Significance
Long-term potentiation (LTP), the communication between neurons that is potentiated upon strong stimulus, is an important cellular mechanism underlying learning and memory. Mechanistically, LTP expression involves AMPA-type glutamate receptors insertion into synapses (initiation) and trapped at the synapses for an extended period of time (maintenance). Here, we show that the amino-terminal domain of GluA1, a subunit of AMPA receptors, selectively interacts with the adhesion protein Np65, and this interaction is required for the synaptic anchoring of AMPA receptors during LTP. Our study reveals a molecular mechanism underlying GluA1-dependent LTP maintenance.
Keywords: amino-terminal domain, GluA1, LTP, AMPA receptors, neuroplastin
Abstract
Long-term potentiation (LTP) has long been considered as an important cellular mechanism for learning and memory. LTP expression involves NMDA receptor-dependent synaptic insertion of AMPA receptors (AMPARs). However, how AMPARs are recruited and anchored at the postsynaptic membrane during LTP remains largely unknown. In this study, using CRISPR/Cas9 to delete the endogenous AMPARs and replace them with the mutant forms in single neurons, we have found that the amino-terminal domain (ATD) of GluA1 is required for LTP maintenance. Moreover, we show that GluA1 ATD directly interacts with the cell adhesion molecule neuroplastin-65 (Np65). Neurons lacking Np65 exhibit severely impaired LTP maintenance, and Np65 deletion prevents GluA1 from rescuing LTP in AMPARs-deleted neurons. Thus, our study reveals an essential role for GluA1/Np65 binding in anchoring AMPARs at the postsynaptic membrane during LTP.
In 1973, Bliss and Lomo published the first observation of long-term potentiation (LTP) in which a tetanic stimulus caused a prolonged enhancement of synaptic transmission in rabbit hippocampus (1). Numerous studies have since demonstrated that LTP contributes to the neuronal mechanisms underlying learning and memory (2–4). The classic NMDA receptor (NMDAR)-dependent LTP is found in many brain regions and is studied mostly in hippocampal CA1 synapses (5, 6). Mechanistically, LTP can be divided into two sequential phases: initiation and maintenance. During LTP initiation, tetanic stimulation activates NMDARs that mediate rapid Ca2+ influx into dendritic spines, resulting in CaMKII activation, which subsequently recruits more AMPA-type glutamate receptors (AMPARs) into synapses, thus strengthening AMPAR-mediated excitatory postsynaptic currents (AMPAR-EPSCs) (7, 8). LTP maintenance is thought to require the newly recruited AMPARs to remain at the postsynaptic membrane for an extended period of time, a process called synaptic trapping (9).
AMPAR cellular trafficking, synapse anchoring, and synaptic function are dependent on the subunit composition of the core functional ion channel, which consists of a tetramer of subunits GluA1-GluA4. Each subunit consists of an amino-terminal domain (ATD, also known as N-terminal domain), a ligand-binding domain, four membrane-spanning segments, and an intracellular C-terminal domain (CTD). In mouse hippocampal CA1 pyramidal neurons, the most common types of AMPAR subunits are GluA1, GluA2, and GluA3 (10). Early studies using virus-based overexpression of the green fluorecent protein (GFP)-tagged AMPAR subunits suggest that GluA1 and GluA2 have differential trafficking capabilities in hippocampal neurons (11, 12); GluA2/A3 heteromers are constitutively trafficked to dendritic spines, while the synaptic cooperation of GluA1-containing AMPARs is dependent on neuronal activity. Thus, a subunit-specific model for AMPAR trafficking has been proposed (13), and LTP is thought to require certain sequences or domains of GluA1 (14). The emergent roles of the GluA1 CTD in synaptic plasticity have been extensively documented (13, 15–17). However, the observation that the CTD-lacking GluA1 is still present at the postsynaptic membrane and mediates LTP (18, 19) challenged the absolute requirement for GluA1 CTD in synaptic transmission and plasticity, indicating that other domains, such as the ATD, might have a previously uncovered role in LTP.
Indeed, recent studies have revealed that the GluA1 ATD is required for synaptic transmission and LTP (20, 21). The ATD, which accounts for nearly half of the AMPAR coding sequence, projects nearly midway into the synaptic cleft where it may dynamically interact with proteins; such interactions might contribute to synaptic plasticity (22, 23). N-cadherin (24), a cell adhesion molecule, and neuronal pentraxins (25), secretory proteins, have been reported to associate with the ATDs of GluA2 and GluA4, respectively. However, whether GluA1 ATD has binding partners in the synaptic cleft remains unclear.
In this study, we aimed to further understand the role of the GluA1 ATD in synaptic transmission and LTP and to investigate the molecular mechanism underlying its function. We found that the ATD is required for GluA1 synaptic function, both under basal conditions and during LTP. Furthermore, we have identified that GluA1 ATD directly interacts with neuroplastin-65 (referred to throughout as Np65), a single-transmembrane protein belonging to the immunoglobulin superfamily of cell adhesion molecules. Interaction of Np65 with the ATD of GluA1 is required for prolonged enhancement in synaptic transmission during LTP. Interestingly, it has been reported that as early as 20 y ago, Np65 antibody treatment causes impairment in LTP maintenance in hippocampal slices (26). Therefore, our results provide a molecular mechanism for GluA1- and Np65-mediated LTP maintenance.
Results
GluA1 ATD Is Required for LTP Maintenance.
To understand the role of GluA1 ATD in LTP, we used a molecular replacement strategy based on CRISPR/Cas9 gene deletion system that was proven to enable the efficient depletion of multiple synaptic proteins simultaneously (27). First, we developed a knockout construct, CRISPR_Gria1-3, containing Cas9-T2A-EGFP (enhanced GFP) and individual single-guide RNAs (sgRNAs) targeting Gria1, Gria2, and Gria3, the genes encoding GluA1, GluA2, and GluA3, respectively. The lentivirus-mediated expression of these sgRNAs in cultured hippocampal neurons dissociated from Cas9–knock-in mice nearly completely depleted the protein expression of all three AMPAR subunits (SI Appendix, Fig. S1A). The time course for the full deletion effect is about 3 wk, and therefore recordings for CRISPR/Cas9-mediated gene deletion or molecular replacement were made 3 wk posttransfection by in utero electroporation (IUE) (SI Appendix, Fig. S1B). In hippocampal slices, somatic outside-out patches excised from CA1 neurons transfected with CRISPR_Gria1-3 exhibited little AMPAR-mediated currents (SI Appendix, Fig. S1C). Simultaneous recording of a CRISPR_Gria1-3–transfected and untransfected (control) neighboring CA1 pyramid neuron showed nearly complete loss of AMPAR-EPSCs in transfected neurons (SI Appendix, Fig. S1D). In contrast, NMDAR-mediated excitatory postsynaptic currents (NMDAR-EPSCs) remained unaltered (SI Appendix, Fig. S1E). LTP, induced with a pairing protocol, was also absent in neurons transfected with CRISPR_Gria1-3 (Gria1-3 null neurons), compared to the robust potentiation in untransfected neurons (SI Appendix, Fig. S1F). The effects of CRISPR_Gria1-3 were not due to Cas9 overexpression because CA1 neurons transfected with Cas9 resulted in intact AMPAR-EPSCs (SI Appendix, Fig. S1G) and NMDAR-EPSCs (SI Appendix, Fig. S1H). Taken together, these data verified the capability of CRISPR_Gria1-3 to delete the endogenous AMPAR subunits in CA1 pyramid neurons.
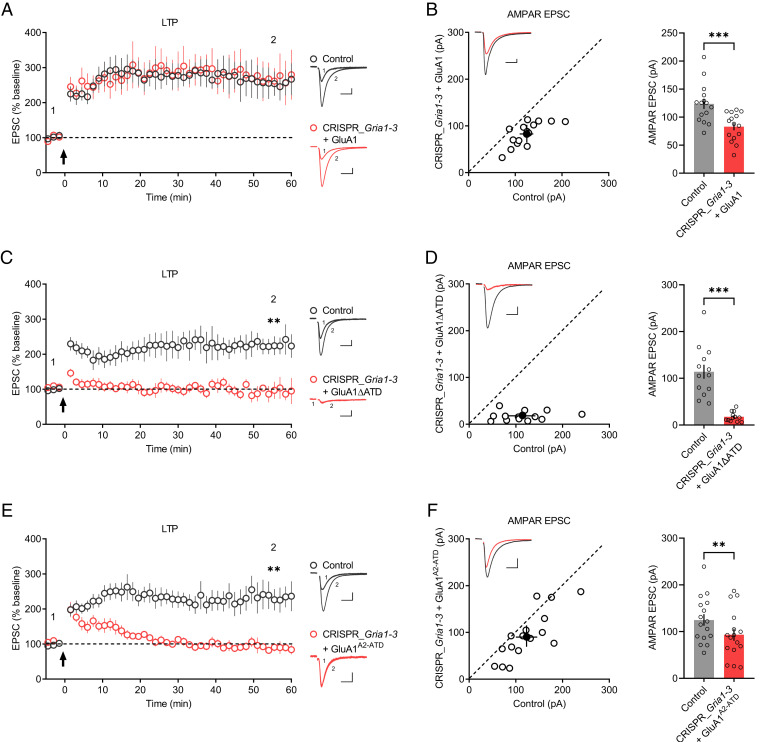
Since the endogenous AMPARs are largely eliminated in CRISPR_Gria1-3–transfected neurons, we then attempt to investigate the role of GluA1 ATD in synaptic transmission and LTP by expressing back GluA1 and GluA1ΔATD, a mutant in which the ATD was deleted. In hippocampal CA1 neurons, overexpression of GluA1 produced significant inward rectifying synaptic responses (SI Appendix, Fig. S2A) and slightly decreased AMPAR-EPSCs (SI Appendix, Fig. S2B). The current-voltage (I-V) curve of the evoked AMPAR-EPSCs was unaltered in GluA1ΔATD-transfected cells compared to untransfected cells (SI Appendix, Fig. S2 C and D), while the somatic outside-out patches from GluA1ΔATD-overexpressing neurons were inward rectifying (SI Appendix, Fig. S2E), indicating that GluA1ΔATD functions at the neuronal somatic surface but not synapses. Moreover, in Gria1-3 null neurons, the expression of GluA1 but not GluA1ΔATD rescued AMPAR-EPSCs (Fig. 1 B and D) and LTP (Fig. 1 A and C), suggesting that GluA1ΔATD has impaired synaptic function under basal conditions and during LTP, in line with a previous study based on a Cre-loxP–mediated recombination system (20).
Fig. 1.
GluA1 ATD is required for LTP. (A, C, and E) Plots show the mean ± SEM. AMPAR-EPSC amplitude normalized to the mean AMPAR-EPSC amplitude before LTP induction (arrow) in control (gray) and transfected (red) neurons expressing (A) GluA1 (control: n = 7; CRISPR_Gria1-3 + GluA1: n = 7; three mice; P = 0.8413 at 55.5 min), (C) GluA1ΔATD (control: n = 8; CRISPR_Gria1-3 + GluA1ΔATD: n = 8; four mice; **P < 0.01 at 55.5 min), or (E) GluA1A2-ATD (control: n = 10; CRISPR_Gria1-3 + GluA1A2-ATD: n = 8; four mice; **P < 0.01 at 55.5 min). Representative AMPAR-EPSC current traces from control (gray) and transfected (red) neurons before (1) and after LTP (2) are shown to the right of the graphs. (Scale bars, 25 pA, 25 ms.) (B, D, and F) Scatterplots show the AMPAR-EPSC amplitudes of single pairs (open circles) of control and transfected neurons; filled circle indicates mean ± SEM amplitude. Insets show sample traces from control (gray) and transfected (red) cells. The bar graphs to the right of the scatterplots show mean ± SEM amplitude of AMPAR-EPSC of untransfected neurons and CRISPR_Gria1-3–transfected neurons expressing (B) GluA1 (control: 124 ± 9.09; CRISPR_Gria1-3 + GluA1: 83.1 ± 6.55; n = 15 pairs; three mice; ***P < 0.001), (D) GluA1ΔATD (control: 114 ± 15.0; CRISPR_Gria1-3 + GluA1ΔATD: 17.7 ± 2.81; n = 13 pairs; four mice; ***P < 0.001), or (F) GluA1A2-ATD (control: 125 ± 12.4; CRISPR_Gria1-3 + GluA1A2-ATD: 93.4 ± 13.0; n = 16 pairs; four mice; **P < 0.01). (Scale bars, 25 pA, 25 ms.)
Next, we investigated whether the inability of GluA1ΔATD to rescue LTP in Gria1-3 null neurons was due solely to deficient AMPAR-EPSCs. We then constructed a chimera GluA1A2-ATD in which the ATD of GluA1 was replaced with GluA2 ATD. GluA1A2-ATD was delivered to synapses, as indicated by the inward rectifying synaptic responses in GluA1A2-ATD-overexpressing neurons (SI Appendix, Fig. S2 F and G), consistent with the previous report (21). In Gria1-3 null neurons, the expression of GluA1A2-ATD resulted in robust AMPAR-EPSCs (Fig. 1F); however, these neurons exhibited only transient potentiation in synaptic transmission upon LTP induction, which returned to basal levels 20–30 min post-LTP induction (Fig. 1E). These results demonstrate that the GluA1 ATD is required for LTP maintenance.
GluA1 ATD Selectively Interacts with Np65.
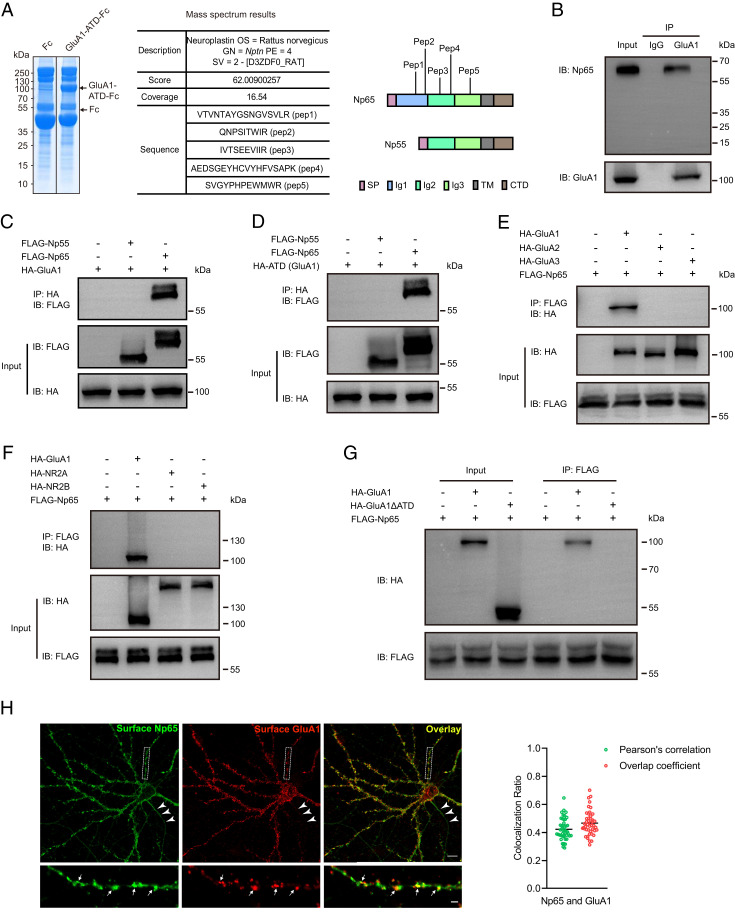
Since GluA1 but not GluA1A2-ATD can rescue LTP in Gria1-3 null neurons, we hypothesized that some molecules in the synaptic cleft may specifically interact with GluA1 ATD and that the interactions are critical for LTP maintenance. To identify GluA1 ATD binding partners in the brain, we purified a recombinant GluA1 ATD protein composed of the ATD of GluA1 fused to the Fc domain of human immunoglobulin (GluA1-ATD-Fc) and then performed affinity chromatography experiments with rat brain lysates followed by mass spectrometry analysis. We found that neuroplastin protein exhibits high score in mass spectrometry results (Fig. 2A and Dataset S1), indicating a high binding affinity between the ATD of GluA1 and neuroplastin. Neuroplastins are cell adhesion proteins encoded by Nptn gene. There are two types of neuroplastins, Np65 and Np55, generated from alternative splicing. Both Np65 and Np55 are single-transmembrane proteins. The longer-isoform Np65 contains three Ig-like domains at the extracellular N-terminal sequences, while Np55 lacks Ig1 (Fig. 2A). Our mass spectrometry analysis revealed that among the five peptide segments, three are shared by Np65 and Np55, and two are Np65-specific (Fig. 2A), implicating that the GluA1 ATD-interacting protein is Np65. Pull-down assay with mouse hippocampus lysates also verified the interaction between GluA1 and Np65 (Fig. 2B).
Fig. 2.
GluA1 ATD selectively interacts with Np65. (A) Coomassie staining of an SDS-PAGE gel containing the recombinant protein (upper arrow). The table shows mass spectrometry results; the diagrams show protein domains of Np65 and Np55; the peptide segments identified by mass spectrometry are tagged. (B) Coimmunoprecipitation of GluA1 and Np65 from adult mouse hippocampal lysates (n = 3 replicates). IP, immunoprecipitation; IB, immunoblotting. (C and D) Coimmunoprecipitation results of (C) HA-tagged GluA1 or (D) HA-tagged GluA1 ATD with FLAG-tagged Np65 and Np55 from HEK293T cell lysates (n = 3 replicates for each experiment). (E) Coimmunoprecipitation results of FLAG-tagged Np65 with HA-tagged AMPAR subunits GluA1, GluA2, and GluA3 from HEK293T cell lysates (n = 3 replicates). (F) Coimmunoprecipitation results of FLAG-tagged Np65 with GluN1 cotransfected HA-tagged NR2A or HA-tagged NR2B from HEK293T cell lysates (n = 3 replicates). (G) Coimmunoprecipitation results of FLAG-tagged Np65 with HA-tagged GluA1 or HA-tagged GluA1ΔATD from HEK293T cell lysates (n = 3 replicates). (H) Representative hippocampal neurons (DIV 17) derived from P0 mouse pups were costained with antibodies targeting Np65 and GluA1. (Bottom) The enlarged areas indicated in the white boxes in Top. Arrows indicate example Np65 and GluA1 colocalization puncta in Bottom; arrowheads in the Top indicate axonal expression of Np65 (n = 36 neurons from three independent cultures). (Scale bars, 10 μm in Top and 2 μm in Bottom.) Right graph, quantification of the colocalization ratio of Np65 and GluA1 at dendritic surface is shown (n = 45 dendritic segments with total length of 1,350 μm from 15 neurons; mean ± SEM).
To study this interaction, we performed coimmunoprecipitation assays in HEK293T cells. We found that GluA1 associated with Np65 but not Np55 (Fig. 2C). The GluA1 ATD alone also interacted with Np65 but not Np55 (Fig. 2D). The GluA1 lacking the ATD did not interact with Np65 (Fig. 2G). The extracellular domain of Np65, Ig123 but not shortened Ig12 efficiently interacted with GluA1 ATD. Ig23, the extracellular domain of Np55, did not interact with GluA1 ATD (SI Appendix, Fig. S3A). We also assessed whether Np65 interacted with other glutamate receptor subunits in HEK293T cells and found that Np65 did not bind the AMPAR subunit GluA2 or GluA3 (Fig. 2E) and NMDAR subunit NR2A or NR2B (Fig. 2F). In primary cultured hippocampal neurons, we found high surface expression of Np65 in dendrites and dendritic spines, where it colocalized with GluA1 (Fig. 2H). We also noticed that Np65 expressed at axons in which GluA1 was absent (Fig. 2H), indicating that Np65 may also express at presynaptic terminals. Besides, in permeabilized condition, GluA1 and Np65 were colocalized intracellularly (SI Appendix, Fig. S4A). Taken together, these results show that GluA1 selectively interacts with Np65 and that the GluA1 ATD is required for this interaction.
Neuroplastin Deletion Reduces AMPAR-Mediated Transmission.
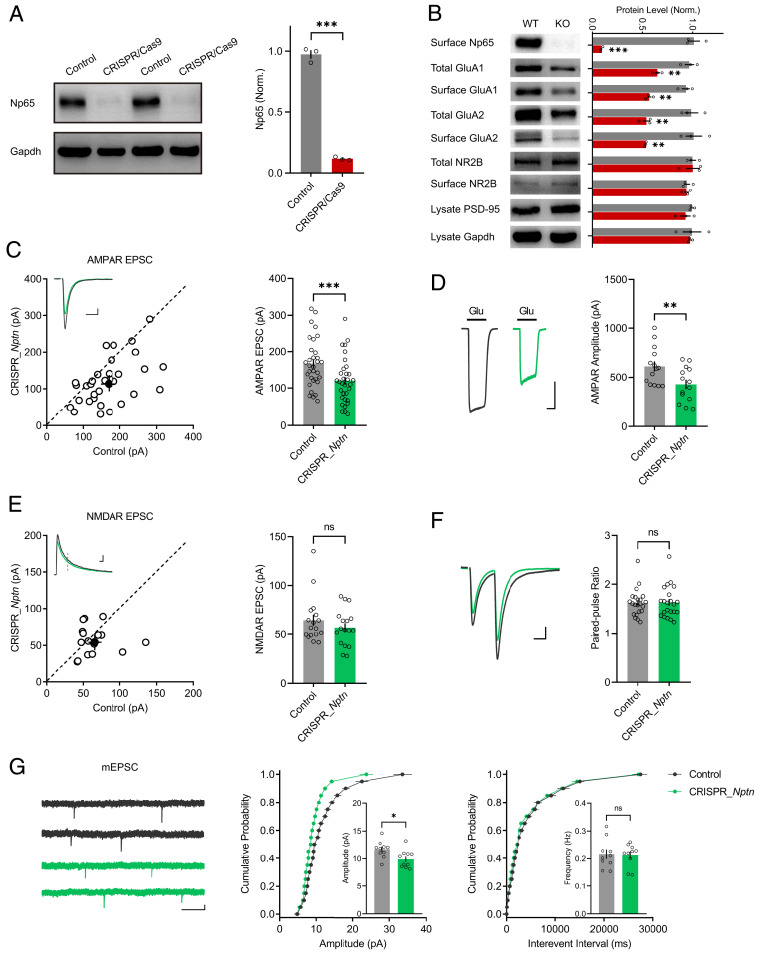
To understand how Np65 is involved in the role of GluA1 in synaptic transmission, we designed a CRISPR_Nptn construct containing both Cas9-T2A-EGFP and sgRNA targeting Nptn. In cultured neurons isolated from hippocampi of the Cas9–knock-in mice, lentivirus-mediated expression of Nptn sgRNA nearly completely depleted Np65 (Fig. 3A). Moreover, both total and surface GluA1 and GluA2 were reduced in these neurons (Fig. 3B), whereas the expressions of NR2B and postsynaptic density (PSD)-95 were unaltered (Fig. 3B). When CRISPR_Nptn was delivered in hippocampal CA1 neurons by IUE, both synaptic (Fig. 3C) and somatic (Fig. 3D) AMPAR-mediated currents were evidently decreased. NMDAR-EPSCs (Fig. 3E) and the paired-pulse ratio (Fig. 3F), a measure of presynaptic release probability, remained unchanged. The amplitude of AMPAR-mediated miniature EPSCs (mEPSCs) was significantly decreased in neurons transfected with CRISPR_Nptn (Fig. 3G), whereas the frequency was unaffected (Fig. 3G). All these observations suggest that neuroplastin is required for synaptic AMPAR function in CA1 pyramid neurons.
Fig. 3.
Np65 deletion reduces AMPAR-mediated transmission. (A) Western blot for Np65 protein expression in primary neuronal cultures infected with Nptn sgRNA-expressing lentivirus and untransfected control (n = 3 replicates; ***P < 0.001). (B) Western blot results of the protein expression of Np65, GluA1, GluA2, NR2B, PSD-95, and Gapdh in Np65-knockout (KO) and untransfected wild-type (WT) neurons. Representative blots and bar graphs of the normalized protein expression levels are shown (mean ± SEM; n = 3 experiments; **P < 0.01; ***P < 0.001). (C and E) Scatterplots show the amplitudes of (C) AMPAR-EPSCs and (E) NMDAR-EPSCs for single pairs (open circles) of untransfected (control) cells and cells transfected with CRISPR_Nptn; the filled circle represents mean ± SEM amplitude. Insets show sample traces from control (gray) and transfected cells (green). The bar graphs to the right of the scatterplots show the mean ± SEM amplitude of (C) AMPAR-EPSC (control: 168 ± 11.9; CRISPR_Nptn: 120 ± 11.0; n = 32 pairs; five mice; ***P < 0.001) and (E) NMDAR-EPSCs (control: 64.5 ± 5.79; CRISPR_Nptn: 56.8 ± 4.57; n = 17 pairs; three mice; P = 0.4586; ns: not significant), (Scale bars, 25 pA, 25 ms.) (D) Representative traces of AMPAR-mediated currents from outside out patches from CRISPR_Nptn-transfected (green) and control (gray) neurons. (Scale bar, 200 pA, 500 ms.) The summary bar graph shows the mean ± SEM amplitude (control: 612 ± 51.5, n = 14; CRISPR_Nptn: 425 ± 47.5, n = 14; three mice; **P < 0.01). (F) Representative traces of paired-pulse ratio (PPR) from neurons transfected with CRISPR_Nptn (green) and control (gray). The bar graph to the right of the scatterplots shows the mean ± SEM of PPR (control: 2 ± 0.06; CRISPR_Nptn: 2 ± 0.07; n = 22 pairs; four mice; P = 0.9493; ns: not significant). (Scale bar, 25 pA, 25 ms.) (G) Representative traces of mEPSCs recorded from CA1 neurons electroporated with CRISPR_Nptn (green) and from untransfected control (gray) neurons. (Scale bars, 5 pA, 1 s.) Cumulative distributions of the mEPSC amplitude and interevent intervals are shown. The bar graphs show the mean ± SEM of mEPSC amplitude (control: 11.64 ± 0.47; CRISPR_Nptn: 9.9 ± 0.49; n = 10 pairs; three mice; *P < 0.05) and frequency (control: 0.22 ± 0.02; CRISPR_Nptn: 0.21 ± 0.01; n = 10 pairs; three mice; P = 0.92; ns: not significant).
We next examined whether Np65 or Np55 overexpression could change AMPAR-mediated transmission in CA1 neurons. We found that Np65 overexpression modestly increased AMPAR-EPSCs (SI Appendix, Fig. S5A) without altering NMDAR-EPSCs (SI Appendix, Fig. S5B). Np55 overexpression did not show any effects on AMPAR-EPSCs or NMDAR-EPSCs (SI Appendix, Fig. S5 C and D).
Neuroplastin Deletion Blocks LTP Maintenance.
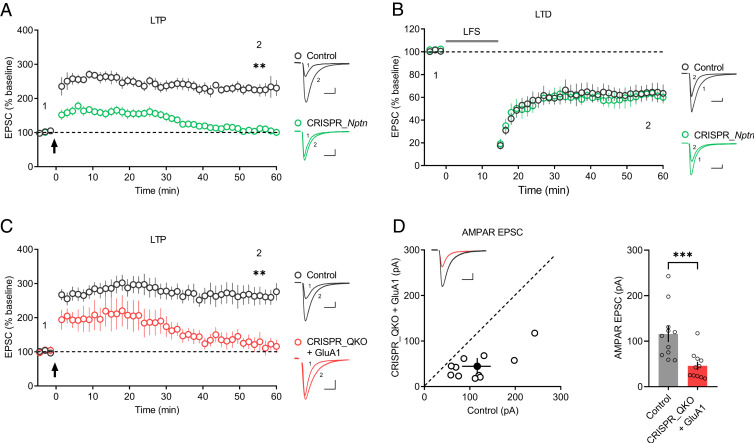
As neuroplastin is required for AMPAR-mediated transmission, and Np65 selectively interacts with GluA1, we then examined whether neuroplastin deletion affects LTP in CA1 pyramid neurons. As expected, neuroplastin deletion severely impaired LTP maintenance, although the potentiation of synaptic transmission was initially induced in CRISPR_Nptn-transfected neurons (Fig. 4A), in line with previous observations (26, 28). We also assessed the effects of neuroplastin depletion on NMDAR-dependent long-term depression (LTD), another type of neural synaptic plasticity, but found no change (Fig. 4B).
Fig. 4.
Neuroplastin deletion blocks LTP maintenance. (A and B) Plots show the mean ± SEM. AMPAR-EPSC amplitude of CRISPR_Nptn-transfected (green) and untransfected (control) (gray) CA1 pyramid neurons normalized to the mean AMPAR-EPSC amplitude before the induction of (A) LTP (arrow) (control: n = 7; CRISPR_Nptn: n = 10; four mice; **P < 0.01 at 55.5 min) and (B) LTD (LFS [low-frequency stimulation]) (control: n = 9; CRISPR_Nptn: n = 9; four mice; P = 0.535 at 55.5 min). Representative AMPAR-EPSC current traces from control (gray) and transfected (green) neurons before and after LTP (A) and LTD (B) are shown to the right of the graphs. (C) Plots show the mean ± SEM. AMPAR-EPSC amplitude of transfected (red) and untransfected (control) (gray) neurons normalized to the mean AMPAR-EPSC amplitude before the induction of LTP (arrow) (control: n = 11; CRISPR_QKO + GluA1: n = 8; five mice; **P < 0.01 at 55.5 min). Representative AMPAR-EPSC current traces from control (gray) and transfected (red) neurons before and after LTP are shown to the right of the graphs. (D) Scatterplots show the AMPAR-EPSC amplitudes of single pairs (open circles) of control and transfected neurons; filled circle represents the mean ± SEM amplitude. Insets show sample traces from control (gray) and transfected cells (red). The bar graph to the right of the scatterplots shows the mean AMPAR-EPSC amplitude ± SEM of neurons transfected with CRISPR_QKO + GluA1 and untransfected neurons (control) (control: 116 ± 17.7; CRISPR_QKO + GluA1: 45.8 ± 8.94; n = 11 pairs; five mice; ***P < 0.001). (Scale bars, 25 pA, 25 ms.)
Based on the above results, we suspected that in the absence of neuroplastin, GluA1 would be unable to rescue LTP in Gria1-3 null neurons. To address this hypothesis, we developed a quadruple knockout construct, CRISPR_QKO, containing Cas9-T2A-EGFP and four individual sgRNAs targeting Gria1, Gria2, Gria3, and Nptn. Neurons transfected with CRISPR_QKO failed to show normal LTP after GluA1 overexpression (Fig. 4C). Moreover, in CRISPR_QKO-transfected neurons, GluA1 only modestly rescued AMPAR-EPSCs, compared to those CRISPR_Gria1-3–transfected neurons (Figs. 4D and 1B). These results demonstrate that the GluA1/Np65 interaction is required for LTP maintenance.
Np65 but Not Np55 Is Required for LTP Maintenance.
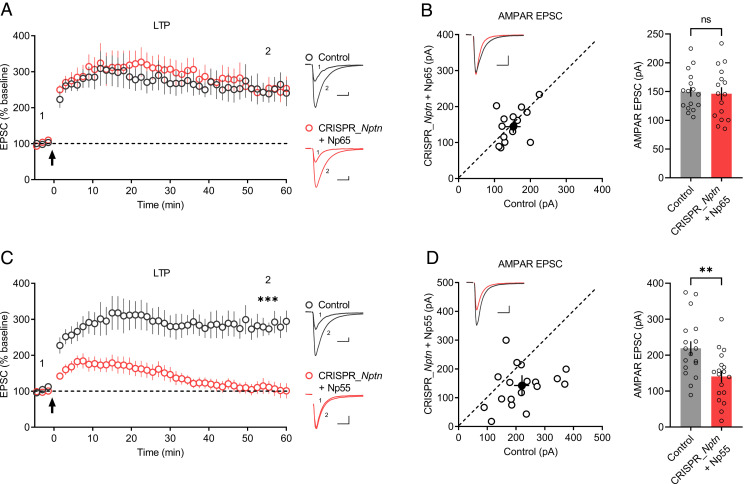
Since Np65 interacts with GluA1, while Np55 does not (Fig. 2C), we predicted that Np65 but not Np55 would rescue the LTP impairment of CRISPR_Nptn-transfected neurons. Indeed, neurons transfected with CRISPR_Nptn and Np65 showed robust LTP (Fig. 5A) and AMPAR-EPSCs (Fig. 5B) similar to those of untransfected neurons. In contrast, when neurons transfected with CRISPR_Nptn and Np55, both LTP (Fig. 5C) and AMPAR-EPSCs (Fig. 5D) remained impaired. These results suggest an isoform-specific requirement for neuroplastin in LTP maintenance.
Fig. 5.
Np65 but not Np55 is required for LTP maintenance. (A and C) Plots show the mean ± SEM. AMPAR-EPSC amplitude of control (gray) and transfected (red) CA1 pyramid neurons normalized to the mean AMPAR-EPSC amplitude before LTP induction (arrow). Experiments were performed in neurons transfected with (A) CRISPR_Nptn + Np65 (control: n = 9; CRISPR_Nptn + Np65: n = 9; four mice; P = 0.8609 at 55.5 min) or (C) CRISPR_Nptn + Np55 (control: n = 11; CRISPR_Nptn + Np55: n = 11; five mice; ***P < 0.001 at 55.5 min). Representative AMPAR-EPSC current traces from control (gray) and transfected (red) neurons before and after LTP are shown to the right of the graphs. (B and D) Scatterplots show the AMPAR-EPSC amplitudes of single pairs (open circles) of control and transfected neurons; filled circle represents the mean ± SEM amplitude. Insets show representative traces from control (gray) and transfected cells (red). The bar graphs to the right of the scatterplots show the mean AMPAR-EPSC amplitude ± SEM of neurons transfected with (B) CRISPR_Nptn + Np65 (control: 150 ± 8.52; CRISPR_Nptn + Np65: 146 ± 11.2; n = 16 pairs; four mice; P = 0.6322; ns: not significant) or (D) CRISPR_Nptn + Np55 (control: 219 ± 19.4; CRISPR_Nptn + Np55: 141 ± 16.4; n = 18 pairs; five mice; **P < 0.01). (Scale bars, 25 pA, 25 ms.)
The Extracellular Domain of Np65 Is Required for LTP Maintenance.
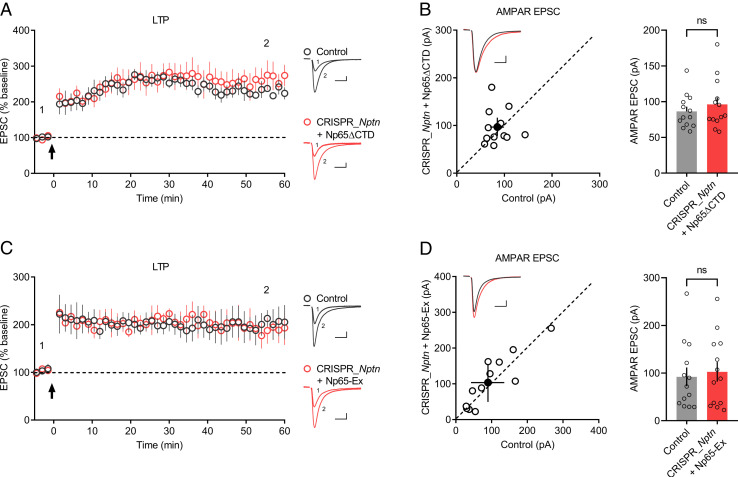
Np65 has a large extracellular domain, a transmembrane helix, and a short intracellular CTD (29); we next investigated which region of Np65 interacts with GluA1 ATD to facilitate LTP maintenance. We first deleted the CTD from Np65, yielding the construct Np65ΔCTD, and tested its interaction with GluA1 ATD and function in neurons. When coexpressed in HEK293T cells, Np65ΔCTD interacted with GluA1 ATD (SI Appendix, Fig. S6A). Overexpression of Np65ΔCTD in primary hippocampal cultures resulted in normal distribution of Np65ΔCTD at the neuronal surface (SI Appendix, Fig. S6B). Moreover, neurons transfected with CRISPR_Nptn and Np65ΔCTD yielded LTP (Fig. 6A) and AMPAR-EPSCs (Fig. 6B) comparable to those of untransfected neurons. These findings indicated that Np65 CTD is not required for AMPAR-mediated synaptic transmission and LTP.
Fig. 6.
The extracellular domain of Np65 is required for LTP maintenance. (A and C) Plots show the mean ± SEM. AMPAR-EPSC amplitude of control (gray) and transfected (red) CA1 pyramid neurons normalized to the mean AMPAR-EPSC amplitude before LTP induction (arrow). Experiments were performed in neurons transfected with (A) CRISPR_Nptn + Np65ΔCTD (control: n = 7; CRISPR_Nptn + Np65ΔCTD: n = 7; three mice; P = 0.400 at 55.5 min) or (C) CRISPR_Nptn + Np65-Ex (control: n = 7; CRISPR_Nptn + Np65-Ex: n = 8; four mice; P = 0.7302 at 55.5 min). Representative AMPAR-EPSC current traces from control (gray) and transfected (red) neurons before and after LTP are shown to the right of the graphs. (B and D) Scatterplots show amplitudes of AMPAR-EPSC for single pairs (open circles) of control and transfected neurons; filled circle represents the mean ± SEM amplitude. Insets show representative traces from control (gray) and transfected cells (red). The bar graphs to the right of the scatterplots show the mean AMPAR-EPSC amplitude ± SEM of neurons transfected with (B) CRISPR_Nptn + Np65ΔCTD (control: 86.4 ± 6.58; CRISPR_Nptn + Np65ΔCTD: 96.2 ± 9.73; n = 13 pairs; three mice; P = 0.4973; ns: not significant) or (D) CRISPR_Nptn + Np65-Ex (control: 92.3 ± 19.7; CRISPR_Nptn + Np65-Ex: 103 ± 20.6; n = 13 pairs; four mice; P = 0.2439; ns: not significant). (Scale bars, 25 pA, 25 ms.)
Then we investigated the role of Np65 extracellular domain in LTP maintenance. We developed a chimeric molecule Np65-Ex by replacing the extracellular domain of Pdgfrα with that of Np65, a strategy that was previously applied to evaluate the role of leucine-rich repeat transmembrane proteins (LRRTMs) in LTP (30, 31). When transfected into HEK293T cells, Np65-Ex but not Pdgfrα was pulled down by GluA1 ATD (SI Appendix, Fig. S6C); moreover, similar to Np65ΔCTD, Np65-Ex was strongly expressed at the neuronal surface in primary hippocampal cultures (SI Appendix, Fig. S6D). More importantly, in CRISPR_Nptn-transfected neurons, the expression of Np65-Ex resulted in LTP (Fig. 6C) and rescued AMPAR-EPSCs (Fig. 6D) identical to those of untransfected neurons. These results indicate that the Np65 extracellular domain is essential for the GluA1 ATD interaction and LTP maintenance.
GluA2(Q)-Mediated LTP Is Independent of the ATD and Np65.
Our study demonstrated that the GluA1/Np65 interaction is essential for LTP expression in CA1 neurons. However, previous studies showed that GluA2(Q) can mediate LTP in AMPARs-deleted neurons (18). Furthermore, GluA2(Q)ΔATD can mediate LTP (20). In order to reconcile these observations, we introduced GluA2(Q) or GluA2(Q)ΔATD in wild-type neurons or Gria1-3 null neurons by IUE. In line with previous studies (20, 21), overexpression of GluA2(Q) in CA1 neurons caused robust decreased rectification index (SI Appendix, Fig. S7A); AMPAR-EPSCs were significantly increased (SI Appendix, Fig. S7B), indicating that GluA2(Q) homomers are able to express and function at the neuronal synapses. Similarly, GluA2(Q)ΔATD also produced inward rectifying synaptic responses (SI Appendix, Fig. S7C), but AMPAR-EPSCs were largely reduced in GluA2(Q)ΔATD-transfected neurons (SI Appendix, Fig. S7D). In Gria1-3 null neurons, both GluA2(Q) and GluA2(Q)ΔATD showed LTP similar to the level of untransfected (control) neurons (SI Appendix, Fig. S8 A and C); GluA2(Q) fully rescued AMPAR-EPSCs (SI Appendix, Fig. S8B), while GluA2(Q)ΔATD exhibited much smaller AMPAR-EPSCs (SI Appendix, Fig. S8D), implying that GluA2 ATD is not required for LTP but is essential for AMPAR-mediated EPSCs, consistent with previous studies (18, 20). We also observed that in CRISPR_QKO neurons, GluA2(Q) showed normal LTP (SI Appendix, Fig. S8E) and largely rescued AMPAR-EPSCs (SI Appendix, Fig. S8F), suggesting that GluA2(Q)-mediated synaptic plasticity is independent of Np65. However, it is important to note that deletion of neuroplastin leads to LTP impairment in CA1 neurons, suggesting that the LTP, supposedly mediated by GluA1/A2 heteromers, is normally based on an Np65-dependent GluA1 mechanism.
Discussion
Our main finding is that the maintenance of LTP depends on the interaction between Np65 and GluA1. Several lines of evidence support this model. First, full length of GluA1 but not GluA1ΔATD rescued LTP in AMPARs-knockout neurons (Gria1-3 null neurons). Second, we showed that Np65 directly interacts with GluA1 ATD and that the deletion of Np65 in CA1 neurons severely impaired LTP maintenance as well as AMPAR-mediated synaptic transmission. Third, the expression of Np65 was sufficient to rescue the deficiency in LTP and AMPAR-EPSCs in neuroplastin-knockout neurons. This model reveals a comprehensive picture of how GluA1 contributes to LTP maintenance via its ATD in hippocampal CA1 pyramid neurons.
In this study, we observed that overexpression of GluA1ΔATD fails to change the synaptic rectification index in CA1 neurons, suggesting that the ATD of GluA1 is necessary for synaptic targeting, in line with the results obtained in hippocampal slice cultures (20, 21). More recently, a study presented on the bioRxiv shows that, although GluA1ΔATD fails to rescue synaptic function, it does target at the PSD, implying that it may have different nanocluster properties compared to GluA1 (32). We also observed that GluA1A2-ATD can mediate synaptic transmission, consistent with a previous study in slice cultures (21), indicating that an ATD is required for GluA1 to efficiently localize at the postsynaptic membrane.
Recent experiments from different laboratories demonstrate that GluA1 ATD is important for LTP expression. Overexpression of GluA1ΔATD in wild-type neurons impairs LTP maintenance (21). In Gria1-3 null neurons, we and others (20) found that GluA1 but not GluA1ΔATD is sufficient to rescue LTP. The deficiency of GluA1ΔATD in rescuing LTP is not solely due to inability in mediating synaptic transmission because the GluA1A2-ATD, which is normal in mediating synaptic transmission, also fails to rescue the long-lasting maintenance of LTP. In contrast, GluA2(Q)ΔATD only modestly rescues AMPAR-EPSCs but expresses robust LTP. These observations suggest that AMPAR-mediated basal synaptic transmissions can be separated from LTP. The properties of nanoclusters formed by GluA1ΔATD but not GluA2ΔATD are changed compared to the full-length receptors at postsynaptic membrane (32). This observation implies that GluA2 ATD may not be involved in shaping the nanocluster property of the receptor; thus replacing with GluA2 ATD may fail to rescue the nanocluster properties of GluA1ΔATD assumably required for LTP. GluA1 ATD may have a subunit-specific role in mediating the organization and stabilization of the nanocluster properties of AMPA receptors, possibly via interaction with other synaptic molecules. Here we report that Np65, a protein that specifically interacts with GluA1 ATD, fulfills this role.
Through proteomic analysis, we identified Np65, the longer splicing isoform of neuroplastins, selectively interacts with GluA1 ATD. Neuroplastins are adhesion molecules belonging to the immunoglobulin superfamily. There are two different neuroplastins encoded by the same gene Nptn, Np65 and Np55, according to their apparent molecular weights. Np65 is selectively expressed in the central nervous system, while Np55 is globally expressed (29). Interestingly, a few lines of evidence have previously suggested that Np65 is involved in cognitive functions in mammal brains (33, 34); altered expression of Np65 in the central nervous system may potentially link to neurodegenerative diseases (35, 36). In hippocampus, Np65 is highly enriched in CA1 neurons (26). Our study reveals that only Np65 can interact with GluA1 and expression of Np65 but not Np55 in neuroplastin-knockout neurons rescued LTP and AMPAR-EPSCs. Np65 antibody treatment or genetic deletion of neuroplastin causes failure in LTP maintenance (26, 28). Np65-Fc, the extracellular sequences of Np65 fused to the Fc segment, also causes similar LTP impairment in hippocampal slices (26). Mechanistically, those observations can be addressed by the impairment in Np65 binding to GluA1; Np65-Fc and Np65 antibody may interfere with GluA1/Np65 interaction.
In Nptn-deleted neurons we observed decreased AMPAR expression and AMPAR-mediated currents. This is consistent with the previous observation that Np65-Fc reduces surface expression of GluA1-containing AMPARs (37), suggesting that GluA1/Np65 interaction might stabilize AMPARs at the neuronal surface. Neuroplastin knockout in mice reduces both amplitude and frequency of mEPSCs (28, 38). However, we only observed decreased mEPSC amplitude in CRISPR_Nptn-transfected neurons. The reduced mEPSC frequency in neuroplastin-knockout mice may reflect the impairment of presynaptic glutamate release probability. Our single-cell genetic manipulation is restricted to the postsynaptic CA1 neurons, thereafter, fails to detect the transsynaptic effects of neuroplastin deletion.
Besides AMPA receptors, neuroplastin is reported to interact with plasma membrane Ca2+-ATPases (PMCAs) via its transmembrane domain and adjacent C-terminal residues (39, 40). In our study, Np65-Ex and Np65ΔCTD can fully rescue LTP in neuroplastin-deleted neurons, suggesting that the binding of neuroplastin/PMCAs is unlikely involved in LTP expression. Np65 is also reported to interact with GABAA receptors and regulate their expression at the inhibitory synapses (38, 41); the exact binding domains are currently unknown.
In addition to Np65, other transmembrane proteins appear to interact with the extracellular domain of AMPARs. Neuropilin-2 interacts with GluA1 via its extracellular CUB domains and mediates semaphorin 3F-dependent homeostatic plasticity in mouse cortical neurons (42). LRRTM2 interacts with AMPAR subunits GluA1 and GluA2 via the extracellular LRR domain to regulate the cell surface expression of GluA1-containing AMPARs (30, 43). However, the domains of AMPARs that mediate the interactions with those proteins have not yet been determined. Our findings of the GluA1 ATD-interacting protein Np65 and the demonstration of this interaction in mediating LTP maintenance, together with recent works (20, 21, 32), highlight the importance of the ATDs in AMPARs’ synaptic anchoring during LTP.
Materials and Methods
Mice.
All animal experiments performed in this study were in accordance with the guidelines of the Institutional Animal Care and Use Committee of Nanjing University. Mice were group-housed with a standard 12-h light/dark cycle and fed ad libitum. For primary hippocampal cultures, P0 pups of both sexes were used. For IUE, 8- to 10-wk-old male and female mice were used for mating, and the day that a vaginal plug was detected was designated pregnancy day E0.5. The generation of the Cas9-knock-in mouse line was performed as previously described (44).
Electrophysiology.
Hippocampal CA1 pyramid neurons were visualized via infrared differential interference contrast microscope. Paired whole-cell recordings were achieved through the simultaneous recording of a GFP- or mCherry-positive neuron and a neighboring untransfected neuron. Acute slices were prepared from E14.5 in utero-electroporated (SI Appendix, Materials and Methods) P17–P25 mice.
Experimental procedures are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This work is supported by grants from the National Key R&D Program of China (2019YFA0801603 and 2017YFA0105201), the National Natural Science Foundation of China (91849112, 31571060, 31900698, 316708142, and 81925011), the Natural Science Foundation of Jiangsu Province (BE2019707), the Beijing Municipal Commission of Science & Technology, and Education (Z181100001518001, CIT & TCD20190334, and KZ201910025025), the Youth Beijing Scholars Program (015), the Kay Realm R&D program of Guangdong Province (2019B030335001), and the Fundamental Research Funds for the Central Universities (0903-14380029).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019194118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Bliss T. V., Lomo T., Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J. Physiol. 232, 331–356 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.J. L. Martinez, Jr, Derrick B. E., Long-term potentiation and learning. Annu. Rev. Psychol. 47, 173–203 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Nabavi S., et al., Engineering a memory with LTD and LTP. Nature 511, 348–352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicoll R. A., A brief history of long-term potentiation. Neuron 93, 281–290 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Lüscher C., Malenka R. C., NMDA receptor-dependent long-term potentiation and long-term depression (LTP/LTD). Cold Spring Harb. Perspect. Biol. 4, a005710 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakazawa K., McHugh T. J., Wilson M. A., Tonegawa S., NMDA receptors, place cells and hippocampal spatial memory. Nat. Rev. Neurosci. 5, 361–372 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Herring B. E., Nicoll R. A., Long-term potentiation: From CaMKII to AMPA receptor trafficking. Annu. Rev. Physiol. 78, 351–365 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Lisman J., Yasuda R., Raghavachari S., Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci. 13, 169–182 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehlers M. D., Heine M., Groc L., Lee M. C., Choquet D., Diffusional trapping of GluR1 AMPA receptors by input-specific synaptic activity. Neuron 54, 447–460 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu W., et al., Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron 62, 254–268 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi S. H., et al., Rapid spine delivery and redistribution of AMPA receptors after synaptic NMDA receptor activation. Science 284, 1811–1816 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Shi S., Hayashi Y., Esteban J. A., Malinow R., Subunit-specific rules governing AMPA receptor trafficking to synapses in hippocampal pyramidal neurons. Cell 105, 331–343 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Kessels H. W., Malinow R., Synaptic AMPA receptor plasticity and behavior. Neuron 61, 340–350 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi Y., et al., Driving AMPA receptors into synapses by LTP and CaMKII: Requirement for GluR1 and PDZ domain interaction. Science 287, 2262–2267 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Diering G. H., Huganir R. L., The AMPA receptor code of synaptic plasticity. Neuron 100, 314–329 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee H. K., et al., Phosphorylation of the AMPA receptor GluR1 subunit is required for synaptic plasticity and retention of spatial memory. Cell 112, 631–643 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Anggono V., Huganir R. L., Regulation of AMPA receptor trafficking and synaptic plasticity. Curr. Opin. Neurobiol. 22, 461–469 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Granger A. J., Shi Y., Lu W., Cerpas M., Nicoll R. A., LTP requires a reserve pool of glutamate receptors independent of subunit type. Nature 493, 495–500 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Díaz-Alonso J., et al., Long-term potentiation is independent of the C-tail of the GluA1 AMPA receptor subunit. eLife 9, e58042 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Díaz-Alonso J., et al., Subunit-specific role for the amino-terminal domain of AMPA receptors in synaptic targeting. Proc. Natl. Acad. Sci. U.S.A. 114, 7136–7141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watson J. F., Ho H., Greger I. H., Synaptic transmission and plasticity require AMPA receptor anchoring via its N-terminal domain. eLife 6, e23024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buonarati O. R., Hammes E. A., Watson J. F., Greger I. H., Hell J. W., Mechanisms of postsynaptic localization of AMPA-type glutamate receptors and their regulation during long-term potentiation. Sci. Signal. 12, eaar6889 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.García-Nafría J., Herguedas B., Watson J. F., Greger I. H., The dynamic AMPA receptor extracellular region: A platform for synaptic protein interactions. J. Physiol. 594, 5449–5458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saglietti L., et al., Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron 54, 461–477 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Pelkey K. A., et al., Pentraxins coordinate excitatory synapse maturation and circuit integration of parvalbumin interneurons. Neuron 85, 1257–1272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smalla K. H., et al., The synaptic glycoprotein neuroplastin is involved in long-term potentiation at hippocampal CA1 synapses. Proc. Natl. Acad. Sci. U.S.A. 97, 4327–4332 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Incontro S., et al., The CaMKII/NMDA receptor complex controls hippocampal synaptic transmission by kinase-dependent and independent mechanisms. Nat. Commun. 9, 2069 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharya S., et al., Genetically induced retrograde Amnesia of associative memories after neuroplastin ablation. Biol. Psychiatry 81, 124–135 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Langnaese K., Beesley P. W., Gundelfinger E. D., Synaptic membrane glycoproteins gp65 and gp55 are new members of the immunoglobulin superfamily. J. Biol. Chem. 272, 821–827 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Bhouri M., et al., Deletion of LRRTM1 and LRRTM2 in adult mice impairs basal AMPA receptor transmission and LTP in hippocampal CA1 pyramidal neurons. Proc. Natl. Acad. Sci. U.S.A. 115, E5382–E5389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soler-Llavina G. J., et al., Leucine-rich repeat transmembrane proteins are essential for maintenance of long-term potentiation. Neuron 79, 439–446 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watson J. F., Pinggera A., Ho H., Greger I. H., AMPA receptor anchoring at CA1 synapses is determined by an interplay of N-terminal domain and TARP γ8 interactions. bioRxiv [Preprint] (2020). 10.1101/2020.07.09.196154 (Accessed 10 July 2020). [DOI]
- 33.Amuti S., et al., Neuroplastin 65 mediates cognitive functions via excitatory/inhibitory synapse imbalance and ERK signal pathway. Neurobiol. Learn. Mem. 127, 72–83 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Herrera-Molina R., et al., Neuroplastin deletion in glutamatergic neurons impairs selective brain functions and calcium regulation: Implication for cognitive deterioration. Sci. Rep. 7, 7273 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ilic K., et al., Hippocampal expression of cell-adhesion glycoprotein neuroplastin is altered in Alzheimer’s disease. J. Cell. Mol. Med. 23, 1602–1607 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beesley P. W., Herrera-Molina R., Smalla K. H., Seidenbecher C., The neuroplastin adhesion molecules: Key regulators of neuronal plasticity and synaptic function. J. Neurochem. 131, 268–283 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Empson R. M., et al., The cell adhesion molecule neuroplastin-65 inhibits hippocampal long-term potentiation via a mitogen-activated protein kinase p38-dependent reduction in surface expression of GluR1-containing glutamate receptors. J. Neurochem. 99, 850–860 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Herrera-Molina R., et al., Structure of excitatory synapses and GABAA receptor localization at inhibitory synapses are regulated by neuroplastin-65. J. Biol. Chem. 289, 8973–8988 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt N., et al., Neuroplastin and basigin are essential auxiliary subunits of plasma membrane Ca2+-ATPases and key regulators of Ca2+ clearance. Neuron 96, 827–838.e9 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Gong D., et al., Structure of the human plasma membrane Ca2+-ATPase 1 in complex with its obligatory subunit neuroplastin. Nat. Commun. 9, 3623 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarto-Jackson I., et al., The cell adhesion molecule neuroplastin-65 is a novel interaction partner of γ-aminobutyric acid type A receptors. J. Biol. Chem. 287, 14201–14214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Q., et al., Neuropilin-2/PlexinA3 receptors associate with GluA1 and mediate Sema3F-dependent homeostatic scaling in cortical neurons. Neuron 96, 1084–1098.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Wit J., et al., LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron 64, 799–806 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J., Du Y., He X., Huang X., Shi Y. S., A convenient Cas9-based conditional knockout strategy for simultaneously targeting multiple genes in mouse. Sci. Rep. 7, 517 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.