Significance
Natural products provide the inspiration for most drugs, and marine natural products, in particular, are emerging as promising new therapeutics with new targets or mechanisms of action. Pharmacological targeting of tubulin dynamics has been a validated strategy for cancer therapy for decades, yielding structurally diverse natural products and derivatives, including paclitaxel, vincristine, maytansine, and eribulin, targeting six known and different binding sites. We discovered a chemical scaffold from marine cyanobacteria that targets a seventh tubulin binding site. We report the entire spectrum of the discovered chemical and biological novelties, including the isolation, structure determination, and chemical synthesis of the natural product, and the investigation of its mechanism of action, target identification, and binding mode elucidation at the atomic level.
Keywords: marine natural product, cyanobacteria, total synthesis, microtubules, tubulin
Abstract
Tubulin-targeted chemotherapy has proven to be a successful and wide spectrum strategy against solid and liquid malignancies. Therefore, new ways to modulate this essential protein could lead to new antitumoral pharmacological approaches. Currently known tubulin agents bind to six distinct sites at α/β-tubulin either promoting microtubule stabilization or depolymerization. We have discovered a seventh binding site at the tubulin intradimer interface where a novel microtubule-destabilizing cyclodepsipeptide, termed gatorbulin-1 (GB1), binds. GB1 has a unique chemotype produced by a marine cyanobacterium. We have elucidated this dual, chemical and mechanistic, novelty through multidimensional characterization, starting with bioactivity-guided natural product isolation and multinuclei NMR-based structure determination, revealing the modified pentapeptide with a functionally critical hydroxamate group; and validation by total synthesis. We have investigated the pharmacology using isogenic cancer cell screening, cellular profiling, and complementary phenotypic assays, and unveiled the underlying molecular mechanism by in vitro biochemical studies and high-resolution structural determination of the α/β-tubulin−GB1 complex.
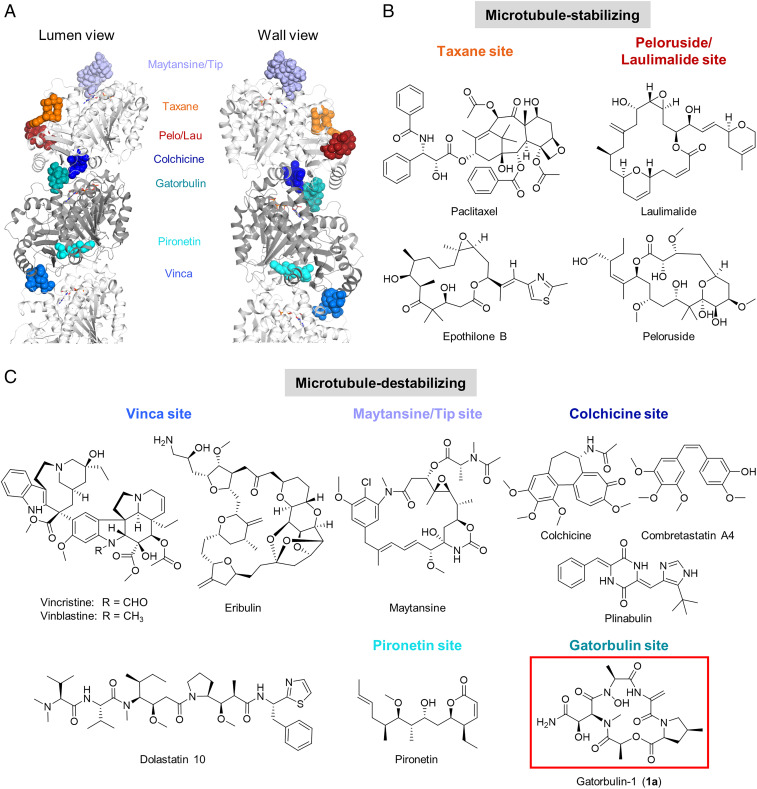
Microtubules are polarized polymers consisting of α/β-tubulin heterodimers involved in cellular structure, motility, proliferation, and intracellular trafficking (1). Pharmacological targeting of tubulin dynamics at different sites (Fig. 1A) has been a validated strategy for cancer therapy for decades and has mostly been linked to the antimitotic effects of these compounds, although increasing evidence has emerged for the importance of nonmitotic effects (1). Natural products targeting tubulin, in particular, have yielded a wealth of chemically diverse agents and provided the basis for several Food and Drug Administration-approved drugs, for both cancer and other pathologies, either alone or as antibody−drug conjugate (ADC), including paclitaxel, vincristine, maytansine, eribulin, and colchicine (Fig. 1 B and C). Compounds can be classified based on their binding to one of the six known binding sites, and, even though they are all targeting tubulin, they have shown distinct pharmacological effects. Therefore, there is a persistent interest in the identification of novel microtubule-targeting agents. Two α/β-tubulin binding sites are associated with microtubule stabilization (taxane and laulimalide/peloruside sites; Fig. 1 A and B), while binding to four other sites causes microtubule destabilization (vinca, maytansine, colchicine, and pironetin sites; Fig. 1 A and C) (1).
Fig. 1.
Binding sites and structures of microtubule-targeting agents. (A) Tubulin heterodimer (α-tubulin in gray and β-tubulin in white) in ribbon representation, where six known binding sites have been highlighted showing representative ligands in sphere representation: maytansine (PDB ID code 4tv8, violet); epothilone (PDB ID code 4o4i, orange); peloruside (PBD ID code 4o4j, red); colchicine (PDB ID code 4o2b, dark blue); pironetin (PDB ID code 5fnv, cyan), and vinblastine (PDB ID code 4eb6, light blue). The gatorbulin binding site has been also included (PDB ID code 7alr, teal). (B and C) Representative compounds targeting tubulin binding sites. (B) Microtubule-stabilizing agents. (C) Microtubule-destabilizing agents, including the structure of GB1 (1a).
Our investigation of marine cyanobacteria as a source of potential anticancer agents has previously yielded the modified peptides dolastatin 10 (Fig. 1C) and dolastatin 15 (2–4), targeting the vinca site (5, 6). Three ADCs with a dolastatin 10 analog (monomethyl auristatin E) as the cytotoxic payload are approved for the treatment of various lymphomas and refractory bladder cancer, while dolastatin 15-based ADCs have advanced to clinical trials (4). We identified both dolastatins 10 and 15 as indirect hypoxia-inducible factor (HIF) inhibitors based on differential cytotoxicity against a panel of isogenic HCT116 colorectal cancer cells (4, 7), which indicated that HIF inhibition is functionally relevant for the mechanisms of action of these compounds. HIF is activated in solid tumors and promotes metastasis, and targeted screening early in the drug discovery process could provide a rapid indication for requisite selectivity for cancer treatment (8–10). Using the same isogenic screening system, we now identified an antiproliferative agent that also possessed preferential activity for oncogenic KRAS-containing and HIF-1α−containing HCT116 cells and is a microtubule-destabilizing cyclodepsipeptide. We named the compound gatorbulin-1 (GB1, 1a; Fig. 1C), in analogy to eribulin (Eisai Research Institute), to symbolically represent the discovery of its unique chemical structure and pharmacological potential at the University of Florida and global Gator Nation partners. Here we report the bioassay-guided isolation, structure determination, synthesis, preliminary structure−activity relationships, mechanism of action, target identification, and binding mode elucidation. Our studies revealed that GB1 represents a unique chemical scaffold targeting a different binding site near the colchicine site and displays distinct pharmacology (Fig. 1A).
Results
Isolation and Structure Determination of GB1.
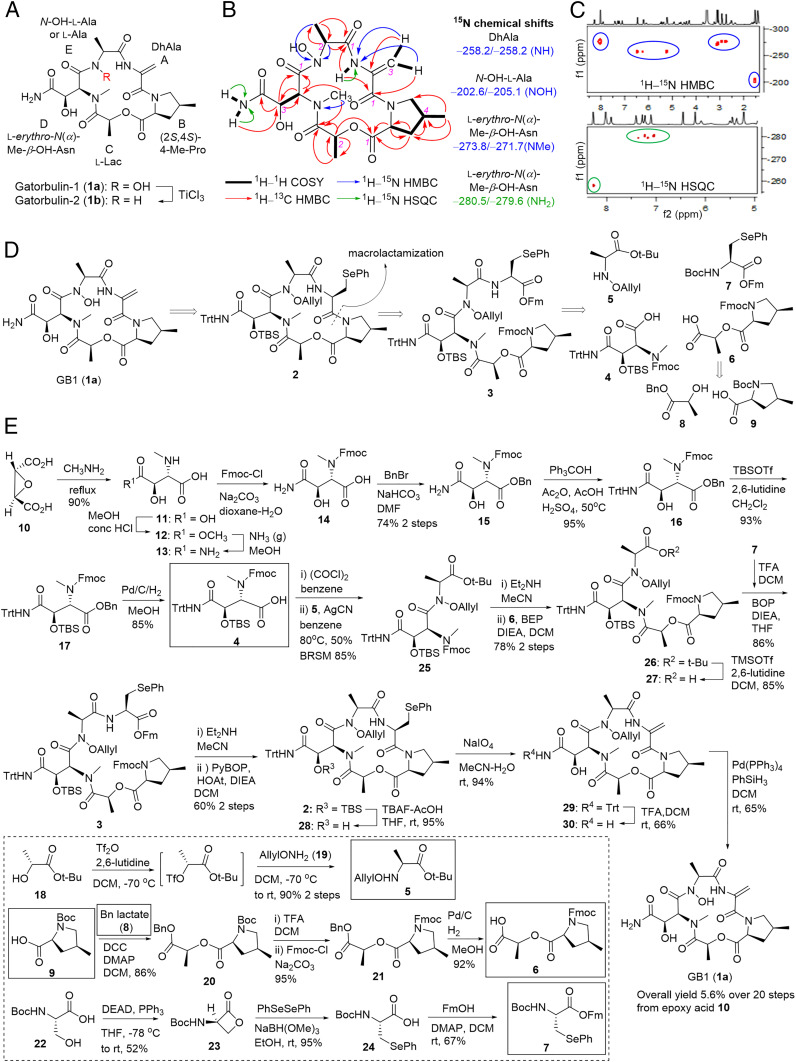
Various collections of the cyanobacterium Lyngbya cf. confervoides during blooms off the coast near Ft. Lauderdale (11) (Broward County) were extracted with EtOAc–MeOH (1:1). The extracts, previously proven to be rich in secondary metabolites and possessing antifeedant activity (12, 13), were either solvent−solvent partitioned first or directly applied onto a Diaion HP-20 column, and then fractions were subjected to reversed-phase high-pressure liquid chromatography (HPLC) to afford GB1 (1a) as an optically active, colorless amorphous solid ([α]20D –84.0 [c 0.10, MeOH]) and as the most antiproliferative extract component by bioassay-guided isolation using colon cancer cells, along with a minor analog, termed gatorbulin-2 (GB2, 1b; Fig. 2A). Doubling of virtually every signal in the 1H NMR spectrum of 1a recorded in DMF-d7 suggested the presence of an asymmetric dimer or the presence of conformers in a ratio of 1:1. This observation coupled with the [M + H]+ ion peak at m/z 484.2043 obtained by simultaneous electrospray ionization and atmospheric pressure chemical ionization high-resolution mass spectrometry (HRMS) and 13C NMR data suggested a molecular formula of C20H29N5O9 (calculated for C20H30N5O9, 484.2044) and consequently the presence of conformers in the NMR solvent. NMR analysis using 1H NMR, 13C NMR, 1H–1H correlation spectroscopy (1H–1H COSY), heteronuclear multiple quantum coherence (HMQC), and 1H–13C heteronuclear multiple bond correlation (HMBC) data were carried out for both conformers, revealing two sets of five spin-coupled systems as part of a pentapeptide structure; one signal set appeared slightly broader (Table 1, Fig. 2B, and SI Appendix, Figs. S1–S7).
Fig. 2.
Structure determination and total synthesis of GB1. (A) Structures of the isolated natural products, GB1 (1a) and its N-deoxy derivative, GB2 (1b). GB1 units are labeled A–E. (B) Homonuclear and heteronuclear 2D NMR correlations for GB1. (C) Selected regions of the 1H–15N HMBC and 1H–15N HSQC spectra of GB1. (D) Retrosynthetic analysis for the total synthesis of GB1 (1a). (E) Forward synthetic route. Longest linear sequence is shown in the main scheme. Building blocks 4 to 9 from the retrosynthesis (in D) are indicated by boxes with solid lines. The synthesis of the building blocks 5 to 7 is provided in the box with the dashed line.
Table 1.
NMR data for GB1 (1a) for both conformers (1:1) in DMF-d7
| Conformer 1 | Conformer 2 | |||||||||
| Unit | C/H no. | δH (J in Hz)* | δC, mult† | δN‡ | COSY* | 1H–13C HMBC* | 1H–15N HMBC§ | δH (J in Hz)* | δC, mult† | δN‡ |
| DhAla | 1 | 165.9, qC | 166.8, qC | |||||||
| (A) | 2 | 136.3, qC | 136.2, qC | |||||||
| 3a | 6.46, s | 101.8, CH2 | H-3b | 1, 2 | 6.22, s | 103.0, CH2 | ||||
| 3b | 5.22, s | H-3ª | 1 | 5.11, s | ||||||
| NH | 8.28, s | −258.2 | 1, 1 (Ala) | H-3a, H-3b | 8.60, s | −258.2 | ||||
| 4-Me-Pro | 1 | 170.8, qC | 170.4, qC | |||||||
| (B) | 2 | 4.42, t (7.0) | 62.6, CH | H-3a, H-3b | 1, 3 | 4.94, dd (9, 8) | 63.8, CH | |||
| 3a (proS) | 2.58, m | 37.0, CH2 | H-2, H-3b, H-4 | 1, 2, 4, 5, 6 | 2.53, m | 40.8, CH2 | ||||
| 3b (proR) | 1.58, m | H-2, H-3a, H-4 | 1, 2, 4, 5, 6 | 1.65, m | ||||||
| 4 | 2.53, m | 34.7, CH | H-3a, H-3b, H-5a, H-5b, H3-6 | 3, 5, 6 | 2.38, m | 32.4, CH | ||||
| 5a (proR) | 4.31, dd (–10.0, 6.0) | 56.9, CH2 | H-4, H-5b | 2, 3, 4, 6 | 3.71, dd (–11.0, 7.0) | 55.6, CH2 | ||||
| 5b (proS) | 3.25, dd (–10.0, 8.5) | H-4, H-5ª | 3, 6 | 3.11, dd (–11.0, 11) | ||||||
| 6 | 1.12, d (7.0) | 18.0, CH3 | H-4 | 3, 4, 5 | 1.11, d (7.0) | 16.9, CH3 | ||||
| Lac | 1 | 169.9, qC | 169.7, qC | |||||||
| (C) | 2 | 5.47, q (7.0) | 72.9, CH | H3-3 | 1, 3, 1 (MePro) | 5.39, q (7.0) | 72.1, CH | |||
| 3 | 1.42, d (7.0) | 17.6, CH3 | H-2 | 1, 2 | 1.41, d (7.0) | 17.3, CH3 | ||||
| N(α)-Me-β-OH-Asn | 1 | 170.4, qC | 169.4, qC | |||||||
| (D) | 2 | 5.51, br d (9.5) | 58.0 (br), CH | H-3, | 1, 2 | 5.85 | Not observed | |||
| 3 | 4.51, dd (9.5, 4.5) | 68.8, CH | H-2, 3-OH | 1, 2, 4 | 4.65, br dd (6, 6) | 69.7, CH | ||||
| 3-OH | 6.05, br d | H-3 | 5.06, br | |||||||
| 4 | 175.1, qC | 174.8 | ||||||||
| N-Me | 3.09, s | 34.0, CH3 | −273.8 | 2, 1 (Lac) | N-Me | 3.14, s | 34.5 (br), CH3 | −271.7 | ||
| 4-NHa | 7.26, s | −280.5 | 4-NHb | 7.39, s | −279.6 | |||||
| 4-NHb | 7.09, s | 4-NHa | 3 | 7.21, s | ||||||
| N-OH-Ala | 1 | 170.5, qC | 170.2, qC | |||||||
| (E) | 2 | 4.29, q (6) | 64.7, CH | H3-3, NH | 1, 3 | 4.71, br q (6) | 60.0, CH | |||
| 3 | 1.54, d (6) | 13.8, CH3 | H-2 | 1, 2 | 1.42, d (6) | 15.6, CH3 | ||||
| N-OH | 11.35, br s | −202.6 | H-2, H3-3 | 10.58, br | −205.1 | |||||
br: broad, s: singlet, d: doublet, q: quartet, dd: doublet of doublets, qC: quaternary carbon.
Recorded at 600 MHz.
Recorded at 150 MHz.
Deduced from 1H–15N HSQC and 1H–15N HMBC (500 MHz).
Recorded at 500 MHz.
For both signal sets, one putative NH singlet each (δH 8.28 and 8.60) showed COSY correlations to sp2-methylene protons (δH 6.46/5.22 for conformer 1, 6.22/5.11 for conformer 2), which also appeared as singlets in the 1H NMR spectrum. Correlations from the NH to the corresponding olefinic methylene carbon (δC 101.8 and 103.0), to a quaternary sp2 hybridized carbon (δC 136.3 and 136.2), and to two carbonyl carbons (δC 165.9/170.5 for conformer 1 and 166.8/170.2 for conformer 2) defined the first residue (unit A; Fig. 2A) as a dehydroalanine (DhAla) residue (Table 1 and Fig. 2B).
The second multiproton spin system consisted of two methylenes (δH 4.31/3.25, 2.58/1.58 for conformer 1), two methines (δH 4.42, 2.53), and one methyl group (δH 1.12 d). COSY analysis established their arrangement supported by HMBC data (Table 1 and Fig. 2B). The terminal methine and methylene carbons of this spin system appeared to be nitrogenated (δC 62.6, 56.9), and the HMBC correlation of one of the methylene protons (δH 4.31) to one of the methine carbons (δC 62.6) clarified that the carbons were joined in a 3-methyl pyrrolidine structure (Fig. 2B), which, upon further analysis of HMBC correlations to a carbonyl carbon (δH 170.8), identified the second residue (unit B; Fig. 2A) as a 4-methylproline unit (4-MePro). In a similar fashion, a signal set corresponding to the second conformer for this unit was unambiguously identified (Table 1).
Analysis of the third spin system was straightforward, and this unit consisted of only one methine and a methyl group and NMR data (Table 1 and Fig. 2B), consistent with an acylated lactic acid (Lac) moiety (unit C; Fig. 2A).
Two singlets of another signal set for heteroatom-bound protons (δH 7.26/7.09 for conformer 1 and δH 7.39/7.21 for conformer 2) showed cross-peaks in the COSY spectrum, suggesting a primary amide for the fourth moiety. Another set of singlets at δH 3.09 and 3.14 was indicative of an N-methyl tertiary amide group; expectedly, these signals exhibited HMBC correlations with a carbonyl carbon of the adjacent residue (δC 169.9 and 169.7) and, for conformer 1, also to the α-carbon of the N-methylated amino acid (δC 58.0). Rigorous two-dimensional (2D) NMR analysis (Table 1 and Fig. 2B) established the fourth residue (unit D; Fig. 2A) as an N(α)-methyl-β-hydroxy-asparagine (N(α)-Me-β-OH-Asn). Even though the significant broadening of all signals for this unit for the second conformer resulted in fewer HMBC correlations, all 1H and 13C NMR resonances could be assigned except for the α-carbon, since its NMR signal was too broad to be observed (Table 1).
The fifth and last moiety (unit E; Fig. 2A) exhibited similarity to a lactic acid or alanine moiety, yet the α-carbon resonated at higher field than the corresponding carbon for lactic acid (δC 64.7 and 60.0) and thus more likely bore a nitrogen atom, which then, in turn, had to bear a substituent that was not accounted for yet. This NMR analysis so far led to the assignment of all atoms except one oxygen and hydrogen based on HRMS analysis. In the 1H NMR spectrum, the only unassigned signal at this point was a signal for an exchangeable proton at δH 11.35 (br s) for conformer 1 and at δH 10.58 (br) putatively for conformer 2, which could not be rationalized by a secondary amide since it did not show a COSY correlation to the nitrogen-bearing methine and also resonated too far downfield. The chemical shifts were consistent with carboxylic acid protons which could exist in a linear structure; however, it would not leave a substituent for the nitrogen atom in the alanine-like moiety. Therefore, a bond between two heteroatoms, nitrogen and oxygen, had to be invoked, which led us to propose an N-hydroxy group in a cyclic hydroxamate (N-OH-Ala); its hydroxy proton was also expected to resonate between δH 10 and 12 as observed (Table 1).
The doubling and overlap of signals for several carbonyl carbons for different conformers slightly complicated the sequencing of the individual units. To ultimately prove the existence of the hydroxamate and to validate the nature of the nitrogen atoms, we carried out a 1H–15N HMBC and 1H–15N heteronuclear single quantum correlation (HSQC) analysis (Table 1 and SI Appendix, Figs. S8 and S9). HMBC correlations of the H-3a/b methylene protons of the DhAla unit to a nitrogen atom resonating at δN –258.2 (relative to external MeNO2, δN 0.0) supported the earlier assignment of a secondary amide (Fig. 2 B and C). The N-Me protons of the N-Me-3-OH-Asn showed HMBC correlations to a nitrogen possessing a chemical shift of δN –273.8, an expected value for a tertiary amide (Fig. 2 B and C). Most importantly and confirmatory for the hydroxamate moiety were two- and three-bond correlations from the α-methine and the β-methyl protons to a signal at δN –202.6 (Fig. 2 B and C), which is in agreement with literature values for hydroxamate nitrogens [δN –199.2 for polyoxypeptin A (14)], further validating the proposed structure for GB1 (1a). The 1H–15N HSQC spectrum, in addition to the secondary amide proton for DhAla (δN –258.2), also showed one-bond correlations for both protons of the primary amide functionality for both conformers (δN –280.5 and –279.6; Fig. 2C). The presence of the hydroxamate functionality was supported analytically by ferric hydroxamate complex formation (15) (SI Appendix).
Further support for the proposed structure was found with the isolation of its N-deoxy derivative, GB2, or N-deoxy-GB1 (1b; Fig. 2A). The 1H NMR spectrum was strikingly similar to the one of 1a, including the presence of conformers in a 1:1 ratio in DMF-d7 (SI Appendix, Table S1 and Figs. S10–S15). The most significant difference appeared to be the lack of the N-OH protons at δH 10 to 12; instead, a new set of doublets appeared in the range for amide protons (δH 8.21 and 8.38 for conformers 1 and 2).
To establish the absolute configuration, we performed acid hydrolysis to liberate the individual units and synthesized all isomers of the amino acid standards for comparative chiral HPLC analysis and advanced Marfey’s analysis. The 4-MePro standards were prepared as described previously (16), and the N(α)-Me-β-OH-aspartic acid stereoisomers were prepared as described in SI Appendix. We detected l-erythro-N(α)-Me-β-OH-Asp, (2S,4S)-4-Me-Pro, and l-Lac in the hydrolyzate of 1a. The structures and identical absolute configurations were confirmed by conversion of 1a into 1b via TiCl3-mediated reduction (Fig. 2A) (17). Upon acid hydrolysis, 1b yielded l-Ala as detected by chiral HPLC analysis, establishing the remaining stereogenic center.
Structure Validation by the Total Synthesis of GB1.
To prove the structure and overcome the supply issue, we embarked on total synthesis. The retrosynthetic analysis of total synthesis of GB1 (1a) is shown in Fig. 2D. The final product 1a could be obtained from the fully masked cyclized precursor 2 by sequential deprotection. The site between 4-MePro and (Se)-phenylselenocysteine [Sec(Ph)] was chosen for macrolactamization. In linear precursor 3, the 9-fluorenylmethoxycarbonyl (Fmoc) and 9-fluorenylmethyl (Fm) pair was designed as the protection groups of amino and carboxy termini, which could be cleaved simultaneously with base to provide the precursor of macrocyclization (18). Sec(Ph) was proposed as the prounit of DhAla (19). Linear compound 3 was disconnected into four building blocks 4 to 7, which could be constructed from commercially available reagents (e.g., 6 from 8 and 9) using established or modified protocols.
Fig. 2E depicts the synthetic route to GB1 (1a). The synthesis of acid 4 was adopted from published procedures (20). The (2R,3R)-epoxysuccinic acid (10) was converted to erythro-N(α)-methyl-3-hydroxy-l-aspartic acid (11) by treatment with methylamine−water under reflux. Then 11 was selectively esterified with acidic methanol under refluxing (21) to provide monoester 12. Without purification, aminolysis of 12 with ammonia (gas) in MeOH provided N(α)-Me-β-OH-Asn (13) (20, 21), which had poor solubility in MeOH, so that pure product could be obtained by simple filtration. Sequential protections of the groups of 13 using standard methods (22) provided full masked compound 17. Finally, acid 4 was obtained from 17 by hydrogenation with palladium catalyst.
The synthesis of the building block of allyloxamine 5 adopted the triflate method (23). Allyl was chosen as the NOH protecting group as it could be selectively removed by Pd(Ph3P)4 in the presence of dehydropeptide. Acid 6 was synthesized from (4S)-N-Boc-4-methyl-Pro (9) and benzyl-l-lactate (8) by standard protocols of esterification, protection, and deprotection. Following established procedures (19), N-Boc-serine (22) was converted to BocSec(Ph) (24) via β-lactone 23, which was esterified with FmOH to provide building block 7. DhAla could be obtained from phenylselenocysteine by oxidative β-elimination.
The fusion of building blocks was initiated by coupling of acid 4 with allyloxamine 5. Acid 4 was activated to the acid chloride, which was then coupled with 5 in the presence of AgCN (24, 25) to provide 25 in 50% isolated yield and 85% yield based on recovered starting materials. This acylation was not successful when other common coupling reagents were used, because of poor nucleophilicity of the nitrogen and high steric hindrance (26). Compound 26 was obtained in 78% yield by 2-bromo-1-ethyl-pyridinium tetrafluoroborate–mediated coupling (27) of acid 6 with the free methylamine generated by selective deprotection of 25. Selective removal of the t-butyl group of 26 by trimethylsilyl trifluoromethanesulfonate/2,6-lutidine (28) afforded acid 27, which was coupled with free amine from 7 using (benzotriazol-1-yloxy)tris(dimethylamino)phosphonium hexafluorophosphate as a coupling reagent to yield the linear compound 3 in 86% yield. The use of the t-butyl protecting group prevented diketopiperazine (29) formation upon coupling of 6 and 25, and ensured that trityl and tert-butyldimethylsilyl (TBS) groups were intact for the generation of acid 27. Both Fmoc and Fm protection groups were removed simultaneously when compound 3 was exposed to Et2NH in MeCN. The macrocyclization was mediated by PyBOP/HOAt to give macrocycle 2 in 60% yield for two steps. Removal of TBS of 2 with the tetrabutylammonium fluoride/HOAc buffer (22, 30) provided 28, and subsequent oxidation of SePh with NaIO4 (19) yielded dehydropeptide 29. Trityl was removed with trifluoroacetic acid in CH2Cl2 (31) to yield primary amide 30. The removal of allyl group by Pd(PPh3)4/PhSiH3 (30, 32) provided final product 1a. The removal sequences for trityl and allyl groups are interchangeable; however, the yield would drop from 66 to 35%. GB1 (1a) was purified by reversed-phase thin-layer chromatography plate (C18). The overall yield over 20 steps from the epoxy acid 10 was 5.6% (see SI Appendix for details and SI Appendix, Figs. S16–S61 for all NMR spectra). The synthetic sample was identical to the isolated natural product, which was verified by NMR, HRMS, and optical rotation (SI Appendix, Figs. S62 and S63).
GB1 Cellular Profiling Identifies the Mechanism of Action.
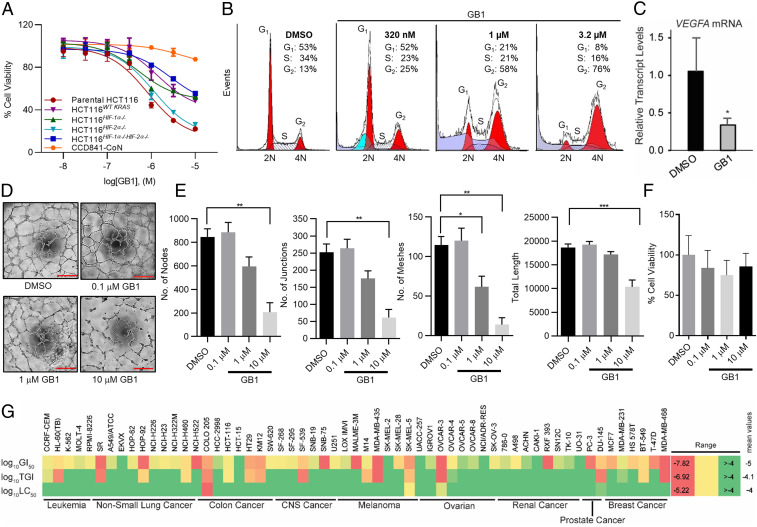
GB1 (1a) was identified as the extract’s active component against colon cancer cells and showed a concentration that inhibits response by 50% (IC50) of 0.80 µM in a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide assay against HCT116 colorectal cancer cells (Fig. 3A), while 1b was inactive at the highest concentration tested (IC50 > 10 µM), indicating that the hydroxamate moiety is indispensable to the antiproliferative activity. Isogenic cell line selectivity screening indicated preferential activity against parental HCT116 compared with the oncogenic KRAS knockout or double knockout of both HIF-1α and HIF-2α transcription factors (Fig. 3A) (7). Deconvolution using single knockouts of HIF-1α and HIF-2α clearly demonstrated that only cells depleted in HIF-1α had reduced susceptibility to GB1, which is consistent with HIF-1α being in the same pathway and activated by oncogenic KRAS. Furthermore, normal epithelial colon cells (CCD841-CoN) were less inhibited, indicating an additional promising level of selectivity (Fig. 3A). The preference for HIF-1α−expressing cells parallels the selectivity profiles for microtubule agents we previously discovered (dolastatins 10 and 15) (4, 7). DNA content analysis revealed the concentration-dependent G2/M cell cycle accumulation characteristic for antimitotic agents (Fig. 3B). A concentration of GB1 (4 × IC50) that results in a complete antiproliferative response initiated a concomitant down-regulation of the HIF-1α target gene VEGFA in parental HCT116 cells (Fig. 3C) that was even more pronounced than for dolastatin 15 (4), which also elevated GB1’s potential as antiangiogenic agent. Consistent with the role of microtubule dynamics in angiogenesis (33), GB1 also inhibited endothelial tube formation without cytotoxicity (Fig. 3 D–F). Endothelial cell tube formation in Matrigel mimics the process of blood vessel formation in vivo (33). Our study suggested that 10 μM GB1 was necessary to block the ability of human umbilical vein endothelial cells (HUVECs) to form tube-like structures within 3 h (Fig. 3D). Quantification of the structural network revealed a significant reduction in the number of nodes, number of junctions, number of meshes, and total length of tubes (Fig. 3E). Importantly, GB1 did not affect HUVEC cell viability, even after 24 h (Fig. 3F). Taken together, this reveals an antiangiogenic potential of the compound in inhibiting the formation and stabilization of a 3D vascular network. The additional VEGF down-regulation in growth factor-secreting cells, which is a driver of angiogenesis, is expected to result in a pronounced antiangiogenic action of GB1 in vivo.
Fig. 3.
Mechanism of action of GB1 through cellular profiling. (A) Antiproliferative activity of GB1 in parental HCT116 colon cells, isogenic HCT116 knockout cells, and CCD-841CoN normal epithelial colon cells (48 h treatment). GB1 showed a marginal effect on the viability of CCD-841CoN normal epithelial colon cells (0.5% DMSO vehicle). Parental HCT116 cells and HCT116HIF-2α−/− were most susceptible, while the potency and efficacy of GB1 were reduced against HCT116HIF-1α−/−HIF-2α−/−, HCT116HIF-1α−/−,, and oncogenic KRAS knockout (HCT116WT KRAS). Cell viability was measured by MTT assay (n = 3). Data are represented as average ± SD. (B) Cell cycle analysis. HCT116 cells were treated with GB1 (320 nM, 1 µM, or 3.2 µM) or vehicle control (0.25% DMSO) for 24 h, and DNA content was assessed by flow cytometry of propidium iodide stained cells. GB1 induced G2/M accumulation. (C) HIF target gene (VEGFA) expression after 16-h exposure of parental HCT116 cells to GB1 (3.2 µM) or vehicle (0.25% DMSO). RNA was isolated, reverse transcribed, and subjected to qPCR using TaqMan analysis. β-actin served as endogenous control. Error bars indicate mean ± SD of three replicates (Student t test, *P < 0.05). (D–F) GB1 inhibits HUVEC tube formation in vitro without toxicity. (D) Representative images of HUVEC tube formation in growth factor-reduced Matrigel upon treatment with DMSO (0.1%) or varying concentrations (10 μM, 1 μM, or 0.1 μM) of GB1 (9 h). (Scale bar: 200 μm.) All images shown are representative, and data are represented as mean ± SEM; one-way ANOVA followed by Tukey's multiple comparisons test; *P < 0.05, **P < 0.01, and ***P < 0.001. (E) Quantification of number of nodes, number of junctions, number of meshes, and total length of tubes (n = 3). (F) GB1 does not affect HUVEC cell viability. HUVEC cell viability was quantified by absorbance at 490 nm using (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt) (MTS assay, 24 h). GB1 treatment did not affect total number of viable HUVEC cells compared to DMSO-treated control (n = 3). For E and F, data are represented as mean ± SEM; one-way ANOVA followed by Tukey’s multiple comparisons test. (G) Heatmap for the performance of GB1 across cell lines in the NCI-60 screen using three different values (growth-inhibitory effect, GI50; cytostatic effect, TGI; cytotoxic effect, LC50; concentration in molars).
The NCI-60 cell line screen data (Fig. 3G), analyzed by the COMPARE algorithm (34), were indicative of a cytotoxicity profile most related to antimitotic/tubulin agents, including paclitaxel, eribulin, colchicine, and vinca bis-indole alkaloid derivatives (P 0.75 to 0.85), suggesting that the biochemical mechanisms of action are related. While GB1 displayed an IC50 of >10 µM against normal mucosal colon cells, it had submicromolar activity against HCT116 cells (GI50 306 nM) and was even more potent against COLO205 cells (GI50 92 nM) based on the NCI-60 data (sulforhodamine B assay). Other susceptible cell types included certain melanoma (SK-MEL-5), ovarian (OVCAR-3), and prostate (DU-145) cancer cells, which correspond to certain cancers where microtubule agents have been successful. Furthermore, cervical and breast cancers are relevant indications, prompting additional studies in these cell types (see below) (35).
Target Identification and GB1 Underlying Molecular Mechanism.
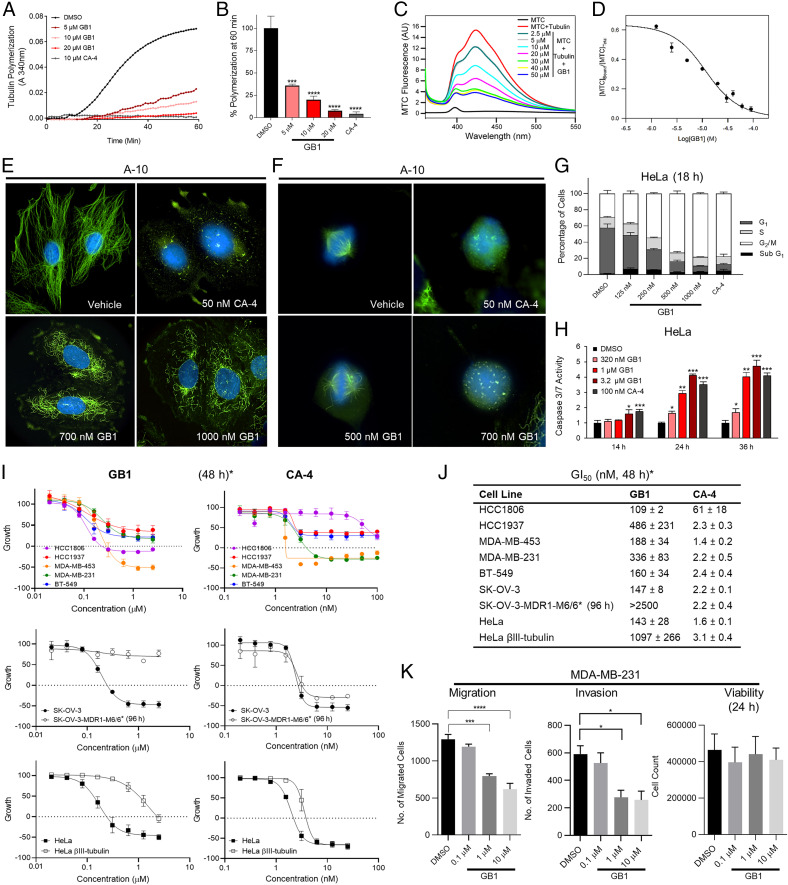
First, using a biochemical assay, we demonstrated that GB1 directly inhibits tubulin polymerization in vitro in an iron-independent manner (Fig. 4 A and B). Second, we tested GB1 binding to commonly targeted sites for microtubule-destabilizing agents, including the vinblastine, maytansine, and colchicine sites (Fig. 1A), by employing fluorescent bona fide probes (36–38). GB1 was unable to displace the fluorescent derivatives of eribulin and maytansine, indicating that it did not bind to these sites. However, GB1 successfully competed with a fluorescent probe of the colchicine site [2-methoxy-5-(2,3,4-trimethoxyphenyl)-2,4,6-cycloheptatrien-1-one (MTC)] (Fig. 4 C and D) with an apparent binding affinity of 1.01 ± 0.08 × 106 M−1, which is below colchicine apparent binding affinity [1.5 × 107 M−1 (39)].
Fig. 4.
Target identification and selectivity profiling of GB1 in comparison with combretastatin A-4 (CA-4). (A and B) Inhibition of tubulin polymerization by GB1. (A) Time course of tubulin polymerization (20 μM) (vehicle: DMSO; 1%). (B) Quantification of tubulin polymerization at 60 min. (C and D) Displacement of MTC. (C) Fluorescence emission spectra of 10 μM MTC in the absence (black line) and the presence (red line) of 10 μM tubulin plus varying concentrations of GB1 (vehicle: DMSO; 1%). (D) Displacement isotherm at 25 °C of MTC by GB1. Line is best fit of the GB1 equilibrium constant, assuming 0.8 sites per tubulin dimer. (E) Microtubule depolymerization GB1 in interphase cells. A-10 cells treated for 18 h (vehicle: DMSO; 0.1%). (Magnification: 1,000×.) (F) GB1 promotes formation of aberrant mitotic spindles in A-10 cells (18 h treatment). (G) GB1 arrest cell cycle in G2/M phase. Cell cycle distribution of HeLa cells in the presence of GB1 (vehicle: DMSO; 0.1%) (H) GB1 causes apoptosis in HeLa cells as measured by Caspase-3/7 activity (vehicle: DMSO; 0.5%). (I and J) Antiproliferative and cytotoxic effects of GB1 and CA-4 in a panel of TNBC cell lines, in parental and Pgp-expressing SK-OV-3 cells, or in parental and βIII-expressing HeLa cells (sulforhodamine B assay, vehicle: DMSO; 0.5%). Asterisk denotes that treatment time was 48 h, except for SK-OV-3-MDR1-M6/6 (96 h). (K) Effects of GB1 on transwell migration and invasion of serum-starved MDA-MB-231 breast cancer cells pretreated with GB1 for 24 h. Pretreated cells were allowed to migrate or invade for 5 h. Quantitative analysis of migrated or invaded cells across a Matrigel layer (vehicle: DMSO; 0.1%). Cell count (24 h) was determined by Trypan Blue exclusion assay. (A, C, E, and F) Representative set from three experiments shown. (B and G–K) Error bars represent ± SEM (B, G, and I–K) or SD (H) from three independent experiments. (B) One-way ANOVA with Dunnett’s post hoc test; ***P < 0.001, ****P < 0.0001. (D and K) n = 3. (E and F) Microtubules visualized using a β-tubulin antibody (green) and DNA was visualized using DAPI (blue). (H) Two-tailed Student t test; *P < 0.05, **P < 0.01, ***P < 0.005. (K) One-way ANOVA followed by Tukey’s multiple comparisons test; *P < 0.05, ***P < 0.001, ****P < 0.0001.
We performed subsequent cell-based studies with GB1 in comparison with combretastatin A4 (CA-4), a known colchicine site binder. We demonstrated that GB1 promotes microtubule depolymerization in A-10 (rat smooth muscle) interphase cells, similar to CA-4, although with lower potency (Fig. 4E). GB1 promoted aberrant mitotic spindle formation at submicromolar concentrations (Fig. 4F). HeLa cervical cancer cells were highly susceptible to GB1; the compound induced accumulation of cells in the G2/M cell cycle phases after 18 h, with low levels of apoptosis as suggested by an emerging sub-G1 population (Fig. 4G). We then measured the caspase-3/7 activity in a concentration- and time-dependent manner, validating that treatment with GB1 induces apoptosis at concentrations that affect tubulin dynamics in cells, especially after 24 h and beyond, similar to CA-4 (Fig. 4H). We then compared the antiproliferative effects of GB1 and CA-4 in a panel of triple-negative breast cancer (TNBC) cells and in drug-resistant models, including parental and Pgp-overexpressing SK-OV-3 ovarian adenocarcinoma cells (48 and 96 h of treatment, respectively) and parental and βIII-expressing HeLa cells (48 h) (40). The compounds showed a different fingerprint. GB1 does not circumvent Pgp-mediated or βIII-tubulin−mediated drug resistance (Fig. 4I), indicating that it was mechanistically distinct from CA-4. Additionally, it has a distinct profile of relative efficacy against a panel of TNBC cell lines as compared to CA-4 (48 h; Fig. 4 I and J). MDA-MB-231 cells pretreated with GB1 for 24 h demonstrated a significant concentration-dependent inhibition of cancer cell migration and invasion abilities across a transwell membrane as compared to dimethyl sulfoxide (DMSO)-treated controls, without significantly affecting cell viability (Fig. 4K). This phenotype is also consistent with the microtubule-targeting mechanism (41). Taken together, the mechanistic biochemical and pharmacological data suggested that GB1 can block ligand binding to the colchicine site but may not fit in the classic pocket, resulting in a distinct profile of cellular activity.
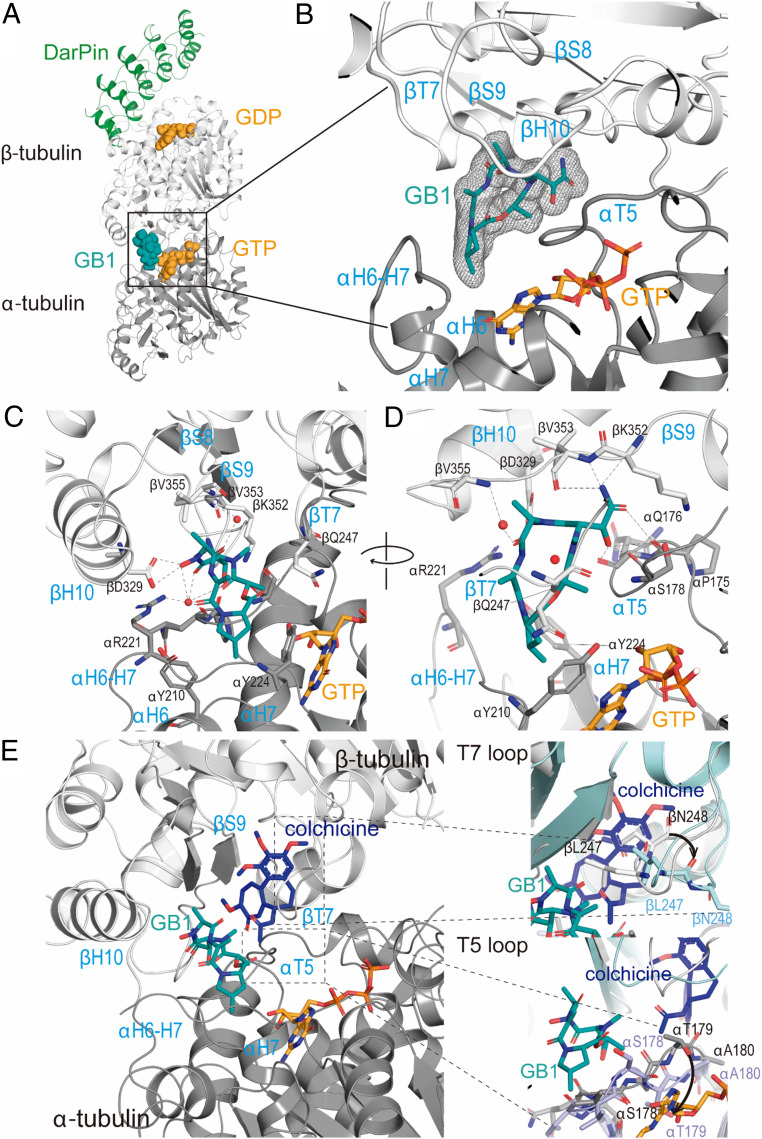
To further probe our hypothesis and identify the specific binding mechanism, we aimed at high-resolution macromolecular crystallography. The T2R-TTL system (two α/β-tubulin dimers in complex with the stathmin-like protein RB3 and tubulin tyrosine ligase) has successfully produced several structures of ligand bound to tubulin using X-ray crystallography (42). However, GB1 soaking experiments did not reveal any difference density related to the ligand. We finally succeeded using the T1-DARPin (T1D) complex that was previously used to solve the structure of colchicine-bound tubulin by serial millisecond crystallography (43). We solved the structure of T1D-GB1 at 1.94-Å resolution (SI Appendix, Table S2), and we unequivocally found ligand density at a site that is close to but clearly different from the colchicine binding site (Fig. 5A). This binding site sits at the intradimer interface between α- and β-tubulins. It is equivalent to the vinca site, but the defined pocket boundaries are reversed (Fig. 5B), meaning that contacts with loop T7, helix H10, and strand S9 are provided by β-tubulin (whereas these are at α-tubulin in the vinca site) and α-tubulin supplies interactions by loops T5 and H6−H7 (which correspond to β-tubulin in the vinca site).
Fig. 5.
Crystal structure of TD1−GB1 complex. (A) Overall view of the TD1−GB1 complex (PDB ID code 7alr). Tubulin (α-tubulin in gray and β-tubulin in light gray) and DarPin (green) are in ribbon, and tubulin-bound nucleotides (orange) and the ligand GB1 (teal) are in sphere representation. (B) Zoom into the composite GB1 site. Simulated annealing omit map of GB1 in the corresponding TD1 complex structure. The mFo-DFc electron density map (gray mesh) is contoured at 3.0σ. The GB1 molecule is shown in stick representation and α- and β-tubulin in ribbon. Secondary structural elements involved in protein–compound interactions are depicted in blue. (C and D) Close-up view of the interaction observed between GB1 (teal, sticks) and tubulin (α-tubulin in gray and β-tubulin in light gray, ribbon). Interacting residues are shown in stick representation and are labeled. D is a rotated view of C. (E) Comparison of GB1 (teal) and colchicine (blue) binding sites, where tubulin is in ribbon, and ligands are in stick representation. Zoom-in panels show α-tubulin loop T5 and β-tubulin loop T7 conformational changes required for colchicine accommodation when compared with colchicine (PDB ID code 5nm5) and GB1 (PDB ID code 7alr) structures. Ligands are in stick representation (GB1, teal; colchicine, blue), and tubulin is in ribbon representation (PDB ID code 5nm5, blue; PDB ID code 7alr, gray). Main residues involved are in stick representation and labeled.
The structure superimposes very well with the T1D apo-structure (Protein Data Bank [PDB] ID code 4drx) with an overall rmsd of 0.39 Å over 771 Cα-atoms, suggesting that the binding of GB1 does not affect the conformation of tubulin. Indeed, there are no major changes on the secondary structural elements surrounding the compound. GB1 is a cyclic peptide that shares a slightly greater buried interface area with α-tubulin (304 Å2) than with β-tubulin (278 Å2), displaying interactions with both tubulin molecules. GB1 4-MePro is sandwiched at the bottom of the pocket within the α-tubulin molecule between αY210 (helix H6) and αY224 (helix H7) (Fig. 5C), while the carbonyl moiety of this residue points toward the lateral chain of βQ247 (loop T7). The next clockwise residue is the DhAla that interacts with α-tubulin through hydrogen bond to the αR221 (loop H6−H7) side chain amide. The same carbonyl additionally co-coordinates a crystallographic water molecule that is shared with other carbonyl and hydroxy groups of the compound. The following residue is an alanine that interacts with β-tubulin through a coordinated crystallographic water molecule to the βV355 (strand S9) amide nitrogen. The hydroxamate of this alanine (N-OH-Ala) hydrogen bonds to βD329 (helix H10) side chain Oδ2 and Oδ1 (Fig. 5C), rationalizing the critical function of the hydroxamate for binding. The hydroxy propanamide residue (N(α)-Me-β-OH-Asn) makes extensive contacts with α- and β-tubulins through loop T5, including the carbonyl moiety of αP175 and αQ176, the carbonyl and the amide of αS178, and strand S9, with βK352 main chain and βV353 amide nitrogen (Fig. 5D). Additionally, the N-Me group of this residue further hydrogen bonds to βV353 carbonyl moiety. The carbonyl of the Lac that closes the ring with 4-MePro further stabilizes contacts with the α-tubulin loop T5 through hydrogen bond to the carbonyl of αQ176.
The tubulin−colchicine (PDB ID code 5nm5) and tubulin−GB1 structures superimpose with an overall rmsd of 0.497 Å over 756 Cα-atoms, with the main differences located at β-tubulin loop T7 and α-tubulin loop T5 (Fig. 5E). These loops imperatively change their conformations upon colchicine binding because, otherwise, the trimethoxy phenyl ring of colchicine (Fig. 1C) would clash with βL247 and βN248 (loop T7) and the amide of ring B would clash with the αT179 (loop T5). The presence of GB1 at the site precludes the simultaneous binding of a second ligand in the colchicine binding site due to GB1 interactions with those loops, thus correlating with the results from our competition assay in which increasing concentrations of GB1 prevent any binding of the MTC fluorescence probe to tubulin (Fig. 4 C and D).
Discussion
We took an integrated approach toward natural products drug discovery by targeting minor, highly bioactive compounds from a chemically prolific cyanobacterium, combining innovative screening and rigorous bioassay-guided isolation and structure determination with chemical synthesis to overcome the supply problem, and achieving in depth mechanistic studies as well as direct target and binding site identification. We advocated for such an approach to fully exploit the proven potential of natural products and increase the value of bioactive natural products (44). The ultimate key for a successful natural product drug discovery campaign is the choice of the source organism. We have been focusing on marine cyanobacteria, which are prolific yet underexplored marine prokaryotes with a tremendous biosynthetic potential. The gatorbulin-yielding sample was derived from a blooming “superproducer” of secondary (specialized) metabolites (natural products) that previously yielded lyngbyastatins 4 to 6, pompanopeptins A and B, tiglicamides, and largamides/largamide D oxazolidine, most of which are noncytotoxic serine protease inhibitors (12, 13, 45–47). Beyond showcasing the biosynthetic capacity of marine cyanobacteria, our discovery of GB1 exemplifies that marine cyanobacterial natural products occupy therapeutically relevant chemical space that could lead to the discovery of new biology, chemical tools, or even drug leads.
GB1 is a small (molecular weight < 500 g/mol) cyclodepsipeptide, unique from most cyanobacterial modified peptides or peptide−polyketide hybrids, which dominate the landscape of bioactive natural products produced by marine cyanobacteria (48). GB1 is densely functionalized, with all amino acids being modified and the presence of one hydroxy acid. Natural products possessing all these unusual structural features of GB1 (the hydroxamate, C-hydroxylated, and dehydroamino acids, and modified proline) have not been reported. The 4-MePro residue is a rare amino acid but has been previously found in cyanobacterial natural products (16). Interestingly, the hydroxamate group that is typical for metal chelators (especially iron siderophores) and present in other antiproliferative compounds (49) plays an iron-independent functional role in GB1’s binding to tubulin as the major mechanism of antiproliferative action. The additional metal binding ability potentially increases the pharmacological complexity of GB1 and remains to be investigated. However, the iron complexing ability due to the hydroxamate functionality was proven but appeared not to play a predominant role with respect to the antiproliferative activity, based on the NCI-60 profile and cell cycle arrest characteristic for tubulin-targeting agents. We found the metal was necessary for neither GB1 binding to tubulin nor the inhibition of tubulin assembly into microtubules in our in vitro assays.
Tubulin-targeting agents strongly affect microtubule dynamics, which is a key feature for the functioning of these filaments in a wide range of biological processes. In fact, many of these compounds have been originally discovered within the extracts from living sources that often have mobility restrictions and, hence, use them as a protection mechanism against consumers. Some compounds promote microtubule stabilization through the interaction with either the taxane site (in the lumen of the microtubule) or the pelo/lau site (wall of the microtubule). The underlying mechanism is under discussion, but these compounds prevent microtubule depolymerization by locking lateral and/or longitudinal contacts. Microtubule-destabilizing compounds are more common and frequently more toxic. These induce rapid microtubule disassembly and prevent tubulin polymerization by two different molecular mechanisms: blockage (50, 51) and wedging (52, 53). Compounds that bind to the tip (maytansine) site (top surface of β-tubulin) or pironetin site (bottom surface of α-tubulin) prevent the incorporation of a new tubulin heterodimer to the microtubule-growing end, due to the inactivation of the contact surfaces. Hence, these follow a blockage mechanism. Meanwhile, compounds interacting with the colchicine or the vinca site block the essential curve-to-straight conformational change in tubulin upon assembly, which necessarily comprise microtubule formation. Our GB1 high-resolution structure revealed a site and contributed to understanding the molecular mechanism underlying the compound’s destabilization effect. GB1 binds to a pocket localized between α- and β-tubulin, close to the colchicine site and equivalent to the vinca site (that sits between two tubulin heterodimers), denoting a common underlying mechanism to destabilize microtubule formation: wedging. The colchicine site is an elongated pocket, where some compounds (colchicine, podophyllotoxin, or noscapine) bind to the first third, interacting with β-tubulin S9 and H7 (some also interact with the α-tubulin T5 loop), whereas other compounds (e.g., nocodazole) bind deeper and also interact with the β-tubulin β sheet of the N-terminal domain (52, 54, 55). None have the extensive interactions with α-tubulin that are observed with GB1.
Most tubulin-targeting agents bind to β-tubulin, which is the active molecule within the heterodimer upon guanosine-5′-triphosphate hydrolysis, with the exception of the pironetin site that is localized in α-tubulin. Tubulin has been revealed as a complex molecule, since it includes two similar targeting sites that comprise interfaces of the two proteins, either within the heterodimer (intradimer interface, gatorbulin site) or between two heterodimers (interdimer interface, vinca site). Interestingly, these sites are equivalent, but contact surfaces are swapped between tubulin subunits. The top surface contains α-tubulin (vinca) or β-tubulin (gatorbulin) residues, whereas the bottom surface contributes with β-tubulin (vinca) or α-tubulin (gatorbulin) amino acids. Since tubulin is a key target for the treatment of cancer, this increases our options for the screening and development of new, safer, and more effective drugs. GB1 is a cyclic depsipeptide that represents a distinct chemotype that differs from other peptides targeting tubulin, such as dolastatin 10, and possesses low toxicity and molecular weight, adding to its promising small-molecule, drug-like properties and translational potential.
Materials and Methods
All experimental details on the chemistry are provided in SI Appendix, including the isolation of the natural products, determination of the absolute configuration, and synthetic procedures. Biological methods and details about cell lines, proteins, and reagents are described in SI Appendix. Detailed descriptions are provided for the cell viability assessments, cell cycle analysis (HCT116 and HeLa cells), RNA isolation and RT-qPCR, indirect immunofluorescence, caspase activity assays, tubulin polymerization, measurement of binding constants, tube formation assays, and migration and invasion assays. SI Appendix, Table S1 contains NMR data for GB2, and SI Appendix, Table S2 contains crystallography data collection and refinement statistics. NMR spectra of the natural products (GB1 and GB2) and all synthetic compounds are also provided as SI Appendix, Figs. S1–S63.
Supplementary Material
Acknowledgments
This research was supported by the NIH, National Cancer Institute Grants R01CA172310 (to H.L.) and R50CA211487 (to R.R.) and National Institute of General Medical Sciences Grant P41GM086210 (to H.L. and V.J.P.), Commercialization Fund Award from University of Florida (UF) Innovate (UF Office of Technology Licensing), and Debbie and Sylvia DeSantis Chair professorship (H.L.). The biochemical and the crystal structure work was supported by grants Ministerio de Ciencia e Innovación PID2019-10454RB-I00/AEI/10.13039/501100011033, Fondo de Investigaciones Sanitarias and COV20/01007E Proyecto Intramural Especial 201920E111 from Consejo Superior de Investigaciones Científicas (to J.F.D.) and European Union H2020-MSCA-ITN-2019 860070 TUBINTRAIN grant (to J.F.D. and A.E.P.). We thank staff of the Smithsonian Marine Station for assistance with sample collection and extraction. We thank Wesley Yoshida (University of Hawaii at Manoa) for assistance with the 15N NMR experiments and Long H. Dang (UF Department of Medicine) for providing the HCT116 knockout cells. We thank Susan L. Mooberry (University of Texas Health Science Center at San Antonio) for support with the pharmacological studies; she acknowledges Greehey Endowment support. We thank John A. Beutler of the Molecular Targets Program at the National Cancer Institute for assistance with COMPARE analysis. We thank Ganadería Fernando Díaz for calf brains supply, and thank staff of beamline XALOC (ALBA, Cerdanyola del Vallès, Spain) for their support. We thank Cora Petersen for her assistance with the cell proliferation studies.
Footnotes
Competing interest statement: The University of Florida has filed a patent application relating to the content of this article.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021847118/-/DCSupplemental.
Data Availability
Atomic coordinates (crystallographic data) have been deposited in Protein Data Bank (PDB ID code 7ALR) (56).
References
- 1.Risinger A. L., Du L., Targeting and extending the eukaryotic druggable genome with natural products: Cytoskeletal targets of natural products. Nat. Prod. Rep. 37, 634–652 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luesch H., Moore R. E., Paul V. J., Mooberry S. L., Corbett T. H., Isolation of dolastatin 10 from the marine cyanobacterium Symploca species VP642 and total stereochemistry and biological evaluation of its analogue symplostatin 1. J. Nat. Prod. 64, 907–910 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Salvador-Reyes L. A., Engene N., Paul V. J., Luesch H., Targeted natural products discovery from marine cyanobacteria using combined phylogenetic and mass spectrometric evaluation. J. Nat. Prod. 78, 486–492 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ratnayake R., et al., Dolastatin 15 from a marine cyanobacterium suppresses HIF-1α mediated cancer cell viability and vascularization. ChemBioChem 21, 2356–2366 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai R. L., Pettit G. R., Hamel E., Binding of dolastatin 10 to tubulin at a distinct site for peptide antimitotic agents near the exchangeable nucleotide and vinca alkaloid sites. J. Biol. Chem. 265, 17141–17149 (1990). [PubMed] [Google Scholar]
- 6.Cruz-Monserrate Z., Mullaney J. T., Harran P. G., Pettit G. R., Hamel E., Dolastatin 15 binds in the vinca domain of tubulin as demonstrated by Hummel-Dreyer chromatography. Eur. J. Biochem. 270, 3822–3828 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Bousquet M. S., et al., Multidimensional screening platform for simultaneously targeting oncogenic KRAS and hypoxia-inducible factors pathways in colorectal cancer. ACS Chem. Biol. 11, 1322–1331 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pouysségur J., Dayan F., Mazure N. M., Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature 441, 437–443 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Semenza G. L., Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3, 721–732 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Zhong H., et al., Overexpression of hypoxia-inducible factor 1α in common human cancers and their metastases. Cancer Res. 59, 5830–5835 (1999). [PubMed] [Google Scholar]
- 11.Paul V. J., Thacker R. W., Banks K., Golubic S., Benthic cyanobacterial bloom impacts the reefs of South Florida (Broward County, USA). Coral Reefs 24, 693–697 (2005). [Google Scholar]
- 12.Sharp K., et al., Phylogenetic and chemical diversity of three chemotypes of bloom-forming Lyngbya species (Cyanobacteria: Oscillatoriales) from reefs of southeastern Florida. Appl. Environ. Microbiol. 75, 2879–2888 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matthew S., et al., Intramolecular modulation of serine protease inhibitor activity in a marine cyanobacterium with antifeedant properties. Mar. Drugs 8, 1803–1816 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umezawa K., Ikeda Y., Kawase O., Naganawa H., Kondo S., Biosynthesis of polyoxypeptin A: Novel amino acid 3-hydroxy-3-methylproline derived from isoleucine. J. Chem. Soc., Perkin Trans. 1 13, 1550–1553 (2001). [Google Scholar]
- 15.Steele A. D., et al., Diverted total synthesis of promysalin analogs demonstrates that an iron-binding motif is responsible for its narrow-spectrum antibacterial activity. J. Am. Chem. Soc. 138, 5833–5836 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luesch H., et al., Biosynthesis of 4-methylproline in cyanobacteria: Cloning of nosE and nosF genes and biochemical characterization of the encoded dehydrogenase and reductase activities. J. Org. Chem. 68, 83–91 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Mattingly P. G., Miller M. J., Titanium trichloride reduction of substituted N-hydroxy-2-azetidinones and other hydroxamic acids. J. Org. Chem. 45, 410–415 (1980). [Google Scholar]
- 18.Kim B., et al., Evaluation of class I HDAC isoform selectivity of largazole analogues. Bioorg. Med. Chem. Lett. 24, 3728–3731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ley S. V., Priour A., Heusser C., Total synthesis of the cyclic heptapeptide Argyrin B: A new potent inhibitor of T-cell independent antibody formation. Org. Lett. 4, 711–714 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Sendai M., et al., Synthesis of carumonam (AMA-1080) and a related compound starting from (2R,3R)-epoxysuccinic acid. Chem. Pharm. Bull. (Tokyo) 33, 3798–3810 (1985). [Google Scholar]
- 21.Guzmán-Martinez A., Vannieuwenhze M. S., An operationally simple and efficient synthesis of orthogonally protected L-threo-b-hydroxyasparagine. Synlett 2007, 1513–1516 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boger D. L., Lee R. J., Bounaud P. Y., Meier P., Asymmetric synthesis of orthogonally protected L-threo-β-hydroxyasparagine. J. Org. Chem. 65, 6770–6772 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Shustov G. V., Chandler M. K., Wolfe S., Stereoselective synthesis of multiply substituted [1,2]oxazinan-3-ones via ring-closing metathesis. Can. J. Chem. 83, 92–103 (2005). [Google Scholar]
- 24.Hale K. J., Manaviazar S., George J., Total synthesis of (+)-A83586C, (+)-kettapeptin and (+)-azinothricin: Powerful new inhibitors of β-catenin/TCF4- and E2F-mediated gene transcription. Chem. Commun. (Camb.) 46, 4021–4042 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Hale K. J., Manaviazar S., George J. H., Walters M. A., Dalby S. M., Total synthesis of (+)-azinothricin and (+)-kettapeptin. Org. Lett. 11, 733–736 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Ottenheijm H. C. J., Herscheid J. D. M., N-hydroxy-α-amino acids in organic chemistry. Chem. Rev. 86, 697–707 (1986). [Google Scholar]
- 27.Li P., Xu J. C., 1-Ethyl 2-halopyridinium salts, highly efficient coupling reagents for hindered peptide synthesis both in solution and the solid-phase. Tetrahedron 56, 8119–8131 (2000). [Google Scholar]
- 28.Smith A. B. 3rd, et al., Design, synthesis, and biological evaluation of EF- and ABEF- analogues of (+)-spongistatin 1. Org. Lett. 12, 1792–1795 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shute R. E., Rich D. H., Prevention of diketopiperazine formation in peptide synthesis by a simultaneous deprotection-coupling procedure: Entrapment of reactive nucleophilic species by in situ acylation. J. Chem. Soc. Chem. Commun., 1155–1156 (1987). [Google Scholar]
- 30.Kogen H., et al., Crystal structure and total synthesis of globomycin: Establishment of relative and absolute configurations. J. Am. Chem. Soc. 122, 10214–10215 (2000). [Google Scholar]
- 31.Bajjuri K. M., Liu Y., Liu C., Sinha S. C., The legumain protease-activated auristatin prodrugs suppress tumor growth and metastasis without toxicity. ChemMedChem 6, 54–59 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaucher-Wieczorek F. S., Maillard L. T., Badet B., Durand P., Fluorous tagged N-hydroxy phthalimide for the parallel synthesis of O-aryloxyamines. J. Comb. Chem. 12, 655–658 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Schwartz E. L., Antivascular actions of microtubule-binding drugs. Clin. Cancer Res. 15, 2594–2601 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paul K. D., Hamel E., Cancer Chemotherapeutic Agents (American Chemical Society Books, Washington, DC, 1995). [Google Scholar]
- 35.Parker A. L., Teo W. S., McCarroll J. A., Kavallaris M., An emerging role for tubulin isotypes in modulating cancer biology and chemotherapy resistance. Int. J. Mol. Sci. 18, 1434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doodhi H., et al., Termination of protofilament elongation by eribulin induces lattice defects that promote microtubule catastrophes. Curr. Biol. 26, 1713–1721 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Menchon G., et al., A fluorescence anisotropy assay to discover and characterize ligands targeting the maytansine site of tubulin. Nat. Commun. 9, 2106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.La Regina G., et al., Arylthioindole inhibitors of tubulin polymerization. 3. Biological evaluation, structure-activity relationships and molecular modeling studies. J. Med. Chem. 50, 2865–2874 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Díaz J. F., Andreu J. M., Kinetics of dissociation of the tubulin-colchicine complex. Complete reaction scheme and comparison to thermodynamic measurements. J. Biol. Chem. 266, 2890–2896 (1991). [PubMed] [Google Scholar]
- 40.Risinger A. L., et al., The taccalonolides: Microtubule stabilizers that circumvent clinically relevant taxane resistance mechanisms. Cancer Res. 68, 8881–8888 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Čermák V., et al., Microtubule-targeting agents and their impact on cancer treatment. Eur. J. Cell Biol. 99, 151075 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Prota A. E., et al., Structural basis of tubulin tyrosination by tubulin tyrosine ligase. J. Cell Biol. 200, 259–270 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinert T., et al., Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons. Nat. Commun. 8, 542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luesch H., MacMillan J. B., Targeting and extending the eukaryotic druggable genome with natural products. Nat. Prod. Rep. 37, 744–746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matthew S., Ross C., Paul V. J., Luesch H., Pompanopeptins A and B, new cyclic peptides from the marine cyanobacterium Lyngbya confervoides. Tetrahedron 64, 4081–4089 (2008). [Google Scholar]
- 46.Matthew S., Paul V. J., Luesch H., Tiglicamides A.-C., Tiglicamides A-C, cyclodepsipeptides from the marine cyanobacterium Lyngbya confervoides. Phytochemistry 70, 2058–2063 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matthew S., Paul V. J., Luesch H., Largamides A.-C., Largamides A-C, tiglic acid-containing cyclodepsipeptides with elastase-inhibitory activity from the marine cyanobacterium Lyngbya confervoides. Planta Med. 75, 528–533 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan L. T., Phyo M. Y., Marine cyanobacteria: A source of lead compounds and their clinically-relevant molecular targets. Molecules 25, 2197 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bertrand S., Hélesbeux J.-J., Larcher G., Duval O., Hydroxamate, a key pharmacophore exhibiting a wide range of biological activities. Mini Rev. Med. Chem. 13, 1311–1326 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Prota A. E., et al., A new tubulin-binding site and pharmacophore for microtubule-destabilizing anticancer drugs. Proc. Natl. Acad. Sci. U.S.A. 111, 13817–13821 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prota A. E., et al., Pironetin binds covalently to αCys316 and perturbs a major loop and helix of α-tubulin to inhibit microtubule formation. J. Mol. Biol. 428, 2981–2988 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Ravelli R. B. G., et al., Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 428, 198–202 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Gigant B., et al., Structural basis for the regulation of tubulin by vinblastine. Nature 435, 519–522 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Oliva M. A., et al., Structural basis of noscapine activation for tubulin binding. J. Med. Chem. 63, 8495–8501 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Wang Y., et al., Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery. FEBS J. 283, 102–111 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Oliva M. A., Díaz J. F., Crystal structure of TD1-gatorbulin1 complex. Protein Data Bank. 10.2210/pdb7ALR/pdb. Deposited 7 October 2020. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates (crystallographic data) have been deposited in Protein Data Bank (PDB ID code 7ALR) (56).