Significance
Lipids of bilayer membranes can segregate laterally into distinct liquid phases of different composition called liquid ordered and liquid disordered, and corresponding in the plasma membrane of living cells to nanodomains called raft and nonraft domains. Using Förster resonance energy transfer and genetically expressible protein probes of lipid domains, we find that several steps of the metabolism of phosphoinositide lipids are concentrated in cholesterol-rich liquid-ordered domains of the plasma membrane. The receptor-mediated breakdown and the restoration of major phosphoinositide pools are faster in the liquid-ordered than in the liquid-disordered domains. Thus, the ordered domains host a key lipid signaling system of the cell.
Keywords: PIP2, GPMV, cyclodextrin, cholesterol, lipid diffusion
Abstract
Possible segregation of plasma membrane (PM) phosphoinositide metabolism in membrane lipid domains is not fully understood. We exploited two differently lipidated peptide sequences, L10 and S15, to mark liquid-ordered, cholesterol-rich (Lo) and liquid-disordered, cholesterol-poor (Ld) domains of the PM, often called raft and nonraft domains, respectively. Imaging of the fluorescent labels verified that L10 segregated into cholesterol-rich Lo phases of cooled giant plasma-membrane vesicles (GPMVs), whereas S15 and the dye FAST DiI cosegregated into cholesterol-poor Ld phases. The fluorescent protein markers were used as Förster resonance energy transfer (FRET) pairs in intact cells. An increase of homologous FRET between L10 probes showed that depleting membrane cholesterol shrank Lo domains and enlarged Ld domains, whereas a decrease of L10 FRET showed that adding more cholesterol enlarged Lo and shrank Ld. Heterologous FRET signals between the lipid domain probes and phosphoinositide marker proteins suggested that phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] and phosphatidylinositol 4-phosphate (PtdIns4P) are present in both Lo and Ld domains. In kinetic analysis, muscarinic-receptor-activated phospholipase C (PLC) depleted PtdIns(4,5)P2 and PtdIns4P more rapidly and produced diacylglycerol (DAG) more rapidly in Lo than in Ld. Further, PtdIns(4,5)P2 was restored more rapidly in Lo than in Ld. Thus destruction and restoration of PtdIns(4,5)P2 are faster in Lo than in Ld. This suggests that Lo is enriched with both the receptor G protein/PLC pathway and the PtdIns/PI4-kinase/PtdIns4P pathway. The significant kinetic differences of lipid depletion and restoration also mean that exchange of lipids between these domains is much slower than free diffusion predicts.
This article concerns signaling in distinct domains of the plasma membrane (PM). It has long been hypothesized that the lipids and proteins of the PM are laterally heterogeneous, being organized in nanodomains or liquid phases of differing composition (1, 2). When distinct liquid phases coexist in membranes, different lipids prefer liquid-ordered (Lo) or liquid-disordered (Ld) phases, sometimes viewed as corresponding to membrane rafts and nonrafts (3, 4). Such domains have been difficult to study in the PM of live cells due to their postulated diffraction-limited size and rapid dynamics (5). The barriers to observation in living cells meant that many early studies used fixed cells or detergent extracts (6–10). However, such processing might itself introduce artifacts, and the cells are no longer living. For example, the nonionic detergent Triton X-100, is considered to extract the lipids of Ld domains selectively, leaving behind the insoluble Lo domains, yet it was also suggested to induce artifactual aggregation of phosphoinositides like phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] in the PM (11, 12). More recent work has used giant plasma membrane vesicles (GPMVs) derived as induced blebs from living cells where macroscopic, visible lipid phase separation can be induced by lowering the temperature (6–10, 13, 14). Lo lipid domains of the PM also are said to segregate some signaling proteins, to nucleate caveolae (15), and even to host the budding (in the “budozone”) and/or entry of some membrane viruses including influenza, hepatitis C, HIV, and coronaviruses (16–19).
PtdIns(4,5)P2, predominantly located at the PM, regulates many essential functions, including vesicle docking and secretion, endocytosis, excitability of neurons, and ion channel activity (20). It is the substrate of receptor-activated phospholipase C (PLC). The submicroscopic distribution of PtdIns(4,5)P2 at the PM has been studied using superresolution microscopy with PtdIns(4,5)P2-specific antibodies (21), fluorescently tagged pleckstrin homology domains (PHPLCδ1) (11, 22), and electron microscopy (11). But the nanodomain distribution of PtdIns(4,5)P2 remains controversial. Many results suggest that PtdIns(4,5)P2 is heterogeneously distributed in the PM (21, 23–25) and favor the notion that it is concentrated in Lo or raft domains. As detergent-insoluble membranes that define rafts could be an artifact of sample preparation, others argue that PtdIns(4,5)P2 may be homogenous at the PM (11, 12, 26).
Because lipid domains of the living cell PM are considered too small to measure with light microscopy, we turned to Förster resonance energy transfer (FRET) in live cells labeled with PM-targeted fluorescent probes showing some selectivity for Lo or Ld domains (27, 28). This live-cell optical method avoids detergent extraction or formation of GPMVs. The Lck-derived L10 probe (L10) with two palmitoylation sites and one myristoylation site segregates predominantly to lipid-ordered domains, whereas the Src-derived S15 probe (S15) with one myristoylation site and a string of basic residues, segregates almost fully to lipid-disordered domains (28) (see Fig. 1A for the structures). We performed control experiments to validate the selectivity of the L10 and S15 probes. Then, using quantitative optical measurements during receptor stimulation, we assessed the spatiotemporal kinetics of phosphatidylinositide metabolism in domains labeled by the probes. This approach allowed us to determine that the receptor-induced degradation and restoration of PtdIns(4,5)P2 show a preference for Lo domains. We also recognized that the lateral exchange of lipids between Lo and Ld domains was far slower than predicted by free diffusion.
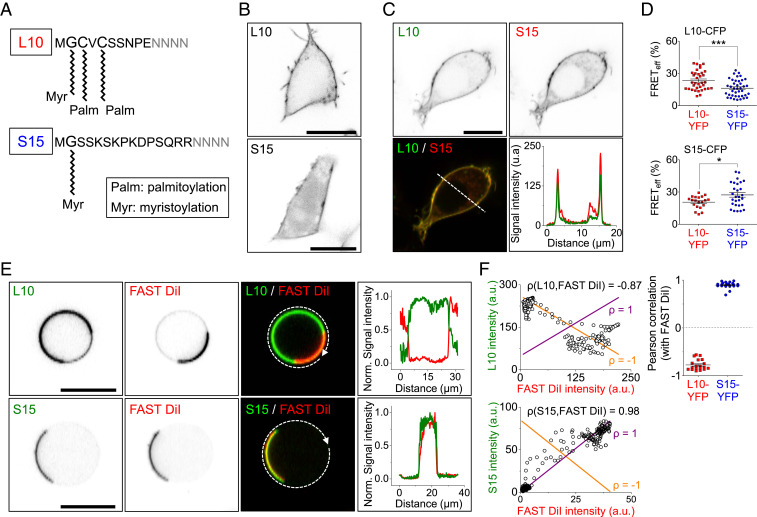
Fig. 1.
Lo and Ld domain targeting of fluorescent-tagged peptide probes. (A) Targeting of specific PM domains by L10 and S15 derived from 10 and 15 N-terminal residues of Lck and Src. Sites of myristoylation and palmitoylation are indicated. (B) Confocal image showing PM localization of the L10 or S15 probes in intact tsA-201 cells. Negative contrast (fluorescence is dark). (C) Two-color confocal images of a cell coexpressing L10-CFP and S15-YFP showing the CFP channel, the YFP channel, and a superposition. The last panel with two line scans along the dashed line in the superimposed image reveals membrane localization. (D) Comparison of homologous and heterologous FRETeff between of L10 and S15. FRETeff of L10-CFP and L10-YFP (n = 35 cells) or S15-YFP (n = 40 cells) and S15-CFP and L10-YFP (n = 20 cells) or S15-YFP (n = 28 cells). (E) Representative confocal images of L10 and S15 show segregated localization in cooled GPMVs. A cooled GPMV expressing L10-YFP (Top) or S15-YFP (Bottom) is loaded with FAST DiI, which targets Ld domains. The superimposed normalized line scans follow a circular curve around the cooled GPMVs. The GPMV is derived from a transfected tsA-201 cell. (F) FAST DiI intensity plotted against L10-YFP intensity (Top Left plot) or S15-YFP (Bottom Left) along with the ρ = −1 (orange line) and ρ = +1 (purple line). Pearson correlations between FAST DiI and L10-YFP or S15-YFP were measured (Top Right) (L10-YFP, n = 19; S15-YFP, n = 22). *P < 0.05, ***P < 0.001, n.s., not significant. (All scale bars, 10 μm.)
Results
Targeting Fluorescence-Tagged Peptides to Lo or Ld Domains of the PM.
We fused CFP and YFP onto short peptides containing sequences for both palmitoylation and myristoylation or for myristoylation alone derived from either the N-terminal 10 residues of Lck or the N-terminal 15 residues of Src (Fig. 1A) (27, 28). When transfected into tsA-201 cells, both L10-YFP and S15-YFP showed good PM localization (Fig. 1B). Although we expected that the two probes would target Lo or Ld domains, respectively, conventional confocal light microscopy showed fully overlapping PM expression (Fig. 1C). This observation could still be consistent with tiny domains below the diffraction limit in intact cells. To resolve a shorter spatial scale, we turned to FRET measured by the three-cube method (29). For YFP and CFP pairs, FRET would be sensitive to length scales below 10 nm. We compared FRET efficiency (FRETeff) of CFP- and YFP-tagged L10 and S15 expressed in cells (Fig. 1D). Homologous FRETeff between CFP- and YFP-tagged L10 molecules was higher than heterologous FRETeff between L10-CFP and S15-YFP, suggesting that L10 localizes near itself more than near S15 (Upper graph). Similarly, homologous FRETeff between CFP- and YFP-tagged S15 molecules was higher than heterologous FRETeff between S15-CFP and L10-YFP, suggesting that S15 localizes near itself more than near L10 (Lower graph). Nevertheless, the separation of L10 and S15 was not complete. Reports by detergent extraction that the Lo:Ld distribution ratio for L10 is 63%:37% and that for S15 is 2%:98% (30) in Jurkat T cells, might suggest why there still could be significant FRET between L10 and S15, perhaps occurring primarily in Ld domains.
Macroscopic PM lipid domains are rarely observed by imaging in live cells, but they have been observed in certain GPMVs derived from cells. GPMVs isolated as induced blebs from the mammalian PM have been an important tool for studying physical properties of physiological membranes (13). To validate preferences of L10 and S15 for liquid-ordered versus -disordered phases in the PM, we expressed the probes in cells and then prepared GPMVs using established protocols involving dithiothreitol (DTT) and paraformaldehyde (PFA) (31) (Fig. 1E). Previous studies on the phase behavior of GPMVs from a number of cell types show that large, phase-segregated domains form when these membranes are cooled to temperatures well below physiological temperature (13) or if some of the membrane components are artificially clustered (32). The fluorescent dye, FAST DiI, with diunsaturated linoleyl tails (C18:2), is considered a selective marker of Ld domains (33, 34). In our GPMV membranes, FAST DiI distributed apparently uniformly at 25 °C (SI Appendix, Fig. S1 A, Upper) but showed clear macroscopic phase separation when the temperature was reduced (SI Appendix, Fig. S1 A, Lower). Whereas GPMVs derived from a rat basophilic leukemia cell line become phase separated around 18 °C (35), we found that GPMVs from tsA-201 cells showed phase separation only below 10 °C (SI Appendix, Fig. S1B). Therefore, we conducted our GPMV experiments at ∼5 °C, designated as “cooled GPMVs.”
Confocal fluorescence microscopy of cooled GPMVs revealed macroscopic phase separation of YFP-tagged L10 and, in a separate experiment, macroscopic phase separation of S15 (first column in Fig. 1E). In the same cooled GPMVs, distinct Ld phase domains were revealed using FAST DiI (second column in Fig. 1E). The third column superimposes the YFP (green) and FAST DiI images (red), and the fourth column plots line scans that followed the dashed curved line around the overlaid images. L10-YFP showed a striking negative spatial correlation with FAST DiI (quantified by a Pearson correlation coefficient, ρ = −0.78 ± 0.03, n = 19 GPMVs), and S15-YFP showed a striking positive correlation (ρ = 0.90 ± 0.01, n = 22 GPMVs; Fig. 1F). In summary, in cooled GPMVs from tsA-201 cells, L10 segregated mainly in Lo domains and S15 segregated in Ld. In our experiments, the Lo domains of GPMVs covered a larger membrane fraction than Ld (L10: 62 ± 4%, FAST DiI: 29 ± 1%, S15: 34 ± 3%) (Fig. 2C). This larger Lo area confirmed previous results on GPMVs derived from NIH 3T3 fibroblasts (36). Similar experiments with intact tsA-201 cells did not detect macroscopic phase separation upon cooling to 5 °C; rather the cells just appeared to shrink (SI Appendix, Fig. S1C).
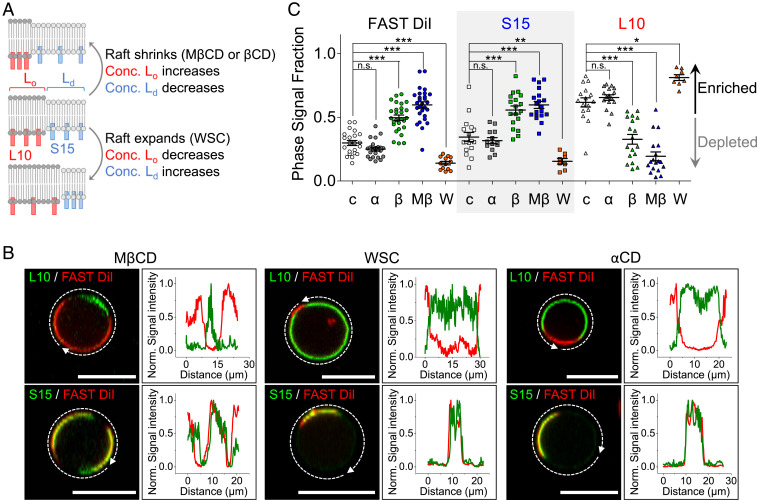
Fig. 2.
Change of domain area by varying cholesterol in the PM. (A) Schematic representation of changes of Lo and Ld size and of the surface density of L10 and S15 during application of cholesterol chelators (MβCD or βCD) or carriers (WSC). (B) Representative GPMV images and line scans in cholesterol-modifying media. L10-YFP (Top) or S15-YFP (Bottom) expressing GPMVs have been chilled to induce PM phase separation. The intensity profiles plot the fluorescence intensity following the white dashed arrows around the GPMVs. (C) Quantification of FAST DiI, S15-YFP, and L10-YFP fractional phase area in GPMVs from three independent experiments. Multiple GPMVs (n) were analyzed for each condition. FAST DiI: (c, control) 0.29 ± 0.01, n = 22; (α, αCD) 0.25 ± 0.01, n = 23; (β, βCD) 0.50 ± 0.02, n = 27; (Mβ, MβCD) 0.60 ± 0.02, n = 28; (W, WSC) 0.14 ± 0.01, n = 13; S15-YFP: (control) 0.34 ± 0.03, n = 17; (αCD) 0.32 ± 0.02, n = 13; (βCD) 0.56 ± 0.03, n = 18; (MβCD) 0.60 ± 0.03, n = 19; (WSC) 0.16 ± 0.02, n = 8; L10-YFP: (control) 0.62 ± 0.04, n = 16; (αCD) 0.66 ± 0.02, n = 16; (βCD) 0.33 ± 0.04, n = 18; (MβCD) 0.20 ± 0.04, n = 18; (WSC) 0.81 ± 0.02, n = 8. *P < 0.05, **P < 0.005, ***P < 0.001, n.s. not significant. Error bars show SEM. (Scale bars, 10 μm.) Conc., concentration.
We performed control intramembrane lateral diffusion experiments with GPMVs to determine the physical state of the cooled membranes. Lateral diffusion coefficients of 0.1 μm2/s are reported for dye-labeled PtdIns(4,5)P2 (with two fatty acid chains) at room temperature in baby hamster kidney cell PM and of 1 μm2/s in macrophages (37). In giant unilamellar membranes at room temperature, diffusion coefficients for a variety of labeled lipids range from 0.1 to 0.7 μm2/s in Lo domains and from 5 to 12 μm2/s in the less rigid Ld domains (33). By contrast, at 4 °C (below a transition temperature) diffusion coefficients in gel-phase dimyristoylphosphatidylcholine multibilayers were below 0.001 μm2/s for a di-C18 dye (38). To measure diffusion in the Lo domains of our cooled GPMVs, we employed fluorescence recovery after photobleaching (SI Appendix, Fig. S1D). A patch of the L10-YFP within an Lo domain was bleached (at arrowhead) with intense light for 2 s, finishing at time 0. Subsequent frames and the sequential line scans (SI Appendix, Fig. S1 D–F) showed spread of the lateral L10-YFP label such that the depressed L10-YFP fluorescence intensity began to even out over 10 s to a new lower baseline. The experiment was done at ∼5 °C. SI Appendix, Fig. S1E plots raw line-scan points from the experiment of SI Appendix, Fig. S1 D and F plots the averaged data from three experiments binned in distance intervals of 0.6 μm to reduce noise. The smooth curves in these two panels are solutions of a one-dimensional diffusion equation with a membrane diffusion coefficient for L10 of D = 0.3 μm2/s in the Lo domain (Materials and Methods). To gauge the sensitivity of our rough analysis, SI Appendix, Fig. S1G compares model curves at 5 s when the assumed diffusion coefficient was increased or decreased threefold. Recalling that L10 (with three fatty acid chains) is linked to a cytoplasmic fluorescent protein, the 0.3 μm2/s diffusion coefficient suggested that the Lo domain of cooled GPMVs retained liquid-like properties at 5 °C rather than acting like a solid or gel phase.
Collectively, the segregated PM domain localizations of fluorescently tagged L10 and S15 in GPMVs validated their use in FRET experiments as markers of Lo and Ld domains, respectively. The C-terminal, CAAX-containing sequences of H-ras and K-ras also have been considered for identifying Lo and Ld, respectively (39). In our hands, they both concentrated in Ld domains of cooled GPMVs (SI Appendix, Fig. S2) and were not studied further.
Controlling PM Domains by Varying the Cholesterol Level.
Cholesterol is a diagnostic component of Lo domains that helps maintain their structure (40, 41). It is more abundant in the outer leaflet of the PM than in the inner leaflet (42, 43). Treatment of cells or GPMVs with cholesterol-chelating methyl-β-cyclodextrin (MβCD) in the medium reduces PM cholesterol (42–44), and treatment with preloaded MβCD-cholesterol complexes (marketed as “water-soluble cholesterol” [WSC]) increases PM cholesterol (45). As diagrammed in Fig. 2A, removing cholesterol from GPMVs by extraction has been observed to change area fraction by shrinking the Lo domains and expanding the Ld domains, and conversely adding cholesterol to the PM expands Lo and shrinks Ld (3, 36). To confirm the effects of cholesterol on Lo and Ld domains, we applied MβCD or WSC to cooled GPMVs from cells expressing the L10 and S15 probes (Fig. 2B). Extraction of cholesterol by 5 mM of active MβCD or active β-cyclodextrin (βCD), shrank the Lo- and expanded the Ld-domain sizes (Fig. 2 B, Left). The addition of cholesterol (1 mg/mL WSC) expanded the Lo- and shrank the Ld-domains (Fig. 2 B, Middle). These dramatic changes in domain size during cholesterol manipulation were quantified by calculating the fraction of the total circumference with normalized intensity greater than 0.5 (Fig. 2C). Thus, we confirmed that the L10-accumulating Lo domains of GPMVs are enriched in cholesterol and change area fraction as cholesterol is varied. As a control experiment, 5 mM of inactive α-cyclodextrin (αCD), which does not chelate cholesterol, did not change Lo domain fractions (Fig. 2 B, Right and Fig. 2C).
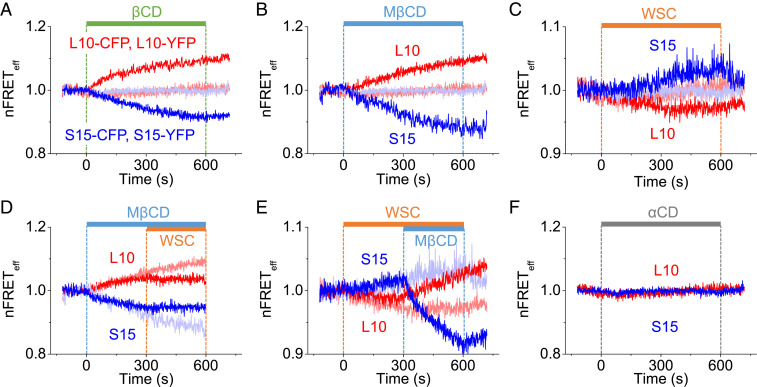
After these successful preliminary experiments in a model system, we could turn to our principal goal, to examine PM domains in intact living tsA-201 cells at room temperature. Since no spatial segregation of our labels was visible in the light microscope, we looked for probe density changes using FRET while manipulating the cholesterol content. If the total area of PM surface did not change, removal of cholesterol should shrink the area fraction of the Lo phase, increasing L10 density, while also increasing the area fraction of the Ld phase, decreasing S15 density, as diagrammed in Fig. 2A. The opposite changes should occur if cholesterol were added. To test whether the total PM surface area changed, we monitored PM electrical capacitance under whole-cell patch clamping during application of αCD and MβCD. There were no significant changes of membrane capacitance (αCD control: before, 14.1 ± 1.0 pF; at 10 min, 14.3 ± 1.0 pF; MβCD: before, 14.0 ± 1.4 pF; at 10 min, 14.8 ± 1.0 pF) meaning that changing the cholesterol did not alter the total area by more than 7.5%. Nevertheless, there were clear changes of FRET that signaled reciprocal changes in surface density of L10 and S15 (Fig. 3). Here we monitored homologous FRET between L10-CFP and L10-YFP, or, in separate experiments, between S15-CFP and S15-YFP in transfected tsA-201 cells. Application of βCD or MβCD gave significant increases in L10-L10 and decreases in S15-S15 homologous FRET as anticipated (Fig. 3 A and B). Similar reciprocal changes were seen with transfected HEK-293 cells (SI Appendix, Fig. S3). Application of WSC gave the opposite changes from MβCD (Fig. 3C). Application of WSC in the middle of MβCD halted the ongoing changes (Fig. 3D), and application of MβCD in the middle of WSC reversed the ongoing changes (Fig. 3E). The effects of adding or removing cholesterol developed with half times of 200 to 300 s and were almost complete in 10 min. As a control, a 10-min application of inactive αCD did not change the density of either probe (Fig. 3F). These results showed that living cells have Lo and Ld domains that are reliably reported by L10 and S15 probes and that Lo domains sequester much of the membrane cholesterol. We can note that the approximate equality in the magnitude of the complementary FRET changes implied that the Lo and Ld domains each initially occupied roughly equal fractions of the total lipid area. Therefore, although their territories were below the resolution of the microscope, neither could be regarded as a minority island in a sea of the other in intact cells.
Fig. 3.
Surface density changes of membrane L10 and S15 during cholesterol manipulation in living cells at room temperature. Averaged time courses of the homologous nFRETeff between CFP- and YFP-labeled L10 (red), and between CFP- and YFP-labeled S15 (blue) in separate tsA-201 cells at room temperature. Normalized FRET traces: (A) 5 mM βCD (L10: n = 6 cells, S15: n = 6 cells); (B) 5 mM MβCD (L10: n = 15 cells and S15: n = 14 cells); and (C) 1 mg/mL WSC (L10: n = 5 cells and S15: n = 5 cells). Pale blue and pink lines are αCD control experiments from F. (D) WSC was applied 5 min after application of MβCD (L10: n = 11 cells and S15: n = 8 cells). (E) MβCD was applied 5 min after application of WSC (L10: n = 6 cells and S15: n = 5 cells). Pale blue and pink in D and E come from B and C. (F) 5 mM αCD control (L10: n = 6 cells and S15: n = 6 cells).
Faster Hydrolysis of PtdIns(4,5)P2 in Lo during Receptor Stimulation.
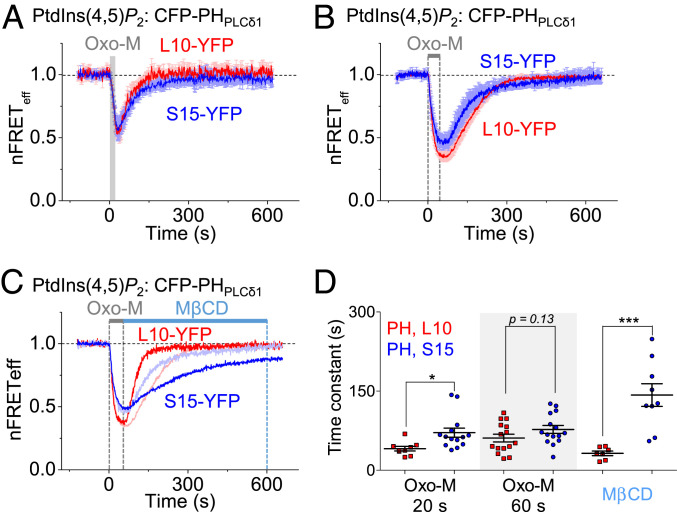
The L10 and S15 probes allowed us to study phosphoinositide metabolism in specific domains of the PM. In these experiments we monitored PM PtdIns(4,5)P2 using CFP-tagged PtdIns(4,5)P2-binding pleckstrin homology (PH) domains of PLCδ1 (CFP-PHPLCδ1) as probes (46), and we stimulated transfected M1 muscarinic receptors (M1R) with the agonist oxotremorine-M (Oxo-M, 10 μM) to activate phospholipase-Cβ (PLCβ) and hydrolyze PM PtdIns(4,5)P2 (Fig. 4A). In order to compare the dynamics of PtdIns(4,5)P2 depletion and restoration in Lo and Ld domains, we measured heterologous FRET between the CFP-PHPLCδ1 lipid probe and either L10-YFP or S15-YFP domain markers in the PM. For the two domain markers the average FRETeff values for the initial 120 s were similar, L10: 18.8% and S15: 17.7%, indicating that PtdIns(4,5)P2 is present in both lipid domains. Fig. 4B shows the time courses of heterologous FRET between CFP-PHPLCδ1 and both domain markers. Activation of M1R by muscarinic agonist Oxo-M (10 μM) promoted depletion of PtdIns(4,5)P2 in both domains, seen as a rapid reduction of FRET. Finally, washout of receptor agonist allowed recovery of PtdIns(4,5)P2 in both lipid domains. Here and in the following experiments, the FRET traces were normalized to the resting FRETeff for the first 120 s, and the absolute resting FRETeff values are given in the legend.
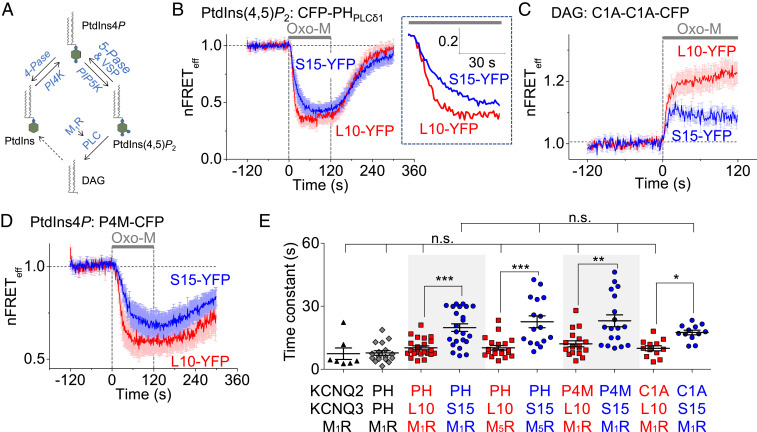
Fig. 4.
Faster PtdIns(4,5)P2 hydrolysis in Lo domains than in Ld. (A) Partial schematic diagram of the phosphoinositide cycle. Blue labels are enzymes or receptors. Abbreviations: 4-Pase, 4-phosphatase; 5-Pase, 5-phosphatase; and VSP, voltage-sensing phosphatase. (B) Averaged time courses of nFRETeff in Lo and Ld domains upon activation of M1R by 10 μM Oxo-M. Heterologous FRET between CFP-PHPLCδ1 and L10-YFP (red) or S15-YFP (blue) in separate tsA-201 cells. Error bars show SEM. The Inset shows the first 60 s expanded (L10: n = 22 cells and S15: n = 23 cells). The absolute resting FRETeff used for normalizations was L10: 18.8 ± 3.4% and S15: 17.7 ± 3.3%. (C) Time courses of nFRETeff between CFP-tagged C1A-C1A domains and L10-YFP or S15-YFP in M1R-expressing cells. CFP-tagged C1A-C1A domains detect DAG at the PM and increase the FRET signal when DAG increases in the labeled domains (L10: n = 12 cells and S15: n = 12 cells). The absolute resting FRETeff used for normalizations was L10: 10.2 ± 1.2% and S15: 10.9 ± 1.2%. (D) Averaged time courses of PtdIns4P in the specified PM domains measured by FRET between the CFP-tagged P4M probe and L10-YFP or S15-YFP in cells (L10: n = 18 cells and S15: n = 19 cells). The absolute resting FRETeff used for normalizations was L10: 11.8 ± 1.2% and S15: 9.3 ± 1.6%. (E) Time constants of lipid depletion during receptor activation in Lo and Ld domains. *P < 0.05, **P < 0.005, ***P < 0.001, n.s., not significant.
Although the decrease and recovery of PM PtdIns(4,5)P2 were qualitatively similar in the two domains, quantitative differences were apparent in closer kinetic analysis. We measured the exponential time constants for the heterologous FRET decrease (τ) representing PtdIns(4,5)P2 depletion during receptor activation. The decrease was twice as fast in Lo domains (L10-YFP, τ = 10 s) as in Ld (S15-YFP, τ = 20 s) (Fig. 4 B and E). Similar results were obtained in cells transfected with M5 muscarinic receptors, showing FRET decreases that were three times faster in Lo (τ = 8 s) than in Ld (τ = 23 s) (SI Appendix, Fig. S4A and Fig. 4E). The domain-specific kinetics during M1R activation could be compared with the more familiar global dynamics of PtdIns(4,5)P2-dependent KCNQ2/3 current and of PH-domain migration reported by homologous FRET between CFP-PHPLCδ1 and YFP-PHPLCδ1. Muscarinic suppression of KCNQ2/3 current developed with a time constant of 7 s (SI Appendix, Fig. S4 B and E), and the decrease of homologous PH domain FRET developed with a time constant of 8 s (SI Appendix, Fig. S4 C and E). Thus these two traditional measures corresponded best to the results with Lo domains, as defined by the L10 domain marker (Fig. 4E), as if the receptor-induced hydrolysis of PtdIns(4,5)P2 occurred preferentially in Lo. To analyze the resting PtdIns(4,5)P2 distribution in intact cells, we monitored homologous FRETeff between CFP-PHPLCδ1 and YFP-PHPLCδ1 (SI Appendix, Fig. S5A) as well as heterologous FRETeff between CFP-PHPLCδ1 and either L10-YFP or S15-YFP (SI Appendix, Fig. S5B) during cholesterol manipulation. None of these FRETeff signals was sensitive to application of MβCD or application of inactive αCD, again as if PtdIns(4,5)P2 is present universally and occupies both the Ld domains and the cholesterol-rich Lo domains of living cells.
As an additional measure of the rate of PtdIns(4,5)P2 breakdown in each domain, we monitored the generation of one of the PtdIns(4,5)P2 hydrolysis products, diacylglycerol (DAG) using CFP-tagged tandem C1A-C1A domains from PKCγ (47). In resting cells, where DAG in the PM was low, the CFP-tagged C1A-C1A probes were localized in the cytoplasm, and L10-YFP and S15-YFP were in the PM producing little heterologous FRET with the DAG probe (Fig. 4C). When PM DAG increased during receptor activation, the cytoplasmic DAG probes were recruited to the L10- or S15-enriched domains increasing the heterologous FRET signals. In principle, the DAG should be produced simultaneously and stoichiometrically with PtdIns(4,5)P2 breakdown (Fig. 4A). As anticipated, we found faster and larger production of DAG in L10-enriched domains (time constant, τ = 10 s) than in S15-enriched domains (τ = 19 s) confirming faster PtdIns(4,5)P2 hydrolysis in Lo domains.
Phosphatidylinositol 4-phosphate (PtdIns4P) is the precursor of PtdIns(4,5)P2 and probably also is a substrate cleaved by the PLC enzyme. The mean resting heterologous FRET between CFP-tagged P4M, a PtdIns4P probe from SidM (48), and L10-YFP or S15-YFP (SI Appendix, Fig. S5B), was 11.8% and 9.3%, respectively, indicating that PtdIns4P is present in both lipid domains at the PM as we found for PtdIns(4,5)P2. We measured the depletion time course of PtdIns4P in the two domains by heterologous FRET during M1R stimulation (Fig. 4 D and E). PtdIns4P decreased in both lipid domains, and again the decrease was twice as fast in Lo domains (11 s) as in Ld domains (24 s). Altogether these data confirm the presence in both domains of a synchronized depletion of PtdIns(4,5)P2 and PtdIns4P that is twice as fast in Lo domains as in Ld during muscarinic M1R and M5R stimulation.
Faster Recovery of PtdIns(4,5)P2 in Lo.
When PLCβ is activated through stimulation of M1R, the pools of PtdIns4P and PtdIns(4,5)P2 decrease rapidly. Subsequent recovery of PtdIns(4,5)P2 is a slower two-step reaction that requires the precursor phosphatidylinositol (PtdIns) and two lipid kinases, PI4-kinase (PI4K) and PIP5-kinase (PIP5K) (Fig. 4A). Are these activities concentrated in specific lipid domains? During agonist application, heterologous FRET between CFP-PHPLCδ1 and L10-YFP or between CFP-PHPLCδ1 and S15-YFP declined and after removal of agonist, recovered, showing the dynamic depletion and restoration of PM PtdIns(4,5)P2. Recovery from a 20 s Oxo-M stimulation was faster in Lo than in Ld (Fig. 5 A and D). The time constant of recovery was 40 ± 4 s with L10 and 71 ± 8 s with S15. When we extended the Oxo-M agonist application time to 60 s, the difference was no longer statistically significant (Fig. 5 B and D). However, a significant difference was revealed by manipulating membrane cholesterol. When we applied MβCD immediately after removing Oxo-M, PtdIns(4,5)P2 recovery was accelerated in Lo domains (32 ± 4 s) and markedly retarded in Ld domains (142 ± 22 s) (Fig. 5 C and D). The differential effects of MβCD application were also seen when it was applied 3 min before Oxo-M (SI Appendix, Fig. S6 A and C), but were less striking when applied 20 min before (SI Appendix, Fig. S6 B and C). Together these data indicate that under many circumstances replenishment of PtdIns(4,5)P2 is demonstrably faster in Lo domains than in Ld domains.
Fig. 5.
Faster PtdIns(4,5)P2 recovery in Lo domains than in Ld after receptor activation. (A) Time courses of averaged heterologous nFRETeff between CFP-PHPLCδ1 and L10-YFP (red) or S15-YFP (blue) during and after 20-s stimulation of the M1 receptor by Oxo-M application (L10: n = 8 cells and S15: n = 14 cells). Error bars show SEM. The absolute resting FRETeff used for normalizations was L10: 20.2 ± 2.8% and S15: 20.5 ± 1.5%. (B) PtdIns(4,5)P2 dynamics during and after 60-s Oxo-M application (L10: n = 15 cells and S15: n = 15 cells). The absolute resting FRETeff used for normalizations was L10: 20.1 ± 2.7% and S15: 21.3 ± 5.2%. (C) PtdIns(4,5)P2 dynamics when 5 mM of MβCD was applied immediately following 60 s of Oxo-M (L10: n = 7 cells and S15: n = 9). The absolute resting FRETeff used for normalizations was L10: 22.4 ± 3.3% and S15: 24.3 ± 4.4%. (D) Quantification of the lipid recovery time constants for different conditions. *P < 0.05, ***P < 0.001.
Contributions of PI4K and PIP5K to Fast Recovery in Lo Domains.
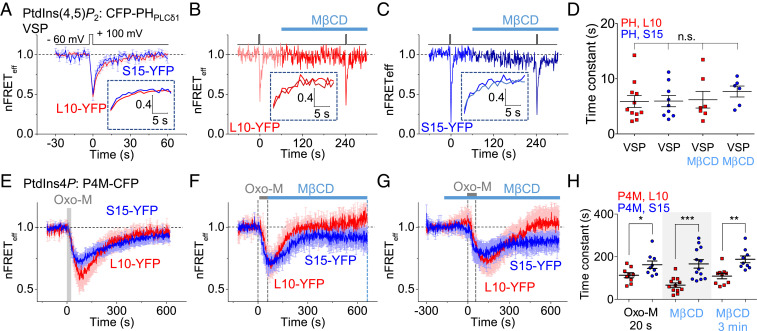
We tested which kinase activity, PI4K, PIP5K, or both, is more active in Lo than in Ld domains. Endogenous PIP5K activity during PtdIns(4,5)P2 recovery, was assessed by measuring the recovery kinetics after a rapid dephosphorylation of PtdIns(4,5)P2 to PtdIns4P at the PM. The Danio rerio voltage-sensing 5-phosphatase (VSP) was expressed in the cells and activated by a short depolarizing voltage step (49) (Figs. 4A and 6A). During this brief activation (2 s at 100 mV), the PtdIns(4,5)P2 in both domains was depleted by conversion to PtdIns4P, and then it recovered with indistinguishable time constants, 5.8 ± 1.1 s in Lo and 5.9 ± 1.0 s in Ld (Fig. 6 A and D). Additionally, we measured PIP5K activity in each domain after shrinking the area fraction of Lo phase and increasing the area fraction of Ld phase by cholesterol extraction (Fig. 6 B and C). The VSP phosphatase was activated twice by voltage steps; the first depolarization served to monitor control recovery, and the second followed 3 min in MβCD. The PtdIns(4,5)P2 recovery rates were not significantly different among the four conditions (Fig. 6D). These experiments indicated that endogenous PIP5K and also overexpressed VSP are distributed in the PM with little preference between the lipid domains.
Fig. 6.
Dissection of phosphoinositide restoration after depletion. (A) Averaged time course of heterologous nFRETeff between PHPLCδ1 and L10 or S15 measuring PtdIns(4,5)P2 recovery in Lo and Ld domains after brief VSP stimulation. A 2-s voltage pulse to 100 mV stimulated VSP to dephosphorylate PtdIns(4,5)P2 to PtdIns4P. The traces in the dashed box expand the first 20 s of recovery after the end of VSP stimulation (L10: n = 11 cells and S15: n = 9 cells). The absolute resting FRETeff used for normalizations was L10: 23.1 ± 4.4% and S15: 21.6 ± 3.8%. (B and C) Paired-pulse stimulation of VSP before and after 3-min treatment with MβCD to measure the sensitivity to change in domain sizes for Lo (B) and Ld (C) (L10: n = 7 cells and S15: n = 6 cells). (D) Summary of PtdIns(4,5)P2 recovery time constants after VSP stimulation. (E) Time course of PtdIns4P recovery after 20-s stimulation of the M1R receptor (L10: n = 9 cells and S15: n = 9 cells). The absolute resting FRETeff used for normalizations was L10: 10.9 ± 2.5% and S15: 12.4 ± 1.5%. (F and G) The same when MβCD was applied immediately after (F) (L10: n = 12 cells and S15: n = 13 cells) or 3 min before (G) (L10: n = 10 cells and S15: n = 9 cells) the Oxo-M. The absolute resting FRETeff used for normalizations was for F, L10: 9.6 ± 1.4% and S15: 9.4 ± 1.4%, and, for G, L10: 11.0 ± 1.4% and S15: 12.8 ± 1.8%. (H) PtdIns4P recovery time constants in Lo and Ld domains after receptor activation. *P < 0.05, **P < 0.005, ***P < 0.001, n.s., not significant.
The similarity of endogenous PIP5K activity in the Lo and Ld domains suggested that we should look for local differences in endogenous PI4K activity instead. We measured the recovery time course of the PI4K product PtdIns4P in each domain after Oxo-M stimulation using heterologous FRET between CFP-tagged P4M probes and L10-YFP or S15-YFP. When the Oxo-M stimulus was short (20 s), the recovery of PM PtdIns4P was faster in Lo domains (τ = 112 ± 12 s) than in Ld (τ = 162 ± 17 s) (Fig. 6 E and H). When the receptor stimulation was lengthened to 60 s, a difference in mean recovery time constant persisted but was no longer significant (SI Appendix, Fig. S7 A and B). As for PtdIns(4,5)P2 recovery experiments, we also tested the effect of MβCD on PtdIns4P recovery after 60-s Oxo-M treatments (Fig. 6 F–H). Shrinkage of the Lo domain immediately following, or 3 min before, receptor stimulation, induced faster recovery of PtdIns4P in the Lo domain than in Ld. The experiments suggested that PtdIns4P recovery is differentially concentrated in Lo domains, but PM PIP5K activity is not. Interestingly, in both domains, PtdIns4P recovery occurred only 70 to 80 s after PtdIns(4,5)P2 had recovered.
Discussion
Using cell-derived GPMVs, we have verified the use of L10 and S15 as marker probes for membrane cholesterol-rich Lo and cholesterol-poor Ld domains, respectively (28, 30). In intact tsA-201 cells, the two exclusive domains each occupy about the same area fraction of the cell membrane surface. Then we used the probes in kinetic studies. We achieved 1-s time resolution because we combined FRET, rapid continuous solution perfusion, and 1-s sampling intervals. Phospholipase activity was clearly faster in Lo domains. We readily distinguished the 10-s time constant for depletion of PtdIns(4,5)P2 during M1 receptor-stimulated PLC activity in Lo from the 20-s time constant in Ld domains. This conclusion was reinforced by measuring PtdIns4P depletion and DAG production and when using the M5 receptor instead of M1. We would agree with previous work that little difference between L10- and S15-marked domains would be evident if one used slower methods. For example Tóth et al. (22) used bioluminescence resonance energy transfer (BRET), manual solution addition, and 15-s sampling intervals and did not detect differences, whereas we find now that discriminating domains with the L10 and S15 probes needs faster methods. One probable reason would be that many membrane molecules might exchange between lipid domains in tens of seconds so that metabolic changes will appear nearly simultaneous and uniform in Lo and Ld domains when sampling on a sufficiently slow time scale or, as we found, when stimulating with agonist or treating with cyclodextrins for longer times.
In intact cells we found that PtdIns(4,5)P2 and PtdIns4P were present in both domains of the plasma membrane. Several authors have favored the view that PtdIns(4,5)P2 is more concentrated in Lo domains (27, 50–52) or in Ld domains (53, 54). Some of these papers show different phenotypes during domain-specific reduction of Ptdins(4,5)P2. For example, reduction of PtdIns(4,5)P2 in Ld domains increases PM ruffling, whereas reduction in Lo domains yields a smooth PM devoid of membrane ruffles and filopodia (27). Some papers indicate that Triton X-100 segregates PtdIns(4,5)P2 in the PM to generate artifactual Lo domains (11). Using FRET as a criterion, we did not find segregation of either PtdIns4P or PtdIns(4,5)P2 into Lo or Ld domains in tsA-201 cells. Further, MβCD did not change homologous FRET between CFP-PHPLCδ1 and YFP-PHPLCδ1 or heterologous FRET between CFP-PHPLCδ1 and L10-YFP or S15-YFP. These results are consistent with prior BRET experiments from Várnai and coworkers (22).
We showed that extraction of cholesterol shrank Lo domains and expanded Ld domains not only in GPMVs as observed before (36) but also in intact cells. Shrinking the area fraction of the Lo phase manifested as increased homologous FRET between L10 probes, presumably by concentrating those protein labels into a reduced area. Shrinking the Lo domains also increased receptor-activated PLC activity, again presumably by concentrating some rate-limiting component(s) of the PLC pathway, which may be receptors, G proteins, PLC, or several of these. Similarly, the restoration of PtdIns(4,5)P2 and PtdIns4P after depletion was faster in Lo domains than in Ld domains, and shrinking the Lo domains by extracting cholesterol augmented this difference. The faster PtdIns(4,5)P2 restoration was not due to higher PIP5K activity in Lo domains. In summary, our observations showed that receptor-mediated phosphoinositide hydrolysis and the subsequent restoration of PtdIns(4,5)P2 occurred preferentially in Lo domains which also segregated much of the membrane cholesterol.
We should note three caveats here. First when we speak of segregation of lipid or protein components, we should always be thinking of a fractional partitioning between domains rather than some absolute exclusion. Second, longer treatments with cyclodextrins can be damaging to cells and may disrupt Lo-related structures like caveolae and signaling complexes (22, 51). We focused on the initial actions of cyclodextrins when the Lo domain is believed to be gradually shrinking rather than falling apart. And third, the probes we relied on here contained fluorescent proteins that Zacharias et al. (55) have shown can induce homodimerization, unlike the monomeric modified version they developed. This could lead to artifacts from excess FRET. However, fortunately here we are primarily comparing FRET between L10 or S15 as FRET acceptors with various other probes as a donor. Since there were profound differences between L10 and S15 signals that used the same versions of YFP, the differences we are reporting are not due to a dimerization artifact. Further, one might suspect that the resting FRET we report from the PHPLCδ1 and P4M probes could be due to dimerization artifacts. However, since activating PLC reduces this FRET signal further (Fig. 4 B and D), a significant portion of the resting FRET reflects actual phosphoinositide rather than artifactual dimerization.
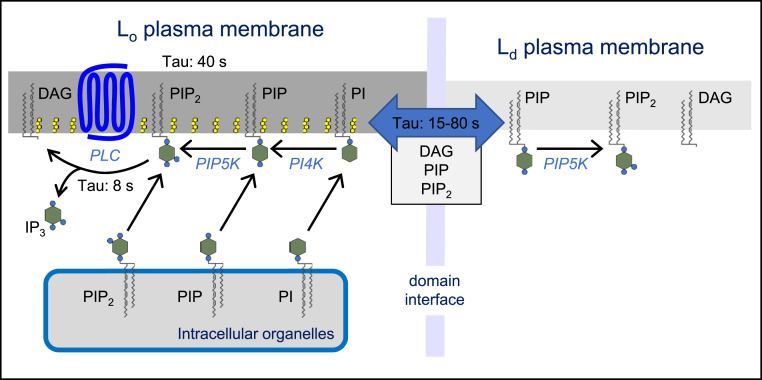
We offer alternative interpretations of our results as questions for future study with an extreme hypothesis drawn in Fig. 7. Our results show that some steps in agonist-induced PtdIns(4,5)P2 breakdown are more concentrated in cholesterol-enriched Lo domains than in Ld as defined by the L10 and S15 probes. What is rate limiting here? Perhaps there are more muscarinic receptors, or better coupling through G proteins, or more PLC activity in Lo.
Fig. 7.
An extreme hypothesis with almost all of phosphoinositide metabolism occurring in Lo/raft domains of the PM. The Lo domain includes receptors, fast PtdIns(4,5)P2 breakdown, and fast restoration of PtdIns(4,5)P2 by local synthesis and/or by transfer or membrane fusion delivering precursors from intracellular membranes. The Ld domain has local PIP5K activity and exchanges DAG, PtdIns4P, and PtdIns(4,5)P2 surprisingly slowly with the Lo domain across the domain interface. PI, PtdIns; PIP, PtdIns4P; and PIP2, PtdIns(4,5)P2.
Similarly the restoration of PtdIns(4,5)P2 is faster in Lo domains than in Ld domains and interestingly the restoration of PM PtdIns(4,5)P2 is faster than that of its precursor PM PtdIns4P, as has been reported for whole-cell phosphoinositides for a long time (56). One likely factor in the latter observation is that the enzyme PIP5K is so fast that any PM PtdIns4P may be immediately converted to PtdIns(4,5)P2 until the PtdIns(4,5)P2 pool is fully replenished. But what makes the PtdIns(4,5)P2 restoration faster in Lo domains? Possible rate-limiting steps to consider in Lo domains are delivery of the precursor PtdIns to the PM where it is at very low concentration (57, 58), activity of PI4K, and delivery of already formed PtdIns4P or PtdIns(4,5)P2 from other organelles. PI4KIIIα is targeted to the PM by the accessory proteins, EFR3B, TTC7, and FAM126 (59–61). Interestingly, the N terminus of EFR3B has three cysteines (C5, C7, and C8) whose palmitoylation is essential for PM targeting. Thus, this signature in ERH3B is likely to localize functional PI4KIIIα complexes in Lo domains much as multiple palmitoylation localizes L10. It would be simple to assume that PI4K is the key limiting enzyme in Lo, but delivery of each of the phosphoinositides by vesicular traffic or by PtdIns transfer proteins at membrane contact sites merits serious evaluation as well (62–64). Lo domains may be favored sites for membrane contact formation and for membrane fusion events. During stimulation, one or several of these factors may become locally accelerated by the presence in Lo of activated receptors and active PLC (64, 65).
Two nonexclusive interpretations are possible for the slower breakdown and restoration in Ld domains (Fig. 7). The Ld domains may contain all of the mechanisms above as well but with only e.g., 50% of the activity. Alternatively and in the extreme, neither cleavage nor resynthesis of Ptdins(4,5)P2 occurs in Ld, and instead the changes of Ptdins(4,5)P2 that we measured in Ld with our probes reflect lateral diffusional exchange of lipid between the domains with actual cleavage and synthesis occurring only in Lo as drawn in Fig. 7. To explain how the depletion and restoration events in Ld lag well behind those in Ld as we observed, such lipid exchange would need to occur with time constants on the order of 10 to 40 s or longer. We consider lipid exchange inevitable for liquid lipid domains as small as is usually postulated in the PM of living cells, but in this case, exchange would have to be unexpectedly slow. For example, to be very conservative, even if this exchange were simply two-dimensional free diffusion with a diffusion constant as low as 0.1 μm2/s (37), it would take less than 1 s for half the molecules to escape from a 0.5 μm disk (the size of the wavelength of light), which is an order of magnitude too fast to account for the observed lag. The implication is that the boundary between Lo and Ld domains has to offer a significant energy barrier that slows lateral movement of phosphoinositide lipids. Is there a protein fence, or are there retaining forces at the lipid phase boundary? [Please recall that this conclusion is based on the lag in Ld phosphoinositide kinetics in intact cells at room temperature (Figs. 4–6) and not on the lateral diffusion measurements within Lo of cooled GPMVs (SI Appendix, Fig. S1).]
To close, we emphasize the segregation of function, the cholesterol dependence, and the rapid dynamics of lipid domains in living cell plasma membranes. With less than 10 min of cyclodextrin treatment, the relative proportion of specific domains was strongly reset. Acute and chronic hypercholesterolemia might quickly alter key plasma membrane signaling pathways simply due to resetting the relative area fractions of Lo and Ld domains. Finally, we promote the use of L10 and S15 as genetically expressible FRET acceptors to probe the lipid and protein dynamics of lipid nanodomains.
Materials and Methods
Cell Culture and Gene Transfection.
All experiments except in SI Appendix, Fig. S3 used the tsA-201 cell line (Sigma) maintained with 5% CO2 at 37 °C in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (Sigma) and 2% penicillin/streptomycin. Cells (passages 15 to 40) were transfected at ∼75% confluency and subsequently plated on poly-l-lysine-coated cover-glass chips (#0; Thomas Scientific). X-tremeGENE 9 (Sigma) and cDNA plasmids were used for transfection.
Plasmids for CFP-PHPLCδ and YFP-PHPLCδ were gifts from Kees Jalink (Netherlands Cancer Institute, Amsterdam, Netherlands) (11); P4M-CFP (single P4M version, not tandem), L10-CFP, L10-YFP, S15-CFP, and S15-YFP from B. C. Suh (49); Venus-K-ras and Venus-H-ras from Peter Várnai (Semmelweis University, Budapest, Hungary) (66); and M5R and C1A-C1A-CFP from Myeong et al. (67, 68). Of the fluorescent protein labels, all were standard versions except for Venus-K-Ras and Venus-H-Ras, which carried the monomeric mutations of Zacharias et al. (55). M1R cDNA was purchased from the cDNA Resource Center. FAST DiI (1,1′-Dilinoleyl-3,3,3′,3′-tetramethylindocarbocyanine, 4-chlorobenzenesulfonate) was from Life Technologies. N-ethylmaleimide (NEM), DTT, PFA, MβCD, and WSC were from Sigma. αCD and βCD were from Santa Cruz Biotechnology. Phosphate-buffered saline (PBS) was from Thermo Fisher Scientific.
Molecular Cloning.
For the generation of various L10 and S15 constructs, we used the one-step sequence- and ligation-independent cloning (SLIC) (69). First, pEYFP-N1 and pECFP-N1 vectors (Clontech) were linearized by KpnI restriction enzyme digestion. The cDNAs encoding FRB were amplified by PCR using primers with L10 or S15 sequences including an 18-bp sequence homologous to each end of the linearized vector. Primers used for L10 and S15 constructs are listed in SI Appendix, Table S1. Second, the linearized vector and PCR fragments were blended and incubated at room temperature for 2.5 min with T4 DNA polymerase (NEB). Third, the DNA mixture was kept on ice for 10 min and then competent Escherichia coli cells were transformed directly. For the deletion of FRB of L10 and S15 constructs: First, L10-FRB-fluorescent protein (YFP and CFP) constructs were amplified by inverse PCR using nPfu-special DNA polymerase (Enzynomics). Second, the PCR product was 5′-phosphorylated by T4 polynucleotide kinase (Enzynomics), and plasmid DNA was digested by DpnI (Agilent Technologies). Finally, the PCR product was ligated by T4 DNA ligase (NEB). The primers used for deletion are listed in SI Appendix, Table S2. All the constructs were verified by DNA sequencing (Macrogen).
Fluorescence and Three-Cube FRET Measurements.
Three-cube FRET was measured between transfected CFP- and YFP-tagged fluorescent proteins (29). For a detailed description of the recording and analysis of FRET signals, see SI Appendix, Supplementary FRET Methods.
Except where indicated, all experiments were done at room temperature (21 to 23 °C).
GPMV Preparation.
Twenty-four hours after transfection, the medium was removed from cells, and cells were washed three times with PBS. FAST DiI (5 μg/mL) was then added to the dishes and incubated at 4 °C for 10 min. Cells were washed five times by GPMV buffer (10 mM Hepes, 15 mM NaCl, 2 mM CaCl2, pH 7.4). GPMV buffer plus 25 mM PFA and 2 mM DTT was added to the dishes, and cells were incubated at 37 °C for 1 to 2 h until vesicles became visible outside the cells. The GPMV-containing supernatant was then decanted into 1.5-mL Eppendorf tubes, and cyclodextrin or WSC was added to the solution for 30 min. A total of 50 μL of GPMV solution was pipetted from the bottom of the Eppendorf tube and sandwiched between two coverslips coated with 0.1% bovine serum albumin (BSA).
Confocal Microscopy.
Cells and GPMVs were imaged with a Zeiss LSM 710 laser-scanning confocal microscope using a 63× oil objective. Fluorophores were excited by argon (for CFP and YFP) and helium-neon (for FAST DiI) lasers. The confocal images were analyzed with ImageJ/Fiji (NIH). For GPMV experiments, the GPMV solution was cooled using a 12 V, 60 W Peltier plate and a DC power supply (Digital Electronics). Temperature was measured with a thermistor but was not regulated by feedback, so we describe the GPMVs simply as “cooled” when the temperature was near 5 ± 2 °C.
Membrane Diffusion Coefficient Analysis.
The mobility of L10-YFP in Lo domains was assessed by fluorescence recovery after photobleaching on the confocal microscope. The spatial spread of YFP fluorescence was approximated by a mathematical model of one-dimensional diffusion in an 8-μm closed diffusion regime with reflecting ends. Space was represented as a series of 0.05-μm-thick compartments, and mass was transferred between them by Fick’s law in time steps of 1 ms by Euler integration. Validity was checked by two tests: 1) verifying that the spread of mass obeyed the one-dimensional Einstein relation (root-mean-square displacement of 2 Dt, where D is the diffusion coefficient and t is time); and 2) verifying that total mass was conserved except during photobleaching. Bleaching for 2 s was simulated as a gradual exponential decay of mass with a rate constant of 2/s operating on the spatial compartments between 2.5 and 5.5 μm (while diffusion continued to occur).
Analysis and Statistics.
The results are presented as mean ± SEM. Statistical significance was determined using the two-tailed Student’s t test, except in Figs. 2C, 4E, and 6D, which used one-way ANOVA, *P < 0.05, **P < 0.005, and ***P < 0.001.
Supplementary Material
Acknowledgments
We thank Lea M. Miller for technical assistance; Drs. Sarah L. Keller and Alexey J. Merz for helpful advice; and Drs. Duk-Su Koh, Donald Hilgemann, Gucan Dai, Tamas Balla, and Pietro De Camilli for insightful comments on the manuscript. This work was supported by NIH Grant R37-NS08174; by the Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Education (2019R1A6A3A03031486); the National Research Foundation of Korea grant funded by the Korea government (Ministry of Science, Information and Communications Technology, and Future Planning) (2019R1A2B5B01070546); and by the Wayne E. Crill Endowed Professorship.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025343118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Bagatolli L. A., Mouritsen O. G., Is the fluid mosaic (and the accompanying raft hypothesis) a suitable model to describe fundamental features of biological membranes? What may be missing? Front Plant Sci 4, 457 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simons K., Ikonen E., Functional rafts in cell membranes. Nature 387, 569–572 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Levental I., Veatch S., The continuing mystery of lipid rafts. J. Mol. Biol. 428, 4749–4764 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rietveld A., Simons K., The differential miscibility of lipids as the basis for the formation of functional membrane rafts. Biochim. Biophys. Acta 1376, 467–479 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Sezgin E., Levental I., Mayor S., Eggeling C., The mystery of membrane organization: Composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 18, 361–374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schroeder R., London E., Brown D., Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins: GPI-anchored proteins in liposomes and cells show similar behavior. Proc. Natl. Acad. Sci. U.S.A. 91, 12130–12134 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuck S., Honsho M., Ekroos K., Shevchenko A., Simons K., Resistance of cell membranes to different detergents. Proc. Natl. Acad. Sci. U.S.A. 100, 5795–5800 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Owen D. M., Williamson D. J., Magenau A., Gaus K., Sub-resolution lipid domains exist in the plasma membrane and regulate protein diffusion and distribution. Nat. Commun. 3, 1256 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Sengupta P., et al., Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat. Methods 8, 969–975 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prior I. A., Muncke C., Parton R. G., Hancock J. F., Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 160, 165–170 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Rheenen J., Achame E. M., Janssen H., Calafat J., Jalink K., PIP2 signaling in lipid domains: A critical re-evaluation. EMBO J. 24, 1664–1673 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heerklotz H., Szadkowska H., Anderson T., Seelig J., The sensitivity of lipid domains to small perturbations demonstrated by the effect of Triton. J. Mol. Biol. 329, 793–799 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Baumgart T., et al., Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc. Natl. Acad. Sci. U.S.A. 104, 3165–3170 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerstle Z., Desai R., Veatch S. L., Giant plasma membrane vesicles: An experimental tool for probing the effects of drugs and other conditions on membrane domain stability. Methods Enzymol. 603, 129–150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Busija A. R., Patel H. H., Insel P. A., Caveolins and cavins in the trafficking, maturation, and degradation of caveolae: Implications for cell physiology. Am. J. Physiol. Cell Physiol. 312, C459–C477 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sviridov D., Miller Y. I., Ballout R. A., Remaley A. T., Bukrinsky M., Targeting lipid rafts-A potential therapy for COVID-19. Front. Immunol. 11, 574508 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J. H., Rhee J. K., Ahn D. G., Kim K. P., Oh J. W., Interaction of stomatin with hepatitis C virus RNA polymerase stabilizes the viral RNA replicase complexes on detergent-resistant membranes. J. Microbiol. Biotechnol. 24, 1744–1754 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Veit M., Thaa B., Association of influenza virus proteins with membrane rafts. Adv. Virol. 2011, 370606 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang S. T., Kiessling V., Simmons J. A., White J. M., Tamm L. K., HIV gp41-mediated membrane fusion occurs at edges of cholesterol-rich lipid domains. Nat. Chem. Biol. 11, 424–431 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickson E. J., Hille B., Understanding phosphoinositides: Rare, dynamic, and essential membrane phospholipids. Biochem. J. 476, 1–23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Richards D. A., Segregation of PIP2 and PIP3 into distinct nanoscale regions within the plasma membrane. Biol. Open 1, 857–862 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tóth J. T., et al., BRET-monitoring of the dynamic changes of inositol lipid pools in living cells reveals a PKC-dependent PtdIns4P increase upon EGF and M3 receptor activation. Biochim. Biophys. Acta 1861, 177–187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hope H. R., Pike L. J., Phosphoinositides and phosphoinositide-utilizing enzymes in detergent-insoluble lipid domains. Mol. Biol. Cell 7, 843–851 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin H. L., Janmey P. A., Phosphoinositide regulation of the actin cytoskeleton. Annu. Rev. Physiol. 65, 761–789 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Laux T., et al., GAP43, MARCKS, and CAP23 modulate PI(4,5)P2 at plasmalemmal rafts, and regulate cell cortex actin dynamics through a common mechanism. J. Cell Biol. 149, 1455–1472 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji C., Zhang Y., Xu P., Xu T., Lou X., Nanoscale landscape of phosphoinositides revealed by specific pleckstrin homology (PH) domains using single-molecule superresolution imaging in the plasma membrane. J. Biol. Chem. 290, 26978–26993 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson C. M., Chichili G. R., Rodgers W., Compartmentalization of phosphatidylinositol 4,5-bisphosphate signaling evidenced using targeted phosphatases. J. Biol. Chem. 283, 29920–29928 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodgers W., Making membranes green: Construction and characterization of GFP-fusion proteins targeted to discrete plasma membrane domains. Biotechniques 32, 1044–1046, 1048, 1050–1051 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Erickson M. G., Alseikhan B. A., Peterson B. Z., Yue D. T., Preassociation of calmodulin with voltage-gated Ca2+ channels revealed by FRET in single living cells. Neuron 31, 973–985 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Chichili G. R., Rodgers W., Clustering of membrane raft proteins by the actin cytoskeleton. J. Biol. Chem. 282, 36682–36691 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Sezgin E., et al., Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat. Protoc. 7, 1042–1051 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Kaiser H. J., et al., Order of lipid phases in model and plasma membranes. Proc. Natl. Acad. Sci. U.S.A. 106, 16645–16650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kusumi A., et al., Defining raft domains in the plasma membrane. Traffic 21, 106–137 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Baumgart T., Hunt G., Farkas E. R., Webb W. W., Feigenson G. W., Fluorescence probe partitioning between Lo/Ld phases in lipid membranes. Biochim. Biophys. Acta 1768, 2182–2194 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levental I., Grzybek M., Simons K., Raft domains of variable properties and compositions in plasma membrane vesicles. Proc. Natl. Acad. Sci. U.S.A. 108, 11411–11416 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levental I., et al., Cholesterol-dependent phase separation in cell-derived giant plasma-membrane vesicles. Biochem. J. 424, 163–167 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Golebiewska U., et al., Evidence for a fence that impedes the diffusion of phosphatidylinositol 4,5-bisphosphate out of the forming phagosomes of macrophages. Mol. Biol. Cell 22, 3498–3507 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derzko Z., Jacobson K., Comparative lateral diffusion of fluorescent lipid analogues in phospholipid multibilayers. Biochemistry 19, 6050–6057 (1980). [DOI] [PubMed] [Google Scholar]
- 39.Niv H., Gutman O., Kloog Y., Henis Y. I., Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. J. Cell Biol. 157, 865–872 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korade Z., Kenworthy A. K., Lipid rafts, cholesterol, and the brain. Neuropharmacology 55, 1265–1273 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simons K., Ehehalt R., Cholesterol, lipid rafts, and disease. J. Clin. Invest. 110, 597–603 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahammad S., Parmryd I., Cholesterol depletion using methyl-β-cyclodextrin. Methods Mol. Biol. 1232, 91–102 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Liu S. L., et al., Orthogonal lipid sensors identify transbilayer asymmetry of plasma membrane cholesterol. Nat. Chem. Biol. 13, 268–274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ilangumaran S., Hoessli D. C., Effects of cholesterol depletion by cyclodextrin on the sphingolipid microdomains of the plasma membrane. Biochem. J. 335, 433–440 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zidovetzki R., Levitan I., Use of cyclodextrins to manipulate plasma membrane cholesterol content: Evidence, misconceptions and control strategies. Biochim. Biophys. Acta 1768, 1311–1324 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harlan J. E., Hajduk P. J., Yoon H. S., Fesik S. W., Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature 371, 168–170 (1994). [DOI] [PubMed] [Google Scholar]
- 47.Tsai F. C., et al., A polarized Ca2+, diacylglycerol and STIM1 signalling system regulates directed cell migration. Nat. Cell Biol. 16, 133–144 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hammond G. R., Machner M. P., Balla T., A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J. Cell Biol. 205, 113–126 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keum D., Kruse M., Kim D. I., Hille B., Suh B. C., Phosphoinositide 5- and 3-phosphatase activities of a voltage-sensing phosphatase in living cells show identical voltage dependence. Proc. Natl. Acad. Sci. U.S.A. 113, E3686–E3695 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pike L. J., Casey L., Localization and turnover of phosphatidylinositol 4,5-bisphosphate in caveolin-enriched membrane domains. J. Biol. Chem. 271, 26453–26456 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Pike L. J., Miller J. M., Cholesterol depletion delocalizes phosphatidylinositol bisphosphate and inhibits hormone-stimulated phosphatidylinositol turnover. J. Biol. Chem. 273, 22298–22304 (1998). [DOI] [PubMed] [Google Scholar]
- 52.Fujita A., Cheng J., Tauchi-Sato K., Takenawa T., Fujimoto T., A distinct pool of phosphatidylinositol 4,5-bisphosphate in caveolae revealed by a nanoscale labeling technique. Proc. Natl. Acad. Sci. U.S.A. 106, 9256–9261 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petersen E. N., Chung H. W., Nayebosadri A., Hansen S. B., Kinetic disruption of lipid rafts is a mechanosensor for phospholipase D. Nat. Commun. 7, 13873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van den Bogaart G., et al., Membrane protein sequestering by ionic protein-lipid interactions. Nature 479, 552–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y., Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Willars G. B., Nahorski S. R., Challiss R. A., Differential regulation of muscarinic acetylcholine receptor-sensitive polyphosphoinositide pools and consequences for signaling in human neuroblastoma cells. J. Biol. Chem. 273, 5037–5046 (1998). [DOI] [PubMed] [Google Scholar]
- 57.Pemberton J. G., et al., Defining the subcellular distribution and metabolic channeling of phosphatidylinositol. J. Cell Biol. 219, e201906130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zewe J. P., et al., Probing the subcellular distribution of phosphatidylinositol reveals a surprising lack at the plasma membrane. J. Cell Biol. 219, e201906127 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakatsu F., et al., PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J. Cell Biol. 199, 1003–1016 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baskin J. M., et al., The leukodystrophy protein FAM126A (hyccin) regulates PtdIns(4)P synthesis at the plasma membrane. Nat. Cell Biol. 18, 132–138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lees J. A., et al., Architecture of the human PI4KIIIα lipid kinase complex. Proc. Natl. Acad. Sci. U.S.A. 114, 13720–13725 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chung J., Nakatsu F., Baskin J. M., De Camilli P., Plasticity of PI4KIIIα interactions at the plasma membrane. EMBO Rep. 16, 312–320 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pemberton J. G., Kim Y. J., Balla T., Integrated regulation of the phosphatidylinositol cycle and phosphoinositide-driven lipid transport at ER-PM contact sites. Traffic 21, 200–219 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Myeong J., et al., Phosphatidylinositol 4,5-bisphosphate is regenerated by speeding of the PI 4-kinase pathway during long PLC activation. J. Gen. Physiol. 152, e202012627 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Falkenburger B. H., Jensen J. B., Hille B., Kinetics of PIP2 metabolism and KCNQ2/3 channel regulation studied with a voltage-sensitive phosphatase in living cells. J. Gen. Physiol. 135, 99–114 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gulyás G., et al., Plasma membrane phosphatidylinositol 4-phosphate and 4,5-bisphosphate determine the distribution and function of K-Ras4B but not H-Ras proteins. J. Biol. Chem. 292, 18862–18877 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Myeong J., et al., Close spatio-association of the transient receptor potential canonical 4 (TRPC4) channel with Gαi in TRPC4 activation process. Am. J. Physiol. Cell Physiol. 308, C879–C889 (2015). [DOI] [PubMed] [Google Scholar]
- 68.Myeong J., et al., Dual action of the Gαq-PLCβ-PI(4,5)P2 pathway on TRPC1/4 and TRPC1/5 heterotetramers. Sci. Rep. 8, 12117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jeong J. Y., et al., One-step sequence- and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Appl. Environ. Microbiol. 78, 5440–5443 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.