Significance
On-target, off-tumor toxicity of anti-CTLA4 checkpoint inhibitors leads to severe adverse events, restricting therapeutic efficacy. We engineered anti-CTLA4 antibodies and generated a new class of antibodies referred to as conditionally active biologic (CAB) antibodies using physiological chemicals (bicarbonate, hydrogen sulfide) as protein-associated chemical switches (PaCS) to reduce binding under normal physiological conditions, while maintaining binding in the tumor. PaCS add a new dimension to drug discovery. PaCS can be applied to a variety of targets and drug formats, improving safety, increasing the number of targets, allowing the development of new therapies, and enhancing the tolerability of existing therapeutics. The PaCS mechanism can be used to tune the binding activity for disease-related microenvironments, including cancer, infection, inflammation, and cellular senescence.
Keywords: conditionally active biologics, protein-associated chemical switches, monoclonal antibodies, immunooncology, CTLA4
Abstract
Anticytotoxic T lymphocyte-associated protein 4 (CTLA4) antibodies have shown potent antitumor activity, but systemic immune activation leads to severe immune-related adverse events, limiting clinical usage. We developed novel, conditionally active biologic (CAB) anti-CTLA4 antibodies that are active only in the acidic tumor microenvironment. In healthy tissue, this binding is reversibly inhibited by a novel mechanism using physiological chemicals as protein-associated chemical switches (PaCS). No enzymes or potentially immunogenic covalent modifications to the antibody are required for activation in the tumor. The novel anti-CTLA4 antibodies show similar efficacy in animal models compared to an analog of a marketed anti-CTLA4 biologic, but have markedly reduced toxicity in nonhuman primates (in combination with an anti-PD1 checkpoint inhibitor), indicating a widened therapeutic index (TI). The PaCS encompass mechanisms that are applicable to a wide array of antibody formats (e.g., ADC, bispecifics) and antigens. Examples shown here include antibodies to EpCAM, Her2, Nectin4, CD73, and CD3. Existing antibodies can be engineered readily to be made sensitive to PaCS, and the inhibitory activity can be optimized for each antigen’s varying expression level and tissue distribution. PaCS can modulate diverse physiological molecular interactions and are applicable to various pathologic conditions, enabling differential CAB antibody activities in normal versus disease microenvironments.
Targeted therapies continue to transform cancer survival rates, as demonstrated by recent successes with immune checkpoint inhibitors, antibody drug conjugates, immune cell-recruiting bispecific antibodies, and chimeric antigen receptor T cell (CAR-T) therapies (1–3). In solid tumors, the tumor-specific expression of a target is rare (4). As a consequence, targets that are in higher abundance on the surface of a tumor cell relative to a normal cell are often selected for the development of immune therapies. The pharmacologic term, therapeutic index (TI), is generally based on the ratio between levels that are therapeutic and levels that are toxic. The differential activity of a ligand of a target molecule in the diseased tissue versus its normal surroundings is of fundamental importance. For example, exploiting the differential binding of an antibody to its target in a tumor versus in the surrounding environment might enhance the TI. Such a strategy could safely maintain antibody potency at higher doses and could enhance tumor-killing mechanisms (e.g., drug conjugates, bispecific antibodies, or CAR-Ts), while avoiding treatment-related toxicities, resulting from systemic, on-target, off-tumor binding on normal cells away from the tumor.
Unfortunately, the low TI that results from the expression of target molecules on normal cells renders most targets problematic or simply undruggable, hindering the development of effective treatments. An extensively studied example is the additive toxicity observed with the clinically validated Programmed Cell Death Protein-1 (PD1) and CTLA4 combination therapy. This combination can be life-threatening to many patients even at a fraction of their doses when given as monotherapy (5, 6). This lower dosing or the reduced ability for patients to remain on therapy attenuates the potential clinical effectiveness of these important immunooncology (IO) treatments (7). Enhanced targeting strategies are needed urgently, since the traditional objective of identifying new targets with high preferential expression on the tumor cell has largely failed for solid tumors (8), forcing the abandonment of otherwise valuable targets. Given that solid tumors represent over 90% of all cancer types (9), there is considerable need for improved tumor targeting and higher potency to breach solid tumor defenses.
The unique microenvironmental differences in diseased tissues such as those found associated with tumor cells, that is, the tumor microenvironment (TME), offer an important opportunity to improve target selectivity (10–13). There are several conditions that could lead to a differential antibody selectivity, including temperature (14, 15), oxygen levels (16–18), pressure (19), pH (20–27), and small-molecule and protein concentrations (28–33). Proteins have the capability to be responsive to these conditions; for example, previously, we developed enzymes to respond to differentially selective conditions (34–37). This conditional activity allows the enzyme to remain safely expressed, but physiologically inactive or active as needed in order to reduce physiological harm. Analogous approaches were utilized previously to increase the useful window of activity in microbes, plants, and humans for a wide range of effects, including processing efficiencies, maintaining yields (34), and improving drug delivery (cited in the acknowledgments of ref. 38).
An extensive body of literature demonstrates the acidic external microenvironment of tumor cells since the original observation that tumor cells are metabolically glycolytic and secreting high levels of lactic acid, even in aerobic conditions (39). Recently, Rohani et al. (40) have demonstrated that over 87% of aerobic tumor cells are acidic (<pH6.5), which is noteworthy considering that not all cells found within a tumor are malignant cells. Extracellular pH (pHe) in tumors can be as low as pH5.8 (21). The acidic nature of tumors contrasts with the highly controlled slightly alkaline pH7.4 of blood and healthy tissues. Although other microenvironmental conditions differ between tumor and normal cells (10), acidity is correlated with tumor aggressiveness (41). The consistency of this acidic microenvironment characteristic is driven, in part, by the need for precursor molecules required for rapid cell replication that are primarily supplied through glycolysis. Studies also demonstrate that metastatic cells maintain an acidic external pH close to the cell membrane even when surrounded by pH7.4 buffer (24). It is hypothesized that the combination of high tumor glycolytic rates combined with the delayed external cell equilibration due, in part, to the ∼500-nm-thick carbohydrate layer on the surface of the tumor cells assists in establishing a pH gradient keeping the tumor cell surface acidic (42, 43). Cell surface proteins are typically less than 100 nm in length, that is, less than 20% of the size of the surrounding carbohydrate layer, and, as a result, proteins reside within the tumor’s most acidic external microenvironment (44).
Here we demonstrate the ability to engineer conditionally and reversibly active CAB proteins that gain binding activity as the pHe becomes acidic when entering the TME. We engineer the conditional activation and conserve the natural structure of the human immunoglobulin by restricting amino acid changes to the human immune system’s mutation-tolerant hypervariable or complementary-determining regions (CDRs). By limiting changes to the hypervariable regions that are tolerant to some mutations without losing the fine specificity of the immunoglobulin, the strategy allows for an improved TI. The strategy also maintains efficient manufacturing characteristics and minimizes the risk of a decreased or negative immune response. Notably, we describe a novel H+ ion-dependent mechanism that blocks binding of the therapeutic molecule at normal conditions through the involvement of a noncovalently bound physiological chemical(s) (e.g., bicarbonate or hydrogen sulfide), acting as a chemical switch. This chemical switch can reversibly inhibit binding of the therapeutic effector antibody to the target antigen depending upon the extracellular pH. We refer to this novel mechanism for generating CABs as utilizing protein-associated chemical switch(es), or PaCS mechanism. PaCS-utilizing proteins or antibodies are developed by including this chemical dimension in protein engineering, evolution, and design. Characteristically, the pH inflection point of a CAB antibody−antigen interaction using a PaCS mechanism is largely established via the pKa of the PaCS chemical, as opposed to the pKa of the protein. PaCS enables pH-dependent binding without relying on the classic key pH-determining amino acid, histidine. This robust PaCS mechanism provides molecular insights into pathogenesis, as well as potentially delivering improved therapies.
Results
Engineering of Tumor-Selective Anti-CTLA4 Monoclonal Antibodies.
The goal of this study was to generate CAB anti-CTLA4 antibodies with reduced binding at normal physiological conditions (pH of ∼7.4), while maintaining binding at TME conditions [pH of ∼6.3 to 6.9 (45) and even lower at the cell membrane (24)]. A CAB anti-CTLA4 antibody which is reversibly active only in the TME, but not under normal physiological conditions, is expected to have reduced peripheral immune-related toxicity and therefore demonstrate a wider safety margin compared to the anti-CTLA4 antibodies currently available for cancer therapy, allowing higher dosing and longer treatments for improved efficacy.
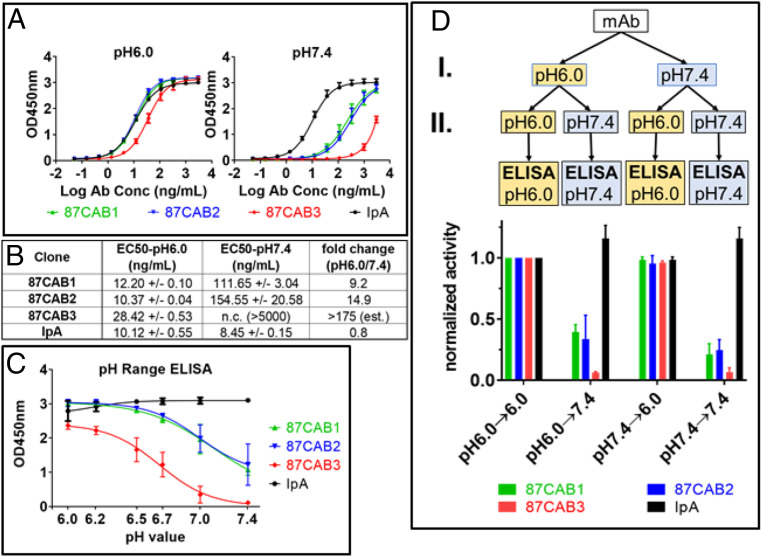
Hallmarks of the TME are a lower extracellular pH due to the increased lactic acid secretion resulting from glycolytic tumor metabolism even under aerobic conditions [i.e., the Warburg effect (39, 46)]. A library of anti-CTLA4 antibody variants [initially based on Ipilimumab (47), including mutations introduced in the six CDR loops and with optimized frameworks] were screened for binding to recombinant human CTLA4 at pH6.0 (TME) and pH7.4 (normal physiological condition). The screening yielded several clones with the desired properties (low binding at pH7.4; high binding at pH6.0). Three of these selected CAB anti-CTLA4 clones are described in this report. The identified variants included two with similar (clones 87CAB1 and 87CAB2) or slightly reduced (clone 87CAB3) binding to CTLA4 at pH6.0 compared to an Ipilimumab analog (IpA), while all had substantially reduced binding at pH7.4 (Fig. 1A). Also, the calculated effective concentration, 50% (EC50) values for clones 87CAB1 and 87CAB2 are similar to the EC50 value of the IpA at pH6.0 (∼10 ng/mL to 12 ng/mL; Fig. 1B), while clone 87CAB3 has a moderately increased EC50 at pH6.0 (∼28 ng/mL; Fig. 1B). IpA shows undesirable stronger binding at pH7.4 (∼8.5 ng/mL) relative to pH6.0, while the binding of clones 87CAB1 and 87CAB2 decreases to only 6 to 11% at pH7.4 relative to pH6.0 (i.e., a 9- to 15-fold change). Clone 87CAB3 has the lowest binding at pH7.4 for the largest binding differential between pH6.0 and pH7.4, with a decrease in EC50 to 0.5% of the binding at pH6.0 (i.e., a 175-fold change as estimated based on the data shown in Fig. 1A).
Fig. 1.
The pH selectivity of anti-CTLA4 variants. CAB anti-CTLA4 antibodies were identified by screening a library of antibody variants for binding to recombinant human CTLA4 in acidic pH, but not in neutral pH, using the PaCS process. (A) Binding activities of CAB anti-CTLA4 variants to human CTLA4 at pH6.0 (Left) and at pH7.4 (Right) were determined using pH affinity ELISA; y axis, optical density (OD) 450 nm; x axis, antibody concentration (log nanograms per milliliter); the starting concentration is 3 µg/mL. The mean OD values from two independent experiments with duplicate reactions are shown. (B) EC50 values of CAB anti-CTLA4 variants binding to human CTLA4 at pH6.0 and at pH7.4 were calculated using the nonlinear fit (variable slope, four parameters) model built into GraphPad Prism software version 7.03. The binding curve for clones 87CAB3 did not reach saturation at pH 7.4, and therefore the EC50 value could not be calculated. (C) Binding activities of CAB anti-CTLA4 variants to human CTLA4 at different pH values were determined by pH range ELISA; y axis, OD 450 nm; x axis, pH of the ELISA mix and wash buffers. The mean OD values from two independent experiments with two replicates for each pH test are shown. Inflection point of the pH curves: Mean OD values (from two independent experiments) at the different pH were plotted against the pH of the buffer using GraphPad Prism software version 7.03. Curve fitting was done using the four-parameter model built into the software. The inflection point of the pH curve (pH with 50% binding activity) equals IC50 of the fitting equation. Binding activity at pH6.0 was set to 100%; 87CAB1 pH inflection point, pH6.92; 87CAB2 inflection point, pH6.95; 87CAB3 pH inflection point, pH6.66. (D) The pH-dependent binding of CAB anti-CTLA4 variants is reversible. The CAB anti-CTLA4 variants (clones 87CAB1, yellow; 87CAB2, purple; and 87CAB3, red) were tested in four different conditions: Antibodies were first diluted from stock solutions (in phosphate-buffered saline [PBS] pH7.4) into pH6.0 or pH7.4 ELISA mix at 250 ng/mL and incubated at room temperature for 30 min (step I in the flowchart). The mAbs were then further diluted to 25 ng/mL in pH6.0 or pH7.4 ELISA mix (step II). Binding to human CTLA4 immobilized in the wells was tested by ELISA. All ELISA steps (blocking, incubation, and washing) were done in either pH6.0 or pH7.4 ELISA mix as indicated in the flowchart. Data from two independent experiments with two replicates were normalized to test condition I (pH6.0→6.0). IpA was used as non-CAB reference in all experiments (black).
IpA showed a similar binding signal across the pH range from pH7.4 down to pH6.5, with slightly lower binding signal between pH6.2 and pH6.0 (Fig. 1C). With the selected antibody−antigen ratio for this analysis, the binding signal for clones 87CAB1 and 87CAB2 is 1/4 to 1/3 of the signal compared to IpA at pH7.4, while clone 87CAB3 has nearly undetectable binding at pH7.4 (Fig. 1C). For all CAB anti-CTLA4 clones analyzed, the signal increased with lower pH. Clones 87CAB1 and 87CAB2 reached the maximum signal at pH6.5, while clone 87CAB3 reached the maximum at pH6.0 (Fig. 1C). The pH inflection point (halfway point between minimum and maximum signal) is ∼pH6.9 for clones 87CAB1 and 87CAB2, while it is ∼pH6.7 for clone 87CAB3.
We then tested whether the exposure to low pH would permanently activate the binding activity of clones 87CAB1, 87CAB2, and 87CAB3 and whether the binding of these clones was reversible (Fig. 1D). Antibodies incubated at pH6.0 and then assayed at pH7.4 showed the same reduced activity as the antibodies which were assayed at pH7.4 throughout the experiment. Conversely, antibodies incubated at pH7.4 and then assayed at pH6.0 retained the same activities as the antibodies which were assayed at pH6.0 throughout the experiment. These data indicate that the binding activities of clones 87CAB1, 87CAB2, and 87CAB3 are pH dependent and reversible.
Identification of PaCS as an Important Modulator of pH-Dependent Antigen Binding and In Vitro Bioactivity.
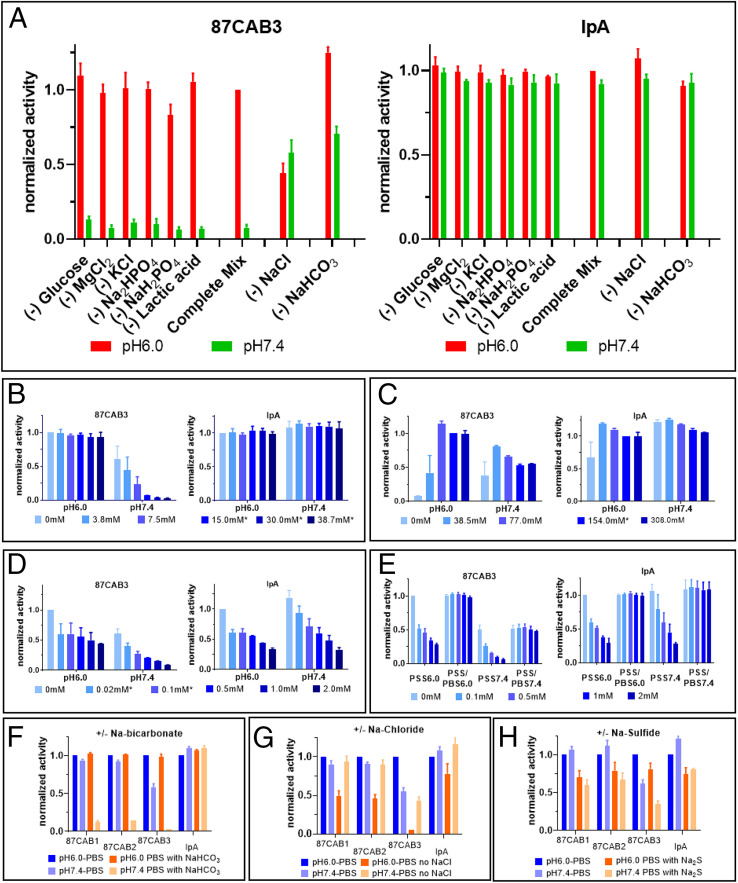
Clone 87CAB3 has the largest pH selectivity (Fig. 1) and was chosen for further characterization. The complete ELISA mix used for screening consists of eight different components. Clone 87CAB3 was tested for binding at pH6.0 and pH7.4 in ELISA mix, with each assay lacking one of the components. Both sodium chloride and bicarbonate affect the binding signal compared to the complete ELISA mix. Without sodium chloride, the binding signal at pH6.0 drops to ∼50% of that observed in the presence of sodium chloride with a fourfold to fivefold increase in binding at pH7.4, reducing overall pH-dependent selectivity (Fig. 2A). Furthermore, in the absence of sodium bicarbonate and in the presence of sodium chloride, clone 87CAB3 exhibited a sixfold loss in pH selectivity (i.e., maintaining high-affinity binding activity at pH6.0 and gaining substantial binding activity at pH7.4). In the presence of both sodium bicarbonate and sodium chloride, clone 87CAB3 demonstrated the strongest, selective pH-dependent binding (high binding activity under the pH6.0 condition, but barely detectable binding activity at pH7.4) (Fig. 2A). Removing any of the components from the ELISA mix did not affect the binding activities of the non-CAB IpA.
Fig. 2.
Influence of chemical compounds on the pH selectivity. (A–E) Normalized binding activities of clone 87CAB3 (Left) and Ipilimumab analogue (IpA, Right) to human CTLA4 at pH6.0 and pH7.4 were determined by ELISA in different buffer conditions. y axis: normalized binding activities; x axis: sample ID. Normalized activity from at least two independent experiments with duplicates are shown. (A) Influence of individual assay components on pH selectivity. Individual chemical components were left out of the complete assay mix as indicated on the graph. Data normalized to the signal at pH6.0 using the complete assay mix. Red bars: assay at pH6.0; green bars: assay at pH7.4. (B) Sodium bicarbonate titration (0 mM to 38.7 mM NaHCO3 sodium bicarbonate). Bicarbonate concentrations within the physiological range (48) are marked with asterisks. Data normalized to pH6.0, 0 mM sodium bicarbonate. (C) Sodium chloride titration (0 mM to 308 mM NaCl). Sodium chloride concentrations within the physiological range (46) are marked with asterisks. Data normalized to pH6.0, 154 mM sodium chloride. (D) Sodium sulfide titration (0 mM to 2 mM Na2S). Physiological concentration of sulfide is less than 0.1 mM (48). Data normalized to pH6.0, 0 mM sodium sulfide. (E) Sodium sulfide induced reduction in binding activity is reversible. Clone 87CAB3 and IpA were first diluted from stock solutions (in PBS pH7.4) into pH6.0 or pH7.4 ELISA assay mix (containing sodium sulfide with various concentrations as indicated) at 1,000 ng/mL and incubated at room temperature for 30 min. The mAbs were then further diluted to 2 ng/mL in pH6.0 or pH7.4 ELISA assay mix with or without sodium sulfide. Binding to human CTLA4 immobilized in the wells was tested by ELISA. All ELISA steps (blocking, incubation, washing) were done either in pH6.0 or pH7.4 assay mix in the presence of sodium sulfide (PSS6.0 or PSS7.4) or without sodium sulfide (PSS/PBS6.0 or PSS/PBS7.4). Data were normalized to pH6.0, 0 mM sodium sulfide. PSS: PBS buffer with sodium sulfide. Influence of sodium bicarbonate (F), sodium chloride (G), and sodium sulfide (H) on other anti-CTLA4 variants. Binding activities of anti-CTLA4 variants (clones 87CAB1, 87CAB2, and 87CAB3) and IpA to human CTLA4 at pH6.0 (dark columns) and pH7.4 (light columns) in the absence (blue) or presence (orange) of the indicated component. y axis: normalized OD 450 nm; x axis: sample ID. Data were normalized to pH6.0, two independent experiments with duplicate reactions. (F) Binding to human CTLA4 in the presence or absence of 30 mM sodium bicarbonate in the assay. (G) Binding to human CTLA4 in the presence or absence of 154 mM sodium chloride in the assay. (H) Binding to human CTLA4 in the presence or absence of 0.1 mM sodium sulfide in the assay.
In addition, we observed bicarbonate concentration-dependent inhibition of the binding activity at pH7.4, while no inhibition was observed at pH6.0 (Fig. 2B). Based on the estimated pKa of 6.35 of bicarbonate under physiological conditions (48), the concentration of the negatively charged bicarbonate ion will decrease at pH6.0, and the equilibrium reaction shifts toward carbonic acid (CO2+H2O↔H2CO3↔HCO3−+H+), while the total CO2 concentration stays the same. This suggests that the negatively charged bicarbonate ion is inhibiting the antibody−antigen interaction at pH7.4.
Titrations with sodium chloride in the absence of bicarbonate showed that the binding of clone 87CAB3 to CTLA4 is dependent on the presence of at least low levels of sodium chloride and requires near-physiological levels [137 mM to 145 mM (49, 50)] for full binding (Fig. 2C). The presence of sodium chloride near physiological levels is also required for the nonbicarbonate-dependent pH selective binding (Fig. 2C). The binding activity of IpA was slightly reduced at pH6.0 in the absence of sodium chloride. Taken together, the data show that sodium chloride concentrations approaching physiological levels are essential for clone 87CAB3′s binding to CTLA4. In addition, bicarbonate and sodium chloride are needed for maximizing the pH selectivity of clone 87CAB3, that is, the inhibition of binding at pH7.4 with bicarbonate and the increased binding at pH6.0. While bicarbonate is the primary contributor in pH selectivity of clone 87CAB3, sodium chloride alone enabled approximately twofold pH selectivity (Fig. 2 A–C).
We noted the disease-relevant pKa of ∼7.04 for the physiologically occurring molecule, hydrogen sulfide (HS−), and therefore we investigated whether it could also influence pH-dependent binding. We observed some general reduction in binding activity for both IpA and 87CAB3 with concentrations even below normal physiological levels [i.e., <∼0.1 mM (51, 52)] of hydrogen sulfide, experimentally provided as sodium sulfide (Fig. 2 D and H). This reduction was observed with all of the proteins shown in this report and is possibly a general phenomenon (Fig. 2H and SI Appendix, Fig. S1C). Importantly, we also noticed that the pH selectivity of clone 87CAB3 is increased at sodium sulfide concentrations of 0.5 mM to 2 mM (Fig. 2D), while the activity of IpA is equally affected at both conditions with no improvement in selectivity. In addition, we observed that the decreased binding in the presence of sodium sulfide is also reversible (Fig. 2E). Clone 87CAB3 and IpA show full binding activity after incubation with sodium sulfide followed by reducing concentration through dilution before assaying. This indicates that hydrogen sulfide can facilitate PaCS activity, even though the original antibody was selected using bicarbonate instead of hydrogen sulfide. The pH selectivity dependence on sodium bicarbonate and sodium sulfide, as well as sodium chloride for binding activity, was not limited to clone 87CAB3. More specifically, we found that bicarbonate also inhibited the binding activity of all three anti-CTLA4 variants at pH7.4, but not at pH6.0 (Fig. 2F). Furthermore, all three anti-CTLA4 variants showed reduced binding at pH6.0 in the absence of sodium chloride (Fig. 2G). This loss of binding activity was most pronounced in clone 87CAB3. Clone 87CAB3 was the only clone showing a reduced binding signal at pH7.4 in the absence of sodium bicarbonate. In the presence of sodium chloride (and the absence of bicarbonate), only clone 87CAB3 maintains some pH-dependent binding (Fig. 2G), but the pH selectivity was not as pronounced as in the presence of bicarbonate (Fig. 2F). We also observed pH selectivity for all three anti-CTLA4 variants when replacing sodium bicarbonate with sodium sulfide (Fig. 2H). Finally, the calculated isoelectric point (pI) of all the selected anti-CTLA4 CAB variants did not differ appreciably from the parent IpA (IpA, pI = 8.54; 87CAB1, pI =7.91; 87CAB2, pI = 8.25; 87CAB3, pI = 8.67).
Lactic acid is highly enriched in the TME as a result of high glycolytic rates even in oxygenated microenvironments, known as the Warburg effect (39, 53). However, lactic acid showed no effect on the binding of IpA and on any of the CAB antibodies we tested. This suggests that the blocking activity of bicarbonate and hydrogen sulfide is not just dependent upon the presence of a negative charge, but may also be a function of their electronegativity, molecular size, and shape. In addition, some limited screening of antibody libraries for pH-dependent activation by lactic acid did not yield CABs utilizing lactic acid as a PaCS chemical.
Generating PaCS Mechanistic CAB Antibodies against Other Tumor Targets.
We also tested the binding activities of conditionally selected antibodies against five different targets (SI Appendix, Fig. S1), which were generated using the PaCS process as described in Materials and Methods for the anti-CTLA4 variants in the presence or absence of bicarbonate, sodium chloride, and sodium sulfide. The results indicate that there are two major classes of CAB antibodies in this set: 1) pH selectivity strictly dependent on the presence of PaCS chemicals (e.g., bicarbonate, hydrogen sulfide) at physiological concentrations and 2) pH selectivity that is independent of the presence of PaCS chemicals (SI Appendix, Fig. S1 A and C). The requirement for sodium chloride for binding (especially at pH6.0) was not universal, but it does frequently modulate binding in this broader set of antibodies (SI Appendix, Fig. S1B). The large number of conditionally active antibodies discovered when using bicarbonate demonstrates that PaCS selection greatly expands the success rate and degree of selectivity observed in pH selective antibodies compared to solely histidine-dependent protein selectivity. Furthermore, the impact of protein-associated endogenous chemicals emphasizes the underappreciated importance of using disease-like assay conditions, instead of normal physiological conditions, for protein discovery and evolution in order to maximize responses and to help ensure effective translation in vivo.
PaCS-Responsive Amino Acid Changes.
The mutations found in the collection of CAB antibodies described in this report are summarized in SI Appendix, Fig. S2. Mostly polar and nonpolar amino acids are replaced by charged amino acids. Over 50% of the introduced mutations are aspartic (D) and glutamic (E) acid. Histidine (H) is introduced only in less than 10% of the molecules, but is also eliminated in ∼5% of the mutations, indicating that histidine does not play a major role in the design of PaCS pH-based CABs. It is also noteworthy that only a few changes are necessary to render an antibody sensitive to PaCS.
Kinetic Analysis of pH-Dependent Binding of Clone 87CAB3.
Binding kinetics of clone 87CAB3 were analyzed by surface plasmon resonance (SPR) SI Appendix, Fig. S3 A and B. Varying antibody concentrations were injected over a sensor surface with immobilized CTLA4 extracellular domain. Experiments were carried out at pH6.0 or pH7.4 with 30 mM sodium bicarbonate in the buffer. The tests showed that clone 87CAB3 has a dissociation constant (Kd) of ∼0.53 nM at pH6.0 and a Kd of ∼2.2 nM at pH7.4 (SI Appendix, Fig. S3C). In addition to the change in Kd, we also observed a sharp drop in the maximum SPR signal from pH6.0 to pH7.4 (from ∼30 resonance units [RU] to ∼10). Simulations of the SPR data showed that this drop was not caused by the approximately fourfold change in Kd. The shape of the SPR curves at pH7.4 indicates that fewer CTLA4 ligands are available for binding, compared to those available at pH6.0. This was also confirmed by SPR simulations (SI Appendix, Fig. S3 A, Bottom). The simulations indicated that, at pH7.4, less than 30% of binding sites are available on the chip compared to those available at pH6.0, which is in good agreement with the observed signals. This reduction in reactive ligand was not caused by protein denaturation after repeated regeneration cycles, as the signal recovered again when the condition was switched back to pH6.0. The data indicate that bicarbonate interacts with CTLA4 at pH7.4, preventing clone 87CAB3 from binding. As predicted from the ELISA tests, IpA was not affected, and its Kd and SPR signal intensity did not change significantly from pH6.0 to 7.4 (SI Appendix, Fig. S3 B and C). As a result, pH selectivity is a product of both the fold change in Kd and the fold change in maximum signal observed (i.e., estimated >12-fold shift in binding).
In Vitro Functional Activities of Clone 87CAB3.
The pH selectivity of clone 87CAB3 was also tested by flow cytometry using mammalian cells expressing human CTLA4 on the cell surface (SI Appendix, Fig. S4A). The high expression levels of the recombinant receptor increase the potential for avidity, which could dampen the selectivity in this assay. Nevertheless, the EC50 for clone 87CAB3 at pH7.4 is about threefold higher compared to pH6.0 in the presence of bicarbonate (SI Appendix, Fig. S4B).
We also compared the functional activity of clone 87CAB3 in tumor-associated acidic pH and normal physiological pH conditions using human lymphocytes stimulated with staphylococcal enterotoxin B (SEB; SI Appendix, Fig. S5). In SEB-stimulated peripheral blood mononuclear cells (PBMC) cultures, the addition of 87CAB3 antibody enhanced IL-2 production over the level observed with the added isotype control antibody in the acidic pH (pH6.2). In contrast, IL-2 levels did not increase with the addition of clone 87CAB3 in the neutral pH (pH7.4). At the concentration of 10 μg/mL, 87CAB3 promoted a 1.5-fold increase in IL-2 production compared to the isotype control in the pH6.2 tumor environment, similar to the increase levels observed with the IpA (SI Appendix, Fig. S5). These results support that clone 87CAB3 is functionally active and selective in vitro (active in acidic pH; inactive in neutral pH).
In Vivo Efficacy and Immunotoxicity of CAB and Non-CAB Anti-CTLA4 Antibodies in Mice and Nonhuman Primates.
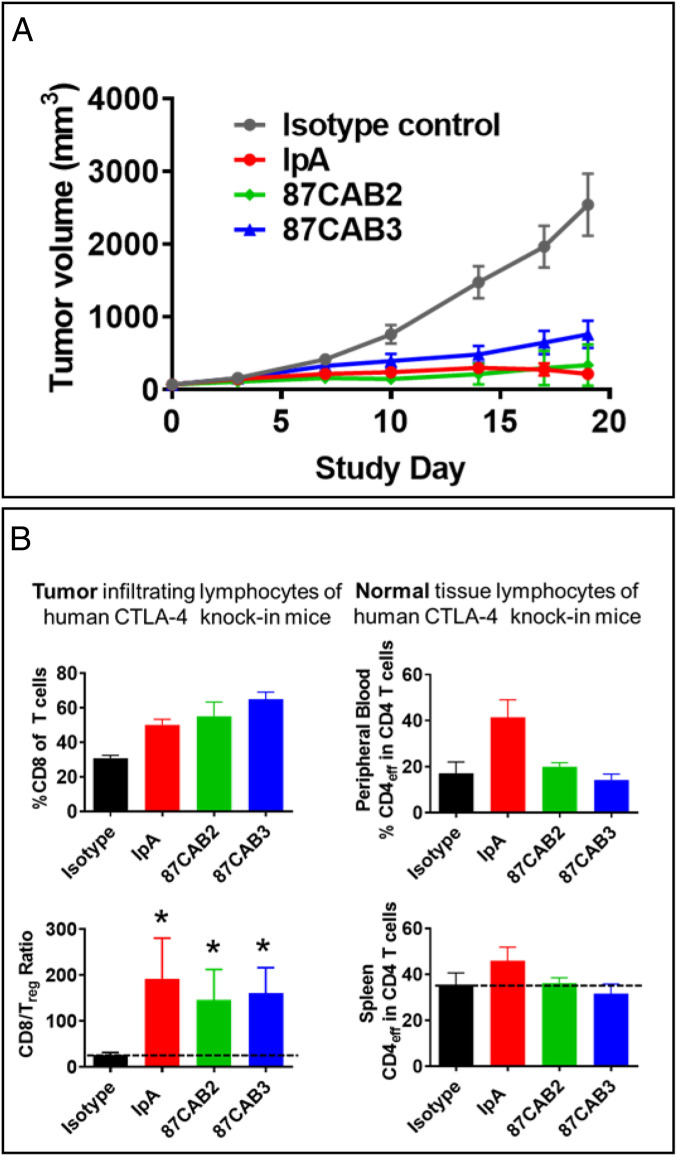
The PaCS process enabled the identification of novel CAB anti-CTLA4 antibodies which would not have been identified using conventional approaches. The CAB anti-human CTLA4 antibodies that differed from their wild-type parental antibodies with respect to pH selectivity were tested for their efficacy and potency. Since the CAB and non-CAB anti-CTLA4 antibodies did not cross-react with the murine ortholog, we used a human CTLA4 knock-in mouse model to assess antitumor efficacy in a syngeneic tumor xenograft model previously shown to be useful in demonstrating the effects of CTLA4 blockade on tumor growth (54). The CTLA4 knock-in mice were xenografted with the mouse MC-38 syngeneic colon adenocarcinoma tumor and then administered CAB and non-CAB anti-CTLA monoclonal antibodies (mAbs) (Fig. 3A). The anti-CTLA4 antibody IpA had potent antitumor activity that led to regressions when given at 3 mg/kg every 4 d for 3 wk. The CAB antibodies 087CAB3 and 87CAB2 had similar activity at the same dosage and dosing frequency. A humanized isotype negative control antibody (anti-HIV gp120) (55) had no effect on tumor growth when administered in the same manner. This indicates that the CAB and non-CAB anti-CTLA4 antibodies had equivalent efficacy. We analyzed the data using quantitative systems pharmacology and determined a correlation between an individual subject’s EC50 and Emax values which suggests that maximal efficacy has not yet been at a maximum with the dose levels tested. In addition, graphical assessment of individually estimated Emax and EC50 values (random effects) showed no apparent difference in the median values, indicating that the CAB anti-CTLA4 variants had the same activity as the non-CAB IpA. We then compared the effect of CAB and non-CAB antibodies on the activity of T cells within the TME and periphery of the treated mice using immunophenotyping (Fig. 3B). Our results indicate that all of the CAB anti-CTLA4 antibodies tested led to increases in T cell activation within the tumor and a reduction in T regulatory (Treg) cell types (Fig. 3 B, Left). Further, while the non-CAB antibodies increased T cell markers in the periphery, the CAB variants did not (Fig. 3 B, Right). This suggests that the CAB anti-CTLA4s did not lead to peripheral activation of T cells that are thought to drive the dangerous immunotoxicity associated with CTLA4 treatments (54, 56, 57). The CAB anti-CTLA4 antibodies have antitumor efficacy equivalent to that of IpA, including with tumor regression against MC38 colorectal tumor in a syngeneic human CTLA4 knock-in mice. Like IpA, CAB anti-CTLA4 antibodies modulate the activity and the numbers of infiltrating T cell subsets within the TME, but, unlike IpA, the CAB anti-CTLA4 antibodies do not increase CD4 effector T cells in the periphery beyond the TME, for potential reduction in systemic immune toxicities impacting normal tissues. Taken together, the data suggest that CAB anti-CTLA4 antibodies provide the opportunity to be dosed to much higher levels to achieve maximal efficacy, yet would have reduced toxicities and improved safety.
Fig. 3.
In vivo efficacy of CAB and non-CAB anti-CTLA4 antibodies in a humanized CTLA4 knock-in mouse tumor xenograft model. The antitumor effects and impact on tumor and peripheral immunophenotype of anti-CTLA4 antibody treatments. (A) Human CTLA4 knock-in mice (huCTLA4-KI) were xenografted with the syngeneic mouse MC-38 colon tumor cell line, then split into four cohorts treated with either IpA (red), CAB anti-CTLA4 antibodies 87CAB2 (green) or 87CAB3 (blue), or humanized IgG isotype control antibody (black) twice weekly for 3 wk at 3 mg/kg by intravenous administration. Tumor volume was measured for 3 wk every 4 d. Plotted are the average growth curves for each cohort consisting of 10 animals each. (B) T cell lymphocyte levels in the tumor and in the periphery were measured by immunohistochemistry (intratumor) and by flow cytometry (peripheral blood and splenocytes) to quantitate T cell and T cell subset levels. The percentage of CD8+ T cells (Upper Left) and the ratio of CD8+ to Treg cells (Lower Left) were analyzed in tumors as described (58). To access the impact of treatments of T cells levels in the periphery, the number of CD4+ cells in the peripheral blood (Upper Right) and spleen (Lower Right) was measured by flow cytometry. *P < 0.05 as indicated above the bars.
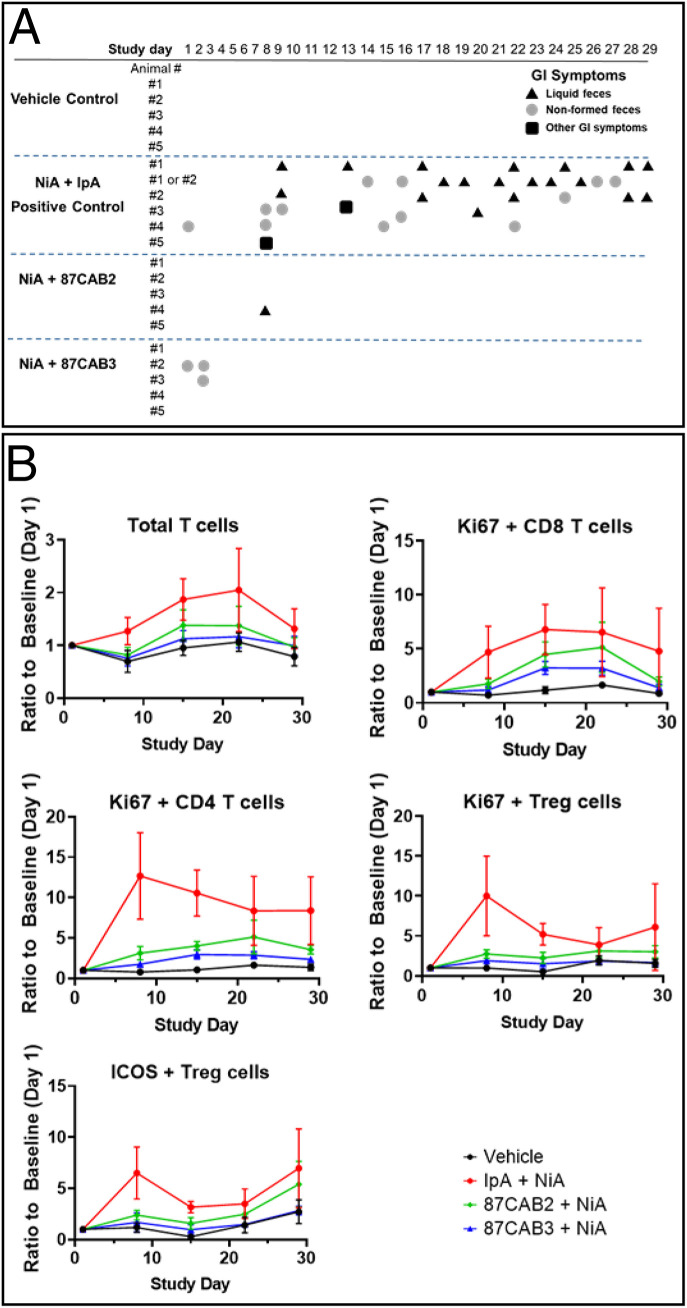
To look more closely at the apparent differences in immunotoxicity associated with non-CAB and CAB anti-CTLA4 antibodies, we chose a suitable nonhuman primate model that is sensitive to anti-CTLA related toxicities (58). Repeated coadministration of either CAB anti-CTLA4 or IpA in combination with an anti-PD1 (nivolumab analog; NiA) into monkeys for 4 wk was performed to access the peripheral systemic and normal tissue effects of combination treatments (Fig. 4A). Combination treatment with both IpA and NiA analogs resulted in increases in T cell proliferation markers in peripheral blood cells, while the CAB anti-CTLA4 plus NiA had normal immunophenotypic patterns (Fig. 4B). All animals in the IpA plus NiA treated groups had significant gastrointestinal (GI) symptoms (diarrhea, loose stools) that presented early, were sustained throughout the treatment period, and were associated with substantial mononuclear infiltration within the intestinal wall. In sharp contrast, the CAB anti-CTLA4 plus NiA treated groups showed no significant GI symptomology nor histopathology. In the cohorts given IpA plus NiA, all the animals showed signs of GI toxicity on at least 1 d, and a majority of the animals suffered GI toxicity on multiple days. In contrast, for 87CAB2, for example, only one animal showed signs of GI toxicity on a single day. The collective analysis of our mouse and monkey studies demonstrated that the TI for 87CAB2 compared to IpA is approximately sixfold higher than IpA. We believe this number is likely an underestimate of the TI, since the levels used did not reach the “no adverse effect level” in nonhuman primates. These data indicate that our CAB anti-CTLA4 molecule may have a superior safety profile when used in combination with PD1 inhibitors and allow increased dosing levels to achieve superior efficacy relative to current anti-CTLA4 therapy as a single agent or in combination with other anticancer therapies, including IO agents.
Fig. 4.
Anti-CTLA4 CABs nonhuman primate toxicity study. (A) Clinical observations of cynomolgus macaques treated in combination with anti-PD1 antibody (NiA) and anti-CTLA4 antibodies (IpA and CABs 87CAB2 and 87CAB3). Gastrointestinal toxicity was monitored as previously described (58) by measuring liquid feces or diarrhea (triangles), loosely formed feces (circles), or other GI symptoms such as vomiting or failure to eat food (squares). In some cases (animals 1 and 2), the source of liquid feces or loose stools could not be determined, as they were cohabitated during the experiment and listed as either 1 or 2. (B) Immunophenotyping of PBMC isolated from blood samples taken during the time course of anti-PD1 and anti-CTLA4 antibody treatments. Day 1 represents pretreatment baseline measurements, and day 29 represents 7 d following the last (fourth) antibody treatment. PBMC samples were isolated from heparinized blood samples by standard density gradient centrifugation using Ficoll−Hypaque medium. PBMCs were analyzed with antibodies that specifically recognize T cells (CD3) or T cell subsets T helper (CD4), T cytotoxic (CD8), or Treg cells (CD3, CD4, CD25, CD127, and FoxP3) as previously described (58). Cell activation state was measured by staining for the nuclear antigen Ki67. Inducible T cell costimulator (ICOS) staining was used as an additional antigen to also determine the level of the peripheral Treg cell activation state. The absolute levels and ratios of cells were compared by measuring the mean fluorescent intensity produced by staining using flow cytometry as previously described (58).
Discussion
We have shown that we can generate antibodies that have conditional binding to their target antigen based on amino acid changes only in hypervariable regions of the antibody. Conditional binding is primarily the result of selection of antibodies in conditions that reflect the unique chemical differences existing between normal and disease microenvironments. In this example, the conditional binding is governed by the characteristic differences in pH between the TME and normal tissue. This novel, broadly applicable conditional binding is the result of selecting antibody variants that are modulated by PaCS utilizing physiological chemicals, including sodium bicarbonate, sodium sulfide, and sodium chloride. By selecting antibodies that are responsive to a PaCS mechanism at pH levels that typify the surface of tumor cells, we can now generate antibodies to important solid tumor targets that selectively act with high binding in the TME. This chemical switch system extends the blocking, as well as enhancement of protein−protein interactions, beyond the inherent ability of a protein’s physiologically relevant pH sensing amino acid, histidine (pKa 6.1). These new pH-sensitive chemical components expand the number and degree of pH-controlled interactions. We evaluated the pH conditionally selected antibodies against different targets in the absence or presence of the PaCS chemical, bicarbonate. We found that many of these antibodies required bicarbonate for their pH selectivity (Fig. 4A). However, there were a few antibodies which have pH selectivity in the absence of bicarbonate or other chemical-dependent binding (SI Appendix, Fig. S1). The large number of antibodies discovered with high selectivity when using bicarbonate demonstrates that PaCS greatly expands the potential applicability of pH selectivity with antibodies, as well as in all proteins. Furthermore, these data underscore the importance of using precise physiological conditions for protein discovery and evolution to maximize responses and to ensure effective translation in vivo. It is notable that antibodies were also developed with both lower and higher pH midpoint inflections with high selectivity (Fig. 1). A range of pH selectivity inflection points is valuable, since the appropriate pH inflection point and selectivity for a given antibody is dependent upon several factors, including cell target density, antibody affinity, antibody effector mechanism, pharmacokinetics, dosing frequency, and disease type.
The ability to engineer CAB antibodies with conditional activity in the TME using the PaCS generation process allows us to target the cell-surface proteins that are relevant cancer targets but yet exist on normal tissues, where antibody binding outside the TME would cause significant toxicity. The CAB/PaCS system can improve the TI for both monotherapies and combination therapies, including combining antibodies to checkpoint inhibitors, which are often not tractable due to toxicity resulting from normal tissue expression. We have generated conditionally active anti-CTLA4 antibodies with restricted activity to the TME. These conditionally active anti-CTLA4 antibodies provide strong anticancer immune responses, while avoiding activity in the peripheral immune system in the blood or lymphoid tissues, thus reducing immune-related toxicity, reflected in an improved TI.
In addition, we also selected one antibody, 87CAB2, that maintains good activity at pH7.0 for potentially enabling activity in the lymph nodes for the recruitment of new T cells, while still avoiding immune-related toxicity via low binding at pH7.4 in normal tissues. CTLA4 is a receptor on T cells that acts as a brake on their activation during immune responses, and, when blocked, can stimulate cancer immune surveillance. The use of anti-CTLA4 antibodies to unleash T cells has demonstrable antitumor efficacy in animal models and in clinical studies (54, 56). However, blockade of CTLA4 as a single agent and in combination with chemotherapy, targeted, and other immunomodulatory therapies has been limited clinically, in part, due to systemic immune-related toxicities. In addition, there is increasing concern that systemic immune activation may lead to sustained autoimmune-related disease in survivors. Our CAB technology generates antibodies that reversibly bind to the target antigen in the context of diseased tissues, but not normal tissues, by taking advantage of the unique cancer microenvironment that is produced largely because of glycolytic tumor metabolism, including the Warburg effect.
In the clinic, immune-related adverse effects (irAEs) present a major obstacle limiting the use of checkpoint inhibitors. Grade III irAEs were observed in 28% of the patients in a recent phase III trial with anti-CTLA4 treatment (59). When combined with anti-PD1 antibodies, the frequency of irAEs can go up to 60%. These disturbing adverse reactions clearly emphasize the currently unmet need for CTLA4 inhibition that is active only in the TME but not under normal physiological conditions. The CAB technology minimizes systemic exposure to anti-CTLA4 treatment.
We have engineered CAB anti-CTLA4 antibodies that reversibly bind human CTLA4 and enhance T cell response to bacterial superantigen in vitro under TME conditions, but not in normal tissue conditions. The CAB anti-CTLA4 antibodies were developed by screening antibody libraries (based, in part, on the Ipilimumab sequence) under normal physiological conditions (characterized by pH7.4) and TME conditions (characterized by pH6.0). Comprehensive positional evolution (CPE) was used to mutate each position of the six CDRs. The CDRs of selected hits were combined with validated human framework libraries and screened again for enhanced pH selectivity. All mutants were expressed as full-length IgGs in mammalian cells using our comprehensive integrated antibody optimization process as outlined in Materials and Methods. We observe that the use of the final molecule format (in this case, IgG1) and the production host (mammalian cells) in all steps of the molecular evolution process is optimal for the identification of clones with the desired characteristics in combination with good manufacturability.
Clone 87CAB3 demonstrates pH selectivity in the absence of sodium bicarbonate; however, this selectivity is further increased in the presence of bicarbonate. The pH selectivity of two other anti-CTLA4 CABs discussed in this report depends on the presence of physiological levels of bicarbonate. This finding supports the observation that bicarbonate is one of the key PaCS chemicals enabling pH-selective binding. All three PaCS molecules presented in this study were present in Earth’s earliest stages of formation and were likely coevolutionary components, enabling greater functionality in early molecules that has been preserved and likely expanded over time. In fact, the bicarbonate molecule is well known as being part of the body’s natural buffer system to maintain the pH at 7.4 (±0.1) (60), NaCl is a ubiquitous charge-shielding and cell-signaling molecule, and H2S has been increasingly implicated in modulating biochemical activity (51, 61–65).
The binding of a PaCS-antibody to its target shifts as a function of the microenvironmental pH and the pKa of the specific PaCS chemical(s) utilized. In the case of the bicarbonate/carbonic acid (pKa is ∼6.4) at pH7.4, the equilibrium is shifted toward negatively charged bicarbonate. In the acidic TME, the bicarbonate level is lower as the equilibrium shifts toward carbonic acid. At physiological conditions (pH7.4), bicarbonate actions are consistent with interacting with the protein surface and directly or indirectly blocking the binding of the CAB antibodies to the antigen. Analysis of the binding kinetics of clone 87CAB3 by surface plasmon resonance showed that the affinity is about fourfold lower at pH7.4 compared to pH6.0 in the presence of bicarbonate. In addition, the maximum SPR signal is only about 1/3 at pH7.4 compared to pH6.0. The change in the shape of the SPR sensorgrams from pH6.0 to pH7.4 implies that, in this situation, the bicarbonate primarily interacts with CTLA4 and not the CAB antibody. This finding is supported by the in silico SPR simulations (SI Appendix, Fig. S3). The PaCS mechanism therefore enables enhanced selectivity from both the antibody and antigen components of the binding reaction. In addition, the two independent mechanisms resulting from the pH-dependent affinity shift and the reduction of the available binding sites between the antigen−antibody interaction result in far greater selectivity. The combination of these two independent effects results in a greater than 12-fold change in selectivity.
These CAB CTLA4 mAbs have antitumor efficacy equal to that of IpA, including tumor regression against MC38 colorectal tumor in a syngeneic human CTLA4 knock-in mouse model. Importantly, CAB CTLA4 antibodies modulate the activity and numbers of infiltrating T cell subsets within the TME, but, unlike IpA, the CAB CTLA antibodies do not increase CD4 effector T cells in the periphery, predicting less systemic immune toxicity impacting normal tissues. Repeated coadministration of either CAB CTLA4 or IpA in combination with an anti-PD1 (NiA) into cynomolgus monkeys for 4 wk was performed to access the peripheral systemic and normal tissue effects of combination treatments. Combination treatment with both IpA and NiA resulted in increases in T cell proliferation markers in peripheral blood cells, while the CAB CTLA4 plus NiA had normal immunophenotypic patterns. All animals in the IpA plus NiA treated groups had significant GI symptoms (diarrhea, loose stools) that presented early, were sustained throughout the treatment period, and were associated with substantial mononuclear infiltration within the intestinal wall. In sharp contrast, the CAB CTLA4 plus NiA treated groups showed no significant GI symptomology or histopathology. These data indicate that CAB CTLA4 molecules may have a superior safety profile when used in combination with PD1 inhibitors and allow increased dosing levels to achieve efficacy superior to current anti-CTLA4 therapy as a single agent or in combination with other anticancer therapies including IO agents.
We have successfully generated CAB antibodies against several different targets including EpCAM, Her2, Nectin-4, CD73, and CD3 shown in addition to CTLA4, as examples in this report. We found that many of these antibodies required PaCS for pH selective binding (SI Appendix, Fig. S1). In the majority of cases studied, PaCS appears to interact with the antigen and block binding at pH7.4. Our data indicate that PaCS interacts with a wide variety of cell-surface proteins. This makes it unlikely that individual PaCS molecules are coordinated on the protein surface by specific arginine and lysine residues as has been reported for human soluble adenylyl cyclase (66). The crystal structure of IpA bound to CTLA4 has been solved (67). No arginine or lysine residues suitable for coordinated binding of the bicarbonate could be identified. In addition, several of the CAB mutations are positioned outside the Ipilimumab\CTLA4 binding interface. It is more likely that PaCS are bound to the (entire) protein surface with a fast on- and off-rate as we observed in our SPR studies and as has been recently described for hydrogen sulfide (68), which allows for rapid reversibility in binding. At pH7.4, PaCS chemicals must be effective at displacing the high-molarity polar water molecules that prevent many weak chemical interactions (69). At the acidic pHe of the TME, fewer PaCS substituent chemicals are bound to the protein surface, exposing positively charged residues on the target for antibody interaction primarily through the negatively charged CAB amino acid substitutions (i.e., aspartic and glutamic acid with pKas lower than PaCS molecules), leading to strong CAB pH selectivity (Fig. 3).
Based on these studies it is expected that there is a potential for other yet to be identified PaCS molecules in disease-related microenvironments, whether controlled through pH, concentration, or other molecular characteristics (intramolecularly or intermolecularly) for enhancing a drug’s TI, is not limited to antibodies, but also includes small molecules, encompassing lipids, sugars, and nucleic acid-based agents or drugs. Further, it is expected that PaCS protein−chemical systems are important naturally occurring regulatory systems linked to a range of disease-related microenvironments, including cancer, inflammation, and cellular senescence. Several groups have sought to improve TI by leveraging the increased presence of variably expressed enzymes circulating in the TME and to exploit these enzymes to clip or remove a mask or protective peptide group from a prodrug in order to activate an antibody near the tumor (70, 71). This prodrug approach is an irreversible process, which generally can occur at a distance from the cancer cell. In contrast, the rapid kinetics of H+ modulation of PaCS chemicals or ions allows for near-homogenous activation and deactivation of the CAB molecules based on the microenvironment near the cancer cell surface, without elevating immunogenicity risk by the addition of foreign sequences. The requirement for precursors from glycolysis for cell growth is a fundamental hallmark of not just tumors but individual cancer cells, with the most rapidly growing cancer cells correlated with greater acidity (24). This ability to design conditional therapeutics with stronger selectivity over narrower pH ranges using the PaCS mechanism offers the ability to greatly enhance both the safety and potency of the future therapies. Beyond antibodies, the PaCS mechanism, especially in view of the antigen interaction, is likely adaptable to peptide and small-molecule drugs. In addition, it is known that intracellular microenvironments are also different between disease and normal cells, potentially further expanding the applicability of the PaCS mechanism.
We see the potential for even broader applications of this technology for targeting antigens in other disease microenvironments. A key characteristic of the TME is acidic pHe (below pH7), resulting, in large part, from increased lactic acid concentrations from glycolytic metabolism (Warburg effect) (46, 72). Our preliminary exploration in other disease areas indicates that PaCS may also underpin the differential binding activities of the different ApoE isoforms (ApE2, ApoE3, and ApoE4) to amyloid-β (1–40) peptide in a pH-dependent manner. These observations suggest the possible involvement of PaCS in the pathogenesis of Alzheimer disease. Further, the inflamed senescent cell populations with senescence-associated secretory phenotype (SASP), implicated in multiorgan disease and shortened lifespan (73–77), are also glycolytic and have acidic microenvironments. The pH selectivity enabled by the PaCS mechanism may lead to many future target opportunities beyond cancer in major disease populations, including cardiovascular disease, dementia, and inflammatory diseases as well as aging.
Materials and Methods
Antibody Engineering and Screening.
Antibody variant libraries of Ipilimumab were built using BioAtla’s proprietary Comprehensive Protein Evolution (CPE) and Comprehensive Protein Synthesis (CPS) processes, shown in detail in SI Appendix.
Animal Model Assessment of Efficacy and Safety.
Tumor xenograft models were performed in human CTLA4 knock-in mice (WuXi Apptec) using the murine syngeneic colon adenocarcinoma cell line MC-38 as previously described (54). Details of the assessments in animal models are shown in SI Appendix.
Supplementary Material
Acknowledgments
We thank the BioDuro Discovery Biology team for their valuable technical assistance and R&D support; Christina Wheeler, Ana Cugnetti, Jing Wang, Monica Sullivan, Carolyn Short, and James Butler for insightful discussions and critical reading of the manuscript; and Dr. Leslie L. Sharp for help with the design and support of the in vivo animal model studies.
Footnotes
Competing interest statement: All authors are shareholders of BioAtla, Inc., which owns the intellectual property rights to CABs- and PaCS-related technologies. H.W.C., G.F., and J.M.S. are inventors on relevant patents. (L.S. is not an inventor on relevant patents.) L.S. and J.M.S. serve as Directors of BioAtla.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020606118/-/DCSupplemental.
Data Availability
All study data are included in the article and SI Appendix.
References
- 1.Murciano-Goroff Y. R., Warner A. B., Wolchok J. D., The future of cancer immunotherapy: Microenvironment-targeting combinations. Cell Res. 30, 507–519 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H., et al., Immune checkpoint blockade and CAR-T cell therapy in hematologic malignancies. J. Hematol. Oncol. 12, 59 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckert F., et al., Beyond checkpoint inhibition—Immunotherapeutical strategies in combination with radiation. Clin. Transl. Radiat. Oncol. 2, 29–35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Junghans R. P., The challenges of solid tumor for designer CAR-T therapies: A 25-year perspective. Cancer Gene Ther. 24, 89–99 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Larkin J., et al., Combined Nivolumab and Ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 373, 23–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larkin J., et al., Five-year survival with combined nivolumab and Ipilimumab in advanced melanoma. N. Engl. J. Med. 381, 1535–1546 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Kaufman H. L., et al., The promise of immuno-oncology: Implications for defining the value of cancer treatment. J. Immunother. Cancer 7, 129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maeda H., Khatami M., Analyses of repeated failures in cancer therapy for solid tumors: Poor tumor-selective drug delivery, low therapeutic efficacy and unsustainable costs. Clin. Transl. Med. 7, 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butcher L., Solid tumors: Prevalence, economics, and implications for payers and purchasers. Biotechnol. Healthc. 5, 20–21 (2008). [PMC free article] [PubMed] [Google Scholar]
- 10.Vaupel P., Metabolic microenvironment of tumor cells: A key factor in malignant progression. Exp. Oncol. 32, 125–127 (2010). [PubMed] [Google Scholar]
- 11.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Kroemer G., Pouyssegur J., Tumor cell metabolism: Cancer’s Achilles’ heel. Cancer Cell 13, 472–482 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Hellmich M. R., Szabo C., Hydrogen sulfide and cancer. Handb. Exp. Pharmacol. 230, 233–241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mansfield C. M., Dodd G. D., Wallace J. D., Kramer S., Curley R. F., Use of heat-sensing devices in cancer therapy. A preliminary study. Radiology 91, 673–678 (1968). [DOI] [PubMed] [Google Scholar]
- 15.Mambou S. J., Maresova P., Krejcar O., Selamat A., Kuca K., Breast cancer detection using infrared thermal imaging and a deep learning model. Sensors (Basel) 18, 2799 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rotin D., Robinson B., Tannock I. F., Influence of hypoxia and an acidic environment on the metabolism and viability of cultured cells: Potential implications for cell death in tumors. Cancer Res. 46, 2821–2826 (1986). [PubMed] [Google Scholar]
- 17.Gatenby R. A., et al., Cellular adaptations to hypoxia and acidosis during somatic evolution of breast cancer. Br. J. Cancer 97, 646–653 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward C., et al., New strategies for targeting the hypoxic tumour microenvironment in breast cancer. Cancer Treat. Rev. 39, 171–179 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Ariffin A. B., Forde P. F., Jahangeer S., Soden D. M., Hinchion J., Releasing pressure in tumors: What do we know so far and where do we go from here? A review. Cancer Res. 74, 2655–2662 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Tannock I. F., Rotin D., Acid pH in tumors and its potential for therapeutic exploitation. Cancer Res. 49, 4373–4384 (1989). [PubMed] [Google Scholar]
- 21.Gerweck L. E., Seetharaman K., Cellular pH gradient in tumor versus normal tissue: Potential exploitation for the treatment of cancer. Cancer Res. 56, 1194–1198 (1996). [PubMed] [Google Scholar]
- 22.Zhang X., Lin Y., Gillies R. J., Tumor pH and its measurement. J. Nucl. Med. 51, 1167–1170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webb B. A., Chimenti M., Jacobson M. P., Barber D. L., Dysregulated pH: A perfect storm for cancer progression. Nat. Rev. Cancer 11, 671–677 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Anderson M., Moshnikova A., Engelman D. M., Reshetnyak Y. K., Andreev O. A., Probe for the measurement of cell surface pH in vivo and ex vivo. Proc. Natl. Acad. Sci. U.S.A. 113, 8177–8181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song C. W., Griffin R., Park H. J., “Influence of tumor pH on therapeutic response” in Cancer Drug Resistance, Teicher B. A., Ed. (Humana, 2006), pp. 21−42. [Google Scholar]
- 26.Cruz-Monserrate Z., et al., Targeting pancreatic ductal adenocarcinoma acidic microenvironment. Sci. Rep. 4, 4410 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corbet C., Feron O., Tumour acidosis: From the passenger to the driver’s seat. Nat. Rev. Cancer 17, 577–593 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Downes H. R., Lactic acid formation in tumor tissue. J. Cancer Res. 13, 268–282 (1929). [Google Scholar]
- 29.Grignani G., Jamieson G. A., Platelets in tumor metastasis: Generation of adenosine diphosphate by tumor cells is specific but unrelated to metastatic potential. Blood 71, 844–849 (1988). [PubMed] [Google Scholar]
- 30.Fraenkl S. A., et al., Plasma citrate levels as a potential biomarker for glaucoma. J. Ocul. Pharmacol. Ther. 27, 577–580 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Mayers J. R., et al., Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat. Med. 20, 1193–1198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma X. J., Dahiya S., Richardson E., Erlander M., Sgroi D. C., Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 11, R7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo Y., Zeng G., Wu S., Identification of microenvironment-related prognostic genes in bladder cancer based on gene expression profile. Front. Genet. 10, 1187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neto A. I., Pinto I. S., “Introduction to marine algae: Overview” in Marine Macro- and Microalgae An Overview, Malcata F. X., Pinto I. S., Guedes A. C., Ed. (CRC, 2019), pp. 1−19. [Google Scholar]
- 35.Richardson T. H., et al., A novel, high performance enzyme for starch liquefaction. Discovery and optimization of a low pH, thermostable alpha-amylase. J. Biol. Chem. 277, 26501–26507 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Palackal N., et al., An evolutionary route to xylanase process fitness. Protein Sci. 13, 494–503 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garrett J. B., et al., Enhancing the thermal tolerance and gastric performance of a microbial phytase for use as a phosphate-mobilizing monogastric-feed supplement. Appl. Environ. Microbiol. 70, 3041–3046 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paladini R. D., et al., Mutations in the catalytic domain of human matrix metalloproteinase-1 (MMP-1) that allow for regulated activity through the use of Ca2+. J. Biol. Chem. 288, 6629–6639 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warburg O., Posener K., Negelein E., Über den Stoffwechsel der Carcinomzelle. Biochem. Z. 152, 309–344 (1924). [Google Scholar]
- 40.Rohani N., et al., Acidification of tumor at stromal boundaries drives transcriptome alterations associated with aggressive phenotypes. Cancer Res. 79, 1952–1966 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Estrella V., et al., Acidity generated by the tumor microenvironment drives local invasion. Cancer Res. 73, 1524–1535 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krähling H., et al., The glycocalyx maintains a cell surface pH nanoenvironment crucial for integrin-mediated migration of human melanoma cells. Pflugers Arch. 458, 1069–1083 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Mitchell M. J., King M. R., Physical biology in cancer. 3. The role of cell glycocalyx in vascular transport of circulating tumor cells. Am. J. Physiol. Cell Physiol. 306, C89–C97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reth M., Matching cellular dimensions with molecular sizes. Nat. Immunol. 14, 765–767 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Chen M., et al., Extracellular pH is a biomarker enabling detection of breast cancer and liver cancer using CEST MRI. Oncotarget 8, 45759–45767 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warburg O., On the origin of cancer cells. Science 123, 309–314 (1956). [DOI] [PubMed] [Google Scholar]
- 47.Korman Alan J., Halk Edward L., Lonberg N., Human Ctla-4 Antibodies and Their Uses (Medarex, 2005). [Google Scholar]
- 48.Middleton P., Kelly A. M., Brown J., Robertson M., Agreement between arterial and central venous values for pH, bicarbonate, base excess, and lactate. Emerg. Med. J. 23, 622–624 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Wardener H. E., He F. J., MacGregor G. A., Plasma sodium and hypertension. Kidney Int. 66, 2454–2466 (2004). [DOI] [PubMed] [Google Scholar]
- 50.He F. J., Markandu N. D., Sagnella G. A., de Wardener H. E., MacGregor G. A., Plasma sodium: Ignored and underestimated. Hypertension 45, 98–102 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Panthi S., Chung H. J., Jung J., Jeong N. Y., Physiological importance of hydrogen sulfide: Emerging potent neuroprotector and neuromodulator. Oxid. Med. Cell. Longev. 2016, 9049782 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karunya R., et al., Rapid measurement of hydrogen sulphide in human blood plasma using a microfluidic method. Sci. Rep. 9, 3258 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Romero-Garcia S., Moreno-Altamirano M. M., Prado-Garcia H., Sánchez-García F. J., Lactate contribution to the tumor microenvironment: Mechanisms, effects on immune cells and therapeutic relevance. Front. Immunol. 7, 52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lute K. D., et al., Human CTLA4 knock-in mice unravel the quantitative link between tumor immunity and autoimmunity induced by anti-CTLA-4 antibodies. Blood 106, 3127–3133 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zwick M. B., et al., Molecular features of the broadly neutralizing immunoglobulin G1 b12 required for recognition of human immunodeficiency virus type 1 gp120. J. Virol. 77, 5863–5876 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leach D. R., Krummel M. F., Allison J. P., Enhancement of antitumor immunity by CTLA-4 blockade. Science 271, 1734–1736 (1996). [DOI] [PubMed] [Google Scholar]
- 57.Liu Y., Zheng P., Preserving the CTLA-4 checkpoint for safer and more effective cancer immunotherapy. Trends Pharmacol. Sci. 41, 4–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Selby M. J., et al., Preclinical development of Ipilimumab and nivolumab combination immunotherapy: Mouse tumor models, in vitro functional studies, and cynomolgus macaque toxicology. PLoS One 11, e0161779 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolchok J. D., et al., Overall survival with combined nivolumab and Ipilimumab in advanced melanoma. N. Engl. J. Med. 377, 1345–1356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwalfenberg G. K., The alkaline diet: Is there evidence that an alkaline pH diet benefits health? J. Environ. Public Health 2012, 727630 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cao X., et al., A review of hydrogen sulfide synthesis, metabolism, and measurement: Is modulation of hydrogen sulfide a novel therapeutic for cancer? Antioxid. Redox Signal. 31, 1–38 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Z., et al., Hydrogen sulfide donor NaHS alters antibody structure and function via sulfhydration. Int. Immunopharmacol. 73, 491–501 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Mao Z., et al., Hydrogen sulfide mediates tumor cell resistance to thioredoxin inhibitor. Front. Oncol. 10, 252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xie Z. Z., Liu Y., Bian J. S., Hydrogen sulfide and cellular redox homeostasis. Oxid. Med. Cell. Longev. 2016, 6043038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Predmore B. L., Lefer D. J., Gojon G., Hydrogen sulfide in biochemistry and medicine. Antioxid. Redox Signal. 17, 119–140 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kleinboelting S., et al., Crystal structures of human soluble adenylyl cyclase reveal mechanisms of catalysis and of its activation through bicarbonate. Proc. Natl. Acad. Sci. U.S.A. 111, 3727–3732 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramagopal U. A., et al., Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab. Proc. Natl. Acad. Sci. U.S.A. 114, E4223–E4232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tan B., et al., New method for quantification of gasotransmitter hydrogen sulfide in biological matrices by LC-MS/MS. Sci. Rep. 7, 46278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Darby J. F., et al., Water networks can determine the affinity of ligand binding to proteins. J. Am. Chem. Soc. 141, 15818–15826 (2019). [DOI] [PubMed] [Google Scholar]
- 70.Polu K. R., Lowman H. B., Probody therapeutics for targeting antibodies to diseased tissue. Expert Opin. Biol. Ther. 14, 1049–1053 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Vasiljeva O., Menendez E., Nguyen M., Craik C. S., Michael Kavanaugh W., Monitoring protease activity in biological tissues using antibody prodrugs as sensing probes. Sci. Rep. 10, 5894 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alfarouk K. O., et al., Glycolysis, tumor metabolism, cancer growth and dissemination. A new pH-based etiopathogenic perspective and therapeutic approach to an old cancer question. Oncoscience 1, 777–802 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coppé J. P., Desprez P. Y., Krtolica A., Campisi J., The senescence-associated secretory phenotype: The dark side of tumor suppression. Annu. Rev. Pathol. 5, 99–118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McHugh D., Gil J., Senescence and aging: Causes, consequences, and therapeutic avenues. J. Cell Biol. 217, 65–77 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Valentijn F. A., Falke L. L., Nguyen T. Q., Goldschmeding R., Cellular senescence in the aging and diseased kidney. J. Cell Commun. Signal. 12, 69–82 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu F., et al., Senescent cardiac fibroblast is critical for cardiac fibrosis after myocardial infarction. PLoS One 8, e74535 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jeon O. H., et al., Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat. Med. 23, 775–781 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.