Since timely determination of nvCT is critical for restricting further infection spread in the population, we have developed a screening assay based on high‐resolution melting (HRM) that can detect C. trachomatis and identify nvCT. Our results showed that the HRM scanning assay is not only able to directly identify C. trachomatis in clinical specimens, but also to correctly differentiate nvCT with mutation C1514T, C1515T and G1523A from the wild‐type.

Summary
Chlamydia trachomatis is the most common sexually transmitted pathogen globally, causing serious health problems and representing a burden on public health. A new variant of C. trachomatis (nvCT) that carries mutations (C1514T, C1515T and G1523A) in the 23S rRNA gene has eluded detection in Aptima Combo 2 assays. This has led to false negatives in diagnostics tests and poses a challenge for C. trachomatis diagnostics on a global level. In this study, we developed a simple and cost‐effective assay to identify C. trachomatis, with a potential application to screen for nvCT. We developed a screening assay based on high‐resolution melting (HRM), targeting the 23S rRNA gene and cryptic plasmid. To evaluate the performance of the assay, 404 archived C. trachomatis DNA specimens and 570 extracted clinical specimens were analysed. Our HRM assay not only identified C. trachomatis in clinical specimens, but also correctly differentiated nvCT carrying C1514T, C1515T and G1523A mutations from the wild‐type. We observed no cross‐reactions with other clinically related agents, and the limit of detection was 11.26 (95% CI; 7.61–31.82) copies per reaction. Implementation of this screening assay could reduce detection times and costs for C. trachomatis diagnoses, and facilitate increased research on the presence and monitoring of nvCT.
Introduction
Worldwide, Chlamydia trachomatis is one of the most prevalent sexually infectious aetiological agents (Menon et al., 2015; Aung et al., 2019; Chi Wai Wong et al., 2019), causing approximately 131 million new infections each year (https://www.who.int/reproductivehealth/publications/stis‐surveillance‐2018/en/). C. trachomatis infections not only can lead to severe reproductive sequelae, including ectopic pregnancy, pelvic inflammatory disease and infertility, but also can facilitate the transmission of human immunodeficiency virus (Xiu et al., 2019; Durukan et al., 2020). However, in some populations, C. trachomatis infections are often asymptomatic, causing significant challenges (underestimations) for detection and epidemiological characterization.
Routine screening and accurate diagnosis for C. trachomatis infections are mainstays for deciding appropriate treatments and interrupting transmission. The current diagnostic algorithms for C. trachomatis are based on nucleic acid amplification tests (NAATs), which are highly sensitive and specific (Turingan et al., 2017). Currently, NAATs come in the form of regulatory‐approved commercially available molecular kits, including Hologic Aptima Combo 2 assay (AC2), Becton Dickinson Molecular Diagnostics BD Max CT/GC/TV, Roche Cobas CT/NG Test, Abbott RealTime CT/NG assay and Cepheid GeneXpert Xpert CT/NG (Hokynar et al., 2019). However, in recent years, continued false‐negative results have been reported during C. trachomatis diagnostics, suggesting the occurrence of intrinsic mutation or recombination events (Turingan et al., 2017), thus complicating diagnoses. One possible cause for the higher number of diagnostic discrepancies in the NAATs results for C. trachomatis may be due to genetic diversity for diagnostic methods with a single target. In 2006, the Swedish new variant of C. trachomatis (S‐nvCT) with a plasmid deletion (Ripa and Nilsson, 2006) was reported to have escaped detection in several commercial C. trachomatis diagnostic kits, including Cobas Amplicor/TaqMan48 (Roche Diagnostics, Basel, Switzerland) and the Abbott m2000 (Abbott Laboratories, Abbott Park, IL, USA; Herrmann et al., 2008). Consequently, the S‐nvCT resulted in thousands of false‐negative test results, resulting in an increase in sexually transmitted diseases across several Nordic countries (Unemo and Clarke, 2011). However, the ongoing evolution of C. trachomatis under the high diagnostic selective pressure (Unemo et al., 2019a) is still occurring.
The Finnish new variant of C. trachomatis (F‐nvCT) was first reported in Finland in mid‐February 2019 (Rantakokko‐Jalava et al., 2019) and contained a C1515T mutation in its 23S rRNA gene. Notably, this mutation was located in the region originally utilized by the NAATs Aptima Combo 2 (AC2) (Hologic Inc., San Diego, CA, USA) to detect C. trachomatis, which were used in many countries worldwide. This new variant of C. trachomatis (nvCT) escaping detection in the AC2 assay has subsequently been detected in other European countries such as Sweden, England, Norway and Denmark (Johansen et al., 2019; Roberts et al., 2019; Unemo et al., 2019b; Hadad et al., 2020). Two additional C. trachomatis variants harbouring C1514T and G1523A mutations in the 23S rRNA target region, also escaped AC2 detection in England, Norway and Denmark (Johansen et al., 2019; Roberts et al., 2019; Hadad et al., 2020). False‐negative results due to the continuously emerging mutants (Unemo et al., 2007; Clarke, 2011) have increased the risk of unrecognized transmission. Timely determination and surveillance of the nvCT mutants are, therefore, crucial for our understanding of presence and monitoring of the spread of the C. trachomatis variants (Johansen et al., 2019; Rantakokko‐Jalava et al., 2019; Roberts et al., 2019; Unemo et al., 2019a,b). Here, we report a new molecular screening assay suitable for the identification C. trachomatis and surveillance of these new variants escaping detection by AC2.
Results
The molecular screening assay
After HRM analysis and standardization, melting curve clusters for different targets were generated. A cluster analysis of melting profiles was used to differentiate targets into two distinct clusters (Fig. 1). These two distinct clusters were used to identify and confirm the presence of C. trachomatis in clinical samples. Furthermore, in the 23S rRNA target region, the HRM assay successfully revealed four different cluster formations for the wide‐type and three different point mutations, involving AC2 diagnostic‐eluding nvCTs, C1514T, C1515T and G1523A. Twenty‐eight wild‐type C. trachomatis and three control plasmids harbouring individual mutations (those escaping detection by the AC2 assay) were used to develop and evaluate this HRM assay. Our assay yielded two clusters for the two targets (23S rRNA and the cryptic plasmid) from all 28 well‐characterized C. trachomatis samples. Our data showed that the HRM assay accurately identified C. trachomatis, demonstrating a high specificity for the primers used in this study. Our optimization data also revealed that HRM analysis correctly differentiated C1514T, C1515T and G1523A mutations from the wide‐type. However, plasmids containing C1515T or G1523A mutations were not distinguished from each other due to their similar melting temperatures (Tm) values (Table 1).
Fig. 1.
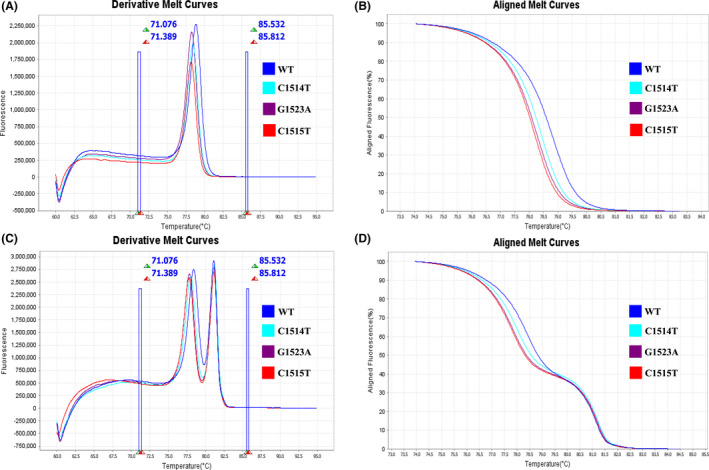
The results of singleplex and multiplex HRM assay. (A) Derivative melt curves of the singleplex HRM assay. (B) Aligned melt curves of the singleplex HRM assay. (C) Derivative melt curves of the multiplex HRM assay. (D) Aligned melt curves of the multiplex HRM assay.
Table 1.
Summary of Tm values for each of the organisms used in the HRM assay.
| Organisms | Tm of target 23S rRNA | Tm of target cryptic plasmid | ||
|---|---|---|---|---|
| Range | Mean ± SD | Range | Mean ± SD | |
| C. trachomatis (WT, n = 28) | 78.40–78.48 | 78.43 ± 0.03 | 81.14–81.16 | 81.14 ± 0.01 |
| Plasmid (C1514T mutation, n = 1) | 78.00–78.06 | 78.02 ± 0.02 | 81.19–81.25 | 81.21 ± 0.02 |
| Plasmid (C1515T mutation, n = 1) | 77.84–77.94 | 77.90 ± 0.04 | 81.17–81.25 | 81.22 ± 0.04 |
| Plasmid (C1523A mutation, n = 1) | 77.84–77.86 | 77.85 ± 0.01 | 81.17–81.20 | 81.19 ± 0.01 |
| Herpes simplex virus types 1 (n = 2) | NA | NA | NA | NA |
| Herpes simplex virus types 2 (n = 2) | NA | NA | NA | NA |
| Neisseria gonorrhoeae (n = 2) | NA | NA | NA | NA |
| Chlamydia pneumonia (n = 2) | 77.69–77.70 | 77.69 ± 0.01 | NA | NA |
| Treponema pallidum (n = 2) | NA | NA | NA | NA |
| Trichomonas vaginalis (n = 2) | NA | NA | NA | NA |
| Mycoplasma hominis (n = 2) | NA | NA | NA | NA |
| Ureaplasma urealyticum (n = 2) | NA | NA | NA | NA |
| Mycoplasma genitalium (n = 2) | NA | NA | NA | NA |
| Ureaplasma parvum (n = 2) | NA | NA | NA | NA |
NA, no amplification was observed; WT, wild‐type.
To assess the potential of the HRM assay to cross‐react with other genetically or clinically related agents, we conducted a specificity test using microbial species DNA extracts from 20 confirmed clinical samples and isolates. No specific melting profiles, consistent with both reference controls, were observed for all organisms across all sample types (Table 1). The average Tm value for each DNA used in the HRM assay is summarized in Table 1.
To investigate the limit of detection (LOD) of our HRM assay, a sensitivity test was performed by testing a dilution series using plasmids containing two target regions at the following concentrations: 200, 100, 50, 20, 10, 5 and 1 gene copy per reaction mixture. Each concentration was tested in 12 replicates, and Probit analysis of the results was carried out using SPSS statistical software package version 17.0 (SPSS Inc., Chicago, IL, USA). Our Probit analysis data revealed a LOD of 11.26 (95% CI, 8.90–19.87) and 9.46 (95% CI, 7.58–14.57) copies per reaction for the C. trachomatis target 23S rRNA gene and the cryptic plasmid at 95% confidence, respectively (Table 2).
Table 2.
Limit of detection of each target gene using regression probit analysis.
| Target | No. of replicates detected at each dilution/total no. of replicated at indicated no. of copies/reaction (% positive rate) | Final LOD, no. of copies/reaction | ||||||
|---|---|---|---|---|---|---|---|---|
| 200 | 100 | 50 | 20 | 10 | 5 | 1 | ||
| 23S rRNA | 12/12 (100) | 12/12 (100) | 12/12 (100) | 12/12 (100) | 10/12 (83.3) | 7/12 (58.3) | 0/12 (0) | 11.26 (95% CI, 8.80–19.87) |
| Cryptic plasmid | 12/12 (100) | 12/12 (100) | 12/12 (100) | 12/12 (100) | 11/12 (91.7) | 8/12 (66.7) | 0/12 (0) | 9.46 (95% CI, 7.58–14.57) |
Evaluation of the performance on clinical samples
A total of 570 DNA samples were first analysed by the HRM assay for the validation of the ability to directly identify C. trachomatis in clinical specimens. Among 570 tested samples, 36 showed similar profiles and aligned with melting curves for both targets (i.e. 23S rRNA and the cryptic plasmid). No melting curves were observed for the remaining negative C. trachomatis specimens (Fig. 2). The test performance characteristics of HRM assay for the detection of C. trachomatis were compared with our reference STI‐MS assay (Table 3). The overall per cent agreement between assays was 98.8% (95% confidence interval [CI], 97.4 to 99.5%), positive per cent agreement was 87.2% (95% CI, 71.8 to 95.2%), and negative per cent agreement was 99.6 (95% CI, 98.5 to 99.9%). When compared with the STI‐MS assay, the HRM assay showed excellent overall and negative agreement. For those positive specimens identified by the developed HRM assay, the results were confirmed using an in house real‐time PCR assay (Mahony et al., 1997). The HRM results for the 36 positive samples were 100% concordant with the results obtained by real‐time PCR. Therefore, our results showed that this HRM assay may be used for the detection and identification of C. trachomatis in clinical samples.
Fig. 2.
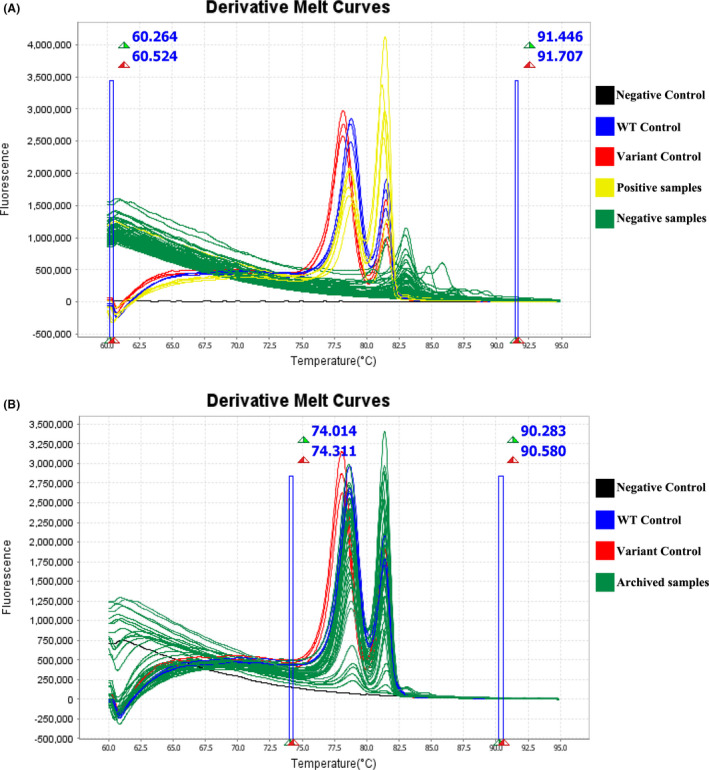
The HRM scanning assay for detecting the nvCT in tested samples. (A) The results of clinical samples. (B) The results of DNA specimens with C. trachomatis positive.
Table 3.
Clinical performance comparison of HRM and STI‐MS assay for the detection of Chlamydia trachomatis.
| HRM assay | STI‐MS assay | % agreement (95% CI) between assays | ||||
|---|---|---|---|---|---|---|
| Positive | Negative | Total | Positive | Negative | Overall | |
| Positive | 34 | 2 | 36 | 87.2 (71.8–95.2) | 99.6 (98.5–99.9) | 98.8 (97.4–99.5) |
| Negative | 5 | 529 | 534 | |||
| Total | 39 | 531 | 570 | |||
Screening samples for the presence of nvCT
To investigate the possible prevalence and spread of nvCT in China, we screened 404 archived DNA specimens that had previously tested positive for C. trachomatis using the Roche Cobas® 4800 CT/NG test. All samples were amplified and resulted in corresponding melting curves, meaning they were positive for C. trachomatis by HRM assay. The nvCT was not identified in any of the samples (Table 4). All tested archived DNA specimens were assigned as wide‐type of C. trachomatis by the HRM assay (Fig. 2). Subsequent DNA sequencing analyses (Hokynar et al., 2019) from 364 specimens were confirmed as wide‐type C. trachomatis. The remaining 40 samples were found to be positive by HRM assay, but their sequences of the 23S rRNA gene were not acquired. We speculate these discrepancies may have been caused by the lower LOD of the HRM assay.
Table 4.
Previous studies and the current study on screening for the nvCT escaping detection in the AC2assay.
| Country | Year a | Type of tested samples | No. retested samples | No. the th nvCT | Mutations in the 23S rRNA gene | Reference |
|---|---|---|---|---|---|---|
| Finland | 2019 | AC2‐negative or equivocal ACT‐positive samples | 180 | 10 | C1515T (n = 10) | Rantakokko‐Jalava et al. (2019) |
| Sweden | 2019 | AC2‐negative or equivocal samples | 70 | 2 | C1515T (n = 2) | Unemo et al. (2019) |
| England | 2019 | AC2/ACT discrepant specimens | 266 | 2 |
C1514T (n = 1) G1523A (n = 1) |
Roberts et al. (2019) |
| Finland | 2018–2019 | AC2‐negative or equivocal ACT‐positive samples | >200 | 39 | C1515T (n = 39) | Hokynar et al. (2019) |
| Norway | 2019 | AC2/ACT discordant samples | 97 | 81 |
C1514T (n = 1) C1515T (n = 79) G1523A (n = 1) |
Johansen et al. (2019) |
| Denmark | 2019 | AC2‐negative or equivocal ACT‐positive samples | 150 | 80 |
C1514T (n = 2) C1515T (n = 2) G1523A (n = 76) |
Hadad et al. (2020) |
| China | 2018 | CT positive samples by Roche Cobas® 4800 CT/NG | 404 | 0 | ND | This study |
AC2, Aptima Combo 2; ACT, Aptima Chlamydia trachomatis; nvCT, AC2 new variant of Chlamydia trachomatis; ND, not detected.
Collection date of retest samples.
Discussion
In the past decade, several PCR‐based molecular diagnostics techniques have been successfully developed and widely applied for the detection of C. trachomatis (Meyer, 2016; Feodorova et al., 2019; Hokynar et al., 2019). However, some of these assays use only one target region for detection. Lessons from the Swedish nvCT (Ripa and Nilsson, 2006; Moller et al., 2010; Unemo et al., 2010; Unemo and Clarke, 2011) and the nvCT discovered in 2019 (Hokynar et al., 2019; Johansen et al., 2019; Rantakokko‐Jalava et al., 2019; Roberts et al., 2019; Unemo et al., 2019a,b; Hadad et al., 2020) have suggested that false‐negative results were generated due to genetic diversity in diagnostic methods using a single target. These findings suggest that the detection of more than one gene from C. trachomatis may be necessary to avoid false‐negative results due to mutation within the target sequences. The implementation of a dual‐target strategy represents an important improvement in molecular screening methods, ensuring the detection of new variants with mutations in one of the target regions (Meyer, 2016). To avoid such false‐negative results, therefore, we used two targets from C. trachomatis to increase the sensitivity and specificity. Previous studies (Jalal et al., 2011; Turingan et al., 2017) reported that the organism load of C. trachomatis in genital swabs ranged from fewer than 10 to more than 106. Our results revealed that our assay was highly sensitive in detecting C. trachomatis to approximately 11.26 (95% CI: 7.61–31.82) copies per reaction. Furthermore, we observed no cross‐reactions during pathogenic microbial species testing. These indicated that the assay had good specificity for detection of the C. trachomatis directly in the clinical specimens.
The mutational events in specific target binding sequences of C. trachomatis can raise the possibility of false‐negative results, which led to increasing the spread of nvCT, especially in high‐frequency transmitting populations (Unemo and Clarke, 2011). Though an updated AC2 assay containing dual‐target detection that detects all known nvCT strains have been FDA‐cleared in the United States and clinically validated (Unemo et al., 2020) and CE‐Marked in Europe, knowledge regarding the spread of the nvCT is relatively limited (Unemo et al., 2019a) because the majority of laboratories are still not able to identify these nvCTs. Enhanced and continued surveillance based on screening C. trachomatis samples (negative results using the AC2 platform and positive results using other detection platforms based on targets without 23S rRNA) by this assay will be very useful in addressing these concerns. On the one hand, our HRM assay reduces the possibility that mutation or recombination results in false‐negative results. On the other hand, this screening assay using HRM analysis allows for the precise assessment of sequences and can be used to identify variants with single point mutations without the need for sequencing, thereby significantly reducing turnaround time (Bernal‐Martinez et al., 2017). Since no clinical samples or nvCT isolates were available, for this study, plasmids containing individual mutations within the 23S rRNA target region for AC2 were constructed as positive controls. Although this is a possible limitation in the development of this assay, it is important to note that the application of this method for nvCT detection would not be affected. A further limitation of this molecular screening assay, which is common to other HRM‐based molecular methods, was that targets with the same GC content were difficult to distinguish. Indeed, we did not correctly distinguish between the C1515T and G1523A mutations. However, the HRM assay correctly differentiated the C1514T, C1515T and G1523A mutations from the wide‐type. As high sequence homology among the 23S rRNA genes is found in all Chlamydia species, a similar melting profile for target 23S rRNA was observed for the C. pneumoniae and C. trachomatis in tested strains. This further illustrates the importance of including dual‐target detection methods for C. trachomatis in the investigation of Chlamydia cases with false‐positive results in PCR targeted exclusively at the C. trachomatis. Even so, the emergence of variants lacking the entire 7.5 kb chlamydial cryptic plasmid (Magbanua et al., 2007; Sweeney et al., 2019) that eludes assay detection may occur. However, the two human species of Chlamydia have a different mode of transmission and that C. pneumoniae is not sexually transmitted. Therefore, the overall utility of this assay for C. trachomatis and nvCT detection is not affected.
Although the nvCT was not been found outside the Europe countries, its potentially wide geographical distribution cannot be excluded (Johansen et al., 2019; Rantakokko‐Jalava et al., 2019; Unemo et al., 2019a). To confirm this, the retrospective analysis of AC2 negative samples all over the world needs to be conducted (Chi Wai Wong et al., 2019; Johansen et al., 2019; Xiu et al., 2019). In addition, data from this small‐scale screening program have shown that this nvCT was not present in our samples. To further investigate whether this nvCT was present and circulating in China, a large‐scale laboratory‐based surveillance program must be incepted. By means of the herein described protocol, it is possible to obtain the results within two hours in less than $ 0.50 for each sample. Therefore, our HRM approach may represent a useful laboratory tool for the identification of C. trachomatis in clinical specimens, with the potential ability to provide clues for distinguishing nvCT from wild‐type. Implementation of such a screening strategy could reduce the time and cost to effectively detect C. trachomatis, thus facilitating a greater understanding of nvCT spread in populations.
Conclusion
The HRM scanning assay is not only able to directly identify C. trachomatis in clinical specimens, but also suitable for distinguishing nvCTs (three different mutations in the 23S rRNA gene) from the wild‐type. Our small‐scale screening study showed that nvCTs were not identified in our sample collection. Thus, further large‐scale investigations must determine whether nvCTs are present and circulating in China.
Experimental procedures
Overview
This molecular screening assay was developed using high‐resolution melting (HRM) analysis technology. HRM is a simple, reliable and cost‐effective method that distinguishes wild‐type and mutant variants by the presence of specific DNA targets (Montgomery et al., 2010; Tong and Giffard, 2012). To avoid false‐negative results, the HRM assay targets two genes (23S rRNA and cryptic plasmid) from C. trachomatis to increase the sensitivity and specificity. The 23S rRNA target was used to identify C. trachomatis and distinguish the nvCT from the wild‐type. The presence of C. trachomatis was then further confirmed by the other target, the cryptic plasmid.
Study samples
The performance of this assay was initially assessed using C. trachomatis samples (n = 28) confirmed using the STI‐MS assay (Xiu et al., 2019; STI‐MS is a detection method combining multiplex‐PCR with MALDI‐TOF analysis). For these 28 samples, the 23S rRNA gene was previously characterized with respect to the wild‐type, using Sanger sequencing. Three control plasmids, bearing individual mutations within the 23S rRNA target region for AC2, were used to develop and evaluate the HRM assay. These recombinant plasmids were directly constructed by Tsingke Biotechnology (Beijing, China). The plasmids bearing the 23S rRNA C1514T, C1515T and G1523A mutations were confirmed by sequencing. Our assay was also validated using 570 urethral/genital swabs, from which DNA was extracted using the MagNA Pure LC DNA Isolation Kit 1 (Roche Diagnostics, Mannheim, Germany) and tested for sexually transmitted infections (STI) agents using the STI‐MS method (Xiu et al., 2019). Moreover, a total of 404 archived DNA specimens extracted from first void urine (FVU) and previously confirmed as positive for C. trachomatis using the Roche Cobas® 4800 CT/NG test (Roche Diagnostics, Indianapolis, IN, USA) were also screened using the HRM assay. These clinical samples were collected from clinical laboratories of different hospitals in the Shenzhen area between 2015 and 2018, obtained as part of the Shenzhen Gonococcal and Chlamydial Intervention Programme (SGCIP). A majority of the patients recruited to the study presented with urinary symptoms or for an asymptomatic sexual health screen and had a higher risk of sexually transmitted infections. No information regarding the sexual orientation of the patient or sexual contacts were available. All study participants older than 18 years who were normally resident in the Shenzhen.
Assay design and reaction conditions
Primers for the two targets (23S rRNA and cryptic plasmid) used in this study were designed by the software package Beacon Designer 7.0 (PremierBiosoft, Palo Alto, USA). All primers were synthesized by Tsingke Biotechnology (Beijing, China). Six specific primer sets were designed on a relatively conserved region of the cryptic plasmid. Six candidate primer pairs were selected to amplify the 23S rRNA region, where the nvCT hotspot mutations were located. The optimum primer pairs were selected using the Primer‐BLAST (https://www.ncbi.nlm.nih.gov/tools/primer‐blast/). The single HRM assay was initially developed and performed using the QuantStudio™ 6 Flex Real‐Time PCR System (Applied Biosciences, Foster City, CA, USA). HRM assay reactions were carried out in a final volume of 20 μl, consisting of 10 μl EvaGreen Master Mix (Biotium, Hayward, CA, USA), 1 μl of each primer and 2 μl of the DNA template. Based on this established single HRM assay, a series of experiments were performed to optimize a multiplex HRM assay, including primer combinations, reagent concentrations and cycling parameters. For multiplex HRM analyses, reaction mixtures containing 10 μl EvaGreen Master, 0.5 μl each corresponding primer to 23S rRNA (HRM‐CT‐23S_F: 5’‐AGTTAAGCACGCGGACGATT‐3’ and HRM‐CT‐23S_R: 5’‐ GCGGATTTGCCTACTAACCG‐3’) and cryptic plasmid (HRM‐CT‐cryP_F: 5’‐CCGGCGGCGGGCCAGCACTCCAATTTCTGAC‐3’ and HRM‐CT‐cryP_R: 5’‐ CGGCGGCCGCCCTCGATGATTTGAGCGTGT‐3’), and 2 μl of bacterial genomic DNA, or extracted DNA from clinical sample. The final reaction volume was adjusted to 20 μl with sterile water. The bases, forming the extra GC‐rich tail were added to adjust and distinguish the melting temperature on the 5’ end of cryptic plasmid primers, are underlined. Cycling conditions consisted of an initial denaturation at 95°C for 10 min, followed by 35 cycles at 95 °C for 15 s and 60 °C for 1 min. The melting analysis for the HRM assay was the same as described previously (Xiu et al., 2020): an initial holding step for 1 min at 60°C, followed by a slow temperature increase at a rate of 0.025°C/s to 95°C with continuous fluorescence signal collection. Melting curves were automatically generated by the QuantStudio 6 and 7 Flex Real‐Time PCR software v1.0 (Applied Biosciences, Foster City, CA, USA) and melting temperature (Tm) values were obtained. Finally, using the Melt Curves (Derivative and/or Aligned Melt Curves), differences in melting curve shapes were analysed by comparing each amplicon curve with the control. This further enabled the clustering of samples into different groups.
Sanger sequencing
The V2 region of the 23S rRNA gene was amplified using conventional PCR methods, as described previously (Hokynar et al., 2019). Sanger sequencing was accomplished at Tsingke Biotechnology (Beijing, China). To confirm the nvCT, the obtained sequences were compared with the 23S rRNA sequences of C. trachomatis E/Bour strain (GenBank accession no. HE601870) and three previously described AC2 diagnostic‐escape nvCTs using the ClustalW algorithms implemented in BioEdit software version 7.0.9 (Ibis Bioscience, Inc., USA).
Statistical analysis
Statistical analyses of the HRM assay’s limit of detection (LOD) were performed using Probit analysis, 95 % confidence interval (CI) was calculated using logistic regression model in SPSS statistical software package version 17.0 (SPSS Inc., Chicago, IL, USA). Overall per cent agreement, positive per cent agreement, negative per cent agreement and associated 95% CI were also calculated.
Conflict of interest
The authors declare no conflicts of interest.
Author contributions
JP, FW and LX were involved in the concept of the study. JP and FW contributed to the study design and formulating the research question. LX performed the experiments, analysed the results and wrote the manuscript. YL conducted the experiments and assisted in writing the manuscript. CZ, YL and YZ conducted the experiments and analysed the results. All authors reviewed and revised the final version of manuscript. All authors approve the work for publication and agree to be accountable for the work.
Ethical approval
All experiments were performed according to the ethical standards of the national research committee and approved by the Institutional Review Boards of the Institute of Pathogen Biology. All samples were obtained under approved ethical protocols and with informed consent from each patient.
Acknowledgements
We would like to thank all members of the Shenzhen Gonococcal and Chlamydial Intervention Programme (SGCIP) for their field support and involvement in sample collection.
Microb. Biotechnol. (2021) 14(2), 668–676
Funding informationThis study was supported by grants from the CAMS Innovation Fund for Medical Sciences (2016‐I2M‐3‐021) and Non‐profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2019PT310029).
Contributor Information
Feng Wang, Email: biowangfeng@163.com.
Junping Peng, Email: pengjp@hotmail.com.
References
- Aung, E.T. , Chow, E.P. , Fairley, C.K. , Hocking, J.S. , Bradshaw, C.S. , Williamson, D.A. , and Chen, M.Y. (2019) International travel as risk factor for Chlamydia trachomatis infections among young heterosexuals attending a sexual health clinic in Melbourne, Australia, 2007 to 2017. Euro Surveill 24: 1900219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal‐Martinez, L. , Gil, H. , Rivero‐Menendez, O. , Gago, S. , Cuenca‐Estrella, M. , Mellado, E. , and Alastruey‐Izquierdo, A. (2017) Development and validation of a high‐resolution melting assay to detect azole resistance in Aspergillus fumigatus . Antimicrob Agents Chemother 61: e01083‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Wai Wong, W. , Tsz Hei Lau, S. , Pui Hang Choi, E. , Tucker, J.D. , Fairley, C.K. , and Saunders, J.M. (2019) A systematic literature review of reviews on the effectiveness of chlamydia screening. Epidemiol Rev 41: 168–175. [DOI] [PubMed] [Google Scholar]
- Clarke, I.N. (2011) Evolution of Chlamydia trachomatis . Ann N Y Acad Sci 1230: E11–18. [DOI] [PubMed] [Google Scholar]
- Durukan, D. , Read, T.R.H. , Bradshaw, C.S. , Fairley, C.K. , Williamson, D.A. , De Petra, V. , et al. (2020) Pooling pharyngeal, anorectal, and urogenital samples for screening asymptomatic men who have sex with men for Chlamydia trachomatis and Neisseria gonorrhoeae . J Clin Microbiol 58: e01969‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feodorova, V.A. , Zaitsev, S.S. , Saltykov, Y.V. , Sultanakhmedov, E.S. , Bakulev, A.L. , Ulyanov, S.S. , and Motin, V.L. (2019) An asymptomatic patient with fatal infertility carried a Swedish strain of Chlamydia trachomatis with additional deletion in the plasmid orf1 that belonged to A different MLST sequence type. Microorganisms 7: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadad, R. , Jensen, J.S. , Westh, H. , Gronbaek, I. , Schwartz, L.J. , Nielsen, L. , et al. (2020) A Chlamydia trachomatis 23S rRNA G1523A variant escaping detection in the Aptima Combo 2 assay (Hologic) was widespread across Denmark in July‐September 2019. APMIS 128: 440–444. [DOI] [PubMed] [Google Scholar]
- Herrmann, B. , Torner, A. , Low, N. , Klint, M. , Nilsson, A. , Velicko, I. , et al. (2008) Emergence and spread of Chlamydia trachomatis variant, Sweden. Emerg Infect Dis 14: 1462–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokynar, Kati , Rantakokko‐Jalava, Kaisu , Hakanen, Antti , Havana, Marika , Mannonen, Laura , Jokela, Pia , et al. (2019) The Finnish new variant of Chlamydia trachomatis with a single nucleotide polymorphism in the 23S rRNA target escapes detection by the Aptima Combo 2 test. Microorganisms 7: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalal, H. , Verlander, N.Q. , Kumar, N. , Bentley, N. , Carne, C. , and Sonnex, C. (2011) Genital chlamydial infection: association between clinical features, organism genotype and load. J Med Microbiol 60: 881–888. [DOI] [PubMed] [Google Scholar]
- Johansen, T.B. , Kløvstad, H. , Rykkvin, R. , Herrfurth‐Erichsen, E.B. , Sorthe, J. , Njølstad, G. , et al. (2019) The 'Finnish new variant of Chlamydia trachomatis' escaping detection in the Aptima Combo 2 assay is widespread across Norway, June to August 2019. Euro Surveill 24: 1900592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magbanua, J.P.V. , Goh, B.T. , Michel, C.‐E. , Aguirre‐Andreasen, A. , Alexander, S. , Ushiro‐Lumb, I. , et al. (2007) Chlamydia trachomatis variant not detected by plasmid based nucleic acid amplification tests: molecular characterisation and failure of single dose azithromycin. Sex Transm Infect 83: 339–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony, J.B. , Jang, D. , Chong, S. , Luinstra, K. , Sellors, J. , Tyndall, M. , and Chernesky, M. (1997) Detection of Chlamydia trachomatis, Neisseria gonorrhoeae, Ureaplasma urealyticum, and Mycoplasma genitalium in first‐void urine specimens by multiplex polymerase chain reaction. Mol Diagn 2: 161–168. [DOI] [PubMed] [Google Scholar]
- Menon, S. , Timms, P. , Allan, J. a , Alexander, K. , Rombauts, L. , Horner, P. , et al. (2015) Human and pathogen factors associated with Chlamydia trachomatis‐related infertility in women. Clin Microbiol Rev 28: 969–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, T. (2016) Diagnostic procedures to detect Chlamydia trachomatis infections. Microorganisms 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller, J.K. , Pedersen, L.N. , and Persson, K. (2010) Comparison of the Abbott RealTime CT new formulation assay with two other commercial assays for detection of wild‐type and new variant strains of Chlamydia trachomatis . J Clin Microbiol 48: 440–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, J.L. , Sanford, L.N. , and Wittwer, C.T. (2010) High‐resolution DNA melting analysis in clinical research and diagnostics. Expert Rev Mol Diagn 10: 219–240. [DOI] [PubMed] [Google Scholar]
- Rantakokko‐Jalava, Kaisu , Hokynar, Kati , Hieta, Niina , Keskitalo, Anniina , Jokela, Pia , Muotiala, Anna , et al. (2019) Chlamydia trachomatis samples testing falsely negative in the Aptima Combo 2 test in Finland, 2019. Euro Surveill 24: 1900298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripa, T. , and Nilsson, P. (2006) A variant of Chlamydia trachomatis with deletion in cryptic plasmid: implications for use of PCR diagnostic tests. Euro Surveill 11: E061109.2. [DOI] [PubMed] [Google Scholar]
- Roberts, D.J. , Davis, G.S. , Cole, M.J. , Naik, D. , Maru, H. , Woodford, N. , et al. (2019) Prevalence of new variants of Chlamydia trachomatis escaping detection by the Aptima Combo 2 assay, England, June to August 2019. Euro Surveill 24: 1900557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney, E.L. , Bletchly, C. , Gupta, R. , and Whiley, D.M. (2019) False‐negative Chlamydia polymerase chain reaction result caused by a cryptic plasmid‐deficient Chlamydia trachomatis strain in Australia. Sex Health 16: 394–396. [DOI] [PubMed] [Google Scholar]
- Tong, S.Y. , and Giffard, P.M. (2012) Microbiological applications of high‐resolution melting analysis. J Clin Microbiol 50: 3418–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turingan, Rosemary S. , Kaplun, Ludmila , Krautz‐Peterson, Greice , Norsworthy, Sarah , Zolotova, Anna , Joseph, Sandeep J. , et al. (2017) Rapid detection and strain typing of Chlamydia trachomatis using a highly multiplexed microfluidic PCR assay. PLoS One 12: e0178653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo, M. , and Clarke, I.N. (2011) The Swedish new variant of Chlamydia trachomatis . Curr Opin Infect Dis 24: 62–69. [DOI] [PubMed] [Google Scholar]
- Unemo, M. , Getman, D. , Hadad, R. , Cole, M. , Thomson, N. , Puolakkainen, M. , and Spiteri, G. (2019a) Letter to the editor: Chlamydia trachomatis samples testing falsely negative in the Aptima Combo 2 test in Finland, 2019. Euro Surveill 24: 1900354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo, M. , Hansen, M. , Hadad, R. , Lindroth, Y. , Fredlund, H. , Puolakkainen, M. , and Sundqvist, M. (2019b) Finnish new variant of Chlamydia trachomatis escaping detection in the Aptima Combo 2 assay also present in Orebro County, Sweden, May 2019. Euro Surveill 24: 1900370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo, M. , Hansen, M. , Hadad, R. , Puolakkainen, M. , Westh, H. , Rantakokko‐Jalava, K. , et al. (2020) Sensitivity, specificity, inclusivity and exclusivity of the updated Aptima Combo 2 assay, which provides detection coverage of the new diagnostic‐escape Chlamydia trachomatis variants. BMC Infect Dis 20: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo, M. , Olcen, P. , Agne‐Stadling, I. , Feldt, A. , Jurstrand, M. , Herrmann, B. , et al. (2007) Experiences with the new genetic variant of Chlamydia trachomatis in Orebro county, Sweden ‐ proportion, characteristics and effective diagnostic solution in an emergent situation. Euro Surveill 12: E5‐6. [DOI] [PubMed] [Google Scholar]
- Unemo, M. , Seth‐Smith, H.M.B. , Cutcliffe, L.T. , Skilton, R.J. , Barlow, D. , Goulding, D. , et al. (2010) The Swedish new variant of Chlamydia trachomatis: genome sequence, morphology, cell tropism and phenotypic characterization. Microbiology 156: 1394–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiu, L. , Zhang, C. , Li, Y. , Wang, F. , and Peng, J. (2019) Simultaneous detection of eleven sexually transmitted agents using multiplexed PCR coupled with MALDI‐TOF analysis. Infect Drug Resist 12: 2671–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiu, L. , Zhang, C. , Li, Y. , Wang, F. , and Peng, J. (2020) High‐resolution melting analysis for rapid detection of the internationally spreading ceftriaxone‐resistant Neisseria gonorrhoeae FC428 clone. J Antimicrob Chemother 75: 106–109. [DOI] [PubMed] [Google Scholar]


