentrations (MIC) were determined as the lowest concentration of antibiotics inhibiting the growth of each strain. Antibiotic susceptibilities of clinical isolates are listed in Table S3.
Phage isolation and characterization
The clinical strain of the multidrug‐resistant A. baumannii AB9 was used as an indicator for phage isolation. Wastewater samples were collected from the Second Hospital of Dalian Medical University. Wastewater was cleared by centrifugation (10 000 × g, 10 min, 4°C) and filtered (0.22‐μm membrane filters). Purified wastewater (10 ml) and mid‐log phase AB9 (1 ml) were mixed with 2× concentrated LB broth (10 ml). Mixtures were cultured at 37°C at 160 r.p.m. for 24 h in order to enrich the phages. These cultures were then centrifuged at 10 000 × g, 4°C for 10 min, and the supernatants were filtered through a 0.22‐μm filter to remove bacteria. A double‐layer agar technique was used to detect and purify phages as described previously (Kutter, 2009). Briefly, 100 μl of filtrate and 200 μl of AB9 (108 cfu ml−1) were mixed with semi‐solid agar medium and mixtures were poured onto LB agar plates. Plaques were incubated at 37°C and left to form on plates overnight. For phage purification, clear, single phage plaques were picked and resuspended in 100 μl of sterilized sodium chloride‐magnesium sulphate (SM) buffer (2% gelatin, 10 mM MgSO4·7 H2O, 10 mM Tris/HCl, 100 mM NaCl, pH 7.5). Then, phage suspensions were plated by the double‐agar layer technique. Single‐plaque isolations were performed in three consecutive rounds in order to obtain the purified phages. All phage lysates were stored at 4°C.
A one‐step growth experiment of the phage was performed at a multiplicity of infection (MOI) of 0.001 as previously described (Pajunen et al., 2000). In brief, phage was mixed with mid‐log phase host culture at MOI of 0.001 and adsorbed at 37°C for 5 min. The mixtures were centrifuged (10 000 × g, 10 min), and pellets were resuspended in fresh LB broth. Resuspensions were cultured at 37°C with shaking. Samples were collected every 10 min for 120 min, and then, the phage titre of each sample was measured by the double‐layer agar technique. All experiments were carried out in triplicate.
To obtain transmission electron micrographs (TEM), phage particles were preliminarily purified through Polyethylene Glycol 8000 (PEG8000) treatments. In brief, 1 M solid NaCl was added to 200 ml of phage lysate, stored at 4°C for 1 h and then centrifuged (10 000 × g, 10 min). To precipitate the phage particles, 10% (w/v) PEG8000 was added to the supernatant and stored overnight on ice. This solution was centrifuged at 10 000 × g for 10 min and pellets were resuspended in 1 ml SM buffer. The condensed phage suspensions were purified by CsCl density gradient centrifugation (CsCl, 0.6 g ml−1, 200 000 × g for 16 h at 4°C) as previously described (Tang et al., 2017). The morphology of the phages was examined using a JEM‐2000EX TEM (JEOL, Tokyo, Japan) (Yang et al., 2010).
Genome sequencing and bioinformatics analysis
Genomic DNA of phage vB_AbaP_D2 was extracted using the Universal Phage Genomic DNA Extraction Kit (Knogen, Guangzhou, China) and stored at −20°C. Sequencing of the whole D2 genome was performed on the Biozeron (China) Illumina Hiseq paired‐end platform. The program GeneMark was used to predict and analyse the potential open reading frames (ORFs) of D2. Functional annotation was assessed using the online platform Blast‐P (http://www.ncbi.nlm.nih.gov/BLAST/). The complete genome sequence of phage D2 has been deposited in the GenBank database and assigned the accession number MH042230.
Sequence analysis of endolysin Abtn‐4
The amino acid sequence of endolysin Abtn‐4 was identified using the online platform Blast‐P. Amino acid sequences of Abtn‐4 and several previously published endolysins were aligned using ClustalW2. The functional domains of Abtn‐4 were analysed using the NCBI Conserved Domain Database (CDD) (Marchler‐Bauer et al., 2015) and InterProScan program (Jones et al., 2014). The structural prediction of Abtn‐4 was performed on the Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index). Protein hydrophobicity and identification of the transmembrane helix was predicted using the following three programs: TMpred (https://embnet.vital‐it.ch/software/TMPRED_form.html), HMMTOP (http://www.enzim.hu/hmmtop/html/submit.html) and TCDB (http://www.tcdb.org/progs/?tool=hydro).
Expression, purification and characterization of Abtn‐4
The putative endolysin Abtn‐4 gene (genetic region 5797–6354 bp) was amplified from the genomic DNA of phage D2 using the following primers: Abtn‐4‐Forward (5‐GCCATATGATGATTCTGACTAAAGACGGG‐3) and Abtn‐4‐Reverse (5‐GCCTCGAGCTATAAGCTCCGTAGAGCGCG‐3), with the restriction endonuclease sites of NdeI/XhoI underlined. PCR fragments and vector pET28a were digested using NdeI/XhoI restriction endonucleases (TaKaRa, Dalian, China) and PCR restriction fragments inserted into pET28a. Plasmids of pET28a‐Abtn‐4 were transformed into E. coli BL21(DE3) and the recombinant proteins were expressed and produced as follows: recombinant cells were incubated in LB broth with 50 µg ml−1 kanamycin at 37°C with shaking. When the OD600 (optical density at 600 nm) reached 0.8, IPTG (isopropyl‐β‐d‐thiogalactopyranoside) was added to a final concentration of 1 mM. Then, cultures were incubated at 16°C for 14 h with shaking. Cells were centrifuged and resuspended in lysis buffer (20 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0) and lysed by sonication on ice. Samples were centrifuged (10 000 g, 4°C for 20 min) and filtered (0.22‐μm membrane filters) to remove debris. For recombinant protein purification, soluble cell‐free extracts were loaded onto a 5 ml Ni‐NTA‐Sefinose columns (Sangon, Shanghai, China) and eluted with 400 mM imidazole buffer solution (Oliveira et al., 2016). Purified recombinant protein was stored at −80°C. Before use, the purified protein was equilibrated by dialysis phosphate‐buffered saline (PBS, 1.8 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl, 10 mM Na2HPO4, pH 7.8).
Antibacterial activity of endolysin Abtn‐4
Acinetobacter baumannii AB9 was used to test the concentration‐dependent effects of Abtn‐4. Mid‐log phase bacterial cultures were centrifuged (6000 × g, 5 min, 4°C), washed three times with sterile PBS and suspended to a final concentration of 108 cfu ml−1. Abtn‐4 was adjusted to different concentrations using PBS; then, 100 µl of each of the different concentrations of Abtn‐4 was added into bacterial suspensions. Final concentrations were 0.5, 2.5, 5 and 7.5 µM. Mixtures were incubated at 37°C, and bacterial counts were done at 0, 20, 40, 60, 80, 100 and 120 min post‐incubation (Yang et al., 2016; Haddad Kashani et al., 2017). For controls, equivalent quantities of PBS were added instead of Abtn‐4.
Strains of AB3, AB7 and AB9 were randomly selected from the listed A. baumannii strains (Table S2) and used to investigate the effect of outer membrane permeabilizers on the antimicrobial activity of Abtn‐4. Briefly, bacterial suspensions that were pretreated with or without an outer membrane permeabilizer (0.1 mM EDTA) were mixed with Abtn‐4 (final concentration was 5 µM) and then were incubated at 37°C for 2 h. For the control, equivalent quantities of PBS were added instead of Abtn‐4. After incubation, bacterial colony‐forming units were counted and the antibacterial activities were expressed as the decrease of viable bacterial counts. All experiments were done in triplicate.
Host range determination
All strains listed in Table S2 were used for the range assay of phage D2 and endolysin Abtn‐4. The host range of the phage was tested by spot testing and efficiency of plating (EOP), but with some modifications (Khan Mirzaei and Nilsson, 2015; Yang et al., 2015). A total of 200 μl of mid‐log phase bacterial cultures were plated on LB agar plates, and phage lysates (109 pfu ml−1) were spotted onto the surface of each plate. Plates were dried at room temperature and incubated overnight at 37°C. After spot formation, individual spots were examined and phage sensitive strains were initially determined. The double‐layer agar technique was performed to determine titres of phage D2 on phage‐sensitive strains. EOP values were calculated as the ratio of titre on the test strain to the titre on the host strain of A. baumannii AB9. EOP values of 0.5 or more were classified as high efficiency ‘+++’; 0.1–0.5 as medium efficiency ‘++’; 0.001–0.1 as low efficiency ‘+’; and 0–0.001 classified as inefficient ‘−’.
To further investigate the host range of phage D2, a modified liquid culture‐based host range assay was performed as described by Xie et al. (2018), with some modifications. Briefly, bacterial strains were cultured overnight at 37°C with shaking. Bacterial cultures were washed twice with sterile PBS and resuspended in fresh TSB to achieve a concentration of 107 cfu ml−1. Each well of the 96‐well microtitre plate was filled with 180 μl of the bacterial suspension, and 20 μl of phage lysate dilution was added at different final concentrations (106, 107 and 108 PFU ml−1). For the controls, 20 μl of PBS was added instead of Abtn‐4. Plates were incubated for 12 h at 37°C without agitation. The OD600 of wells were measured every hour using the microplate spectrophotometer (Multiskan Go, Thermo Scientific). All experiments were done in triplicate.
To test the lytic spectrum of Abtn‐4, Abtn‐4 was added to each mid‐log phase bacterial suspension with the final concentration of 5 µM. The same volume of PBS was added as control. After 2 h incubation at 37°C, samples were serially diluted in PBS and plated for counting of colony‐forming units. All experiments were done in triplicate.
Biofilm assay
The biofilm preventative effects of Abtn‐4 were evaluated by the 96‐well microtitre plate method using the crystal violet technique (Stepanovic et al., 2000; Guo et al., 2017a,b; Glowacka‐Rutkowska et al., 2018). Briefly, 20 µl of overnight grown bacterial cultures and 180 µl of growth media were added to all wells of the 96‐well plates. Wells only containing growth media were used as negative controls. After 48 h incubation at 37°C in static conditions, the supernatants were removed with a pipette and wells were washed twice with sterile PBS to remove non‐adhered cells. Afterwards, wells were fixed with 200 µl of 99% methanol for 30 min, drained and air‐dried at room temperature. A total of 200 µl of 1% (w/v) crystal violet was added to each well and plates were incubated for 30 min at room temperature. Subsequently, wells were washed gently with sterile PBS and left to air‐dry. Dyes were dissolved with 200 µl of 33% vol/vol glacial acetic acid. The OD values were measured at 600 nm using the microplate spectrophotometer. Biofilm formation capabilities of the strains were classified as follows: OD ≤ ODc, non‐biofilm formation; ODc < OD ≤ 2× ODc, weak; 2 × ODc < OD ≤4 × ODc, moderate; 4 × ODc < OD, strong. The optical density cut‐off value (ODc) was defined as three standard deviations above the mean OD600 of the negative control wells. All experiments were done in triplicate.
Biofilm challenge
The biofilm reduction ability of endolysin Abtn‐4 was evaluated as previously described (Zhang et al., 2018a,b), with some modifications. Briefly, 20 μl of fresh log‐phase bacterial culture and 180 µl of growth media were added to each well in the 96‐well plates. Then, Abtn‐4 (5 µM) was added to the two growth stages of the biofilms, namely the early stage (12 h after incubation) and the pre‐mature stage (36 h after incubation). For controls, PBS was added instead of Abtn‐4. After 48 h incubation, the supernatant was removed with a pipette and wells were washed twice with PBS to remove the non‐adhered cells. Fixation, staining and measurement of the biofilms was carried out as described above. Biofilm reduction rates of Abtn‐4 were obtained using the following equation: (N 0‐N 1)/N 0 × 100%, where N 0 = the OD600 in the PBS treatment group and N 1 = the OD600 in the Abtn‐4 treatment group. All experiments were done in triplicate.
Scanning electron micrographs (SEM) were obtained to investigate biofilm formation. Briefly, coverslips were placed into wells of a 12‐well plate with bacterial suspensions (200 µl) and LB broth (1.8 ml). The plate was incubated at 37°C under static conditions. Then, Abtn‐4 (5 µM) was added as described above. After 48 h incubation, the supernatant was removed with a pipette and the coverslips were washed twice with PBS to remove the non‐adhered cells. Coverslips for SEM were immobilized with 2.5% (w/v) glutaraldehyde and dehydrated through a graded ethanol series (50%, 70%, 90%, 100%). All specimens were coated with gold for 3 min, and micrographs were obtained using a NOVA NanoSEM 450 (Japan).
Phage‐resistant bacterial mutants
Phage resistance always occurs in the bactericidal process of lytic phages. Thus, we investigated the effect of Abtn‐4 on phage‐resistant bacterial mutants. The phage‐resistant mutants of the phage D2 host strains (AB9, AB10, AB11, AB15 and AB16) were obtained from double‐layer agar plates (Yu et al., 2018). After 48 h incubation at 37°C, double‐layer agar plates of high titre phage formed single colonies. Colonies were picked from plates and placed into 5 ml of LB broth. After culturing overnight at 37°C with shaking, we used the cross streak assay to look for resistant bacterial cultures. Briefly, 30 µl of phage lysates was spotted on LB agar plates using horizontal phage streaks. Then, 20 µl of the bacterial culture was spotted on the left side of the phage streak, crossing the phage streak to form a vertical bacterial streak on the plate. Bacterial cultures that grew across the phage streak zone were considered as phage‐resistant bacterial mutants. One phage‐resistant mutant of each host strain was randomly selected and named as AB9R, AB10R, AB11R, AB15R and AB16R, and was used for antibacterial activity testing of Abtn‐4. Briefly, the mixtures of Abtn‐4 (5 µM) and the bacterial suspensions (104 cfu ml−1) were added into 96‐well microplates and incubated for 360 min at 37°C. Equal volume of PBS was added instead of Abtn‐4 as the control. The OD600 was measured by a microplate reader at 40‐min intervals.
Statistical analysis
Student’s t‐test was used to evaluate significance and considered values of P < 0.05 to be statistically significant.
Nucleotide sequence accession numbers
The complete genome sequence of phage vB_AbaP_D2 has been deposited in the GenBank database and assigned the accession number MH042230.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Supporting information
Fig. S1. The liquid culture‐based host range assay of phage vB_AbaP_D2. Acinetobacter baumannii strains (107 cfu ml−1) were infected with phages at 106, 107, and 108 pfu ml−1. Optical densities were measured at 600 nm. Error bars represent standard deviations of three replicates.
Table S1. Predicted protein function of phage vB_AbaP_D2.
Table S2. List of strains used in this study.
Table S3. Antibiotic susceptibility of clinical isolates.
Acknowledgements
We would like to thank Daping Nie for the kindness of providing wastewater samples.
Microbial Biotechnology (2021) 14(2), 403–418
Funding information
This work was funded by the Fundamental Research Funds for the Central Universities (DUT20GJ217), National Natural Science Founfation of China (31701719) and LiaoNing Revitalization Talents Program (XLYC1907085).
References
- Abdi‐Ali, A. , Hendiani, S. , Mohammadi, P. , and Gharavi, S. (2014) Assessment of biofilm formation and resistance to imipenem and ciprofloxacin among clinical isolates of Acinetobacter baumannii in Tehran. Jundishapur J Microbiol 7: e8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasaudi, S.B. (2018) Acinetobacter spp. as nosocomial pathogens: epidemiology and resistance features. Saudi J Biol Sci 25: 586–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonova, N.P. , Vasina, D.V. , Lendel, A.M. , Usachev, E.V. , Makarov, V.V. , Gintsburg, A.L. , et al. (2019) Broad bactericidal activity of the Myoviridae bacteriophage lysins LysAm 24, LysECD7, and LysSi3 against Gram‐negative ESKAPE pathogens. Viruses 11: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginska, N. , Pichlak, A. , Gorski, A. , and Jonczyk‐Matysiak, E. (2019) Specific and selective bacteriophages in the fight against multidrug‐resistant Acinetobacter baumannii . Virol Sin 34: 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briers, Y. , Walmagh, M. , and Lavigne, R. (2011) Use of bacteriophage endolysin EL188 and outer membrane permeabilizers against Pseudomonas aeruginosa . J Appl Microbiol 110: 778–785. [DOI] [PubMed] [Google Scholar]
- Briers, Y. , Walmagh, M. , Van Puyenbroeck, V. , Cornelissen, A. , Cenens, W. , Aertsen, A. , et al. (2014) Engineered endolysin‐based “Artilysins” to combat multidrug‐resistant gram‐negative pathogens. MBio 5: e01379‐01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, F. , Wang, X. , Wang, L. , Li, Z. , Che, J. , Wang, L. , et al. (2015) Evaluation of the efficacy of a bacteriophage in the treatment of pneumonia induced by multidrug resistance Klebsiella pneumoniae in mice. Biomed Res Int 2015: 752930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa Luciano, C. , Olson, N. , Tipple, A.F. , and Alfa, M. (2016) Evaluation of the ability of different detergents and disinfectants to remove and kill organisms in traditional biofilm. Am J Infect Control 44: e243–e249. [DOI] [PubMed] [Google Scholar]
- Defraine, V. , Schuermans, J. , Grymonprez, B. , Govers, S.K. , Aertsen, A. , Fauvart, M. , et al. (2016) Efficacy of artilysin Art‐175 against resistant and persistent Acinetobacter baumannii . Antimicrob Agents Chemother 60: 3480–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, H. , Zhu, C. , Chen, J. , Ye, X. , and Huang, Y.P. (2015) Antibacterial activity of Stenotrophomonas maltophilia endolysin P28 against both Gram‐positive and Gram‐negative bacteria. Front Microbiol 6: 1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- During, K. , Porsch, P. , Mahn, A. , Brinkmann, O. , and Gieffers, W. (1999) The non‐enzymatic microbicidal activity of lysozymes. FEBS Lett 449: 93–100. [DOI] [PubMed] [Google Scholar]
- Flach, J. , Pilet, P.E. , and Jolles, P. (1992) What's new in chitinase research? Experientia 48: 701–716. [DOI] [PubMed] [Google Scholar]
- Gilmer, D.B. , Schmitz, J.E. , Euler, C.W. , and Fischetti, V.A. (2013) Novel bacteriophage lysin with broad lytic activity protects against mixed infection by Streptococcus pyogenes and methicillin‐resistant Staphylococcus aureus . Antimicrob Agents Chemother 57: 2743–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka‐Rutkowska, A. , Gozdek, A. , Empel, J. , Gawor, J. , Zuchniewicz, K. , Kozinska, A. , et al. (2018) The ability of lytic Staphylococcal Podovirus vB_SauP_phiAGO1.3 to coexist in equilibrium with its host fcilitates the selection of host mutants of attenuated virulence but does not preclude the phage antistaphylococcal activity in a nematode infection model. Front Microbiol 9: 3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, M. , Feng, C. , Ren, J. , Zhuang, X. , Zhang, Y. , Zhu, Y. , et al. (2017a) A novel antimicrobial endolysin, LysPA26, against Pseudomonas aeruginosa . Front Microbiol 8: 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z. , Huang, J. , Yan, G. , Lei, L. , Wang, S. , Yu, L. , et al. (2017b) Identification and characterization of Dpo42, a novel depolymerase derived from the Escherichia coli phage vB_EcoM_ECOO78. Front Microbiol 8: 1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad Kashani, H. , Fahimi, H. , Dasteh Goli, Y. , and Moniri, R. (2017) A novel chimeric endolysin with antibacterial activity against methicillin‐resistant Staphylococcus aureus . Front Cell Infect Microbiol 7: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, L.E. , and Sillanpaa, J. (2014) Enterococcal cell wall components and structures. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Gilmore, M.S. , Clewell, D.B. , Ike, Y. , and Shankar, N. (eds). Boston, MA: Massachusetts Eye and Ear Infirmary. [PubMed] [Google Scholar]
- He, X. , Lu, F. , Yuan, F. , Jiang, D. , Zhao, P. , Zhu, J. , et al. (2015) Biofilm formation caused by clinical Acinetobacter baumannii isolates is associated with overexpression of the AdeFGH efflux pump. Antimicrob Agents Chemother 59: 4817–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiby, N. , Bjarnsholt, T. , Moser, C. , Bassi, G.L. , Coenye, T. , Donelli, G. , et al. (2015) ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect 21(Suppl. 1): S1–S25. [DOI] [PubMed] [Google Scholar]
- Huang, G. , Shen, X. , Gong, Y. , Dong, Z. , Zhao, X. , Shen, W. , et al. (2014) Antibacterial properties of Acinetobacter baumannii phage Abp1 endolysin (PlyAB1). BMC Infect Dis 14: 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jado, I. , Lopez, R. , Garcia, E. , Fenoll, A. , Casal, J. , Garcia, P. , et al. (2003) Phage lytic enzymes as therapy for antibiotic‐resistant Streptococcus pneumoniae infection in a murine sepsis model. J Antimicrob Chemother 52: 967–973. [DOI] [PubMed] [Google Scholar]
- Jones, P. , Binns, D. , Chang, H.Y. , Fraser, M. , Li, W. , McAnulla, C. , et al. (2014) InterProScan 5: genome‐scale protein function classification. Bioinformatics 30: 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Mirzaei, M. , and Nilsson, A.S. (2015) Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 10: e0118557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter, E. (2009) Phage host range and efficiency of plating. Methods Mol Biol 501: 141–149. [DOI] [PubMed] [Google Scholar]
- Lai, M.J. , Lin, N.T. , Hu, A. , Soo, P.C. , Chen, L.K. , Chen, L.H. , and Chang, K.C. (2011) Antibacterial activity of Acinetobacter baumannii phage varphiAB2 endolysin (LysAB2) against both gram‐positive and gram‐negative bacteria. Appl Microbiol Biotechnol 90: 529–539. [DOI] [PubMed] [Google Scholar]
- Loessner, M.J. , Wendlinger, G. , and Scherer, S. (1995) Heterogeneous endolysins in Listeria monocytogenes bacteriophages: a new class of enzymes and evidence for conserved holin genes within the siphoviral lysis cassettes. Mol Microbiol 16: 1231–1241. [DOI] [PubMed] [Google Scholar]
- Magiorakos, A.P. , Srinivasan, A. , Carey, R.B. , Carmeli, Y. , Falagas, M.E. , Giske, C.G. , et al. (2012) Multidrug‐resistant, extensively drug‐resistant and pandrug‐resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18: 268–281. [DOI] [PubMed] [Google Scholar]
- Mah, T.F. (2012) Biofilm‐specific antibiotic resistance. Future Microbiol 7: 1061–1072. [DOI] [PubMed] [Google Scholar]
- Marchler‐Bauer, A. , Derbyshire, M.K. , Gonzales, N.R. , Lu, S. , Chitsaz, F. , Geer, L.Y. , et al. (2015) CDD: NCBI's conserved domain database. Nucleic Acids Res 43: D222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X. , Shi, Y. , Ji, W. , Meng, X. , Zhang, J. , Wang, H. , et al. (2011) Application of a bacteriophage lysin to disrupt biofilms formed by the animal pathogen Streptococcus suis . Appl Environ Microbiol 77: 8272–8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, M. , Tanji, Y. , Mizoguchi, K. , Soejima, A. , Orito, Y. , and Unno, H. (2001) Antibacterial activity of Bacillus amyloliquefaciens phage endolysin without holin conjugation. J Biosci Bioeng 91: 469–473. [DOI] [PubMed] [Google Scholar]
- Musafer, H.K. , Kuchma, S.L. , Naimie, A.A. , Schwartzman, J.D. , Al‐Mathkhury, H.J. , and O'Toole, G.A. (2014) Investigating the link between imipenem resistance and biofilm formation by Pseudomonas aeruginosa . Microb Ecol 68: 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D. , Loomis, L. , and Fischetti, V.A. (2001) Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc Natl Acad Sci USA 98: 4107–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.C. , Schmelcher, M. , Rodriguez‐Rubio, L. , Klumpp, J. , Pritchard, D.G. , Dong, S. , and Donovan, D.M. (2012) Endolysins as antimicrobials. Adv Virus Res 83: 299–365. [DOI] [PubMed] [Google Scholar]
- Nikaido, H. (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67: 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, H. , Melo, L.D. , Santos, S.B. , Nobrega, F.L. , Ferreira, E.C. , Cerca, N. , et al. (2013) Molecular aspects and comparative genomics of bacteriophage endolysins. J Virol 87: 4558–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, H. , Thiagarajan, V. , Walmagh, M. , Sillankorva, S. , Lavigne, R. , Neves‐Petersen, M.T. , et al. (2014) A thermostable Salmonella phage endolysin, Lys68, with broad bactericidal properties against gram‐negative pathogens in presence of weak acids. PLoS ONE 9: e108376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, H. , Pinto, G. , Oliveira, A. , Oliveira, C. , Faustino, M.A. , Briers, Y. , et al. (2016) Characterization and genome sequencing of a Citrobacter freundii phage CfP1 harboring a lysin active against multidrug‐resistant isolates. Appl Microbiol Biotechnol 100: 10543–10553. [DOI] [PubMed] [Google Scholar]
- Oliveira, H. , Costa, A.R. , Konstantinides, N. , Ferreira, A. , Akturk, E. , Sillankorva, S. , et al. (2017) Ability of phages to infect Acinetobacter calcoaceticus‐Acinetobacter baumannii complex species through acquisition of different pectate lyase depolymerase domains. Environ Microbiol 19: 5060–5077. [DOI] [PubMed] [Google Scholar]
- Pajunen, M. , Kiljunen, S. , and Skurnik, M. (2000) Bacteriophage phiYeO3‐12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J Bacteriol 182: 5114–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, S.Y. , You, R.I. , Lai, M.J. , Lin, N.T. , Chen, L.K. , and Chang, K.C. (2017) Highly potent antimicrobial modified peptides derived from the Acinetobacter baumannii phage endolysin LysAB2. Sci Rep 7: 11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Bano, J. , Marti, S. , Soto, S. , Fernandez‐Cuenca, F. , Cisneros, J.M. , Pachon, J. , et al. (2008) Biofilm formation in Acinetobacter baumannii: associated features and clinical implications. Clin Microbiol Infect 14: 276–278. [DOI] [PubMed] [Google Scholar]
- Schmelcher, M. , Donovan, D.M. , and Loessner, M.J. (2012) Bacteriophage endolysins as novel antimicrobials. Future Microbiol 7: 1147–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuch, R. , Pelzek, A.J. , Nelson, D.C. , and Fischetti, V.A. (2019) The PlyB endolysin of bacteriophage vB_BanS_Bcp1 exhibits broad‐spectrum bactericidal activity against Bacillus cereus sensu lato isolates. Appl Environ Microbiol 85: e00003–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y. , Koller, T. , Kreikemeyer, B. , and Nelson, D.C. (2013) Rapid degradation of Streptococcus pyogenes biofilms by PlyC, a bacteriophage‐encoded endolysin. J Antimicrob Chemother 68: 1818–1824. [DOI] [PubMed] [Google Scholar]
- Stepanovic, S. , Vukovic, D. , Dakic, I. , Savic, B. , and Svabic‐Vlahovic, M. (2000) A modified microtiter‐plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods 40: 175–179. [DOI] [PubMed] [Google Scholar]
- Swift, S.M. , Rowley, D.T. , Young, C. , Franks, A. , Hyman, P. , and Donovan, D.M. (2016) The endolysin from the Enterococcus faecalis bacteriophage VD13 and conditions stimulating its lytic activity. FEMS Microbiol Lett 362: fnw216. [DOI] [PubMed] [Google Scholar]
- Tang, K. , Lin, D. , Zheng, Q. , Liu, K. , Yang, Y. , Han, Y. , and Jiao, N. (2017) Genomic, proteomic and bioinformatic analysis of two temperate phages in Roseobacter clade bacteria isolated from the deep‐sea water. BMC Genom 18: 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thandar, M. , Lood, R. , Winer, B.Y. , Deutsch, D.R. , Euler, C.W. , and Fischetti, V.A. (2016) Novel engineered peptides of a phage lysin as effective antimicrobials against multidrug‐resistant Acinetobacter baumannii . Antimicrob Agents Chemother 60: 2671–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummeepak, R. , Kitti, T. , Kunthalert, D. , and Sitthisak, S. (2016) Enhanced antibacterial activity of Acinetobacter baumannii bacteriophage OABP‐01 endolysin (LysABP‐01) in combination with colistin. Front Microbiol 7: 1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres‐Barcelo, C. (2018) Phage therapy faces evolutionary challenges. Viruses 10: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, D. , Ackermann, H.W. , Kropinski, A.M. , Lavigne, R. , Sutton, J.M. , and Reynolds, D.M. (2017) Comparative analysis of 37 Acinetobacter bacteriophages. Viruses 10: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar, S. , Biswas, I. , and Veeraraghavan, B. (2019) Accurate identification of clinically important Acinetobacter spp.: an update. Future Sci OA 5: FSO395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Gu, J. , Lv, M. , Guo, Z. , Yan, G. , Yu, L. , et al. (2017) The antibacterial activity of E. coli bacteriophage lysin lysep3 is enhanced by fusing the Bacillus amyloliquefaciens bacteriophage endolysin binding domain D8 to the C‐terminal region. J Microbiol 55: 403–408. [DOI] [PubMed] [Google Scholar]
- Wittebole, X. , De Roock, S. , and Opal, S.M. (2014) A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 5: 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M. , Hu, K. , Xie, Y. , Liu, Y. , Mu, D. , Guo, H. , et al. (2018) A novel phage PD‐6A3, and its endolysin Ply6A3, with extended lytic activity against Acinetobacter baumannii . Front Microbiol 9: 3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , Wang, R. , Xu, M. , Liu, Y. , Zhu, X. , Qiu, J. , et al. (2019) A novel polysaccharide depolymerase encoded by the phage SH‐KP152226 confers specific activity against multidrug‐resistant Klebsiella pneumoniae via biofilm degradation. Front Microbiol 10: 2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Y. , Wahab, L. , and Gill, J.J. (2018) Development and validation of a microtiter plate‐based assay for determination of bacteriophage host range and virulence. Viruses 10: 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Liang, L. , Lin, S. , and Jia, S. (2010) Isolation and characterization of a virulent bacteriophage AB1 of Acinetobacter baumannii . BMC Microbiol 10: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M. , Du, C. , Gong, P. , Xia, F. , Sun, C. , Feng, X. , et al. (2015) Therapeutic effect of the YH6 phage in a murine hemorrhagic pneumonia model. Res Microbiol 166: 633–643. [DOI] [PubMed] [Google Scholar]
- Yang, H. , Bi, Y. , Shang, X. , Wang, M. , Linden, S.B. , Li, Y. , et al. (2016) Antibiofilm activities of a novel chimeolysin against Streptococcus mutans under physiological and cariogenic conditions. Antimicrob Agents Chemother 60: 7436–7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L. , Wang, S. , Guo, Z. , Liu, H. , Sun, D. , Yan, G. , et al. (2018) A guard‐killer phage cocktail effectively lyses the host and inhibits the development of phage‐resistant strains of Escherichia coli . Appl Microbiol Biotechnol 102: 971–983. [DOI] [PubMed] [Google Scholar]
- Yuan, Y. , Qu, K. , Tan, D. , Li, X. , Wang, L. , Cong, C. , et al. (2019a) Isolation and characterization of a bacteriophage and its potential to disrupt multi‐drug resistant Pseudomonas aeruginosa biofilms. Microb Pathog 128: 329–336. [DOI] [PubMed] [Google Scholar]
- Yuan, Y. , Wang, L. , Li, X. , Tan, D. , Cong, C. , and Xu, Y. (2019b) Efficacy of a phage cocktail in controlling phage resistance development in multidrug resistant Acinetobacter baumannii . Virus Res 272: 197734. [DOI] [PubMed] [Google Scholar]
- Yuniati, Y. , Hasanah, N. , Ismail, S. , Anitasari, S. , and Paramita, S. (2018) Antibacterial activity of dracontomelon dao extracts on methicillin‐resistant S. aureus (Mrsa) and E. coli multiple drug resistance (Mdr). Afr J Infect Dis 12: 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Xu, L.L. , Gan, D. , and Zhang, X. (2018a) In vitro study of bacteriophage AB3 endolysin LysAB3 activity against Acinetobacter baumannii biofilm and biofilm‐bound A. baumannii . Clin Lab 64: 1021–1030. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Cheng, M. , Zhang, H. , Dai, J. , Guo, Z. , Li, X. , et al. (2018b) Antibacterial effects of phage lysin LysGH15 on planktonic cells and biofilms of diverse Staphylococci . Appl Environ Microbiol 84: e00886‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.L. , Zhou, Y.H. , Chen, J.Q. , Huang, Q.Y. , Han, Q. , Liu, B. , et al. (2015) Quantitative proteomic analysis of sub‐MIC erythromycin inhibiting biofilm formation of S. suis in vitro. J Proteomics 116: 1–14. [DOI] [PubMed] [Google Scholar]
- Zhou, H. , Yao, Y. , Zhu, B. , Ren, D. , Yang, Q. , Fu, Y. , et al. (2019) Risk factors for acquisition and mortality of multidrug‐resistant Acinetobacter baumannii bacteremia: A retrospective study from a Chinese hospital. Medicine (Baltimore) 98: e14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The liquid culture‐based host range assay of phage vB_AbaP_D2. Acinetobacter baumannii strains (107 cfu ml−1) were infected with phages at 106, 107, and 108 pfu ml−1. Optical densities were measured at 600 nm. Error bars represent standard deviations of three replicates.
Table S1. Predicted protein function of phage vB_AbaP_D2.
Table S2. List of strains used in this study.
Table S3. Antibiotic susceptibility of clinical isolates.


