Significance
Infants are born without an established gut microbiota, which develops rapidly after birth and is shaped by the maternal microbiota. However, how the maternal microbiota, through shaping the neonatal microbiota, would affect the establishment of a strong immune system in neonates remains unclear. Here, we show mechanistically how the maternal microbiota regulates the de novo production of neonatal IgA.
Keywords: maternal microbiota, neonatal IgA, Lactobacillus reuteri, T cells, type 3 innate lymphoid cells
Abstract
Infants are prone to enteric infections due to an underdeveloped immune system. The maternal microbiota, through shaping the neonatal microbiota, helps establish a strong immune system in infants. We and others have observed the phenomenon of enhanced early neonatal immunoglobulin A (IgA) production in preweaning immunocompetent mice nursed by immunodeficient dams. Here, we show that this enhancement of IgA in neonates results from maternally derived microbiota. In addition, we have found that the neonatal IgA production can be induced by Lactobacillus reuteri, which is enriched in the milk of immunodeficient dams. Moreover, we show that while the production of neonatal IgA is dependent on neonatal T cells, the immunodeficient maternal microbiota-mediated enhancement of neonatal IgA has a T cell–independent component. Indeed, this enhancement may be dependent on type 3 innate lymphoid cells in the neonatal small intestinal lamina propria. Interestingly, maternal microbiota-induced neonatal IgA does not cross-react with common enteric pathogens. Future investigations will determine the functional consequences of having this extra IgA.
An infant’s gut microbiota is established by the maternal microbiota and develops rapidly after birth (1, 2). Commensal microbe colonization in neonates dramatically decreases the incidence of neonatal infections (3–5). However, how the maternal microbiota, through shaping the neonatal microbiota, would affect the establishment of a strong immune system in neonates remains unclear. The lack of such information has prevented identification of maternal microbiota dysbiosis as a risk factor for neonatal infections.
IgA is mostly produced in the gut, and adult human plasma cells residing in the intestinal lamina propria (LP) excrete about 50 mg/kg of immunoglobulin A (IgA) per day (6). IgA is transcytosed through epithelial cells from the basolateral surface by the polymeric immunoglobulin receptor (pIgR). On the apical side, IgA is released into the gut lumen, where it interacts with commensals and drives gastrointestinal homeostasis. In turn, commensal bacteria can induce IgA, a notion supported by early evidence comparing animals housed under germ-free (GF) and specific-pathogen-free (SPF) conditions (7, 8). Two hypotheses regarding mucosal IgA induction resulted from these studies: 1) T-dependent generation in the germinal centers of Peyer’s patches (PPs) and 2) T-independent generation in intestinal LP and isolated lymphoid follicles (ILFs) (9). The first hypothesis follows well-studied germinal center interactions between B and T cells that results in highly specific antibodies generated by the processes of somatic hypermutation and clonal selection. T-independent reactions in the LP, on the other hand, necessitate innate immune pathways and result in polyreactive antibodies. However, most studies on microbiota and induction of IgA utilized adult animals, making it unclear how IgA production is initiated in neonates. We aimed to address this by focusing on the contribution of maternal microbiota.
Here, we show an interesting phenomenon that de novo synthesis of neonatal IgA in preweaning immunocompetent mouse pups is significantly enhanced by the milk of immunodeficient mothers. Such enhancement of IgA production in neonates is mediated by the maternal microbiota and by at least one specific bacterium, Lactobacillus reuteri, in the maternal microbiota. Furthermore, we show that maternal microbiota-mediated enhancement of neonatal IgA is dependent on T cells and type 3 innate lymphoid cells (ILC3s) in the neonatal small intestinal LP. As this extra IgA does not appear to cross-react with two common enteric pathogens, the physiological function of maternal microbiota-induced neonatal IgA remains to be explored.
Results
Neonatal Mice Nursed by Immunodeficient Dams Have Enhanced De Novo IgA Production.
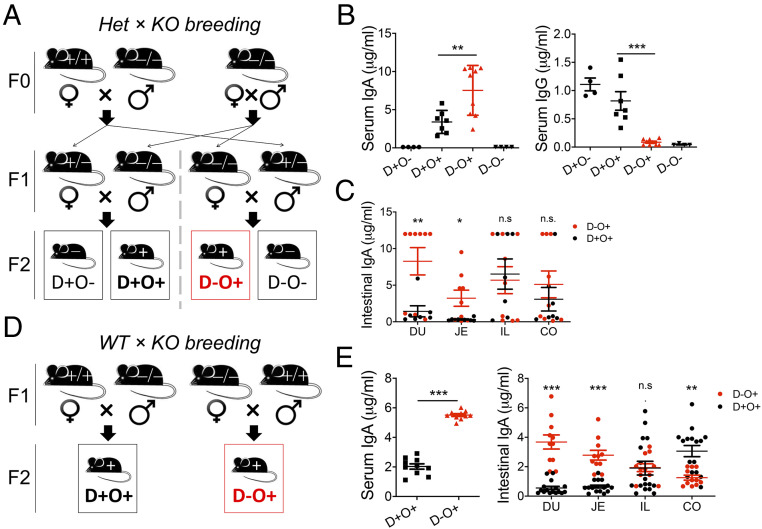
Adult female gut bacteria, which can translocate to the mammary gland via the lymphatic and blood circulation (10), differ in composition and diversity in the presence vs. absence of adaptive immunity (11). We thus used female mice deficient in recombination-activating gene 1 (Rag1−/−) vs. Rag1-sufficient animals to study the neonatal IgA response to diversified maternal microbiota. Using a heterozygous × homozygous knockout breeding strategy, we generated immunodeficient and immunocompetent offspring nursed by immunodeficient vs. immunocompetent dams (Fig. 1A). As anticipated, immunodeficient offspring did not have detectable serum IgA regardless of the type of the dam (Fig. 1A). Strikingly, however, immunocompetent pups nursed by immunodeficient dams had a significantly higher level of serum IgA than those nursed by immunocompetent dams (Fig. 1B). Such an increase of antibody titers was not observed for serum IgG. As the pups reared by immunodeficient dams had smaller spleens (SI Appendix, Fig. S1A), we asked whether de novo synthesis of IgA from the neonatal intestine contributed to the difference in serum IgA levels. Indeed, IgA production from the duodenum and jejunum of the immunocompetent offspring was significantly enhanced when they were nursed by immunodeficient dams (Fig. 1C). Therefore, we predicted that the agent responsible for IgA induction was orally introduced. It is noteworthy that maternal IgA from the breast milk did not contribute to the level of neonatal serum IgA, as immunodeficient offspring reared by immunocompetent dams did not have a detectable level of serum IgA (Fig. 1B). The same results were obtained with a wild type × homozygous knockout breeding strategy (Fig. 1 D and E and SI Appendix, Fig. S1B). Enhancement of IgA induction in the offspring with immunodeficient rearing, hereinafter referred to as “superinduction” of neonatal IgA, was evident at 16 and 21 d of age but not 12 d (SI Appendix, Fig. S1C). We focused on 16 d of age, before the consumption of solid food.
Fig. 1.
Neonatal mice nursed by immunodeficient dams have enhanced de novo IgA production. (A) Breeding scheme. Het, heterozygous; KO, knockout; D, dam; O, offspring; +, immunocompetent; −, immunodeficient. (B) Serum IgA and IgG levels in pups at 16 d of age (n ≥ 4). Results are representative of five independent experiments. (C) Level of de novo synthesized IgA from intestinal segments of pups at 16 d of age (n ≥ 4). DU, duodenum; JE, jejunum; IL, ileum; CO, colon. (D) Breeding scheme. WT, wild type. (E) Serum IgA and level of de novo synthesized IgA from intestinal segments of pups at 16 d of age (n ≥ 10). One-way ANOVA and two-tailed Student’s t test were used as appropriate. Error bars show SEM. *P < 0.05, **P < 0.01, ***P < 0.001; n.s., not statistically significant.
Similar phenomena were described in other immunodeficient models (12, 13). Recently, Koch and colleagues attributed a heightened T cell and germinal center response to the absence of commensal-targeting IgG and IgA from milk (14). However, systemic IgG does not contribute to our phenomenon, as IgG injection did not prevent the induction of IgA synthesis in pups nursed by immunodeficient moms (12). Additionally, pups nursed by IgA−/− mothers did not have accelerated IgA synthesis (12), suggesting that a maternally derived factor other than milk IgA is responsible.
Postnatal Maternal Factors Mediate the Superinduction of IgA in Neonates.
Acquisition of microbiota before birth has been under rigorous debate over the past decade (15), and the placenta was recently determined as sterile except in the case of invading pathogens (16). We used a cross-fostering approach to investigate the contribution of prenatal maternal factors (P) vs. postnatal maternal factors (M) on neonatal IgA production (Fig. 2A). Prenatal maternal factors include placental transfer of antibodies and vaginal microbiota acquired during birth. Postnatal maternal factors include maternal milk containing microbiota and antibodies and contamination from maternal skin and gut microbiota during nursing. When the Rag1-sufficient and Rag1-deficient dams were nonlittermates, the strongest difference in serum IgA was observed between the group receiving immunocompromised prenatal + postnatal factors compared with the group receiving Rag1-sufficient prenatal + postnatal factors (P−M− vs. P+M+, Fig. 2B), suggesting contributions from both factors. Notably, with the same Rag1-deficient postnatal factors (M−), immunocompromised prenatal factors significantly increased IgA production compared with Rag1-sufficient prenatal factors (Fig. 2 B and C). Employment of a littermate strategy, which would equalize the maternal microbiota (17), neutralized most of the observed differences in neonatal IgA production (Fig. 2 D and E). However, the significant difference of neonatal IgA synthesis between types of prenatal factors, which include placentally transferred antibodies, remained when the Rag1-deficient postnatal factors (M−) were kept constant (Fig. 2D). This indicates that the differences related to prenatal factor are independent of maternal microbiota. In contrast, the difference related to postnatal factors is maternal microbiota dependent, as evidenced by the comparison between Fig. 2 B and D, where P+M− vs. P+M+ is significantly different under the nonlittermate condition (Fig. 2B) but not in the littermate condition (Fig. 2D). We thus decided to further investigate the impact of postnatal maternal factors, which include the maternal microbiota, on neonatal IgA production.
Fig. 2.
Postnatal maternal factors mediate the superinduction of IgA in neonates. (A) Breeding scheme to distinguish the effects of P vs. M. (B) Serum IgA level of pups at 16 d of age (n ≥ 10). (C) Fecal IgA and IgG levels in 16-d-old pups without postnatal maternal factors (n ≥ 11). (D) Serum IgA level of 16-d-old pups from littermate dams (n ≥ 18). (E) Fecal IgA and IgG levels in 16-d-old pups from littermate dams (n ≥ 8). In B and D, groups with different letters were significantly different based on one-way ANOVA. In C and E, **P < 0.01, ***P < 0.001. Error bars show SEM.
Superinduction of Neonatal IgA Relies on the Maternal Microbiota Vertically Transferred during and after Birth.
We first characterized the neonatal gut microbiota by using 16S rRNA sequencing. The neonatal gut microbiota should cover all the maternal microbiotas vertically transferred during and after birth, including vaginal, milk, skin, and gut microbiotas acquired from the dam. Both the diversity (Fig. 3A) and composition (Fig. 3 B and C) of microbiotas were different in 16-d-old offspring nursed by immunodeficient vs. immunocompetent dams. The compositions of Rag1+/+ and Rag1−/− milk also differed (SI Appendix, Fig. S2 A and B). Rag1+/+ milk had a higher relative abundance of Bacteroidales, whereas the Rag1−/− milk was overwhelmingly dominated by Lactobacillales. To determine whether the differences in maternal microbiotas led to superinduction of IgA, we again generated littermate dams (Fig. 3D), which equalized the neonatal gut microbiota in the offspring’s stomach at 16 d of age (Fig. 3 E and F). Strikingly, neonatal serum and intestinal IgA levels were not different when the dams were littermates (Fig. 3G and SI Appendix, Fig. S2C). Similar results were obtained when nonlittermate dams were treated with antibiotics that largely removed the maternal microbiota (Fig. 3H and SI Appendix, Fig. S2D). This suggests that IgA superinduction depends on maternal microbiota vertically transferred during and after birth. Importantly, the littermate strategy does not equalize the antibody differences between the two types of milk; thus, the phenomenon of IgA superinduction is independent of milk antibodies.
Fig. 3.
Superinduction of neonatal IgA relies on the maternal microbiota vertically transferred during and after birth. (A) Shannon diversity of microbiota in different sites of the neonatal gastrointestinal tract at 16 d of age (n = 4). ST, stomach. (B) The relative abundance at the order level in 16-d-old pups (n = 4). (C) Principal coordinates analysis of the microbiota in 16-d-old pups (n = 4). (D) Breeding scheme to generate littermate dams. (E) Shannon diversity of the microbiota in the stomach of 16-d-old pups nursed by littermate dams (n ≥ 10). (F) Principal coordinates analysis of the microbiota in the stomach of 16-d-old pups nursed by littermate dams (n ≥ 10). (G) Serum IgA level of 16-d-old pups nursed by littermate dams (n ≥ 21). (H) Serum IgA level of 16-d-old pups nursed by antibiotic-treated dams (n ≥ 7). (I) Scheme showing microbiota transplantation experiments in GF C57BL/6 wild-type mice. The donor material was the stomach contents of 3-d-old immunocompetent pups nursed by SPF Rag1+/+ (WT) or Rag1−/− (KO) dams. (J) Serum IgA levels of 16-d-old pups from GF dams receiving PBS (no), WT, or KO microbiota transplantation (n ≥ 16). One-way ANOVA and two-tailed Student’s t tests were used as appropriate. Error bars show SEM. *P < 0.05, ***P < 0.001; n.s., not statistically significant.
In addition to using a loss-of-function approach, we performed a gain-of-function experiment where pregnant, GF Rag1+/+ females were transplanted with the stomach contents of 3-d-old pups nursed by Rag1+/+ or Rag1−/− dams (Fig. 3I). Pups born to GF dams transplanted with the Rag1−/−-reared stomach contents had a higher level of serum IgA (P = 0.06) than pups born to GF dams transplanted with the Rag1+/+-reared stomach contents (Fig. 3J). While the microbiota from the neonatal stomach contents may not be the same as the maternal microbiota in terms of anaerobic and/or nutritional environment, results of this experiment suggest that IgA superinduction depends on differences in the maternal microbiota.
Lactobacillus reuteri Enhances IgA Production in Neonates.
To determine specific microbial contributions to IgA production, we focused on differences in the stomach contents of 3-d-old pups. A significantly lower overall bacterial load was present in the stomach contents of pups nursed by immunodeficient moms (SI Appendix, Fig. S3A). Looking closer at different bacterial taxa (Fig. 4A), the Bacteriodales family S24-7 was significantly increased with immunodeficient rearing. Although this taxon is not well characterized (18), recent studies have associated it with type 2 innate lymphoid cell (ILC2)-driven IgA production in the stomach (19). While Odoribacter was similarly significantly increased in the stomach content of pups nursed by immunodeficient moms, Lactobacillus reuteri and Lactobacillus casei were by far more abundant. We resorted to qPCR to confirm enrichment of these two species. Unfortunately, the primers designed to determine classification as L. casei were not species specific (SI Appendix, Fig. S3B), so we were not certain of the high abundance of this species in the stomach content of pups nursed by immunodeficient dams. Therefore, we decided to focus on L. reuteri, which was specifically confirmed with qPCR analysis. Interestingly, L. reuteri was significantly enriched in both relative and absolute levels in neonates’ stomachs (Fig. 4 B and C) but not colons (SI Appendix, Fig. S3 C and D). In addition, L. reuteri was equalized in pups stomach contents nursed by littermate dams (Fig. 4D), consistent with the absence of IgA superinduction under this condition. Notably, IgA production was significantly enhanced by 16 d but not 12 d (SI Appendix, Fig. S1C), whereas the absolute abundance of L. reuteri was significantly higher at both time points (Fig. 4C). Putatively, L. reuteri may play a role in early immunological events that consequentially lead to increases in IgA production.
Fig. 4.
Lactobacillus reuteri enhances IgA production in neonates. (A) Volcano plot of 3-d-old stomach microbiotas. Data points with significant differences (P < 0.05) are in red. (B and C) Relative (B) and absolute (C) abundance of L. reuteri in the stomach of pups at 12 and 16 d of age (n ≥ 4) after nursing on nonlittermate dams. (D) Absolute abundance of L. reuteri in the stomach of 16-d-old pups nursed by littermate dams (n = 6). (E) Serum IgA levels of 16-d-old pups from GF dams monocolonized with L. oris (LO) or L. reuteri (LR) (n ≥ 10). (F) Serum IgA from 16-d-old SPF B6 pups neonatally gavaged with LO or LR (n ≥ 8). A two-tailed Student’s t test was used. Error bars show SEM. *P < 0.05, **P < 0.01, ***P < 0.001; n.s., not statistically significant.
We evaluated whether L. reuteri contributes to induction of neonatal IgA by employing GF and SPF mouse models using the L. reuteri strain CF48-3A, which is derived from human breast milk and known to colonize the mouse gut (20). Exposure of GF C57BL/6 (B6) dams to L. reuteri CF48-3A during gestation resulted in significant elevation of neonatal serum IgA compared with a related species not relevant to gastrointestinal colonization, L. oris (Fig. 4E). While exposure of SPF B6 dams to the same strain of L. reuteri did not significantly increase IgA levels in the serum of pups (SI Appendix, Fig. S3E), gavage of neonatal SPF B6 pups starting at 2 to 3 d postbirth significantly induced IgA over L. oris (Fig. 4F). In contrast, serum IgG remained unaltered by neonatal gavage (SI Appendix, Fig. S3F). Furthermore, intestinal IgA was significantly elevated in all sections of the intestinal tract (SI Appendix, Fig. S3G). No sex difference in neonatal IgA levels was observed (SI Appendix, Fig. S3H). Together, these results suggest that the presence of L. reuteri in the immunocompromised maternal microbiota contributes to the superinduction of neonatal IgA.
T Cells and ILC3s Are Required for Maternal Microbiota–Driven Neonatal IgA Induction.
Next, we investigated the immunological mechanism of neonatal IgA induction. Sites of IgA synthesis in the gut include PPs and ILFs (6, 21). Formation of PPs starts during embryogenesis (22, 23) and requires the interaction between hematopoietic cells and stromal cells. The hematopoietic cells produce lymphotoxin (LT) β that signals through its receptor on stromal cells, leading to the migration of lymphoid tissue-inducer (LTi) cells, vital cells for the generation of the lymphoid tissue, into the PP (24). The development of LTi cells depends on transcription factors Id2 and RORγt (25, 26). ILFs, on the other hand, develop upon birth from a structure called cryptopatches, which also consist of RORγt-expressing LTi cells and dendritic cells (27–29). When B and T cells are recruited, cryptopatches become ILFs (30). Therefore, in addition to the absence of all lymph nodes, mice deficient in the Rorc gene fail to develop either PPs or ILFs. To determine whether these lymphoid tissues played a role in maternal microbiota–driven neonatal IgA induction, we cross fostered littermate Rorc+/− and Rorc−/− pups to either Rag1+/+ or Rag1−/− dams (SI Appendix, Fig. S4A). Rorc deficiency in pups completely abrogated the induction of IgA seen in Rorc-sufficient littermate pups when both were cross fostered to Rag1−/− dams, suggesting that lymphoid tissues, including PPs and ILFs, are required.
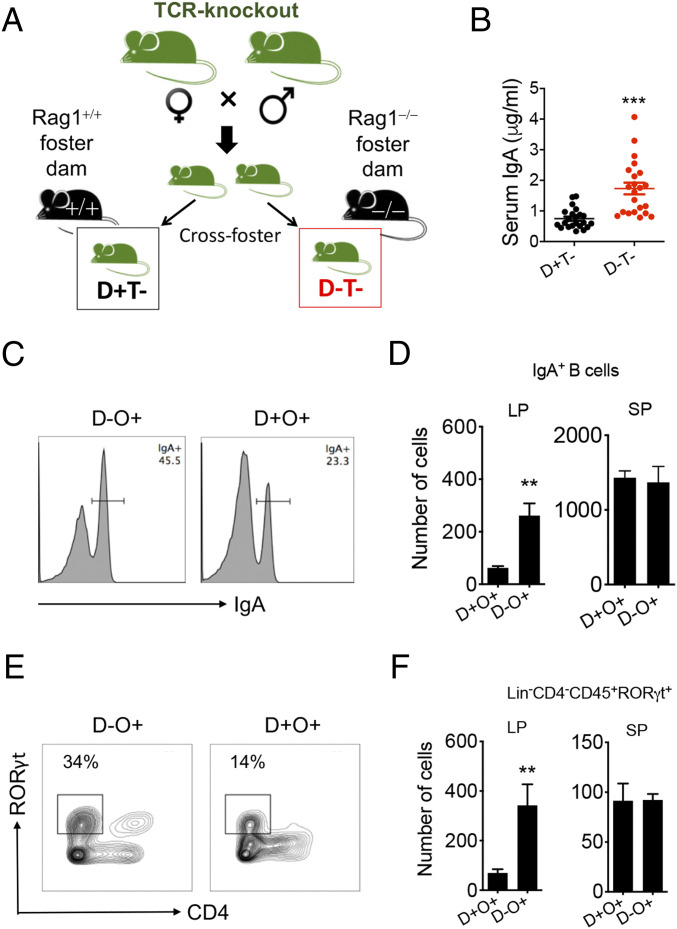
As Rorc-deficient mice are not capable of producing IgA through either T-dependent (25) or T-independent (31) mechanisms, we asked whether neonatal T cells played a role. Indeed, in the neonatal small intestinal LP, 10 to 15% of RORγt+ cells were T cells (SI Appendix, Fig. S4B; and most of them were CD3+CD4−CD8−). Thus, we cross fostered T cell–deficient pups to either Rag1+/+ or Rag1−/− dams (Fig. 5A and SI Appendix, Fig. S4C). Overall, the offspring’s serum IgA levels were lower, indicating T cell–dependent induction of neonatal IgA (Fig. 5B; compared with Fig. 1 B and E and SI Appendix, Fig. S4A). Importantly, the difference in serum IgA between two types of nursing moms was still present (Fig. 5B). This suggests that the superinduction phenomenon by Rag1−/− rearing has a T cell–independent component. The percentages of follicular T helper (Tfh) cells in the small intestine were the same regardless of the type of nursing mom (SI Appendix, Fig. S4D), also suggesting that neonatal IgA superinduction may be independent of T cells.
Fig. 5.
T cells and ILC3s are required for maternal microbiota–driven IgA induction. (A) Breeding scheme showing the cross-fostering experiment where T cell–deficient (Tcrb−Tcrd−) pups were nursed by Rag1+/+ vs. Rag1−/− dams. D−, Rag1−/− nursing dams; D+, Rag1+/+ nursing dams; T−, T cell–deficient pups. (B) Serum IgA level of T cell–deficient pups at 16 d of age (n ≥ 21). (C) FACS analysis of IgA+ cells after gating on B220+ cells in the small intestinal LP of pups at 16 d of age. (D) Absolute number of IgA+ B cells in the small intestinal LP and spleen (SP) of 16-d-old pups (n ≥ 5). (E) FACS analysis of Lin−CD4−CD45+RORγt+ ILC3-like cells in the small intestinal LP of 16-d-old pups. (F) Absolute number of ILC3s in the small intestinal LP and SP of pups at 16 d of age (n ≥ 5). A two-tailed Student’s t test was used. Error bars show SEM. **P < 0.01, ***P < 0.001.
Recently, neonates were shown to have heightened expression of toll-like receptor 5 (TLR5) by the intestinal epithelium relative to adults (32). We thus questioned whether TLR5 signaling led to IgA superinduction. Indeed, the level of flagellin protein, FLiC, was significantly higher in the milk of Rag1−/− dams, and the difference in milk FLiC was eliminated with littermate dams and with antibiotic treatment (SI Appendix, Fig. S4E). However, pups deficient in TLR5 still exhibit superinduction of IgA (SI Appendix, Fig. S4F), suggesting that TLR5 is not required.
Interestingly, nursing B6 pups on immunodeficient dams significantly increased small intestinal LP IgA+ B cells (Fig. 5 C and D) and CD45+CD4−RORγt+ ILC3-like populations (Fig. 5 E and F and SI Appendix, Fig. S4 G and H). Significant differences were present in the small intestinal LP but not the spleen. This suggests that maternal microbiota–stimulated ILC3s may be a critical driver for IgA+ B cell differentiation and neonatal IgA superinduction. Together, our data suggest that neonatal IgA may be catalyzed by maternally derived microbiota through a mechanism dependent on T cells (induction) and ILC3s (superinduction).
Maternal Microbiota–Induced Neonatal IgA Does Not Cross-React with Salmonella or Enterohemorrhagic Escherichia coli.
Finally, we examined the physiological function of maternal microbiota–induced neonatal IgA. IgA is protective against enteric infections as secretory IgA in the gut lumen, so we determined the level of IgA in the luminal contents obtained from upper small intestines. Because much of the IgA in the neonatal gut comes from breast milk, we focused on pups born to different moms but nursed by Rag1−/− dams (Fig. 6A). We were able to detect IgA in the small intestinal contents of these pups, suggesting that some of the de novo synthesized IgA was transported to mucosal secretions. However, the induced neonatal IgA did not cross-react with S. typhimurium or enterohemorrhagic Escherichia coli (EHEC) (Fig. 6B), suggesting a lack of protection against two common pathogens. In contrast, neonatal IgG from the same pups (Fig. 6C) as well as adult IgA and IgG from mice with autoimmune lupus (Fig. 6D) were cross-reactive with both Salmonella and EHEC. These results suggest that the maternal microbiota–induced neonatal IgA may not be protective against enteric infections.
Fig. 6.
Maternal microbiota–induced neonatal IgA does not cross-react with Salmonella or EHEC. (A) Level of IgA in the upper small intestinal contents. Both groups were nursed by Rag1−/− dams (M−) but born to either Rag1+/+ (P+) or Rag1−/− (P−) dams. Error bars show SEM. The Mann–Whitney U test showed P = 0.1375. (B) Cross-reactivity of neonatal IgA with Salmonella or EHEC (n = 5 pooled samples from five different experiments; each experiment had at least nine mice/group). Respective antisera were used as positive controls. OD, optical density. (C) Cross-reactivity of neonatal IgG with Salmonella or EHEC (n = 1 pooled sample from at least 10 mice/group). (D) Cross-reactivity of adult IgA and IgG with Salmonella or EHEC (n = 1 pooled sample from three lupus-prone MRL/lpr mice).
Discussion
In the neonatal gut, IgA limits epithelial penetration of colonizing bacteria, thus preventing inflammation (12). This highlights the importance of IgA’s role in protecting offspring from infections. Yet the underpinning mechanisms that govern initiation of IgA in neonates are incompletely understood. Using a model of early enhanced neonatal IgA production, we found that prenatal maternal factors significantly contribute to elevated IgA induction. However, the contributions of prenatal maternal factors are not as deterministic as postnatal maternal factor–driven IgA induction. Enrichment of certain maternal microbiota–derived bacterial taxa leads to early enhanced IgA production in the intestines, or what we refer to as IgA superinduction. In particular, L. reuteri was identified as a specific microbe derived from the maternal microbiota that colonizes the neonatal gastrointestinal tract to induce IgA. However, L. reuteri may not be the only bacteria with IgA-inducing capability in neonates. It will be intriguing to check other possible candidates, for example, S24-7. At the same time, it is also possible that certain bacterial species, which were enriched in pups nursed by immunocompetent dams, can inhibit IgA synthesis or degrade secreted IgA by bacterial IgA proteases (33).
In the absence of a critical transcription factor RORγt, induction of neonatal IgA is significantly impaired regardless of differences in the maternal microbiota source. In adult mice, RORγt+ ILC3s can regulate gut IgA production in different ways, for example, by controlling B cell class switching to IgA in PPs (34) and by regulating Tfh cells residing in mesenteric lymph nodes as antigen-presenting cells (35). However, in neonates, PPs and lymph nodes are underdeveloped. Thus, it is possible that RORγt+ ILC3s exert IgA regulation in the neonatal gut through a different manner at early age compared with adults. We hypothesize that the maternal microbiota, or, specifically L. reuteri, could induce the formation of ILFs in neonates, leading to differentiation of IgA-producing plasma cells. Further investigation will focus on the complete understanding of how these RORγt+ ILC3s develop and function, with the existence of an IgA-inducing microbiota, to enhance IgA production at an early age.
In this study, we do not have evidence supporting a protective role of maternal microbiota–induced neonatal IgA. The main function of IgA on mucosal surfaces is to protect against pathogenic infections (6, 21), but certain specificities may ultimately interfere with gut homeostasis and induce local inflammation (36). This warrants future investigation of the specificities of neonatal IgA induced by the maternal microbiota. It is likely that this extra IgA is polyreactive or even autoreactive. The challenge is to determine whether application of microbe-based strategies (e.g., L. reuteri as a probiotic) to enhance immune function early in life leads to desired protection and facilitation of healthy immune development, or whether there are caveats for people at risk for developing autoimmune disorders.
Materials and Methods
Mice.
C57BL/6J (Rag1+/+, stock number 000664), B6.129S7-Rag1tm1Mom/J (Rag1−/−, stock number 002216), B6.129P-Tcrbtm1MomTcrdtm1Mom/J (stock number 002121), B6.129S1-Tlr5tm1Flv/J (stock number 008377), and B6.129P2-Rorctm1Litt/J (stock number 007571) were purchased from Jackson Laboratory and bred and maintained in an SPF facility at Virginia Tech, according to the requirements of the Institutional Animal Care and Use Committee. GF B6 mice were purchased from National Gnotobiotic Rodent Resource Center and maintained in the Gnotobiotic Rodent Facility at the Virginia Tech College of Veterinary Medicine. Timed mating was established, and breeder males were removed after 10 d of contact with breeder females. For microbiota equalization (littermate) experiments, two breeder females from the same litter were cohoused after weaning. When the females were 8 wk old, a single male mouse of the correct genotype was introduced to set up a breeding trio. The two pregnant females were separated 3 to 4 d before labor. For gut microbiota removal (antibiotic) experiments, breeder mice were given a mixture of antibiotics (1 g/L ampicillin, 1 g/L neomycin, 0.5 g/L metronidazole, and 1g/L vancomycin) in the drinking water from the day of mating. For gestational gavage, females were orally gavaged with 200 μL of bacteria culture on days 0, 5, 10, 15, and 20 postbreeding. Gavage of neonatal mice was performed as previously described (37). For the GF microbiota transplantation experiments, pregnant GF females were orally gavaged with 200 μL of stomach content suspension or bacteria culture twice at the 10th and 15th day after mating, respectively. For cross-fostering experiments, all switches of pups were finished within 24 h of birth.
Bacterial Culture.
L. reuteri CF48-3A and L. oris F0423 were obtained from BEI Resources. Bacteria were cultured overnight in 5 mL of deMan Rogasa and Sharpe (MRS) broth for 12 to 15 h. The following morning, subcultures were established at 1:100 in 30 mL MRS and grown to exponential phase. Cultures were harvested by centrifugation, washed, and resuspended at desired concentration in sterile phosphate buffered saline (PBS) or for neonatal gavage (5% dextrose saline solution).
Microbiota Sampling and Analysis.
Autoclaved foil, tweezers, and scissors were used to collect microbiota samples. Milk was collected through two methods. For the first method (38), pups (3 to 5 d of age) were removed from dams and euthanized. Eight hours later, dams were injected intraperitoneally with oxytocin that acted on the mammary glands of lactating females to stimulate the release of milk. Immediately after, they were anesthetized with ketamine/xylazine that would sedate the mice for ∼20 to 30 min. During this time, the mammary tissue was gently massaged and squeezed to manually express milk. Another researcher would then use a sterile pipette tip to suck up the expressed milk and transfer to a 1.5 mL sterile centrifuge tube. Using this method, we were able to collect about 200 μL of milk per dam. For the second method, milk was collected from stomachs of 3-d-old pups. For microbiota sampling of 12- and 16-d-old pups, intestinal contents were collected from different sites. To avoid cage effects, samples in each group were collected from at least two different cages. Samples were frozen at −80 °C until processed at the same time. Sample homogenization, cell lysis, DNA extraction, and PCR were performed. qPCR was used to calculate absolute abundance of Lactobacillus species and total bacteria as we previously published (11). Standards for determining absolute abundance were generated by performing DNA extraction on a known concentration of L. reuteri culture. Relative abundance was determined through qPCR by taking the ratio of absolute abundance of Lactobacillus to total bacteria or 16S rRNA sequencing. The primer sequences were 1) total bacteria, forward 5′-ACT CCT ACG GGA GGC AGC AGT-3′, reverse 5′-ATT ACC GCG GCT GCT GGC-3′; 2) L. reuteri, forward 5′-GCG TTG ATG TTG TTG AAG GAA TGA GCT TTG-3′, reverse 5′-CAT CAG CAA TGA TTA AGA GAG CAC GGC C-3′; and 3) L. casei (not specific), forward 5′-GCC CTT AAG TGG GGG ATA AC-3′, reverse 5′-TAG AGT TTG GGC CGT GTC TC-3′. For 16S rRNA sequencing, purified amplicons of the V4 region were sequenced bidirectionally on an Illumina MiSeq at Argonne National Laboratory. Sequences were analyzed by a bioinformatician as we previously published (11).
ELISA.
Serum samples were stored at −20 °C until processing. One-centimeter intestinal segments from different sites were collected during dissection. After thorough washes with PBS, the intestinal segments were cultured in complete cell culture media (RPMI 1640 supplemented with 10% fetal bovine serum, 1 mM sodium pyruvate, 1% minimum essential medium nonessential amino acids, 10 mM Hepes, 55 μM 2-mercaptoethanol, 2 mM l-glutamine, 100 U/mL penicillin–streptomycin [Life Technologies]) for 5 to 7 d. The supernatant was harvested after centrifugation and frozen at −20 °C. During mouse dissection, colonic contents were collected and weighed. The contents were homogenized in PBS, and the supernatant was saved after centrifugation. Milk, serum, intestinal culture supernatant, and supernatant of homogenized intestinal contents were used for ELISA following the instructions of ELISA kits: mouse IgA and IgG (Bethyl Laboratories) and flagellin for mouse samples (MyBiosource).
Cell Isolation and Flow Cytometry.
Spleens were collected and mashed in 70 μm cell strainers with complete culture media. Red blood cells were lysed with RBC lysis buffer (eBioscience). To isolate LP cells, the small intestine was cut into pieces and digested by a buffer with Dispase, DNase, and Collagenase. Intestinal epithelial cells were removed, and Percoll was used for the isolation of lymphocytes. For surface staining, cells were blocked with anti-mouse CD16/32 (eBioscience), stained with fluorochrome-conjugated antibodies, and analyzed with a BD FACSAria II flow cytometer (BD Biosciences). For intracellular staining, a Foxp3 Fixation/Permeabilization kit (eBioscience) was used. Anti-mouse antibodies used in this study include APC anti-CD3, PE-Cy7 anti-CD4, FITC anti-CD8, PerCP-Cy5.5 anti-CXCR5, APC-Cy7 anti-PD-1, PE anti-B220, FITC anti-CD38, APC anti-GL7, FITC anti-IgA, PerCP-Cy5.5 anti-CD45, “Lin”: biotin anti-CD3 and biotin anti-CD19, a Zombie Aqua Fixable Viability Kit (Biolegend), RORγt-PE (BD Biosciences), and APC anti-biotin (Miltenyi Biotec). Flow cytometry data were analyzed with FlowJo.
Salmonella and EHEC Cross-Reactivity Whole-Cell ELISA.
Serum samples were assessed for reactivity to S. typhimurium and EHEC by whole-cell ELISA as described (39). Briefly, Salmonella and EHEC cells were fixed with 1% formalin for 2 h at room temperature (RT). About 108 colony forming units of fixed cells in 100 μL coating buffer (50 mM sodium carbonate) were added into each well of the Nunc Maxisorp plate (ThermoFisher, 44-2404-21) and incubated overnight at 4 °C. The plate was blocked with 4% nonfat milk in PBS-T (PBS with 0.05% Tween-20) for 2 h at RT. Then 50 μL of serially diluted antiserum or mouse serum samples were added into the wells for 1 h incubation at RT. E. coli O157:H7 antiserum (Seracare) was diluted from 1:10,000. Salmonella H antiserum I (1:80,000), O antiserum factors 4,5 (1:80,000), and O antiserum group D1 (1:64) (BD Difco) were combined before serial dilution. Next, wells were incubated for 1 h at RT with the following detection antibodies: horseradish peroxidase-conjugated anti-rabbit, anti-goat, and anti-mouse IgA (SouthernBiotech) and anti-mouse IgG (Bethyl). Super AquaBlue ELISA substrate (Thermo Fisher, 00-4203-58) was used for color development. The plate was read at optical density at 405 nm using SpectraMax M5 (Molecular Devices).
Statistics.
For the comparison of two groups, the two-tailed Student’s t test was used. For the comparison of more than two groups, one-way ANOVA and Tukey’s posttest were used. Results were considered statistically significant when P < 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). All analyses were performed with Prism GraphPad.
Supplementary Material
Acknowledgments
We thank Husen Zhang for microbiota analysis and Melissa Makris for flow cytometry analysis. We also thank Balfour Sartor (University of North Carolina at Chapel Hill) and the National Gnotobiotic Rodent Resource Center for providing GF animals. Microbiota sequencing was performed by Sarah Owens at Argonne National Laboratory. This work was partially supported by the Internal Research Competition (IRC) award from the Office of Research and Graduate Studies, Virginia-Maryland College of Veterinary Medicine.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015691118/-/DCSupplemental.
Data Availability
The 16S rRNA sequence data have been deposited in National Center for Biotechnology Information (NCBI) Sequence Read Archive (PRJNA638971).
Change History
August 13, 2021: The Acknowledgments have been updated.
References
- 1.Pantoja-Feliciano I. G., et al., Biphasic assembly of the murine intestinal microbiota during early development. ISME J. 7, 1112–1115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominguez-Bello M. G., et al., Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U.S.A. 107, 11971–11975 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madan J. C., et al., Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch. Dis. Child. Fetal Neonatal Ed. 97, F456–F462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mai V., et al., Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS One 8, e52876 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matamoros S., Gras-Leguen C., Le Vacon F., Potel G., de La Cochetiere M. F., Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. 21, 167–173 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Suzuki K., Fagarasan S., How host-bacterial interactions lead to IgA synthesis in the gut. Trends Immunol. 29, 523–531 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Benveniste J., Lespinats G., Adam C., Salomon J. C., Immunoglobulins in intact, immunized, and contaminated axenic mice: Study of serum IgA. J. Immunol. 107, 1647–1655 (1971). [PubMed] [Google Scholar]
- 8.Benveniste J., Lespinats G., Salomon J., Serum and secretory IgA in axenic and holoxenic mice. J. Immunol. 107, 1656–1662 (1971). [PubMed] [Google Scholar]
- 9.Fagarasan S., Kawamoto S., Kanagawa O., Suzuki K., Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu. Rev. Immunol. 28, 243–273 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Le Doare K., Holder B., Bassett A., Pannaraj P. S., Mother’s milk: A purposeful contribution to the development of the infant microbiota and immunity. Front. Immunol. 9, 361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H., Sparks J. B., Karyala S. V., Settlage R., Luo X. M., Host adaptive immunity alters gut microbiota. ISME J. 9, 770–781 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris N. L., et al., Mechanisms of neonatal mucosal antibody protection. J. Immunol. 177, 6256–6262 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Kramer D. R., Cebra J. J., Early appearance of “natural” mucosal IgA responses and germinal centers in suckling mice developing in the absence of maternal antibodies. J. Immunol. 154, 2051–2062 (1995). [PubMed] [Google Scholar]
- 14.Koch M. A., et al., Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell 165, 827–841 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aagaard K., et al., The placenta harbors a unique microbiome. Sci. Transl. Med. 6, 237ra265 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Goffau M. C., et al., Human placenta has no microbiome but can contain potential pathogens. Nature 572, 329–334 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robertson S. J., et al., Comparison of Co-housing and littermate methods for microbiota standardization in mouse models. Cell Rep. 27, 1910–1919.e2 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Ormerod K. L., et al., Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome 4, 36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satoh-Takayama N., et al., Bacteria-induced group 2 innate lymphoid cells in the stomach provide immune protection through induction of IgA. Immunity 52, 635–649.e4 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Frese S. A., et al., The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet. 7, e1001314 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki K., Nakajima A., New aspects of IgA synthesis in the gut. Int. Immunol. 26, 489–494 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Randall T. D., Carragher D. M., Rangel-Moreno J., Development of secondary lymphoid organs. Annu. Rev. Immunol. 26, 627–650 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishikawa S., Honda K., Vieira P., Yoshida H., Organogenesis of peripheral lymphoid organs. Immunol. Rev. 195, 72–80 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Veiga-Fernandes H., et al., Tyrosine kinase receptor RET is a key regulator of Peyer’s patch organogenesis. Nature 446, 547–551 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Eberl G., et al., An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 5, 64–73 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Yokota Y., et al., Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature 397, 702–706 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Kanamori Y., et al., Identification of novel lymphoid tissues in murine intestinal mucosa where clusters of c-kit+ IL-7R+ Thy1+ lympho-hemopoietic progenitors develop. J. Exp. Med. 184, 1449–1459 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamada H., et al., Identification of multiple isolated lymphoid follicles on the antimesenteric wall of the mouse small intestine. J. Immunol. 168, 57–64 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Pabst O., et al., Cryptopatches and isolated lymphoid follicles: Dynamic lymphoid tissues dispensable for the generation of intraepithelial lymphocytes. Eur. J. Immunol. 35, 98–107 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Eberl G., Littman D. R., Thymic origin of intestinal alphabeta T cells revealed by fate mapping of RORgammat+ cells. Science 305, 248–251 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Tsuji M., et al., Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity 29, 261–271 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Fulde M., et al., Neonatal selection by Toll-like receptor 5 influences long-term gut microbiota composition. Nature 560, 489–493 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Wang L., et al., Bacterial IgA protease-mediated degradation of agIgA1 and agIgA1 immune complexes as a potential therapy for IgA Nephropathy. Sci. Rep. 6, 30964 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reboldi A., et al., IgA production requires B cell interaction with subepithelial dendritic cells in Peyer’s patches. Science 352, aaf4822 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melo-Gonzalez F., et al., Antigen-presenting ILC3 regulate T cell-dependent IgA responses to colonic mucosal bacteria. J. Exp. Med. 216, 728–742 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maes M., et al., Increased IgA responses to the LPS of commensal bacteria is associated with inflammation and activation of cell-mediated immunity in chronic fatigue syndrome. J. Affect. Disord. 136, 909–917 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Francis F., et al., Probiotic studies in neonatal mice using gavage. J. Vis. Exp. 143, e59074 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Willingham K., et al., Milk collection methods for mice and Reeves’ muntjac deer. J. Vis. Exp. 89, e51007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng W., et al., Microbiota-targeted maternal antibodies protect neonates from enteric infection. Nature 577, 543–548 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA sequence data have been deposited in National Center for Biotechnology Information (NCBI) Sequence Read Archive (PRJNA638971).