Summary
Polyethylene terephthalate (PET) is a mass‐produced synthetic polyester contributing remarkably to the accumulation of solid plastics waste and plastics pollution in the natural environments. Recently, bioremediation of plastics waste using engineered enzymes has emerged as an eco‐friendly alternative approach for the future plastic circular economy. Here we genetically engineered a thermophilic anaerobic bacterium, Clostridium thermocellum, to enable the secretory expression of a thermophilic cutinase (LCC), which was originally isolated from a plant compost metagenome and can degrade PET at up to 70°C. This engineered whole‐cell biocatalyst allowed a simultaneous high‐level expression of LCC and conspicuous degradation of commercial PET films at 60°C. After 14 days incubation of a batch culture, more than 60% of the initial mass of a PET film (approximately 50 mg) was converted into soluble monomer feedstocks, indicating a markedly higher degradation performance than previously reported whole‐cell‐based PET biodegradation systems using mesophilic bacteria or microalgae. Our findings provide clear evidence that, compared to mesophilic species, thermophilic microbes are a more promising synthetic microbial chassis for developing future biodegradation processes of PET waste.
Promising bioremediation strategies for plastics waste are of great importance and requirements. In our study, we constructed a recombinant Clostridium thermocellum strain expressing a secretory cutinase (LCC) as a thermophilic whole‐cell biocatalyst to degrade PET under high‐temperature condition (60°C). To our knowledge, this biocatalysis system demonstrates the highest PET degradation efficiency compared to reported whole‐cell‐based systems and also enjoys a low‐cost advantage over the free enzyme‐based process.
Introduction
Plastic pollution has become a global concern in recent years as a result of the extensive production, use and lack of proper recycling provisions and technologies. The tremendous amount of virgin plastics with a total of over 8300 million metric ton (Mt) has been estimated to be produced by humankind since 1950, out of which over 90% are derived from fossil hydrocarbons and 79% are simply landfilled or carelessly disposed in the natural environments (Geyer, et al., 2017). Consequently, humankind is facing a huge environmental crisis caused by the highly recalcitrant plastic materials as their degradation under natural conditions is extremely slow (Rochman, et al., 2013). As an application sector with the shortest product life cycle length, plastic packaging contributes considerably to the rapid accumulation of solid plastic waste (PlasticsEurope, 2018). In addition to polyolefins and polystyrene, polyethylene terephthalate (PET) plays a pivotal role in the global packaging market, especially for fresh food, water and soft drinks (Ellen MacArthur Foundation, 2016). Moreover, PET contributes more than 70% to the global production of synthetic fibres which are majorly used for the textile industry (Geyer, et al., 2017) and can end up as microplastics pollution in the natural environments (Deng, et al., 2019; Zhang, et al., 2019).
Polyethylene terephthalate is a colourless thermoplastic material with a variety of excellent polymer processing properties (Webb, et al., 2013). Despite the fact that the backbone of PET consists of only ester bonds formed between a diol (ethylene glycol, EG) and an aromatic di‐acid (terephthalic acid, TPA), only a few hydrolytic enzymes have been reported to catalyse the polymer chain breakdown, releasing TPA and EG as monomer degradation products (Wei & Zimmermann 2017a,2017b). In 2016, Yoshida et al. identified at a PET‐bottle recycling site a bacterium, Ideonella sakaiensis, capable of degrading and completely assimilating highly amorphous PET as a major carbon source at a mesophilic temperature of 30°C (Bornscheuer, 2016; Yoshida, et al., 2016). When cultivated in a fed‐batch culture containing a low concentration of yeast extract, this bacterium could completely degrade a small PET film (shown as approximately 60 mg with a dimension of 15 x 20 x 0.2 mm) within 42 days (6 weeks). Two key enzymes involved in the hydrolysis of PET by I. sakaiensis were identified: an extracellular PETase (Han, et al., 2017; Austin, et al., 2018) able to cleave the PET polymers into the intermediates bis‐ and mono(2‐hydroxyethyl) terephthalic acid (BHET, MHET respectively), which could possibly be taken up by the cell and subsequently hydrolysed by the second enzyme, the intracellular MHET hydrolase (Palm, et al., 2019), into TPA and EG (Yoshida, et al., 2016). Moreover, Yoshida et al. (2016) also hypothesized a potential metabolic mechanism of PET and its monomer TPA based on primitive transcriptomic analysis. Very recently, Moog, et al. (2019) genetically engineered the photosynthetic microalgae Phaeodactylum tricornutum to resemble an I. sakaiensis‐like cell factory capable of secreting a single‐site mutant of PETase into the extracellular culture medium. The cell‐free culture supernatant has been shown able to degrade the amorphous copolymer polyethylene terephthalate glycol (PETG) following 7 days of incubation at 30°C, as evidenced by scanning electron microscopy (SEM) and high‐performance liquid chromatography (HPLC) analysis towards the aromatic degradation products. When the engineered diatom was cultivated in a salt water‐based environment at 21°C in the presence of industrially shredded PET particles, the secretory expression of the PETase variant and the simultaneous degradation of PET could be successfully verified by HPLC analysis. This finding provided a proof‐of‐concept of using engineered whole‐cell biocatalyst in plastic degradation. Compared to enzymatic hydrolysis using purified free enzymes, the whole‐cell biocatalysis is a ‘one‐pot’ strategy combining the enzyme production and hydrolysis in a single step. The whole‐cell biocatalyst can produce the functional enzymes and degrade PET simultaneously under a consistent reaction condition. Thus, PET degradation based on whole‐cell biocatalyst can show great advantages over using free enzymes by omitting the time‐consuming and material‐intensive protein purification process (de Carvalho, 2017).
Nevertheless, the polymer properties of semi‐crystalline PET with a glass transition temperature (Tg) of >70°C require effective enzymatic hydrolysis at a temperature significantly higher than 30°C (Wei & Zimmermann 2017a, 2017b; Wei, et al., 2019b). For instance, LC‐cutinase (LCC) isolated from a leaf‐branch compost, which was assumed to provide a thermophilic habitat for microbes (Sulaiman, et al., 2012), was shown to have an optimal temperature for PET hydrolysis at 70°C (Sulaiman, et al., 2014; Wei, et al., 2019b), which is in the range of Tg for PET. At this temperature, the mobile amorphous fraction of PET polymers can be preferentially accessed by hydrolases and converted to soluble oligomeric and monomeric feedstocks (Barth, et al., 2015; Wei, et al., 2019a). High temperatures can not only promote plastic decomposition but also reduce the risk of contamination. Therefore, although the plastic degrading activity of natural thermophiles able to grow at >60°C has not been reported so far, engineering thermophilic microbes by employing thermophilic hydrolases is regarded as a more promising strategy for the development of future bioremediation processes for plastic waste.
However, compared to mesophilic microbes, thermophilic microorganisms are usually difficult to be genetically manipulated due to inadequate genetic tools. One exception is Clostridium thermocellum, a typical thermophilic anaerobe with an optimal growth condition at 60°C (Ng, et al., 1977). C. thermocellum is considered one of the most promising species for highly efficient and low‐cost lignocellulose bioconversion (Lynd, et al., 2005), and mature genetic manipulation platforms have been constructed for this thermophile (Tripathi, et al., 2010; Olson and Lynd, 2012; Mohr, et al., 2013, Olson, et al., 2015; Zhang, et al., 2017). By using the developed tools, C. thermocellum recombinant strains have been constructed and employed as whole‐cell biocatalysts in lignocellulose saccharification and biofuel production (Argyros, et al., 2011; Lin, et al., 2015; Liu, et al., 2019). Thus, C. thermocellum has the potential to be used as the microbial chassis for the degradation of plastic waste at high temperature, e.g. with the secretory expression of thermal stable PET hydrolase LCC.
Results
Construction of a recombinant C. thermocellum strain expressing a secretory LC‐cutinase
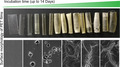
A thermophilic cutinase (LCC) was originally isolated from a plant compost metagenome (Sulaiman, et al., 2012) and has high activity in PET degradation at up to 70°C (Sulaiman, et al., 2014; Wei, et al., 2019b). Thus, LCC was chosen to be expressed in C. thermocellum. C. thermocellum is known for its ability in the highly efficient degradation of lignocellulose by producing an extracellular multi‐protein structure, the cellulosome (Yoav, et al., 2017). Because the exoglucanase Cel48S is the most abundant component in the cellulosome of C. thermocellum (Liu, et al., 2018), the signal peptide sequence of Cel48S was used for the secretory production of LCC. In addition, a strong promoter from the constitutively expressed gene Clo1313_2638 (Olson, et al., 2015) was selected to obtain high‐level expression of LCC in C. thermocellum. As shown in Fig. 1A, the plasmid pHK‐LCC was constructed after PCR verification and was then transformed to C. thermocellum DSM1313 to construct the target strain DSM1313::pHK‐LCC.
Fig. 1.
Construction of LCC‐expressing plasmid pHK‐LCC and extracellular expression of active LCC in C. thermocellum.
A. Construction of pHK‐LCC. SP, signal peptide, P2638, promoter sequence from gene Clo1313_2638, Ter, terminator, Rep Ori, the origin of replicon of E. coli, RepB, replicon origin of C. thermocellum, CAT, thiamphenicol resistant gene. The insertion of the construct ‘P2638‐SP‐lcc’ was verified by PCR with the primers seqF/R and the theoretical size of the PCR product was 1.3 kb. The gel analysis of the PCR product is shown on the right.
B. SDS‐PAGE analysis of cell lysates and extracellular proteins of DSM1313 and DSM1313:: pHK‐LCC stained with Coomassie Brilliant Blue.
C. Fast‐Red dye staining analysis of the protein gel against 1‐naphthyl acetate to visualize esterolytic activity. Lanes 1 and 2, culture supernatant of DSM1313 and DSM1313::pHK‐LCC respectively; lanes 3 and 4, cell lysate of DSM1313 and DSM1313::pHK‐LCC respectively. Ten µg protein was loaded in each lane.
Both the engineered strain and the wild type strain DSM1313, which served as a negative control, were cultivated in GS‐2 medium to investigate the expression and secretion of the LCC protein. According to the Fast‐Red dye‐based staining results (Fig. 1C), active LCC was visible as a violet protein band at approximately 28 kDa in the supernatant of DSM1313::pHK‐LCC, but not detected in the corresponding cell lysate, indicating the produced LCC protein was successfully secreted extracellularly in the supernatant of the culture. In contrast, no LCC activity was observed in either intra‐ or extracellular proteins of the wild type strain. These results indicated the functional expression of LCC as fully secretory protein by C. thermocellum DSM1313::pHK‐LCC.
Influence of PET and PET hydrolysates on the growth of C. thermocellum
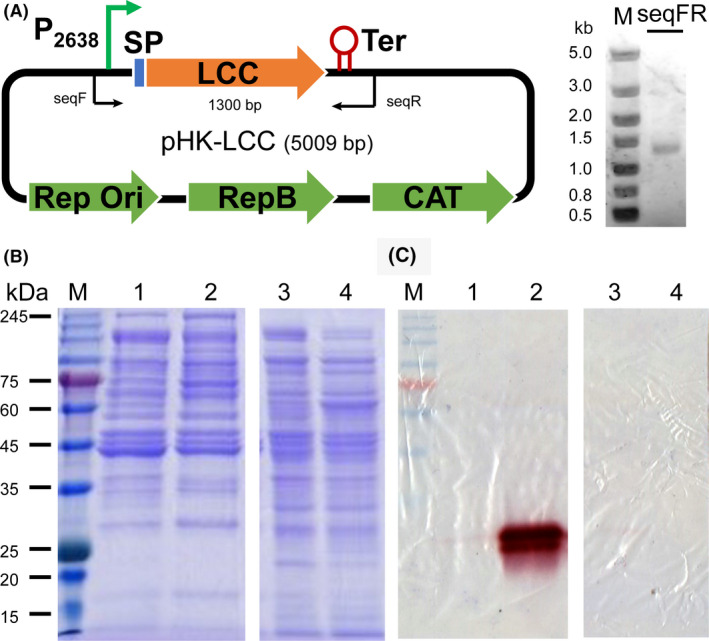
Clostridium thermocellum has no native capability of PET degradation and assimilation based on its genome information. The enzymatic degradation of PET will release TPA and EG as monomeric feedstocks from PET polymers (Wei & Zimmermann 2017a, 2017b) which can potentially influence the growth of C. thermocellum. In our experimental set‐up, we used 50 mg of PET films in an anaerobic glass tube containing a culture medium of 10 ml in total. Therefore, theoretical maximum yield of TPA and EG was estimated to be 25.9 mM according to the molecular mass of one repeating unit of PET of 192.2 g mol−1. Accordingly, we determined the cell growth in terms of optical density at 600 nm (OD600) in the presence of 20‐mM TPA, EG, both of them, or PET films within the first 32 h cultivation time. As shown in Fig. 2, C. thermocellum DSM1313 exhibited a similar shape of growth curve independent of the presence of any PET‐related compounds. Briefly, a lag phase was observed until 12 h after the inoculation followed by a rapid growth until 20 h of cultivation time with a maximum OD600 of approximately 2.2. Afterwards, there was a clear trend of gradually decreasing OD600 until 32 h to reach a final value in the range of 1.6–1.8, suggesting possible cell lysis without further addition of any nutrient. Additionally, no significant difference was observed in the time courses of pH change during cultivation with or without amorphous PET films (Fig. 2).
Fig. 2.
Cell growth and pH of C. thermocellum DSM1313::pHK‐LCC. Cultivation was carried out at 60°C under the agitation of 200 rpm for up to 32 h. The cell growth in the presence of PET film and its degradation products at a constant concentration of 20 mM was determined by monitoring the optical density of cultures at 600nm (OD600). Dashed lines indicate the pH values determined along with the cell growth with or without PET films. Error bars indicate the standard deviations determined from at least triplicate measurements.
Degradation of amorphous PET films using engineered C. thermocellum whole‐cell biocatalyst
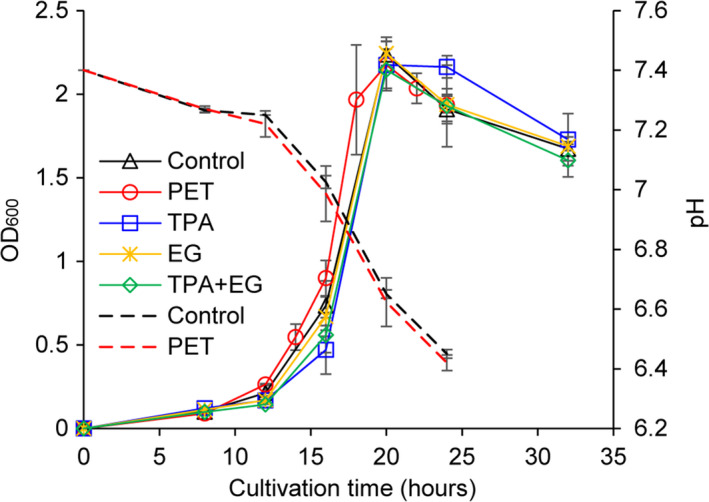
The engineered C. thermocellum strain DSM1313::pHK‐LCC was used to degrade a PET film with an initial weight of approximately 50 mg. The incubation lasted for 14 days at 60°C. The time course of PET weight loss was shown in Fig. 3A. Evident weight loss can be monitored even after 2 days of incubation with a roughly continuously increasing trend until 14 days to achieve a maximum weight loss of approximately 62% (≈ 31 mg).
Fig. 3.
Degradation of amorphous PET films by C. thermocellum DSM1313::pHK‐LCC.
A. Time course of the relative weight loss determined gravimetrically. About 50 mg of PET films were supplemented initially.
B. Time course of the released UV‐absorbing degradation products determined by HPLC in the culture supernatant (stacked columns) as well as calculated sum values based on the weight loss determination (dashed curve, from the same dataset with panel A). The specific concentrations of TPA and MHET are listed in Table S1.
C. Time courses of volumetric esterolytic activity against p‐NPB and pH changes determined in the culture supernatant. Error bars indicate the standard deviation derived from at least triplicate determinations.
D. Specific esterolytic activity analysis determined by Fast‐Red staining. SDS‐PAGE of the culture supernatants (10 μg protein loaded in each lane) removed after cultivation for different time periods following the esterolytic staining with the Fast‐Red dye against 1‐naphthyl acetate as the substrate. The intensities of the violet bands calculated by the Quantity One software indicate the amount of residual specific activities of extracellular LCC.
Culture supernatants removed at an interval of 2 days were subjected to HPLC analysis to identify the soluble UV‐absorbing degradation products. Retention times of standards, as well as a calibration curve for the HPLC analysis, were prepared using commercially purchased pure TPA (Fig. S2). As shown in Fig. 3B, both TPA and MHET were detectable in the culture supernatant after more than 2 days of cultivation. Based on the molecular mass of a PET repeating unit of 192.2 g mol−1, the weight loss curve shown in Fig. 3A was converted into a dashed curve exhibiting the corresponding amount of UV‐absorbing products released (Fig. 2B, Table S1), which is in a good quantitative agreement with the determined sum of TPA and MHET. Slightly lower sum values of the HPLC determined UV‐absorbing degradation products were expected due to the presence of a small fraction of di‐ethylene glycol in the PET samples used in this study, as described previously (Wei, et al., 2019a). The change of pH was also determined in the culture supernatant with a rapid drop in the first 2 days from 7.4 to 6.5 followed by a slower continuous decrease to approximately 6.2 after 14 days of cultivation (Fig. 3C). Because the pH is a logarithmic measure of the present concentration of H3O+, this result showed a good agreement with the determined TPA amounts as indicated by a high correlation coefficient of 0.914 (data not shown).
As shown in Fig. 3D, the intensity of the LCC band was considered to reflect the specific activity because an equal amount of 10 µg of total protein was loaded in each lane. Similar LCC band intensities were observed in the culture supernatants after 1, 2 and 4 days of incubation, and the 8‐day band still showed 88.2% intensity of that obtained with the 1‐day band. Correspondingly, the volumetric esterolytic activity in the culture supernatant increased along with the cell growth in the first day of incubation (Fig. 2), maintained its maximum value (1.01 U ml−1) in the second day of the incubation, and gradually decreased to 0.39 U ml−1 after 8‐day incubation which was 38.9% of the maximal activity (Fig. 3C), although maintained LCC specific activity was detected in the degradation system until 8 days (Fig. 3D). The decreased LCC activity might be caused by the thermal deactivation or released proteases from the lysed cells after long‐term incubation. In line with the degradation performance, as shown in Fig. 3A‐B, which was visible from the second day of the cultivation, the decreased esterolytic activity in the culture supernatant might also be contributed by the mass transfer from free LCC to tightly absorbed enzyme at the surface of PET films, where they catalysed the hydrolysis of the polyesters.
Figure 4 exhibited drastic changes in the surface morphology of PET films after 5‐day cultivation in the presence of the engineered C. thermocellum strain. Cavities and holes appeared primarily as an initial sign for the surface erosion, similarly to the SEM captures of biologically degraded PET films shown in previous studies (Ronkvist, et al., 2009; Moog, et al., 2019; Wei, et al., 2019a). Afterwards, the discrete erosion holes developed to a continuous rugged surface with increasing amounts of irregular crevices as a result of further enzymatic hydrolysis catalysed by the secreted LCC in the culture supernatant until the final cultivation time of 14 days. In contrast to the SEM captures shown by Yoshida et al. (2016), which indicated clearly the I. sakaiensis cell attachment to the surface of PET films, engineered C. thermocellum cells did not attach to the PET film surface (Fig. S3).
Fig. 4.

SEM analysis of PET films during biodegradation using C. thermocellum whole‐cell biocatalyst at 60°C. The PET films were washed and dried by lyophilization before analysis. Cavities and holes appeared as an initial sign for the surface erosion. The number of holes increased along with the incubation time. From day 8, cracks and crevices appeared and became larger as a result of extensive enzymatic hydrolysis.
Discussion
The genetically engineered C. thermocellum constructed in this study is the first thermophilic strain so far able to express a thermophilic hydrolase (LCC) at 60°C. LCC was assumed to have a thermophilic habitat since it was first isolated from the core of a leaf‐branch compost (Sulaiman, et al., 2012). Its optimal temperature for PET hydrolysis was subsequently proved to be 70°C (Sulaiman, et al., 2014; Wei, et al., 2019b), which is in the range of Tg for PET. At such elevated temperature, PET polymer chains in the mobile amorphous fraction will gain higher flexibility and thus can be more easily accessed for enzymatic hydrolysis (Wei, et al., 2019a; Wei, et al., 2019b). Indeed, thermostable enzyme and variants have shown markedly better degradation performance on various PET materials in comparison with their mesophilic or less thermostable counterparts (Ronkvist, et al., 2009; Then, et al., 2016; Wei, et al., 2019b). These thermostable PET hydrolases were so far reported to be recombinantly expressed in E. coli (Herrero Acero, et al., 2011; Roth, et al., 2014), Bacillus subtilis (Wei, et al., 2019a), B. megaterium (Yang, et al., 2007), and Pichia pastoris (Gamerith, et al., 2017; Shirke, et al., 2018). But none of the host species can survive near the optimal temperature of the recombinant enzyme in the PET hydrolysis. Here, the constructed C. thermocellum recombinant strain allowed the simultaneous expression of LCC and biodegradation of PET at 60°C approaching its optimal condition. Although the highest bacterial growth was achieved within the first day of incubation (Fig. 2) and the maximum hydrolytic activity in culture supernatant maintained in the first 2 days of incubation (Fig. 3C), a continuous release of degradation products and a nearly constant increasing of the weight loss of PET films was observed until 14 days of incubation as a result of enzymatic hydrolysis (Fig. 3A‐B).
Compared to PET biodegradation processes using free enzymes which generally require time‐consuming and cost‐intensive protein purifications, whole‐cell biocatalyst based degradation processes profits from the advantages of low‐cost and simple operation process (de Carvalho, 2017). Selected mesophilic microorganisms with a preferential growing temperature at lower than 40°C have been used for the whole‐cell catalysed degradation of various PET materials. For example, I. sakaiensis which can intrinsically express an extracellular PETase and an intracellular MHETase have completely degraded a small piece of amorphous PET material (with a size of 15 x 20 x 0.2mm shown as 60 mg) following fed‐batch incubation at 30°C after 42 days by completely exchanging the medium every week (Yoshida, et al., 2016). This degradation performance was significantly better than using the purified recombinant PETase expressed in E. coli revealed in the same study, suggesting that the presence of a helper enzyme (MHETase) provided by I. sakaiensis whole‐cell system is favourable for accelerating the plastic degradation rate. Recently, an engineered diatom P. tricornutum that can produce a mutant of the I. sakaiensis PETase has been reported to degrade industrially shredded PET particles at 21°C as evidenced by monitoring the PET monomer feedstocks in the culture supernatant by HPLC analysis (Moog, et al., 2019). However, the releasing rate of degradation products by the engineered microalga was markedly lower than the former I. sakaiensis whole‐cell biocatalysis, possibly due to the less favoured temperature for PETase (optimum at 40°C) (Yoshida, et al., 2016), a lower biodegradability of shredded PET particles, as well as the lack of helper enzyme such as MHETase.
Due to the available genetic tools that have been developed previously (Tripathi, et al., 2010, Olson and Lynd, 2012, Mohr, et al., 2013, Olson, et al., 2015, Zhang, et al., 2017), the anaerobic thermophile C. thermocellum was chosen as the chassis to construct a thermophilic whole‐cell biocatalyst for PET degradation in this study. As an anaerobic microorganism, the growth of C. thermocellum requires no aeration and less agitation compared to aerobic microorganisms (Ng, et al., 1977) and thus minimizing the operation cost. Using the previously described pHK plasmid (Mohr, et al., 2013), the modified C. thermocellum DSM1313::pHK‐LCC strain was shown to express functional LCC, and consequently served as a whole‐cell biocatalyst for PET degradation at 60°C. Over 60% weight loss with an amorphous PET film of 50 mg was detected after a 14‐day incubation, corresponding to a degradation rate of > 2.2 mg day−1. This is notably higher than that of previously described mesophilic whole‐cell systems (e.g. > 1.4 mg day−1 for I. sakaiensis biocatalysis) (Yoshida, et al., 2016). Additionally, both reported mesophilic whole‐cell biocatalyses were carried out using fed‐batch cultures with regular supplementation of nutrients at <7 days intervals (Yoshida, et al., 2016; Moog, et al., 2019). By contrast, in this study, the C. thermocellum whole‐cell biocatalyst was batch‐cultivated anaerobically at 60°C in a 10‐ml tube without supplementation of nutrients and exchange of medium. The nutrients initially provided were shown to be adequate to guarantee an optimal cell growth up to 20 h, while the pH value decreased significantly from 7.4 to 6.4 after one‐day cultivation (Fig. 2) although the release of TPA was not yet detectable (Fig. 3B). This decreased pH was an undesirable condition for the growth of C. thermocellum (Whitham, et al., 2018). Under these circumstances, the PET degradation efficiency was considered to be solely dependent on the LCC proteins produced in the first day but without further replenishment from day 2 to day 14. Moreover, due to the cell lysis gradually occurring from 20 h after inoculation, the expressed LCC proteins might be degraded by the potentially released proteases (Zverlov, et al., 2005, Schwarz and Zverlov, 2006). The PET degradation efficiency of the C. thermocellum‐based whole‐cell biocatalyst could be thus further enhanced using a fed‐batch culture with pH control.
Although the whole‐cell biocatalysis strategy has the advantage of simultaneous enzyme production and plastic degradation compared to free enzyme‐based approach, the degradation performance of the secretory LCC‐based whole‐cell biocatalyst was still not comparable to that of purified recombinant LCC at 70°C so far (Sulaiman, et al., 2014; Wei, et al., 2019b). This might be explained by the less favourable temperature, i.e. 60°C, which was selected to match the growth conditions of C. thermocellum, although 70°C was considered a preferred temperature for LCC activity and polymer chain accessibility. Besides, cutinase‐catalysed PET degradation has been shown to be strongly dependent on the surrounding buffer conditions and ion strengths (Barth, et al., 2015; Schmidt, et al., 2016). Further improvements would focus on developing LCC variants with improved PET hydrolytic activity and stability under present conditions to enhance PET plastic degradation efficiency. We detected the accumulation of MHET during the PET degradation, which can act as an inhibitor for LCC in the PET depolymerization as described before (Barth, et al., 2016). Analogously, the construction of a multi‐enzyme system in C. thermocellum able to coexpress LCC and other helper enzymes (e.g. the thermophilic MHET‐hydrolysing carboxyl esterase from Thermobifida fusca) would mitigate the product inhibition and consequently further stimulate the PET degradation. For constructing an engineered cell factory able to solely feeding on PET, the introduction of, e.g. TPA or EG metabolic pathways will be also of interest and envisaged in future works.
In conclusion, our study provides a proof‐of‐concept for the more efficient PET degradation using an engineered thermophilic whole‐cell biocatalyst. This is for sure a promising strategy for the development of effective bioremediation processes of PET waste. Moreover, C. thermocellum is characterized by its ability to efficiently degrade cellulose by producing the cellulosome, an extracellular multi‐protein complex (Bayer, et al., 2008; Artzi, et al., 2017). It is known that the cellulosome system employs assembly modules (cohesin and dockerin) of the scaffoldins and enzymes to form the functional enzyme complex and can interact with the cellulosic substrate using specific carbohydrate‐binding modules (CBM) (Boraston, et al., 2004; Bayer, et al., 2008; Artzi, et al., 2017). Compared to free enzymes, the cellulosome system enjoys the advantage of synergistic interactions due to enzyme proximity (Hong, et al., 2014; Yoav, et al., 2017). Indeed, fusion of a CBM has been shown to notably increase the degradation performance of PET using a homologous enzyme to LCC (Ribitsch, et al., 2013). Thus, it is a promising strategy and also of high scientific interest to develop cellulosome‐like enzyme complexes containing assembled PET degrading enzymes harbouring CBM to promote the overall efficiency of PET degradation. As thermophilic PET hydrolases can also effectively degrade synthetic PET fibres (Wei, et al., 2016), the C. thermocellum‐based whole‐cell biocatalysis strategy is expected to demonstrate great potential for application in the bio‐recycling of mixed textile waste containing both cellulose and polyester fractions.
Experimental procedures
Bacterial strains and cultivation
The bacterial strains used in this study are listed in Table 1. Escherichia coli strains were cultivated aerobically at 37°C in Luria–Bertani (LB) liquid medium by shaking at 200 rpm or on solid LB plate with 1.5% agar. 30 μg ml−1 chloramphenicol was added for screening. C. thermocellum strains were grown anaerobically at 60°C in a modified GS‐2 medium (Cui, et al., 2012) with 5 g l−1 cellobiose. 3 μg ml−1 thiamphenicol (Tm), 20 mM TPA, and 20 mM EG were supplemented to the medium when necessary. Due to the poor solubility of TPA, a stock solution was prepared by dissolving 3.32 g of TPA in 100 ml of 450 mM NaOH and this was adjusted to pH 6.5 afterwards. The pH value of the GS‐2 medium was adjusted to 7.4 after the addition of TPA and EG. The growth curve of the C. thermocellum strains was determined by monitoring the cell optical density at 600 nm (OD600) every 2–8 h.
Table 1.
Bacterial strains and plasmids used in this study.
| Strains/plasmids | Relevant characteristic | Sources |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | f80dlacZΔM15, Δ(lacZYA‐argF)U169, deoR, recA1, endA1, hsdR17(rk−, mk+), phoA, supE44, l−, thi‐1, gyrA96, relA1 | Transgen |
| BL21(DE3) | ompT gal dcm lon hsdSB(rB − mB−) l (DE3 [lacI lacUV5‐T7 gene 1 ind1 sam7 nin5]) | Transgen |
| C. thermocellum | ||
| DSM1313 | Wild type strain | DSMZ |
| DSM1313::pHK‐LCC | Derived from DSM1313, with the plasmid pHK‐LCC | This work |
| Plasmids | ||
| pHK | pNW33N derivative, E. coli‐C. thermocellum shuttle vector, CmR/TmR | Zhang et al. (2017) |
| pHK‐LCC | pHK derivative for expression of LCC in DSM1313 | This work |
Plasmid construction
The recombinant plasmid pHK‐LCC was constructed using the E. coli DH5α strain as the host for LCC expression in C. thermocellum DSM1313. The gene lcc encoding the mature LCC protein (AEV21261.1 omitting the signal peptide) was synthesized with an optimized encoding sequence to adapt the codon usage of C. thermocellum (Genewiz, Suzhou, China) as shown in Fig. S1. The lcc sequence, a signal peptide (SP) sequence of a cellulase Cel48S, and a constitutive promoter of gene Clo1313_2638 (P2638) were amplified with the primer set lccF/R, sigF/R and proF/R, respectively, with the synthesized construct or genome of C. thermocellum DSM1313 as the template, and were ligated together as a whole construct ‘P2638‐SP‐lcc’ using a seamless cloning kit (Vazyme Biotech, Beijing, China) following the principle of overlap PCR. Then, the vector pHK (Mohr, et al., 2013) linearized by reverse PCR using the primer set pHKF/R was ligated with the construct ‘P2638‐SP‐lcc’ by overlap PCR (Fig. 1). The homology regions for overlap PCR were obtained from the designed primers (Table 2). The insertion of the construct was verified by PCR using the primer seqF/R (Fig. 1) and sequencing. A universal PCR condition was used for all amplification experiments, including a pre‐denaturation step at 96°C for 3 min, 35 reaction cycles (95°C for 0.5 min, 55°C for 1.5 min, and 72°C for 1 min), and a complete extention step at 72°C for 5 min. The constructed plasmid was stored at −20°C for further experiment.
Table 2.
Primers used in this study.
| Primers | Sequence (5’‐3’) |
|---|---|
|
lccF |
AGAGTTATTTTTATTTTACTCGAGCTGGCAGTGTC |
|
lccR |
CAAAGGCACCTACAAAAGATAACCCGTATCAGAGAGGACC |
|
sigF |
GGTCCTCTCTGATACGGGTTATCTTTTGTAGGTGCCTTTG |
|
sigR |
GTAAGGAGGAATTTGTTATGATGGTAAAAAGCAGAAAGAT |
|
proF |
ATCTTTCTGCTTTTTACCATCATAACAAATTCCTCCTTAC |
|
proR |
TTCCGGCTCGTATGTGATAAACAAAGGACGGTT |
|
seqF |
TTTATGCTTCCGGCTCGTAT |
|
seqR |
CAATGCTTAATCAGTGAGGC |
|
pHKF |
AATAAAAATAACTCTGTAGAAT |
|
pHKR |
ACATACGAGCCGGAAGCATAAAG |
Electrotransformation and screening of C. thermocellum
The pHK‐LCC plasmid was transformed into E. coli BL21(DE3) to remove the Dcm methylation (Guss, et al., 2012), and transformed into C. thermocellum DSM1313 as previously described (Hong, et al., 2014). Briefly, 100 μl of C. thermocellum competent cells and 10 μl of plasmid DNA were added to a 0.1 cm electroporation cuvette. A series of 40 square pulses were applied, each with an amplitude of 1.5 kV and a duration of 50 ms at 500 ms intervals. Cells were then recovered for 24 h at 51°C in 4 ml of fresh GS‐2 medium before screening on solid medium containing Tm. After growing on GS‐2 solid medium for 5‐7 days, the obtained colonies were screened by colony PCR using the primers seqF/R and sequenced. The colonies verified as the target recombinant strain DSM1313::pHK‐LCC were cultivated in GS‐2 medium and stored at –80°C for further experiments.
Whole‐cell‐based degradation of amorphous PET films
The whole‐cell‐based degradation of PET films was performed according to previously reported PET film degradation experiments using free thermophilic enzymes as summarized by ((Wei & Zimmermann 2017a,2017b;). In detail, amorphous PET film with a thickness of 250 μm was purchased from Goodfellow Ltd. (Bad Nauheim, Germany, product number ES301445), and cut into small pieces with a size of about 2 × 0.8 cm2 and an initial weight of 49.77 ± 0.52 mg. PET films were soaked in 75% ethanol overnight at room temperature and air‐dried under a sterile atmosphere before being added to autoclaved GS‐2 medium containing 5 g l−1 of cellobiose. Each 10 ml culture with one PET film in an anaerobic tube was inoculated with 1% (v/v) DSM1313::pHK‐LCC preculture. The wild type strain DSM1313 was used as a negative control. The cultivation and degradation experiments were performed at 60°C under the agitation of 170 rpm until 14 days. At least three tubes were randomly chosen and removed from the shaker every 24 h to extract the residual PET film, which was washed sequentially using 0.1% SDS and 70% alcohol. The weight loss of the PET film was determined gravimetrically after drying the residual PET films at 60°C for at least 12 h (Ronkvist, et al., 2009; Sulaiman, et al., 2014; Wei, et al., 2019b). Selected culture supernatants were subject to esterolytic activity determination (see below), pH measurement, and HPLC analysis towards the UV‐absorbing degradation products.
Esterolytic activity analyses
To analyse the secretory expression of LCC, SDS polyacrylamide gel electrophoresis (SDS‐PAGE) was carried out by loading 10 µg cell lysate or supernatant protein in each lane followed by an esterolytic activity staining using the Fast‐Red TR salt (Meilunbio, Guangzhou, China) as a dye and 1‐naphthyl acetate (Solarbio, Beijing, China) as a substrate as described previously (Billig, et al., 2010). For selected samples, the intensity of protein bands with hydrolytic activities was calculated using the Quantity One software (version 4.6.2; Bio‐Rad, Shanghai, China) as previously described (Li, et al., 2018). For comparison, polyacrylamide gels loaded by the same supernatant samples were partly also stained with Coomassie Brilliant Blue. Esterolytic activity determination was also performed using p‐nitrophenyl butyrate (p‐NPB) as substrate according to a previous report (Billig, et al., 2010) One unit of esterolytic activity was defined as the amount of enzyme required to hydrolyse 1 μmol of p‐NPB per minute. To obtain cell lysates, cells were harvested by centrifugation at 7000 g for 2 min at room temperature, resuspended in fresh GS‐2 medium and lysed by ultrasonication. The protein concentration was quantified by the Bradford method (Bradford, 1976).
HPLC
The UV‐absorbing hydrolysis products released from PET films in the culture supernatant were analysed similarly as described before (Eberl, et al., 2009; Barth, et al., 2015). A Waters 1525 HPLC system (Waters Cooperation, Milford, CT, USA) equipped with a UV detector (2998 PDA) and an Agilent Eclipse XDB‐C18 column (Agilent Cooperation, Santa Clara, CA, USA) was used with a constant flow rate of 1 ml min−1. The HPLC gradient elution method was used with acetonitrile and phosphate buffer (200 mM, pH 6.0) as the mobile phase. During the process of gradient elution, the proportion of acetonitrile in the mobile phase increased from 10% to 50% within 21 min and then decreased from 50% to 10% within 1 min. The culture supernatant was diluted with the mobile phase to obtain results within the calibration range followed by centrifugation to remove any precipitation. The injection volume was 10 µl. Separated products were detected at a wavelength of 242 nm. Standard curves of TPA were prepared in a concentration range from 0.05 to 1 mM using the TPA stock solution prepared as described above.
Scanning electron microscopy
The surface morphology of PET samples was determined by SEM using a field emission scanning electron microscope (S‐4800; Hitachi, Tokyo, Japan) according to a previously reported method (Hong, et al., 2014). The PET films were extensively washed with distilled water and dried by lyophilization unless stated otherwise.
Conflict of interest
None declared.
Supporting information
Fig. S1. Analysis of the lcc sequences. AEV21261.1_mature is the published LCC sequence omitting the signal peptide (MDGVLWRVRTAALMAALLALAAWALVWASPS‐VEAQ). The sequence was optimized to adapt the codon usage of C. thermocellum. Both sequences encoded the same amino acid sequences except for the change of the first amino acid from Ser to Met
Fig. S2. HPLC analysis of PET hydrolysates degraded by the engineered C. thermocellum whole‐cell biocatalyst. (A), Standard curve for TPA and MHET quantification. (B), HPLC analysis of commercially purchased pure TPA (upper panel) and PET hydrolysates of C. thermocellum DSM1313::pHK‐LCC (lower panel) before and after 14‐day incubation. The peak with the retention time of 2.3 min refers to TPA. The peak with a longer retention time compared to TPA may be the intermediate MHET (Yoshida, et al., 2016, Salvador, et al., 2019).
Fig. S3. SEM analysis of PET films after 3‐day incubation with the C. thermocellum whole‐cell biocatalyst at 60 °C. The PET films were air‐dried without washing. Medium residuals were detected on the surface of the films but no biofilm formation was observed.
Table S1. The concentrations of the released TPA and MHET during PET film degradation shown in Fig. 3B.
Acknowledgements
This work was supported by QIBEBT, Dalian National Laboratory for Clean Energy (DNL), the ‘Transformational Technologies for Clean Energy and Demonstration’, Strategic Priority Research Program of the Chinese Academy of Sciences (grant number XDA 21060201); the National Natural Science Foundation of China (grant number 31570029); QIBEBT and Dalian National Laboratory For Clean Energy (DNL), CAS (Grant number QIBEBT I201905); the Key Technology Research and Development Program of Shandong (grant number 2018GSF116016); and the Major Program of Shandong Provincial Natural Science Foundation (grant number ZR2018ZB0208). The authors R.W. and U.T.B. would like to acknowledge the financial support of the MIX‐UP project received from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 870294.
Microbial Biotechnology (2021) 14(2), 374–385
Funding Information
This work was supported by QIBEBT, Dalian National Laboratory for Clean Energy (DNL), the ‘Transformational Technologies for Clean Energy and Demonstration’, Strategic Priority Research Program of the Chinese Academy of Sciences (grant number XDA 21060201); the National Natural Science Foundation of China (grant number 31570029); QIBEBT and Dalian National Laboratory For Clean Energy (DNL), CAS (Grant number QIBEBT I201905); the Key Technology Research and Development Program of Shandong (grant number 2018GSF116016); and the Major Program of Shandong Provincial Natural Science Foundation (grant number ZR2018ZB0208). The authors R.W. and U.T.B. acknowledge the financial support of the MIX‐UP project received from the European Union’s Horizon 2020 research and innovation programme (grant number 870294).
[Correction added on 08 May 2020 after first online publication: The list of affiliations has been reordered to correct the affiliation details of the first author in this version.]
Contributor Information
Ren Wei, Email: ren.wei@uni-greifswald.de.
Qiu Cui, Email: cuiqiu@qibebt.ac.cn.
Ya‐Jun Liu, Email: liuyj@qibebt.ac.cn.
References
- Argyros, D.A. , Tripathi, S.A. , Barrett, T.F. , Rogers, S.R. , Feinberg, L.F. , Olson, D.G. , et al. (2011) High ethanol titers from cellulose using metabolically engineered thermophilic, anaerobic microbes. Appl Environ Microbiol. 77: 8288–8294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artzi, L. , Bayer, E.A. , and Morais, S. (2017) Cellulosomes: bacterial nanomachines for dismantling plant polysaccharides. Nat Rev Micro 15: 83–95. [DOI] [PubMed] [Google Scholar]
- Austin, H.P. , Allen, M.D. , Donohoe, B.S. , Rorrer, N.A. , Kearns, F.L. , Silveira, R.L. , et al. (2018) Characterization and engineering of a plastic‐degrading aromatic polyesterase. Proc Natl Acad Sci USA 115: E4350–E4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth, M. , Wei, R. , Oeser, T. , Then, J. , Schmidt, J. , Wohlgemuth, F. , and Zimmermann, W. (2015) Enzymatic hydrolysis of polyethylene terephthalate films in an ultrafiltration membrane reactor. J Membr Sci 494: 182–187. [Google Scholar]
- Barth, M. , Honak, A. , Oeser, T. , Wei, R. , Belisário‐Ferrari, M.R. , Then, J. , et al. (2016) A dual enzyme system composed of a polyester hydrolase and a carboxylesterase enhances the biocatalytic degradation of polyethylene terephthalate films. Biotechnol J 11: 1082–1087. [DOI] [PubMed] [Google Scholar]
- Bayer, E.A. , Lamed, R. , White, B.A. , and Flint, H.J. (2008) From cellulosomes to cellulosomics. Chem Rec 8: 364–377. [DOI] [PubMed] [Google Scholar]
- Billig, S. , Oeser, T. , Birkemeyer, C. , and Zimmermann, W. (2010) Hydrolysis of cyclic poly(ethylene terephthalate) trimers by a carboxylesterase from Thermobifida fusca KW3. Appl Microbiol Biotechnol 87: 1753–1764. [DOI] [PubMed] [Google Scholar]
- Boraston, A.B. , Bolam, D.N. , Gilbert, H.J. , and Davies, G.J. (2004) Carbohydrate‐binding modules: fine‐tuning polysaccharide recognition. Biochem J 382: 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornscheuer, U.T. (2016) Feeding on plastic. Science 351: 1154–1155. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- de Carvalho, C.C.C.R. (2017) Whole cell biocatalysts: essential workers from Nature to the industry. Microb Biotechnol 10: 250–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, G.Z. , Hong, W. , Zhang, J. , Li, W.L. , Feng, Y. , Liu, Y.J. , and Cui, Q. (2012) Targeted gene engineering in Clostridium cellulolyticum H10 without methylation. J Microbiol Methods 89: 201–208. [DOI] [PubMed] [Google Scholar]
- Deng, H. , Wei, R. , Luo, W. , Hu, L. , Li, B. , Di, et al. (2019) Microplastic pollution in water and sediment in a textile industrial area. Environ Pollut 258: 113658. [DOI] [PubMed] [Google Scholar]
- Eberl, A. , Heumann, S. , Bruckner, T. , Araujo, R. , Cavaco‐Paulo, A. , Kaufmann, F. , et al. (2009) Enzymatic surface hydrolysis of poly(ethylene terephthalate) and bis(benzoyloxyethyl) terephthalate by lipase and cutinase in the presence of surface active molecules. J Biotechnol 143: 207–212. [DOI] [PubMed] [Google Scholar]
- Ellen MacArthur Foundation . (2016) The new plastics economy: rethinking the future of plastics. Cologny/Geneva, Switzerland: World Economic Forum and McKinsey & Company. [Google Scholar]
- Gamerith, C. , Vastano, M. , Ghorbanpour, S.M. , Zitzenbacher, S. , Ribitsch, D. , Zumstein, M.T. , et al. (2017) Enzymatic degradation of aromatic and aliphatic polyesters by P. pastoris expressed cutinase 1 from Thermobifida cellulosilytica . Front Microbiol 8: 938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer, R. , Jambeck, J.R. , and Law, K.L. (2017) Production, use, and fate of all plastics ever made. Sci Adv 3: e1700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guss, A.M. , Olson, D.G. , Caiazza, N.C. , and Lynd, L.R. (2012) Dcm methylation is detrimental to plasmid transformation in Clostridium thermocellum . Biotechnol Biofuels 5: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, X. , Liu, W. , Huang, J.‐W. , Ma, J. , Zheng, Y. , Ko, T.‐P. , et al. (2017) Structural insight into catalytic mechanism of PET hydrolase. Nat Commun 8: 2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero Acero, E. , Ribitsch, D. , Steinkellner, G. , Gruber, K. , Greimel, K. , Eiteljoerg, I. , et al. (2011) Enzymatic Surface hydrolysis of PET: effect of structural diversity on kinetic properties of cutinases from thermobifida. Macromolecules 44: 4632–4640. [Google Scholar]
- Hong, W. , Zhang, J. , Feng, Y. , Mohr, G. , Lambowitz, A.M. , Cui, G.‐Z. , et al. (2014) The contribution of cellulosomal scaffoldins to cellulose hydrolysis by Clostridium thermocellum analyzed by using thermotargetrons. Biotechnol Biofuels 7: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R. , Feng, Y. , Liu, S. , Qi, K. , Cui, Q. , and Liu, Y.‐J. (2018) Inducing effects of cellulosic hydrolysate components of lignocellulose on cellulosome synthesis in Clostridium thermocellum . Microb Biotechnol 11: 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, P.P. , Mi, L. , Morioka, A.H. , Yoshino, K.M. , Konishi, S. , Xu, S.C. , et al. (2015) Consolidated bioprocessing of cellulose to isobutanol using Clostridium thermocellum . Metab Eng 31: 44–52. [DOI] [PubMed] [Google Scholar]
- Liu, Y.‐J. , Liu, S. , Dong, S. , Li, R. , Feng, Y. , and Cui, Q. (2018) Determination of the native features of the exoglucanase Cel48S from Clostridium thermocellum . Biotechnol Biofuels 11: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Liu, Y.‐J. , Feng, Y. , Li, B. , and Cui, Q. (2019) Construction of consolidated bio‐saccharification biocatalyst and process optimization for highly efficient lignocellulose solubilization. Biotechnol Biofuels 12: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynd, L.R. , van Zyl, W.H. , McBride, J.E. , and Laser, M. (2005) Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotechnol 16: 577–583. [DOI] [PubMed] [Google Scholar]
- Mohr, G. , Hong, W. , Zhang, J. , Cui, G.‐Z. , Yang, Y. , Cui, Q. , et al. (2013) A targetron system for gene targeting in thermophiles and its application in Clostridium thermocellum . PLoS ONE 8: e69032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moog, D. , Schmitt, J. , Senger, J. , Zarzycki, J. , Rexer, K.‐H. , Linne, U. , et al. (2019) Using a marine microalga as a chassis for polyethylene terephthalate (PET) degradation. Microb Cell Fact 18: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, T.K. , Weimer, P.J. , and Zeikus, J.G. (1977) Cellulolytic and physiological properties of Clostridium thermocellum . Arch. Microbiol. 114: 1–7. [DOI] [PubMed] [Google Scholar]
- Olson, D.G. , and Lynd, L.R. (2012) Transformation of Clostridium thermocellum by electroporation. Methods Enzymol 510: 317–330. [DOI] [PubMed] [Google Scholar]
- Olson, D.G. , Maloney, M. , Lanahan, A.A. , Hon, S. , Hauser, L.J. , and Lynd, L.R. (2015) Identifying promoters for gene expression in Clostridium thermocellum . Metab Eng Comm 2: 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm, G.J. , Reisky, L. , Böttcher, D. , Müller, H. , Michels, E.A.P. , Walczak, M.C. , et al. (2019) Structure of the plastic‐degrading Ideonella sakaiensis MHETase bound to a substrate. Nat Commun 10: 1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PlasticsEurope . (2018) Plastics – The Facts 2017. Brussels, Belgium: PlasticsEurope. [Google Scholar]
- Ribitsch, D. , Yebra, A. O. , Zitzenbacher, S. , Wu, J. , Nowitsch, S. , Steinkellner, G. , et al. (2013) Fusion of binding domains to Thermobifida cellulosilytica cutinase to tune sorption characteristics and enhancing PET hydrolysis. Biomacromol 14: 1769–1776. [DOI] [PubMed] [Google Scholar]
- Rochman, C.M. , Browne, M.A. , Halpern, B.S. , Hentschel, B.T. , Hoh, E. , Karapanagioti, H.K. , et al. (2013) Classify plastic waste as hazardous. Nature 494: 169. [DOI] [PubMed] [Google Scholar]
- Ronkvist, Å.M. , Xie, W. , Lu, W. , and Gross, R.A. (2009) Cutinase‐catalyzed hydrolysis of poly(ethylene terephthalate). Macromolecules 42: 5128–5138. [Google Scholar]
- Roth, C. , Wei, R. , Oeser, T. , Then, J. , Föllner, C. , Zimmermann, W. , and Sträter, N. (2014) Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca . Appl Microbiol Biotechnol 98: 7815–7823. [DOI] [PubMed] [Google Scholar]
- Salvador, M. , Abdulmutalib, U. , Gonzalez, J. , Kim, J. , Smith, A.A. , Faulon, J.L. , et al. (2019) Microbial genes for a circular and sustainable Bio‐PET economy, Genes (Basel) 10: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, J. , Wei, R. , Oeser, T. , Belisário‐Ferrari, M.R. , Barth, M. , Then, J. , and Zimmermann, W. (2016) Effect of Tris, MOPS, and phosphate buffers on the hydrolysis of polyethylene terephthalate films by polyester hydrolases. FEBS Open Bio 6: 919–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, W.H. , and Zverlov, V.V. (2006) Protease inhibitors in bacteria: an emerging concept for the regulation of bacterial protein complexes? Mol Microbiol 60: 1323–1326. [DOI] [PubMed] [Google Scholar]
- Shirke, A.N. , White, C. , Englaender, J.A. , Zwarycz, A. , Butterfoss, G.L. , Linhardt, R.J. , and Gross, R.A. (2018) Stabilizing leaf and branch compost cutinase (LCC) with glycosylation: mechanism and effect on PET hydrolysis. Biochemistry 57: 1190–1200. [DOI] [PubMed] [Google Scholar]
- Sulaiman, S. , Yamato, S. , Kanaya, E. , Kim, J.J. , Koga, Y. , Takano, K. , and Kanaya, S. (2012) Isolation of a novel cutinase homolog with polyethylene terephthalate‐degrading activity from leaf‐branch compost by using a metagenomic approach. Appl Environ Microbiol. 78: 1556–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman, S. , You, D.J. , Kanaya, E. , Koga, Y. , and Kanaya, S. (2014) Crystal structure and thermodynamic and kinetic stability of metagenome‐derived LC‐cutinase. Biochemistry 53: 1858–1869. [DOI] [PubMed] [Google Scholar]
- Then, J. , Wei, R. , Oeser, T. , Gerdts, A. , Schmidt, J. , Barth, M. , and Zimmermann, W. (2016) A disulfide bridge in the calcium binding site of a polyester hydrolase increases its thermal stability and activity against polyethylene terephthalate. FEBS Open Bio 6: 425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi, S.A. , Olson, D.G. , Argyros, D.A. , Miller, B.B. , Barrett, T.F. , Murphy, D.M. , et al. (2010) Development of pyrF‐based genetic system for targeted gene deletion in Clostridium thermocellum and creation of a pta mutant. Appl Environ Microbiol 76: 6591–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, H. , Arnott, J. , Crawford, R. , and Ivanova, E. (2013) Plastic degradation and its environmental implications with special reference to poly(ethylene terephthalate). Polymers 5: 1. [Google Scholar]
- Wei, R. , and Zimmermann, W. (2017a) Biocatalysis as a green route for recycling the recalcitrant plastic polyethylene terephthalate. Microb Biotechnol 10: 1302–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, R. , and Zimmermann, W. (2017b) Microbial enzymes for the recycling of recalcitrant petroleum‐based plastics: how far are we? Microb Biotechnol 10: 1308–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, R. , Oeser, T. , Schmidt, J. , Meier, R. , Barth, M. , Then, J. , and Zimmermann, W. (2016) Engineered bacterial polyester hydrolases efficiently degrade polyethylene terephthalate due to relieved product inhibition. Biotechnol Bioeng 113: 1658–1665. [DOI] [PubMed] [Google Scholar]
- Wei, R. , Breite, D. , Song, C. , Gräsing, D. , Ploss, T. , Hille, P. , et al. (2019a) Biocatalytic degradation efficiency of postconsumer polyethylene terephthalate packaging determined by their polymer microstructures. Adv Sci 6: 1900491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, R. , Song, C. , Gräsing, D. , Schneider, T. , Bielytskyi, P. , Böttcher, D. ,et al. (2019b) Conformational fitting of a flexible oligomeric substrate does not explain the enzymatic PET degradation. Nat Commun 10: 5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham, J.M. , Moon, J.‐W. , Rodriguez, M. , Engle, N.L. , Klingeman, D.M. , Rydzak, T. , et al. (2018) Clostridium thermocellum LL1210 pH homeostasis mechanisms informed by transcriptomics and metabolomics. Biotechnol Biofuels 11: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Malten, M. , Grote, A. , Jahn, D. , and Deckwer, W.D. (2007) Codon optimized Thermobifida fusca hydrolase secreted by Bacillus megaterium . Biotechnol Bioeng 96: 780–794. [DOI] [PubMed] [Google Scholar]
- Yoav, S. , Barak, Y. , Shamshoum, M. , Borovok, I. , Lamed, R. , Dassa, B. , et al. (2017) How does cellulosome composition influence deconstruction of lignocellulosic substrates in Clostridium (Ruminiclostridium) thermocellum DSM 1313? Biotechnol Biofuels 10: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S. , Hiraga, K. , Takehana, T. , Taniguchi, I. , Yamaji, H. , Maeda, Y. , et al. (2016) A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351: 1196–1199. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Liu, S. , Li, R. , Hong, W. , Xiao, Y. , Feng, Y. , et al. (2017) Efficient whole‐cell‐catalyzing cellulose saccharification using engineered Clostridium thermocellum . Biotechnol Biofuels 10: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Wang, L. , Halden, R. U. , and Kannan, K. (2019) Polyethylene terephthalate and polycarbonate microplastics in sewage sludge collected from the United States. Environ Sci Technol Lett 6: 650–655. [DOI] [PubMed] [Google Scholar]
- Zverlov, V.V. , Kellermann, J. , and Schwarz, W.H. (2005) Functional subgenomics of Clostridium thermocellum cellulosomal genes: identification of the major catalytic components in the extracellular complex and detection of three new enzymes. Proteomics 5: 3646–3653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Analysis of the lcc sequences. AEV21261.1_mature is the published LCC sequence omitting the signal peptide (MDGVLWRVRTAALMAALLALAAWALVWASPS‐VEAQ). The sequence was optimized to adapt the codon usage of C. thermocellum. Both sequences encoded the same amino acid sequences except for the change of the first amino acid from Ser to Met
Fig. S2. HPLC analysis of PET hydrolysates degraded by the engineered C. thermocellum whole‐cell biocatalyst. (A), Standard curve for TPA and MHET quantification. (B), HPLC analysis of commercially purchased pure TPA (upper panel) and PET hydrolysates of C. thermocellum DSM1313::pHK‐LCC (lower panel) before and after 14‐day incubation. The peak with the retention time of 2.3 min refers to TPA. The peak with a longer retention time compared to TPA may be the intermediate MHET (Yoshida, et al., 2016, Salvador, et al., 2019).
Fig. S3. SEM analysis of PET films after 3‐day incubation with the C. thermocellum whole‐cell biocatalyst at 60 °C. The PET films were air‐dried without washing. Medium residuals were detected on the surface of the films but no biofilm formation was observed.
Table S1. The concentrations of the released TPA and MHET during PET film degradation shown in Fig. 3B.


