Significance
Many diseases involve the propagation of misfolded proteins as in prion diseases. Compounds that can inhibit conversion into misfolded conformers exist, but their mechanism of action remains poorly understood. By observing the effects of the polymeric antiprion pharmacological chaperone pentosan polysulfate on the folding of single prion protein molecules, we find that interactions with partially and fully unfolded states play an important role. A comparison to previous results for a chaperone with very different chemical properties suggests that antiprion agents help to prevent conversion by interacting with the protein when it unfolds from the native structure during the conversion process. Unfolded states may, therefore, be fruitful targets for developing more effective therapeutic inhibitors of prion and prion-like conversion.
Keywords: protein misfolding, pharmacological chaperone, optical tweezers, energy landscape
Abstract
Prion and prion-like diseases involve the propagation of misfolded protein conformers. Small-molecule pharmacological chaperones can inhibit propagated misfolding, but how they interact with disease-related proteins to prevent misfolding is often unclear. We investigated how pentosan polysulfate (PPS), a polyanion with antiprion activity in vitro and in vivo, interacts with mammalian prion protein (PrP) to alter its folding. Calorimetry showed that PPS binds two sites on natively folded PrP, but one PPS molecule can bind multiple PrP molecules. Force spectroscopy measurements of single PrP molecules showed PPS stabilizes not only the native fold of PrP but also many different partially folded intermediates that are not observed in the absence of PPS. PPS also bound tightly to unfolded segments of PrP, delaying refolding. These observations imply that PPS can act through multiple possible modes, inhibiting misfolding not only by stabilizing the native fold or sequestering natively folded PrP into aggregates, as proposed previously, but also by binding to partially or fully unfolded states that play key roles in mediating misfolding. These results underline the likely importance of unfolded states as critical intermediates on the prion conversion pathway.
The propagation of misfolded proteins is a hallmark of a wide variety of diseases, including Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis, and the tauopathies (1). In misfolding diseases like these, nonnative conformers of disease-linked proteins can convert natively folded molecules into additional copies of the misfolded conformers, propagating the misfolding (2). The archetypal misfolding diseases, where propagated misfolding was first discovered, are the prionopathies, such as Creutzfeldt-Jakob disease (CJD) in humans, scrapie in sheep, “mad-cow” disease, and chronic wasting disease in cervids. In prion diseases, the helical fold of native cellular prion protein, PrPC, is converted to a β-rich misfolded isoform, PrPSc, that is both toxic to neurons and acts as an infectious agent to propagate the disease (3).
Because misfolding and conversion are central to prion and prion-like diseases, one therapeutic strategy has been to search for small molecules that can act as pharmacological “chaperones” to inhibit the misfolding (4), analogous to the chaperone proteins that help suppress misfolding in the cell (5). Even though the structure of PrPSc and the mechanism by which it converts PrPC remain controversial (3, 6), a variety of chemically distinct compounds with promising potential against prions have been found (7, 8). The mechanisms by which these putative chaperones act are typically quite poorly understood, however, in part because it is challenging to observe the effects of antiprion compounds on PrP folding directly. As a result, it has proven difficult to develop effective therapeutics or indeed to use the effects of antiprion compounds to gain insight into the misfolding mechanism.
Single-molecule methods are well suited to address this challenge because they can observe individual molecules directly as they change conformation and distinguish subpopulations in a heterogeneous mixture (9, 10), and they have been applied to study misfolding in several disease-related proteins (11–16), including PrP (17–19). Such approaches have also been used to study the mechanisms of cellular chaperones (20), but the effects of pharmacological chaperones have been little explored at the single-molecule level. The only study to date (21) examined the changes in the folding of PrP caused by binding of an antiprion iron tetrapyrrole, Fe-TMPyP, finding that it not only stabilized the native fold as proposed based on ensemble studies (22) but, more interestingly, that it also inhibited the interactions that drove the formation of stable misfolded aggregates, similar to the action of cellular chaperones (23). Owing to the paucity of single-molecule studies of pharmacological chaperones, most of the existing proposals for their mechanism of action are based on the limited information available from ensemble assays, and it remains unclear how they achieve their effect.
Here, we examine how the antiprion compound pentosan polysulfate (PPS) changes the folding of PrP. PPS belongs to a different class of pharmacological chaperone from Fe-TMPyP and other similar molecules: Instead of having a compact, planar structure with cationic charge and low molecular weight, PPS has the opposite character—it is a large, linear polymer with a heparin-like structure (Fig. 1 A, Inset) and anionic charge. It has been found to inhibit the accumulation of PrPSc in prion-infected cells and prolong the lifespan of infected animals (24), and it has even had some success increasing the survival period of patients with CJD, although it is difficult to administer, as it does not penetrate the blood–brain barrier (25–28). PPS is known to interact with PrP (7, 29) and has been proposed to sequester PrPC in nontoxic aggregates so that it cannot be converted to PrPSc (7, 30), but its mechanism of action remains uncertain. To study the interaction between PPS and PrP in more detail, we used a combination of isothermal titration calorimetry (ITC), measuring the thermodynamics of the interaction, and single-molecule force spectroscopy (SMFS), observing the dynamics of a single PrP molecule as it was unfolded and refolded while held under tension by optical tweezers in the presence of PPS. We found very heterogeneous binding: not only binding to two independent sites on natively folded PrP, where each PPS molecule interacted with multiple PrP molecules, but also strong binding to partially and fully unfolded states. The fact that two chemically distinct antiprion agents both interact strongly with unfolded states suggests a central role for unfolded PrP in the prion conversion mechanism.
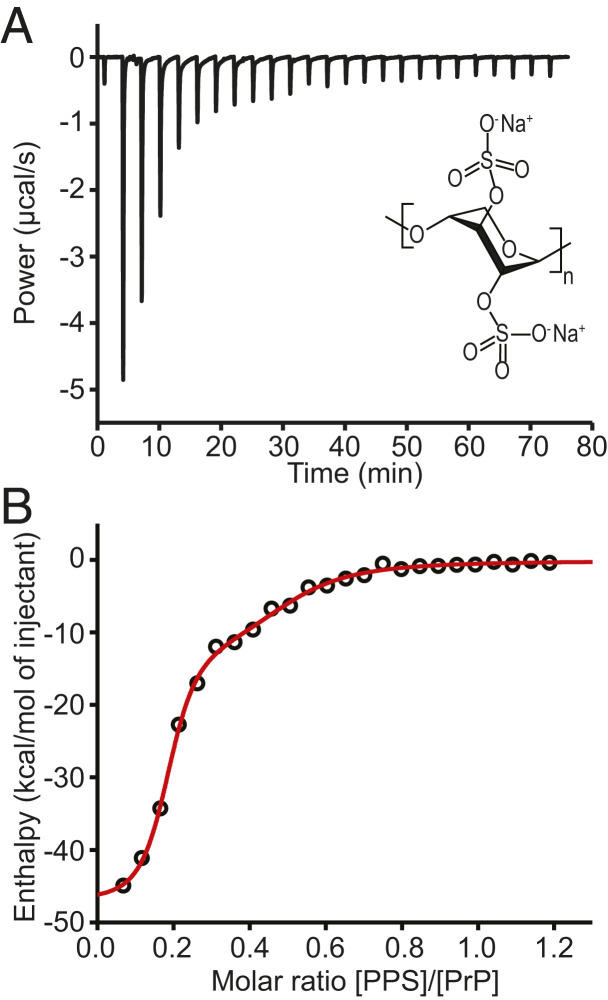
Fig. 1.
ITC of PPS binding to PrPC. (A) ITC thermogram for binding of PPS to hamster PrPC at pH 7. (Inset) Structure of PPS. (B) Binding isotherm (black) and fit to model with two independent binding sites (red).
Results
We first characterized the binding of PPS to natively folded PrP by measuring the rate of heat produced from the binding while PPS was titrated into a sample of recombinant Syrian hamster PrP (see Methods) using ITC. The resulting thermogram (Fig. 1A) was integrated to generate a binding isotherm (Fig. 1B, black). Fitting the shape of the isotherm to the expectations for different binding models, we found that single-site models were unable to account for the observed isotherm (SI Appendix, Fig. S1); instead, a model with two independent binding sites fit better (Fig. 1B, red). One site had dissociation constant KD,1 = 0.19 ± 0.04 μM, binding enthalpy ΔH1 = −47 ± 2 kcal/mol, and binding entropy ΔS1 = −128 ± 3 cal/mol ∙ K, whereas the other was noticeably weaker, with KD,2 = 8 ± 2 μM, ΔH2 = −15 ± 3 kcal/mol, and ΔS2 = −25 ± 13 cal/mol ∙ K (all errors represent SEM). Although binding of PPS to natively folded PrP was detected using surface plasmon resonance previously, the binding affinity and thermodynamics were not reported (31).
The binding stoichiometry from the fitting was particularly interesting: each PPS molecule bound 5.6 ± 0.9 PrP molecules at the stronger binding site and 2.3 ± 0.4 PrP molecules at the weaker binding site. Such a result with a single ligand bound to multiple protein molecules is unusual but possible because PPS is a sizable linear polymer (containing in this case ∼15 monomer repeats) rather than a compact ligand. Furthermore, because each PrP molecule can bind a second PPS molecule, each of which binds to multiple PrP molecules, chains of PPS-linked PrP are possible.
PPS binding to PrP is proposed to be mediated by electrostatic interactions between the negative charge of PPS and the net positive charge of PrP. Since the isoelectric point of PrP is 8.9, the protein will be more positively charged at acidic pH, which should increase the binding affinity. Measuring the binding with ITC at pH 4.5 (SI Appendix, Fig. S2), we found that the stoichiometry of the two binding sites remained the same within error, with each PPS molecule binding 5 ± 1 PrP molecules at the stronger binding site and 2.0 ± 0.2 PrP at the weaker site. As expected, however, the affinity was higher, with KD,1 = 9 ± 5 nM, ΔH1 = −11.3 ± 0.4 kcal/mol, and ΔS1 = −1 ± 1 cal/mol ∙ K for the stronger binding site and KD,2 = 0.4 ± 0.1 μM, ΔH2 = −8.7 ± 0.3 kcal/mol, and ΔS2 = 0 ± 1 cal/mol ∙ K for the weaker binding site. These results confirmed that electrostatic interactions play a central role in the binding of PPS to PrP.
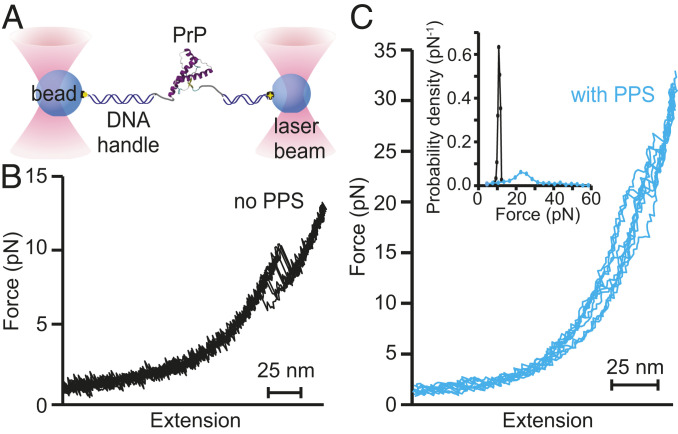
Next, to probe the effects of PPS binding on the folding of PrP, we attached recombinant Syrian hamster PrP covalently to kilobase-long DNA handles bound to beads held in optical traps (Fig. 2A). We then moved the traps apart at constant speed to ramp up the force on the protein and induce unfolding, reversed the trap motion to ramp the force back down and induce refolding, and waited 5 s at near-zero force before repeating the pulling–relaxing cycle, generating sets of force-extension curves (FECs) that show the force applied to the molecule as a function of the molecular extension. In the absence of PPS, unfolding FECs revealed a single “rip” with an abrupt extension increase and force decrease where the protein molecule unfolded (Fig. 2B). As reported previously (18, 19), the contour length change upon unfolding (34.3 ± 0.4 nm) was consistent with the 34.3-nm value expected for complete unfolding of the structured domain of hamster PrP (32), no intermediates were observed, and the unfolding force was narrowly distributed around an average of 10.2 ± 0.2 pN. In the presence of saturating PPS (5 µM), in contrast, the unfolding FECs changed qualitatively in several ways. Only ∼30% of FECs contained rips, indicating that in most of the FECs, the protein did not actually change structure. In those FECs that did contain rips, the unfolding forces were generally both much higher and more broadly distributed than without PPS (Fig. 2C). These FECs also showed the presence of intermediate states, which were never observed for hamster PrP in the absence of PPS, and diverse patterns of unfolding and refolding.
Fig. 2.
FECs of PrP bound to PPS. (A) Schematic of force spectroscopy experiment: A single PrP molecule is attached to beads held in optical traps by DNA handles. (B) Without PPS present, PrP unfolds with a characteristic contour length change of 34.3 nm in a narrow range of forces near 10 pN. (C) With PPS present, PrP unfolds heterogeneously, showing a variety of different contour lengths and unfolding forces. (Inset) The unfolding force distribution with PPS present (cyan) is much broader than without PPS (black).
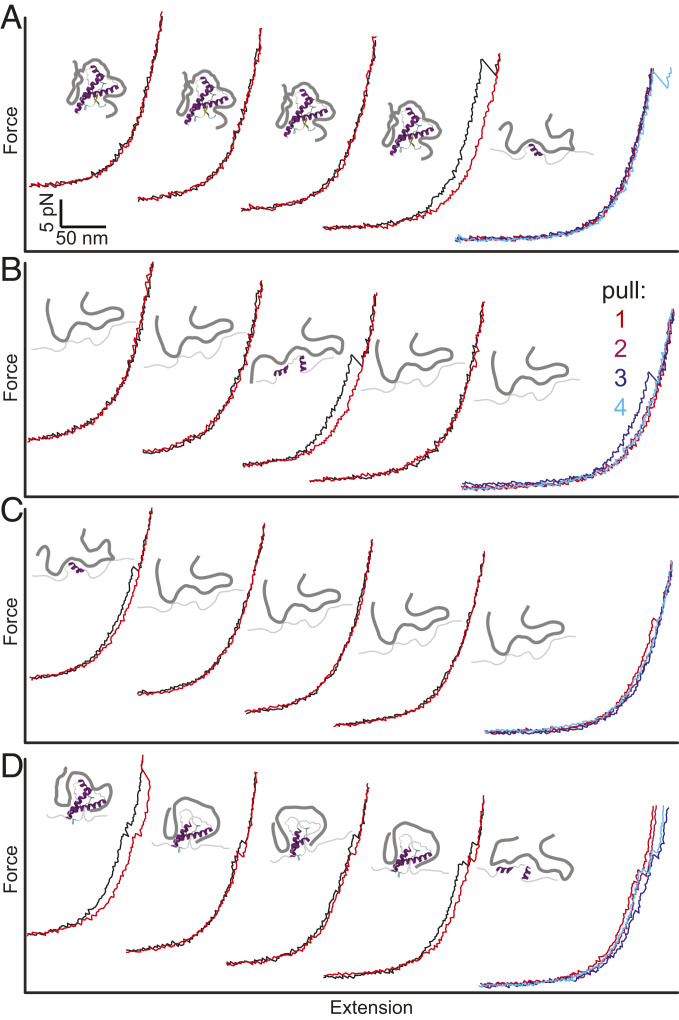
Examining a series of successive pulling–relaxing cycles for a given molecule illustrates this diversity clearly (Fig. 3). In some cases, multiple pulling–relaxing cycles occurred without any transitions before the contour length was seen to increase, indicating unfolding of a state that was strongly stabilized by PPS (Fig. 3A). In other cases, cycles without transitions preceded a decrease in the contour length, indicating that PPS binding had stabilized the protein in a partially or fully unfolded state and the protein subsequently refolded (Fig. 3B). Sometimes, an initial unfolding event was followed by several cycles without further unfolding or refolding (Fig. 3C), suggesting the formation of a stable PPS-bound unfolded state; at other times, the protein unfolded through a series of intermediate states over the course of multiple pulling–relaxing cycles, indicating that PPS acted as a ratchet, binding and stabilizing a sequence of ever-more–unfolded intermediates (Fig. 3D).
Fig. 3.
Heterogeneous unfolding of PPS-bound PrP. Four sets of successive pulling (black) and relaxing (red) curves of PrP with PPS bound show various behaviors. (Far Right) Unfolding curves for the four successive pulls. (A) PrP remains folded through the first three cycles and unfolds on the fourth. (B) PrP remains mostly unfolded but refolds between the second and third cycles. (C) PrP remains mostly unfolded after unfolding in the first cycle. (D) PrP unfolds progressively through a series of intermediates in each cycle. Insets between cycles show cartoons of PPS bound to PrP in different states.
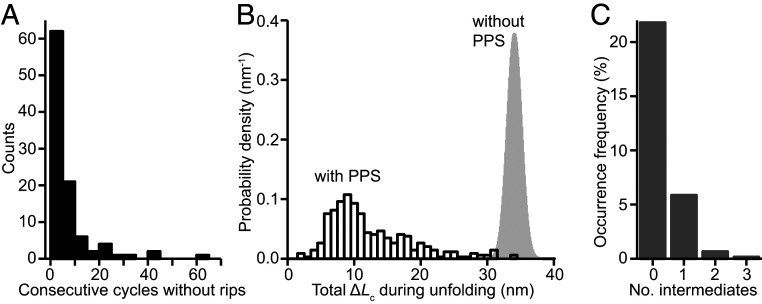
To quantify this behavior, we analyzed the number of cycles without transitions when PPS was present as well as the length changes, unfolding forces, and number of intermediates in FECs that did show transitions. The number of sequential pulling–relaxing cycles without transitions was broadly distributed (Fig. 4A), highlighting the heterogeneity of the behavior. The total contour-length change seen in each unfolding FEC containing a transition, ΔLctot (Fig. 4B, black), was also broadly distributed, ranging from ∼3 to 34 nm but with most transitions at 5 to 20 nm. Almost all (>99%) of these transitions had ΔLctot shorter than the value for complete unfolding of the native state, indicating that partially folded states played a central role with PPS present. Indeed, the importance of intermediates was reinforced by the fact that a significant fraction (∼25%) of the unfolding trajectories containing rips included more than one transition event and thus passed through at least one intermediate (Fig. 4C). In the ratchet-like unfolding events, the net contour-length change per unfolding–refolding cycle ranged from 2 to 22 nm with an average of ∼10 nm, but it had a strong peak at ∼8 nm (the value seen in half of all cases, SI Appendix, Fig. S3A), corresponding to sequestration of ∼20 amino acids when PPS reconfigured to bind the unfolded portion of the protein. The distribution of forces seen in unfolding transitions (Fig. 2 C, Inset, cyan) was also very different from that found previously in the absence of PPS (Fig. 2 C, Inset, black), with an average (25.2 ± 0.5 pN) that was over twice as large and an SD over 16 times as large (∼10.6 pN compared to 0.65 pN without PPS).
Fig. 4.
Characteristics of FECs with PPS bound to PrP. (A) Many FECs showed no unfolding or refolding transitions. (B) The total contour-length change in FECs exhibiting transitions (black) was broadly distributed and almost always shorter than the length observed without PPS (gray). (C) A varying number of intermediates were seen for unfolding with PPS; intermediates were never seen without PPS.
Lastly, we examined the effects of PPS binding on the energy landscape that underlies PrP folding, making use of the ability of SMFS to probe energy landscapes quantitatively (33). To characterize the changes in the landscape induced by PPS binding, we examined the distribution of unfolding forces. Whereas the force distribution in the absence of PPS was previously found to fit well to the expectation for two-state unfolding from a homogenous initial state (34), returning a distance from the folded state to the transition state of Δx‡ = 9 ± 1 nm (18), the distribution with PPS present did not fit well to this model (Fig. 5A, blue), consistent with the observation that the binding induced heterogeneity. A long tail of events at high unfolding forces extended far beyond the sharp cutoff expected for unfolding of a single well-defined state (Fig. 5A, blue); these events could not be explained by the instrumental force noise of <0.3 pN but rather indicated an alternative model was needed to fit the data. We therefore used a model that allows for heterogeneous initial states with different barriers and kinetics, quantifying the heterogeneity in the initial state via a dimensionless parameter Δ that is greater than zero when there are multiple initial states that interconvert at rates slower than the unfolding rates (with larger Δ indicating greater heterogeneity) (35). This model fit the data well (Fig. 5A, red; R2 = 0.85 for fit), returning Δx‡ = 1.1 ± 0.2 nm and a value for the heterogeneity parameter of Δ = 1.7 ± 0.8.
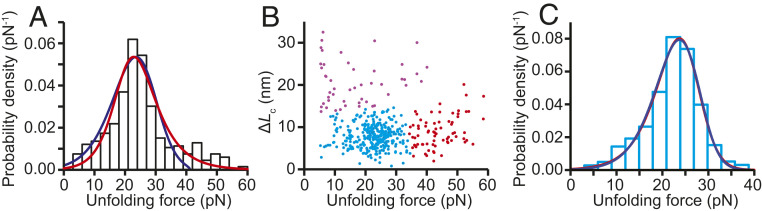
Fig. 5.
Unfolding force distribution. (A) The distribution of all unfolding forces (black) does not fit well to a model that assumes a single well-defined initial state (blue), which cannot account for the long tail at high forces, but it is described well by a model allowing for heterogenous initial states (red). (B) Most of the unfolding events belong to a single cluster (cyan) defined by an unfolding force below ∼30 pN and contour length below ∼12 nm. (C) The unfolding force distribution for the dominant cluster in B is well fit by both models used in A.
These results indicate that PPS binding increased the mechanical rigidity of the system (by decreasing Δx‡) while introducing a moderate amount of heterogeneity. In an attempt to isolate unfolding events arising from a more homogeneous subpopulation of states, we also examined the scatterplot of contour length change versus unfolding force (Fig. 5B), performing a cluster analysis using a variational Gaussian mixture algorithm (36) validated by silhouette index analysis (37). We found that a large majority (∼73%) of the unfolding events belonged to a single cluster (Fig. 5B, cyan), for which the force distribution was well fit by both the heterogeneous-state model (Fig. 5C, red) and the model with a single initial state (Fig. 5C, blue). The two fits returned similar Δx‡ values (1.1 ± 0.1 nm and 1.5 ± 0.3 nm, respectively), but the heterogeneous-state model returned a value for Δ that was reduced significantly from that for the full force distribution, to 0.4 ± 0.2, consistent with the notion that the cluster represented a more homogeneous population. However, Δ remained sufficiently large to suggest that the FECs may not represent a single population. Indeed, the distribution of ΔLc values for this cluster (SI Appendix, Fig. S4) had an SD ∼2.5-fold larger than what was seen previously for unfolding a single well-defined state (16, 18), supporting the notion that this cluster contains a few distinct states, even though they have similar properties. Fitting the force distribution for the cluster at high unfolding force (SI Appendix, Fig. S5) yielded similarly small Δx‡, at 1.3 ± 0.3 nm, but this cluster remained quite heterogeneous, with Δ = 2.2 ± 0.9; the cluster at high ΔLc had too few entries distributed over too wide a force range for reliable fitting. Notably, the unfolding events seen when first pulling on each PrP molecule had similar heterogeneity to the results from subsequent FECs (SI Appendix, Fig. S6), indicating that the heterogeneity was not induced by the unfolding–refolding cycles but was already present at the start of the measurement.
Discussion
PPS has been previously proposed to inhibit misfolding of PrP by sequestering PrPC in aggregates (7, 30). Our ITC results are consistent with this picture and explain how such aggregation could occur: In addition to multiple PrP molecules binding to a single PPS molecule, multiple PPS chains can be cross-linked via the secondary binding sites available on the native fold, generating a web of PPS chains binding natively folded PrP. Using docking simulations to explore the location of the PPS binding sites (SI Appendix), we found that the most likely sites were the middle of helix 2 and the cleft between helices 1 and 3 near localized pockets of positive charge on the surface (SI Appendix, Fig. S7), suggesting that steric hindrance would not prevent multiple PPS molecules binding to a single PrP. Given that previous NMR studies found that PPS can also bind to PrP in the disordered octapeptide repeat region (29), which is absent in the truncated PrP constructs used in our measurements, full-length PrPC must have at least three binding sites for PPS, providing additional opportunities to cross-link multiple PrP and PPS molecules and form aggregates.
Crucially, however, the SMFS measurements showed that PPS interacts with PrP in many other ways, too: It bound not only to the native fold but also to many other conformational states with varied levels of unfolding of the native structure, leading to exceptional heterogeneity that contrasts starkly with the homogeneity of the unfolding and refolding of PrP on its own (17, 18). We note that observing such interactions with unfolded conformers of a normally ordered domain is challenging, as it requires the use of a method like SMFS that destabilizes protein structure without otherwise altering the solution and environmental conditions upon which ligand binding depends. Normally, partially folded intermediates cannot be detected in unfolding or refolding FECs of hamster PrP monomers (17, 18), but with PPS bound, numerous intermediates were populated and stabilized. Intriguingly, in some cases, PrP was seen to unfold in multiple stages over several pulling curves in a ratchet-like manner, with a portion of the protein unfolding each time but not refolding (Fig. 3D). The bound PPS was thus able to reconfigure rapidly and sequester the portions of the protein exposed after each partial unfolding event, displaying a strong affinity for the unfolded protein in addition to the native fold. Although the time required for PPS to reconfigure and bind the exposed regions of PrP could not be detected directly from FECs, it had to be faster than 0.5 s, the minimum time elapsed between unfolding and the point at which refolding occurs in the absence of PPS. The unfolding transitions during ratchet-like events were also somewhat more homogeneous than the overall population, with most transitions falling within the main cluster from Fig. 5B (SI Appendix, Fig. S3B); the net length change per ratchet cycle was particularly regular (SI Appendix, Fig. S3A), suggesting there might be patterns in the rebinding, although the small number of events makes interpretation uncertain.
The heterogeneous binding of PPS to PrP seen by SMFS is consistent with the proposal that PPS interacts nonspecifically with PrP (7, 29), with the ITC results supporting the notion that binding is primarily through electrostatic interactions. However, the full picture of the effects of PPS on PrP is clearly more complex than suggested simply from the interactions of PPS with natively folded PrP, as assumed previously. Indeed, the effects of PPS binding on PrP folding/unfolding dynamics share some important similarities with the effects of the only other antiprion chaperone studied to date at the single-molecule level, Fe-TMPyP. The latter, like PPS, was found to bind to unfolded regions of PrP (stabilizing partially unfolded, nonnative conformations) as well as the native fold, and its binding also made PrP more rigid by decreasing the distance to the transition state from 9 nm to ∼1 nm (21). These similarities are particularly striking because PPS differs from Fe-TMPyP in fundamental ways: It is an extended linear polymer instead of a compact, planar porphyrin; it is negatively charged, as opposed to positively charged; its binding to PrP depends significantly on electrostatic interactions instead of only weakly, as in the case of Fe-TMPyP (22); and it binds nonspecifically at two sites without any obvious binding pocket instead of nestling into a single specific pocket, as does Fe-TMPyP (22).
The fact that PPS rigidifies the native fold of PrP and binds to unfolded regions of PrP, similar to Fe-TMPyP, is very suggestive in terms of possible common mechanisms of action for these pharmacological chaperones. Stabilization of native structures has long been accepted as an important mechanism for inhibiting conversion, but interactions with unfolded states have generally been overlooked (7, 38). Whereas it might be a coincidence that a single antiprion compound interacts strongly with unfolded states, there is no reason to expect a priori that two such chemically distinct compounds as PPS and Fe-TMPyP should do so, suggesting that these interactions are no coincidence but rather are essential to the antiprion activity. We propose that protection against conversion could arise as follows: In order to convert PrPC into PrPSc, the native helical structure has to be unfolded to allow reconfiguration into PrPSc, which has no native helical content (39) and is likely a β-solenoid (6). Antiprion ligands could then interfere with prion conversion and inhibit propagation of misfolding by binding to the partially or fully unfolded states that must presumably be present as transient but obligate intermediates during conversion, over and above any effect they have on reducing the likelihood of unfolding PrPC by stabilizing and rigidifying it. Notably, these interactions with unfolded states are similar to the mechanisms of a variety of cellular chaperones, which interact with partially or fully unfolded conformations of client proteins to inhibit the interactions that drive misfolding and/or aggregation (20, 40, 41).
These results suggest that partially or fully unfolded states may be important targets for inhibiting prion conversion that have been neglected in the search for antiprion therapeutics. The same may possibly hold true for other proteins that undergo prion-like conversion, in which misfolding is mediated by partially unfolded states (16). Testing for binding to unfolded states can be challenging, but it could prove fruitful to do so, and even worth revisiting other antiprion compounds to test if they, too, interact with unfolded states. Finally, we speculate that the heterogeneous interactions and multiplicity of modes of action available to PPS, as well as to Fe-TMPyP, may account for their ability to inhibit structurally distinct prion strains (24, 42), in contrast to the strain-specificity of other antiprion agents like quinacrine (43). This hypothesis could be tested in future work comparing the effects of strain-specific and nonspecific antiprion agents at the singe-molecule level.
Methods
Sample Preparation.
Samples of Syrian hamster PrP truncated to residues 90 to 231 (forming the protease-resistant core of PrPSc) were expressed recombinantly in Escherichia coli, purified, and refolded as described previously (17). Sample purity was assessed with gel electrophoresis and Western blotting, and refolding into the native structure was confirmed using CD spectroscopy (19).
Isothermal Titration Calorimetry.
ITC measurements were made using a MicroCal ITC200. For measurements at neutral pH, 5 kDa PPS at a concentration of 1 mM was titrated into 200 μL 0.1-mM PrP (both reagents in 50 mM MOPS buffer pH 7), using an initial injection of 0.2 to 0.5 μL followed by 1-μL injections with 2-s duration at intervals of 180 to 250 s while stirring at 1,000 rpm. Five separate measurements were performed at pH 7 and the results averaged. For measurements at acidic pH, 1-mM PPS was titrated into 200 μL 0.1-mM PrP (both reagents in 10 mM sodium acetate buffer pH 4.5), using an initial 0.2-μL injection followed by 2-μL injections with 4-s duration at intervals of 200 s while stirring at 1,000 rpm. Three separate measurements were performed at pH 4.5 and the results averaged. For all measurements, the temperature was maintained at 25 °C. The Bayesian information criterion (44) and Akaike information criterion (45) were used to determine that the model with two sets of independent binding sites fit better than the one-binding-site model for both measurement conditions (ΔBIC and ΔAIC, respectively, 27 and 25 at pH 7 and 15 and 8 at pH 4.5).
SMFS Measurements and Analysis.
SMFS measurements were made using custom-built optical tweezers described previously (46). Syrian hamster PrP (90-231) with a Cys residue added to each terminus was conjugated to sulfhydryl-labeled double-stranded DNA handles as described previously (17) after activating the terminal cysteines on PrP with 2,2′-dithiodipyridine (47). Handles were attached specifically to avidin- and anti-digoxigenin–labeled polystyrene beads (600- and 820-nm diameter, respectively) held in traps with stiffness of 0.37 and 0.56 pN/nm, respectively. The traps were moved apart at a constant speed of ∼170 to 250 nm/s to generate FECs, sampling data at 20 kHz and filtering online at the Nyquist frequency. FECs were measured in 50 mM MOPS buffer pH 7, 200 mM KCl, in the presence of 5 μM PPS. Measurements from 18 molecules yielded 1,276 curves for pulling and the same for relaxation. Measurements without PPS were reported previously (17–19).
Contour-length changes in the FECs were determined by fitting the branches of the FECs before and after each structural transition to worm-like chain polymer models (48, 49). The unfolding force distributions were analyzed using models that assume either unfolding from a single initial state (34) or unfolding from multiple initial states (35). Unfolding events were clustered by a variational Gaussian mixture algorithm implemented in Mathematica; the clustering was validated by silhouette index analysis (37), returning a global silhouette index of 0.41. Additional details are provided in the SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by the Alberta Prion Research Institute, Alberta Innovates, and National Research Council Canada.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010213118/-/DCSupplemental.
Data Availability
Single-molecule force spectroscopy data (50) have been deposited in Excel format in Figshare (DOI: 10.6084/m9.figshare.13404719).
References
- 1.Chiti F., Dobson C. M., Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Walker L. C., Jucker M., Neurodegenerative diseases: Expanding the prion concept. Annu. Rev. Neurosci. 38, 87–103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colby D. W., Prusiner S. B., Prions. Cold Spring Harb. Perspect. Biol. 3, a006833 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sweeney P., et al., Protein misfolding in neurodegenerative diseases: Implications and strategies. Transl. Neurodegener. 6, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartl F. U., Bracher A., Hayer-Hartl M., Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Vázquez-Fernández E., et al., The structural architecture of an infectious mammalian prion using electron cryomicroscopy. PLoS Pathog. 12, e1005835 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamatari Y. O., Hayano Y., Yamaguchi K., Hosokawa-Muto J., Kuwata K., Characterizing antiprion compounds based on their binding properties to prion proteins: Implications as medical chaperones. Protein Sci. 22, 22–34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barreca M. L., Iraci N., Biggi S., Cecchetti V., Biasini E., Pharmacological agents targeting the cellular prion protein. Pathogens 7, 27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu H., Dee D. R., Woodside M. T., Single-molecule approaches to prion protein misfolding. Prion 7, 140–146 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann A., Neupane K., Woodside M. T., Single-molecule assays for investigating protein misfolding and aggregation. Phys. Chem. Chem. Phys. 15, 7934–7948 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Ferreon A. C. M., Gambin Y., Lemke E. A., Deniz A. A., Interplay of α-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc. Natl. Acad. Sci. U.S.A. 106, 5645–5650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hervás R., et al., Common features at the start of the neurodegeneration cascade. PLoS Biol. 10, e1001335 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cremades N., et al., Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell 149, 1048–1059 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neupane K., Solanki A., Sosova I., Belov M., Woodside M. T., Diverse metastable structures formed by small oligomers of α-synuclein probed by force spectroscopy. PLoS One 9, e86495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iljina M., et al., Arachidonic acid mediates the formation of abundant alpha-helical multimers of alpha-synuclein. Sci. Rep. 6, 33928 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sen Mojumdar S., et al., Partially native intermediates mediate misfolding of SOD1 in single-molecule folding trajectories. Nat. Commun. 8, 1881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu H., et al., Direct observation of multiple misfolding pathways in a single prion protein molecule. Proc. Natl. Acad. Sci. U.S.A. 109, 5283–5288 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu H., et al., Energy landscape analysis of native folding of the prion protein yields the diffusion constant, transition path time, and rates. Proc. Natl. Acad. Sci. U.S.A. 109, 14452–14457 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu H., et al., Protein misfolding occurs by slow diffusion across multiple barriers in a rough energy landscape. Proc. Natl. Acad. Sci. U.S.A. 112, 8308–8313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mashaghi A., Kramer G., Lamb D. C., Mayer M. P., Tans S. J., Chaperone action at the single-molecule level. Chem. Rev. 114, 660–676 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Gupta A. N., et al., Pharmacological chaperone reshapes the energy landscape for folding and aggregation of the prion protein. Nat. Commun. 7, 12058 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicoll A. J., et al., Pharmacological chaperone for the structured domain of human prion protein. Proc. Natl. Acad. Sci. U.S.A. 107, 17610–17615 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mashaghi A., et al., Reshaping of the conformational search of a protein by the chaperone trigger factor. Nature 500, 98–101 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Doh-ura K., et al., Treatment of transmissible spongiform encephalopathy by intraventricular drug infusion in animal models. J. Virol. 78, 4999–5006 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leveugle B., et al., Heparin oligosaccharides that pass the blood-brain barrier inhibit β-amyloid precursor protein secretion and heparin binding to β-amyloid peptide. J. Neurochem. 70, 736–744 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Stewart L. A., Rydzewska L. H. M., Keogh G. F., Knight R. S. G., Systematic review of therapeutic interventions in human prion disease. Neurology 70, 1272–1281 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Terada T., et al., Less protease-resistant PrP in a patient with sporadic CJD treated with intraventricular pentosan polysulphate. Acta Neurol. Scand. 121, 127–130 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Newman P. K., et al., Postmortem findings in a case of variant Creutzfeldt-Jakob disease treated with intraventricular pentosan polysulfate. J. Neurol. Neurosurg. Psychiatry 85, 921–924 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taubner L. M., Bienkiewicz E. A., Copié V., Caughey B., Structure of the flexible amino-terminal domain of prion protein bound to a sulfated glycan. J. Mol. Biol. 395, 475–490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caughey B., et al., Prions and transmissible spongiform encephalopathy (TSE) chemotherapeutics: A common mechanism for anti-TSE compounds? Acc. Chem. Res. 39, 646–653 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Brimacombe D. B., et al., Characterization and polyanion-binding properties of purified recombinant prion protein. Biochem. J. 342, 605–613 (1999). [PMC free article] [PubMed] [Google Scholar]
- 32.James T. L., et al., Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc. Natl. Acad. Sci. U.S.A. 94, 10086–10091 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodside M. T., Block S. M., Reconstructing folding energy landscapes by single-molecule force spectroscopy. Annu. Rev. Biophys. 43, 19–39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dudko O. K., Hummer G., Szabo A., Intrinsic rates and activation free energies from single-molecule pulling experiments. Phys. Rev. Lett. 96, 108101 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Hinczewski M., Hyeon C., Thirumalai D., Directly measuring single-molecule heterogeneity using force spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 113, E3852–E3861 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hastie T., Tibshirani R., Friedman J., The Elements of Statistical Learning (Springer, ed. 2 2009). [Google Scholar]
- 37.Rousseeuw P. J., Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 20, 53–65 (1987). [Google Scholar]
- 38.Yamasaki T., Suzuki A., Hasebe R., Horiuchi M., Comparison of the anti-prion mechanism of four different anti-prion compounds, anti-PrP monoclonal antibody 44B1, pentosan polysulfate, chlorpromazine, and U18666A, in prion-infected mouse neuroblastoma cells. PLoS One 9, e106516 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smirnovas V., et al., Structural organization of brain-derived mammalian prions examined by hydrogen-deuterium exchange. Nat. Struct. Mol. Biol. 18, 504–506 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bechtluft P., et al., Direct observation of chaperone-induced changes in a protein folding pathway. Science 318, 1458–1461 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Koldewey P., Stull F., Horowitz S., Martin R., Bardwell J. C. A., Forces driving chaperone action. Cell 166, 369–379 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Massignan T., et al., A cationic tetrapyrrole inhibits toxic activities of the cellular prion protein. Sci. Rep. 6, 23180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghaemmaghami S., et al., Continuous quinacrine treatment results in the formation of drug-resistant prions. PLoS Pathog. 5, e1000673 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwarz G., Estimating the dimension of a model. Ann. Stat. 6, 461–464 (1978). [Google Scholar]
- 45.Akaike H., A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19, 716–723 (1974). [Google Scholar]
- 46.Neupane K., Yu H., Foster D. A. N., Wang F., Woodside M. T., Single-molecule force spectroscopy of the add adenine riboswitch relates folding to regulatory mechanism. Nucleic Acids Res. 39, 7677–7687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cecconi C., Shank E. A., Dahlquist F. W., Marqusee S., Bustamante C., Protein-DNA chimeras for single molecule mechanical folding studies with the optical tweezers. Eur. Biophys. J. 37, 729–738 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang M. D., Yin H., Landick R., Gelles J., Block S. M., Stretching DNA with optical tweezers. Biophys. J. 72, 1335–1346 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petrosyan R., Improved approximations for some polymer extension models. Rheol. Acta 56, 21–26 (2017). [Google Scholar]
- 50.Petrosyan R., Patra S., Rezajooei N., Garen C., Woodside M. T., Single-molecule force spectroscopy measurements of pentosan polysulfate interacting with hamster prion protein. Figshare. 10.6084/m9.figshare.13404719. Deposited 7 February 2021. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-molecule force spectroscopy data (50) have been deposited in Excel format in Figshare (DOI: 10.6084/m9.figshare.13404719).