Abstract
The pathogenic consequences of 369 unique human HsMLH1 missense variants has been hampered by the lack of a detailed function in mismatch repair (MMR). Here single-molecule images show that HsMSH2-HsMSH6 provides a platform for HsMLH1-HsPMS2 to form a stable sliding clamp on mismatched DNA. The mechanics of sliding clamp progression solves a significant operational puzzle in MMR and provides explicit predictions for the distribution of clinically relevant HsMLH1 missense mutations.
Keywords: mismatch repair, Lynch syndrome, HNPCC, single molecule, sliding clamp
The common cancer predisposition Lynch syndrome or hereditary nonpolyposis colorectal cancer (LS/HNPCC) is primarily caused by mutations of the human DNA mismatch repair (MMR) genes HsMSH2, HsMSH6, HsMLH1, and HsPMS2 (1). LS/HNPCC genetic diagnostics has identified definitive genomic mutations as well as single-nucleotide variants (SNV) that result in a missense protein variant (MPV) (2). The clinical significance of MPVs is based on an extensive family history that includes cosegregation of the SNV with cancer (3). In the absence of this information, predicting the consequences of an MPV requires complicated functional analysis (4).
MMR maintains genomic stability primarily by excising replication misincorporation errors (5). The LS/HNPCC causative genes are highly conserved members of the MutS homolog (MSH) and MutL homolog (MLH/PMS) proteins that play central roles in directing accurate MMR excision (5). Decades-old studies showed that MSH proteins recognize mismatched nucleotides (6, 7), which triggers adenosine 5′-triphosphate (ATP) binding and the formation of a sliding clamp that randomly diffused along the mismatched DNA (8–11). However, the detailed mechanics of the MLH/PMS proteins has remained a significant puzzle (5). Reconstitution of Escherichia coli (Ec) MMR suggested that EcMutL forms a homodimer and functions as an intermediary between mismatch recognition by the EcMutS homodimer and downstream excision processes that require the EcMutH endonuclease and EcUvrD helicase (12). Recent real-time single-molecule imaging showed how this progression might operate by demonstrating that EcMutS and EcMutL formed cascading sliding clamps on the mismatched DNA (13, 14). These studies revealed that the EcMutS sliding clamp provided a platform for binding one of the N-terminal domains of the EcMutL (8, 15), which then activated a distinctively different ATP-binding activity that coupled both of the EcMutL N-terminal domains after the remaining peptide segments wrapped around the mismatched DNA (13). The EcMutL sliding clamp was exceedingly stable and together with EcMutS enhanced the diffusion-mediated EcMutH search and incision of hemimethylated GATC sites on the error-containing strand (13) as well as acted as a processivity factor for EcUvrD to efficiently displace the error-containing strand (14). These observations suggested that similar functional insights into the human MMR components might help to illuminate the clinical significance of MPVs in LS/HNPCC.
Results and Discussion
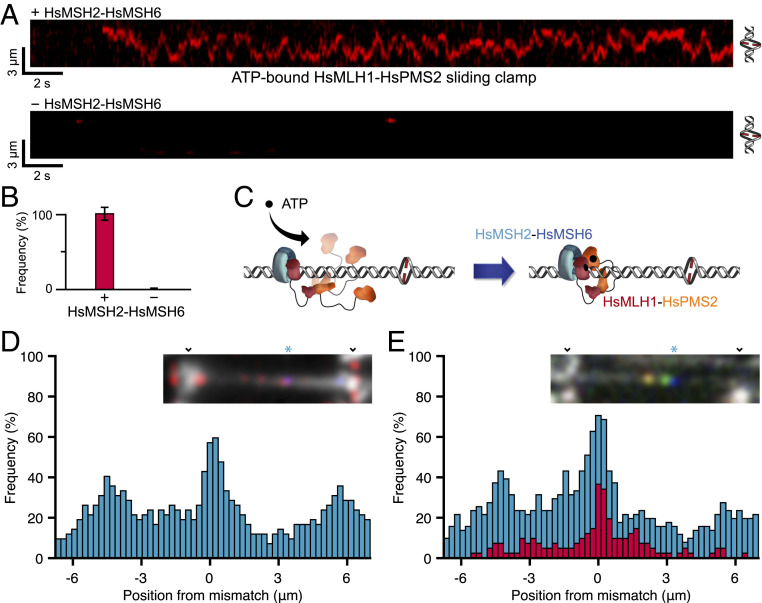
Utilizing single-molecule total internal reflection fluorescence (smTIRF) (13) and DNA SkyBridge surface interference-free light-sheet imaging (16), we found that the principal human (Hs) MSH heterodimer HsMSH2-HsMSH6 was required to load the major human MLH/PMS heterodimer HsMLH1-Cy3HsPMS2 onto a DNA containing a single mismatch (Fig. 1 A and B). The resulting HsMLH1-Cy3HsPMS2 sliding clamp diffused rapidly along the entire length of an 18.4-kilobase (kb) smTIRF or 48.5-kb SkyBridge DNAs (DHsMLH1-HsPMS2 = 0.39 ± 0.09 μm2/s, n = 43, mean ± SE; Fig. 1 A and B) and never stalled at or near the mismatch. These results mirrored the E. coli studies and suggested that HsMSH2-HsMSH6 provided an analogous platform for HsMLH1-HsPMS2 to wrap around the mismatched DNA, forming a stable sliding clamp (Fig. 1C). Tracking HsMSH2-Cy5HsMSH6 revealed molecules that initially bound to the mismatch and then randomly diffused (DHsMSH2-HsMSH6 = 0.036 ± 0.017 μm2/s, n = 52) to points along the entire length of the 18.4-kb or 48.5-kb DNA substrates during a 100-s observation (nDNA = 42; Fig. 1D and SI Appendix, Extended Methods). Inclusion of excess unlabeled HsMLH1-HsPMS2 (100 nM) did not change the widespread localization of HsMSH2-Cy5HsMSH6 (Fig. 1E, blue, nDNA = 51). HsMSH2-Cy5HsMSH6 transiently colocalized with HsMLH1-Cy3HsPMS2, which slowed their diffusion (DHsMSH2-HsMSH6/HsMLH1-HsPMS2 = 0.022 ± 0.0049 μm2/s, n = 35) similar to EcMutS-EcMutL (13). However, the MSH–MLH/PMS complexes ultimately occupied points along the complete length of the 48.5-kb SkyBridge DNA over the 100-s imaging period (Fig. 1E, red, nDNA = 41). These real-time single-molecule observations support the conclusion that the human MMR proteins are only fleetingly stationary, randomly diffuse along the entire mismatched DNA both individually and together, and do not induce detectable MLH/PMS polymerization (5). Such dynamic motions significantly contrast with the static and/or collapsed properties of the MSH–MLH/PMS complexes that aggregated near the mismatch as projected by the Erie, Weninger, and Hingorani as well as the Wang and Li groups (17, 18).
Fig. 1.
Real-time imaging of HsMSH2-HsMSH6 and HsMLH1-HsPMS2 on mismatched DNA. (A) Representative kymographs of a stable ATP-bound HsMLH1-Cy3HsPMS2 (2 nM) sliding clamp on an 18-kb mismatched DNA (see ref. 13). HsMLH1-Cy3HsPMS2 particles were visualized by smTIRF in the presence (Top) or absence (Bottom) of unlabeled HsMSH2-HsMSH6 (8 nM). Helix (Right) approximates the 18-kb mismatch DNA limits on which the HsMLH1-Cy3HsPMS2 clamp is sliding. Time and length reference bars are shown (left corner; 1 μm ∼3,900 bp; see ref. 13). (B) The frequency of HsMLH1-Cy3HsPMS2 sliding clamps on mismatched DNA in the presence (nDNA = 596) and absence (nDNA = 421) of HsMSH2-HsMSH6 (8 nM; mean ± SEM). (C) An illustration of the thermal wrapping mechanism for the formation of an HsMLH1-HsPMS2 sliding clamp. Mismatch recognition by HsMSH2-HsMSH6 provokes ATP binding and the formation of a sliding clamp (blue) that randomly diffuses along the DNA. The HsMSH2-HsMSH6 sliding clamps provides a platform for binding a single N-terminal domain of HsMLH1-HsPMS2, which permits wrapping of the unbound peptide domains around the DNA followed by dimerization and a distinctly different ATP binding process by the N-terminal HsMLH1 and HsPMS2 domains. (D) Position histogram of HsMSH2-Cy5HsMSH6 (50 nM) on a 48.5-kb SkyBridge DNA (see ref. 16) relative to Alexa488-marked mismatch (nDNA = 42). (Inset) Representative DNA SkyBridge molecule showing multiple HsMSH2-Cy5HsMSH6 particles (red) relative to Alexa488-localized mismatch (*, blue). Parallel SkyBridge platforms are marked (v) with mismatched DNA between (light gray). Single molecule positions (nDNA = 42) were established every 20 s for 100 s. (E) Blue histogram: location of HsMSH2-Cy5HsMSH6 (10 nM) in the presence of unlabeled HsMLH1-HsPMS2 (100 nM) on a 48.5-kb SkyBridge DNA (see ref. 16) relative to Alexa488-marked mismatch similar to C (nDNA = 51). Red histogram: location of colocalized HsMSH2-Cy5HsMSH6/HsMLH1-Cy3HsPMS2 (50 nM each) on a 48.5-kb SkyBridge DNA (see ref. 16) relative to Alexa488-marked mismatch similar to panel C (nDNA = 41). (Inset) Representative DNA SkyBridge molecule showing a colocalized HsMSH2-Cy5HsMSH6/HsMLH1-Cy3HsPMS2 (yellow), a solitary HsMLH1-Cy3HsPMS2 (green), and the Alexa488-marked mismatch (*, blue) (nDNA = 35). Parallel SkyBridge platforms are marked (v) with mismatched DNA between (light gray). Single molecules’ positions were established every 20 s for 100 s.
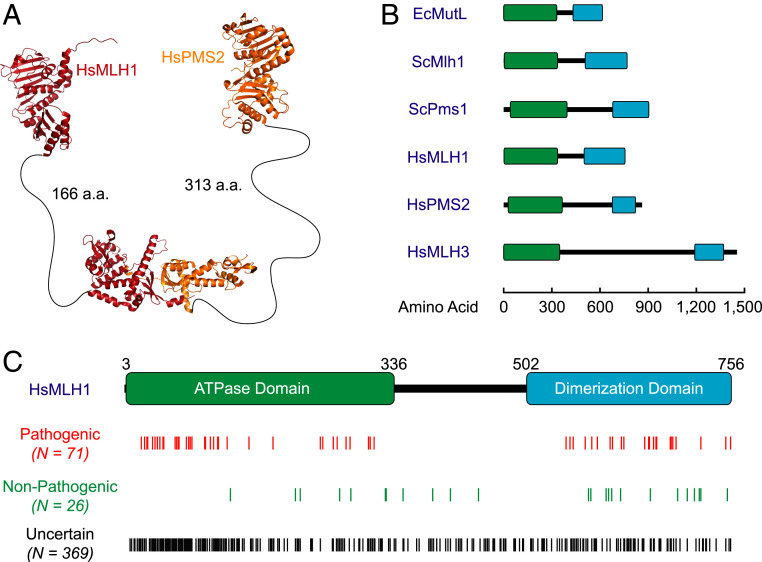
The structure of the N-terminal ATP-binding and C-terminal dimerization domains of several MLH/PMS have been solved (19–21) (Fig. 2A and SI Appendix, Extended Methods). Interestingly, the linker region that connects these two domains is disordered and unresolved and varies in length from relatively short for EcMutL (100 amino acids [aa]) to extremely long for HsMLH3 (840 aa) (Fig. 2B and SI Appendix, Extended Methods). Increasing linker domain deletions of the EcMutL and Sacchromyces cerevisae (Sc) ScMlh1 or ScPms1 (the Sc homolog of HsPMS2) initially prevents their ability to transit roadblocks on the DNA and eventually completely inhibits MMR (22, 23). The most straightforward interpretation of these observations is that MLH/PMS sliding clamps mimic a very flexible donut that encircles the DNA within a relatively large donut hole (Fig. 1C). The flexibility and size of the donut hole is correspondingly controlled by the length of the disordered linkers. Reducing the linker length (size of the donut hole) would inhibit the ability of MLH/PMS sliding clamps to transit roadblocks, while deleting the entire linker domain would inhibit the ability of MLH/PMS to thermally wrap around the DNA when bound to an MSH sliding clamp (Fig. 1C), ultimately inhibiting MMR.
Fig. 2.
The role of HsMLH1-HsPMS2 linker domains in cancer predisposition. (A) The N-terminal structures of HsMLH1 and HsPMS2 (Protein Data Bank [PDB] ID codes 4P7A and 1H7S) with the C-terminal dimerization structure of ScMlh1-ScPms1 (PDB ID code 4E4W) joined by human linker lengths. (B) The locations of N-terminal ATPase (green), linker (central black), and C-terminal dimerization (blue) domains of MutL homologs. (C) The number (N) and locations of unique HsMLH1 pathogenic (n = 71), nonpathogenic (n = 26), and uncertain (n = 369) MPVs, where 220 C-terminal (aa 3 to 336), 49 linker (aa 337 to 501), and 100 N-terminal (aa 502 to 756) unique MPVs were identified from 3,736 total MPVs.
One prediction of the disordered linker-flexibility hypothesis is that any sequence of amino acids could in principle occupy this domain as long as it remained malleable and capable of wrapping around the DNA. As a consequence, there should be few, if any, LS/HNPCC pathogenic MPVs within the HsMLH1 linker domain since it should tolerate most, if not all, amino acid substitutions. We examined 466 unique MPVs located within the coding sequence of HsMLH1 from the InSiGHT database (https://insight-database.org/genes/MLH1). Of these, 71 have been confirmed as pathogenic, 26 have been found to be nonpathogenic, and 369 have been classified as uncertain (Fig. 2C and SI Appendix, Extended Methods) (3). When these MPVs were positioned onto the HsMLH1 backbone a clear pattern emerged in which none of the pathogenic variants were located within the linker domain (aa 337 to 501; Fig. 2C). In contrast, the nonpathogenic variants were located along the entire length of HsMLH1, including the linker domain (Fig. 2C). Of the 369 uncertain MPVs, 49 are located in the HsMLH1 linker domain and unlikely to alter the natural disorder of the HsMLH1 linker domain, making it doubtful that any of these MPVs would be pathogenic. We note that there are 137 reported unique MPVs located within the HsPMS2 gene, including five that have been identified as pathogenic. While none of the pathogenic HsPMS2 MPVs are located in the linker domain, the small number precludes an experimental generalization. Nevertheless, these observations further support dynamic disordered and flexible MLH/PMS linkers, which differs considerably from deduced ATP-dependent ordering of the HsMLH1-HSPMS2 linkers (17, 24). Ultimately the studies reported here should further enhance the understanding of MMR mechanics and lead to clinical discussions of what protein domain defects may constitute a causal mutation for LS/HNPCC.
Materials and Methods
Protein purification, labeling, single-molecule imaging, and analysis have been described previously with some modifications (13, 14). HsMLH1 MPVs were retrieved from the InSiGHT database. See SI Appendix, Extended Methods.
Supplementary Material
Acknowledgments
We thank laboratory members for helpful discussions; this work was supported by the Global Research Laboratory Program National Research Foundation of Korea Information and Communications Technology 2017K1A1A2013241 (to J.-B.L.) and CA067007 (to R.F.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019215118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix. Some study data are available upon request.
References
- 1.Win A. K., et al., Prevalence and penetrance of major genes and polygenes for colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 26, 404–412 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giardiello F. M., et al., Guidelines on genetic evaluation and management of Lynch syndrome: A consensus statement by the US multi-society task force on colorectal cancer. Am. J. Gastroenterol. 109, 1159–1179 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Thompson B. A., et al., Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat. Genet. 46, 107–115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heinen C. D., Juel Rasmussen L., Determining the functional significance of mismatch repair gene missense variants using biochemical and cellular assays. Hered. Cancer Clin. Pract. 10, 9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fishel R., Mismatch repair. J. Biol. Chem. 290, 26395–26403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fishel R. A., Siegel E. C., Kolodner R., Gene conversion in Escherichia coli. Resolution of heteroallelic mismatched nucleotides by co-repair. J. Mol. Biol. 188, 147–157 (1986). [DOI] [PubMed] [Google Scholar]
- 7.Su S.-S., Modrich P., Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc. Natl. Acad. Sci. U.S.A. 83, 5057–5061 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acharya S., Foster P. L., Brooks P., Fishel R., The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol. Cell 12, 233–246 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Cho W. K., et al., ATP alters the diffusion mechanics of MutS on mismatched DNA. Structure 20, 1264–1274 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gradia S., Acharya S., Fishel R., The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell 91, 995–1005 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Gradia S., et al., hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell 3, 255–261 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Lahue R. S., Au K. G., Modrich P., DNA mismatch correction in a defined system. Science 245, 160–164 (1989). [DOI] [PubMed] [Google Scholar]
- 13.Liu J., et al., Cascading MutS and MutL sliding clamps control DNA diffusion to activate mismatch repair. Nature 539, 583–587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J., et al., MutL sliding clamps coordinate exonuclease-independent Escherichia coli mismatch repair. Nat. Commun. 10, 5294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groothuizen F. S., et al., MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA. eLife 4, e06744 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim D., et al., DNA skybridge: 3D structure producing a light sheet for high-throughput single-molecule imaging. Nucleic Acids Res. 47, e107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradford K. C., et al., Dynamic human MutSα-MutLα complexes compact mismatched DNA. Proc. Natl. Acad. Sci. U.S.A. 117, 16302–16312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ortega J., Lee G. Sanghee, Gu L., Yang W., Li G.-M., Mispair-bound human MutS-MutL complex triggers DNA incision and activates mismatch repair. Cell Res., 10.1038/s41422-021-00468-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guarné A., Junop M. S., Yang W., Structure and function of the N-terminal 40 kDa fragment of human PMS2: A monomeric GHL ATPase. EMBO J. 20, 5521–5531 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu S., et al., CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu H., et al., Structure of the human MLH1 N-terminus: Implications for predisposition to Lynch syndrome. Acta Crystallogr. F Struct. Biol. Commun. 71, 981–985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y., Furman C. M., Manhart C. M., Alani E., Finkelstein I. J., Intrinsically disordered regions regulate both catalytic and non-catalytic activities of the MutLα mismatch repair complex. Nucleic Acids Res. 47, 1823–1835 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mardenborough Y. S. N., et al., The unstructured linker arms of MutL enable GATC site incision beyond roadblocks during initiation of DNA mismatch repair. Nucleic Acids Res. 47, 11667–11680 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sacho E. J., Kadyrov F. A., Modrich P., Kunkel T. A., Erie D. A., Direct visualization of asymmetric adenine-nucleotide-induced conformational changes in MutL alpha. Mol. Cell 29, 112–121 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix. Some study data are available upon request.