Significance
Human cytomegalovirus (HCMV) reactivation is a major cause of posttransplant morbidity/mortality. One approach toward reducing this is purging the transplant donor and/or recipient of latently infected cells prior to stem-cell or organ harvest/engraftment. Our findings show the involvement of host bromodomain (BRD) proteins in regulation of HCMV latency and reactivation. Bromodomain and extra-terminal inhibitor (I-BET) treatment of latently infected cells causes reactivation of lytic gene expression by release of transcription activator P-TEFb (CDK9/CycT1) from BRD4-associated repressive complexes, with subsequent recruitment via host superelongation complex (SEC). This results in immune targeting and T cell-mediated killing of these otherwise latently infected cells and provides a therapeutic “shock and kill” strategy that could reduce HCMV-mediated disease in the transplant setting.
Keywords: cytomegalovirus, latency, epigenetics, bromodomain proteins, I-BET
Abstract
Reactivation of human cytomegalovirus (HCMV) from latency is a major health consideration for recipients of stem-cell and solid organ transplantations. With over 200,000 transplants taking place globally per annum, virus reactivation can occur in more than 50% of cases leading to loss of grafts as well as serious morbidity and even mortality. Here, we present the most extensive screening to date of epigenetic inhibitors on HCMV latently infected cells and find that histone deacetylase inhibitors (HDACis) and bromodomain inhibitors are broadly effective at inducing virus immediate early gene expression. However, while HDACis, such as myeloid-selective CHR-4487, lead to production of infectious virions, inhibitors of bromodomain (BRD) and extraterminal proteins (I-BETs), including GSK726, restrict full reactivation. Mechanistically, we show that BET proteins (BRDs) are pivotally connected to regulation of HCMV latency and reactivation. Through BRD4 interaction, the transcriptional activator complex P-TEFb (CDK9/CycT1) is sequestered by repressive complexes during HCMV latency. Consequently, I-BETs allow release of P-TEFb and subsequent recruitment to promoters via the superelongation complex (SEC), inducing transcription of HCMV lytic genes encoding immunogenic antigens from otherwise latently infected cells. Surprisingly, this occurs without inducing many viral immunoevasins and, importantly, while also restricting viral DNA replication and full HCMV reactivation. Therefore, this pattern of HCMV transcriptional dysregulation allows effective cytotoxic immune targeting and killing of latently infected cells, thus reducing the latent virus genome load. This approach could be safely used to pre-emptively purge the virus latent reservoir prior to transplantation, thereby reducing HCMV reactivation-related morbidity and mortality.
Human cytomegalovirus (HCMV) is a ubiquitous beta-herpesvirus that infects between 50 and 90% of the world’s population. Primary infection with HCMV usually results in lifelong carriage of the virus in a latent state, in the absence of infectious virus production, within cells of the myeloid lineage. Subsequent virus reactivation events are usually asymptomatic in a normal healthy individual. However, severe disease is seen following HCMV reactivation in immunocompromised individuals, such as AIDS and transplant patients (1). Reactivation in the transplant recipient or reinfection via reactivation from the donor graft can lead to severe morbidity, including organ rejection and, in some cases, mortality (2). The serostatus of both donor and recipient leads to a graded risk of reactivation and clinical outcome during immunosuppression after solid organ transplantation (SOT), which is highest with seropositive donor to seronegative recipient (D+/R‒) (3). The rate of HCMV reactivation also varies depending on the type of transplantation, with allogeneic SOT and hematopoietic stem-cell transplantation (HSCT) having HCMV-associated disease as high as 56 and 80%, respectively, in highly immune-suppressed seropositive patients (4). Given that kidney, lung, and HSCT transplants alone now globally exceed 200,000 annually (www.transplant-observatory.org), a very high number of patients will experience HCMV reactivation events and potential HCMV-mediated disease unless effective intervention strategies can be devised.
While advancements have been made in designing a vaccine against HCMV, no candidate is currently effective (5). Additionally, while both prophylactic and pre-emptive antiviral therapies can be efficacious for many, for a significant number of patients these can be both ineffective and detrimental; despite improvements with the recently licensed letermovir, drugs such as (val)gancylcovir are poorly bioavailable and can involve high toxicity, resulting in neutropenia (6). Moreover, a major issue is the development of drug resistance, examples of which are already on the rise even with letermovir (7), while therapies such as adoptive T cell transfer can aggravate graft versus host disease with HCMV reactivation (3).
These issues have more recently led to investigations into the potential of directly targeting the latent reservoir; reducing the number of latently infected cells of both seropositive donors and recipients prior to transplantation could reduce the chances of HCMV reactivation posttransplant and provide a better clinical outcome (8). Studies have shown that targeting HCMV latently infected cells, namely myeloid lineage CD14+ monocytes and their CD34+ progenitors (9), can be employed; findings include HCMV-driven down-regulation of multidrug resistance protein-1, which allows specific killing of latent cells with the chemotherapy drug vincristine (10). Work from our group has also shown that targeting HCMV-specific proteins expressed on the surface of latently infected cells, such as US28, using fusion toxin proteins also allows killing of both experimentally and naturally latent HCMV-infected cells in vitro (11).
An alternative approach is to use immune-mediated killing of latently infected cells by enabling recognition through host T cells. While cells undergoing lytic infection can be recognized by naturally present host cytotoxic T cells (CTLs), HCMV latently infected cells do not efficiently present these lytic antigens (8). As such, drugs that induce latently infected cells to express viral lytic proteins should lead to recognition and elimination of latent cells by circulating HCMV-specific CTLs. Commonly, this has become known as “shock and kill” therapy and has led to various latency reversal agents (LRAs), including histone deacetylase inhibitors (HDACis), being trialled with HIV (12). Latency of HCMV in the myeloid lineage is maintained through chromatin-mediated epigenetic repression of the major immediate early promoter (MIEP) (13), with activation of the MIEP usually driven by differentiation of myeloid cells to macrophage or dendritic cells (DCs) (14). We have previously shown that, using HDACis such as valproic acid (VPA) and MC1568, transient HCMV lytic gene expression can be induced, which allows CTL recognition and killing of both experimentally and naturally latent infected cells (15). However, these particular HDACis only caused maximal induction of immediate early gene expression in around 25% of latently infected cells, and HDACis have been shown elsewhere to impair the activity of T cells when targeting HIV reactivation (16).
Newer-generation HDACis with higher potency and lower side effects are now available, as well as inhibitors to a variety of epigenetic modifying enzymes possibly having a role in regulating expression from the HCMV MIEP (17, 18). While targeting epigenetic enzymes in vitro, such as histone demethylases (HDMs), has resulted largely in decreases in HCMV gene expression and virus yield (19, 20), as well as restriction of replication and reactivation of other herpesviruses (21), inhibiting the histone methyltransferase (HMT) component of polycomb-repressive complex-2 (PRC2), EZH2, across two different studies has both activated lytic gene expression (22) and, conversely, suppressed infection by inducing an antiviral cellular state (23). With this in mind, we hypothesized that investigating a range of epigenetic inhibitors for their ability to target possible host regulators of HCMV lytic gene expression might cause increased numbers of latently infected cells to become visible to the host immune system without causing full virus reactivation and avoiding induction of HCMV immune evasion genes, which could protect latently infected cells from CTL recognition.
Here, we show that continuous treatment of latently infected cells with certain HDACis causes not just activation of HCMV gene expression but full reactivation and lytic cascade of the virus. In contrast, through the release of the cellular transcriptional activating complex positive-transcription elongation factor-b (P-TEFb) from repressive BRD4-associated aggregates, and recruitment via the superelongation complex (SEC) to viral promoters, inhibitors of the BET family of bromodomain (BRD)-containing histone acetylation mark reader proteins (I-BETs) (24, 25) induce dysregulated HCMV gene expression. This allows production of virus lytic immunogens, including immunodominant IE72, pp65, and gB, while restricting the expression of many viral immunomodulating proteins (e.g., MHC class I/II downmodulators US2, 3, 6, 8, and 11) and, most importantly, virus DNA replication machinery (e.g., UL44, UL112). Subsequently, these inhibitors allow efficient killing of both experimentally and naturally infected HCMV latent cells by host CTLs in the absence of virus productive infection and, ultimately, reduce the presence of HCMV viral DNA within ex-vivo–treated peripheral blood cells of HCMV seropositive individuals.
Results
BET Inhibitors Induce High MIEP Activity in Cell-Line Models of HCMV Latency.
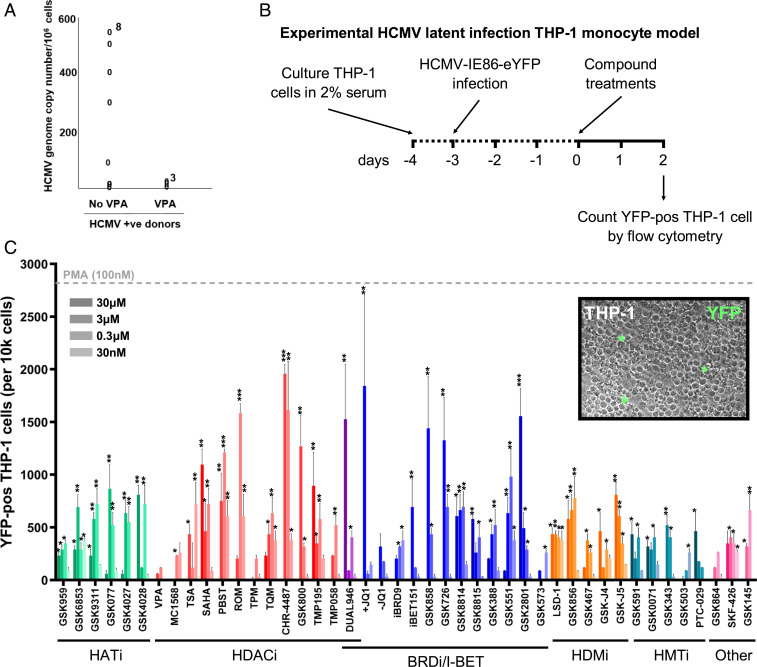
The HDACi VPA is used for the clinical treatment of epilepsy and, in our previous study (15), we showed that treatment of HCMV latently infected cells with VPA induced immediate early (IE) gene expression and made these cells a target for HCMV-specific CTLs. Analysis of HCMV genome carriage in peripheral blood samples by digital droplet PCR (ddPCR) shows that, in comparison to HCMV seropositive patients not receiving HDACi, individuals being treated with VPA had a substantial reduction in their HCMV latent load (although not statistically significant due to available patient numbers) (Fig. 1A). However, since newer generation HDACis have been shown to have higher potency with fewer off-target effects, and it is possible that other epigenetic inhibitors might also act as LRAs, we decided to investigate inhibitors of further targets that might have a superior ability to induce viral gene expression in HCMV latent cells, while not having a significant effect on T cell function or inducing full HCMV reactivation.
Fig. 1.
HDAC and BET inhibitors induce high numbers of IE86-expressing cells in the THP-1 cell line model of HCMV latency. (A) Peripheral blood CD14+ monocytes from three HCMV seropositive epilepsy patients being treated with VPA and from eight controls were analyzed for HCMV copy number by ddPCR. (B and C) Monocytic THP-1 cells infected with HCMV TB40e IE86-eYFP for 3 d were treated with inhibitors of histone acetyltransferases (HATi, green), histone deacetylases (HDACi, red), bromodomain proteins (BRDi/I-BET, blue), histone demethylases (HDMi, orange), histone methyltransferases (HMT inhibitor, teal), as well as other epigenetic modifiers (Other, pink) across a range of concentrations (30 μM to 30 nM) for 48 h before the number of YFP-positive cells was analyzed by flow cytometry in comparison to DMSO (negative control, set to 0) and PMA (100 nM, gray dashed line) (mean + SEM, n = 3; *P < 0.05, **P < 0.01, ***P < 0.001). (Inset) An example image of cells counted (20× magnification).
Using a broad panel of experimental epigenetic inhibitors produced by GlaxoSmithKline (GSK), as well as other commercially available compounds (17, 26) (SI Appendix, Table S1), we first determined their ability to induce HCMV gene expression from the MIEP of a HCMV-TB40e-IE86-eYFP virus latently infecting THP-1 monocytes. After treatment for 48 h and subsequent analysis of YFP expression by flow cytometry (Fig. 1B), all compounds (across a broad range of 30 μM to 30 nM) produced YFP-positive cell numbers above that of dimethylsulfoxide (DMSO) control (Fig. 1C), largely in the absence of any toxicity-directed effects (SI Appendix, Fig. S1). However, induction of IE86 expression with some groups of inhibitors was seen at the same level, if not greater, with negative control molecules (e.g., GSK4028 for HATi and GSK-J5 for HDM inhibitors), suggesting likely nonspecific effects of these inhibitor groups here. Notwithstanding this finding, the two groups of inhibitors with the greatest effects were HDACis and compounds targeting BRD-containing proteins (BRDis). Assessment of MIEP activation in isolation from the virus by these inhibitors in a GFP-expressing THP-1 cell reporter line (27) confirmed that newer generation HDACis (e.g., SAHA, PBST, ROM) and compounds specifically targeting the BET family (BRD2/3/4/T) bromodomains (I-BETs) caused the greatest increase of MIEP activity in comparison with differentiation of monocytes with a phorbol ester (PMA), which was used as positive control (SI Appendix, Fig. S2). In addition, pretreatment of human fetal foreskin fibroblasts (HFFFs) lytically infected for 24 h with HCMV-TB40e-IE86-eYFP with HDACis and I-BETs also broadly caused increased numbers of YFP-positive cells (SI Appendix, Fig. S3A) and YFP expression (SI Appendix, Fig. S3B) in comparison with DMSO controls, although some cytotoxicity was apparent (SI Appendix, Fig. S3C). These data confirmed that newer-generation HDACis had a more potent effect on inducing activity from the MIEP than previously employed HDACis, including VPA, but also that I-BETs caused induction of HCMV major IE gene expression.
BET Inhibitors Induce Virus Gene Expression while Restricting Full Reactivation from Ex Vivo HCMV Latency Models.
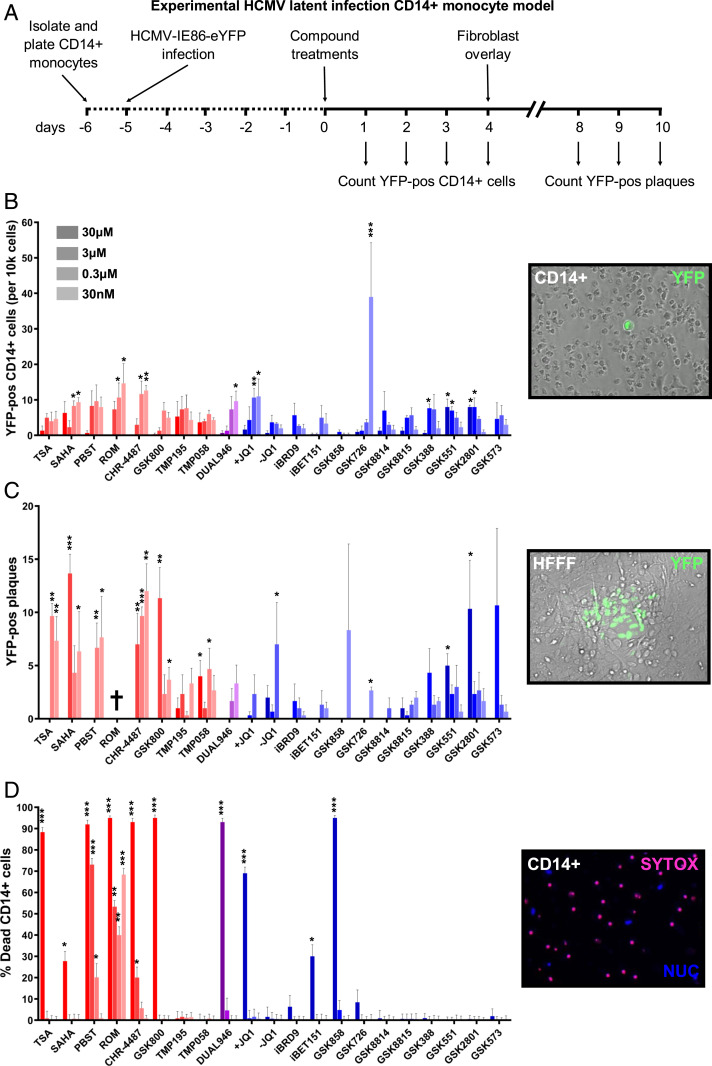
Since HDACis and I-BETs consistently up-regulated HCMV IE gene expression across a range of in vitro models, we next tested their ability to induce HCMV gene expression in our ex vivo model of experimental HCMV latency (Fig. 2). CD14+ monocytes isolated from either peripheral blood of healthy donors or blood donor apheresis cones were infected with HCMV-TB40e-IE86-eYFP (Fig. 2A), with latency established after 5 d as determined by suppression of IE gene expression, maintained expression of latency-associated UL138 transcription and viral genome copy number (SI Appendix, Fig. S4 A–C). Monocytes were then incubated with the range of HDACis and BRDis continuously for 4 d, and the level of HCMV IE induction was assessed by counting YFP-positive CD14+ monocytes (Fig. 2B). Treatment of latently infected primary monocytes with the majority of HDACis and BRDis resulted in YFP-positive cells becoming visible, with highest induction seen with I-BET GSK726 (28). We observed consistent I-BET results with established CD34+ cell-line models of HCMV latency (SI Appendix, Fig. S5). Treated CD14+ monocytes were then overlaid with HFFFs in order to determine if full reactivation of HCMV had caused release of lytic virus. The results clearly show that the majority of HDACis tested led to the production of virus plaques, in contrast to most I-BETs that did not stimulate production of infectious particles, i.e., full reactivation (Fig. 2C). Additionally, many HDACis caused a high level of CD14+ monocyte death, in contrast to I-BETs (Fig. 2D). We compared the least- cytotoxic HDACi that still stimulated virus gene expression (CHR-4487, a myeloid-selective HDACi) with the most potent I-BET (GSK726) over an extended range of concentrations and found that, even with dilution of HDACi and absence of cytotoxicity, HCMV was still fully reactivated (SI Appendix, Fig. S6). In contrast, I-BET GSK726 continued to induce IE gene expression significantly above background down to 30 pM, while causing only minimal plaque formation after HFFF overlay. Interestingly, this finding was phenocopied at higher compound concentrations using a bromodomain-2 (BD2)–selective I-BET, RVX-208 (apabetalone) (29), but not the BD1-selective I-BET, ZL0580 (30) (SI Appendix, Fig. S7).
Fig. 2.
BET inhibitors induce virus gene expression while restricting full reactivation from ex vivo HCMV latency models. (A–C) CD14+ monocytes isolated from apheresis cones were infected with HCMV TB40e IE86-eYFP for 5 d before treatment with HDACi (red bars), BRDi/I-BET (blue bars), or a dual inhibitor (purple bars) across a range of concentrations (30 μM to 30 nM) for 72 h (A). The number of YFP-positive cells per well was then counted (B) before monocytes were overlaid with HFFFs and the resulting plaques were counted after 7 d (C) (mean + SEM, n = 3). (D) CD14+ monocytes isolated from apheresis cones were treated with inhibitors for 72 h before being stained with SYTOX (dead cells, red stain) and Hoechst stain (nucleus, blue stain) and counted (mean + SEM, n = 3) († = total cell death). (Insets) Example images of each condition counted (20× magnification). (*P < 0.05, **P < 0.01, ***P < 0.001.)
BET Inhibitors Dysregulate HCMV Transcription from Latency and Restrict Viral DNA Replication.
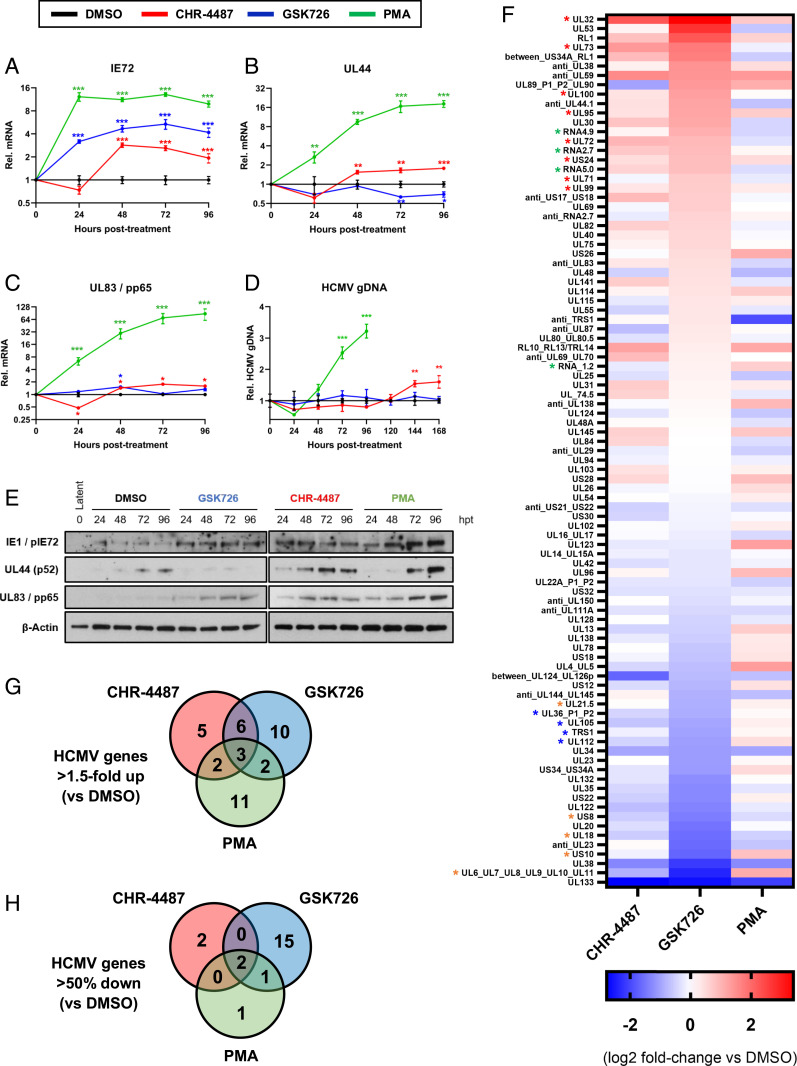
In order to understand why HDACis induce full lytic virus replication but not I-BETs, HCMV gene expression profiles induced by both treatments were analyzed. Compound-treated CD14+ monocytes latently infected with HCMV-TB40e-WT were harvested over a 4-d time course to examine messenger RNA (mRNA) expression of three viral genes by RT-qPCR (Fig. 3 A–C). Results show that I-BET treatment (GSK726) induced both HCMV IE (IE72) and late pp65 (UL83) gene expression in the absence of UL44 production, the HCMV DNA polymerase processivity subunit, in comparison with DMSO-treated cells and in line with known effects on cellular targets (31) (SI Appendix, Fig. S8). In contrast, HDACi CHR-4487 induced all three genes. Immunoblot analysis for IE, UL44, and pp65 (UL83) proteins confirmed mRNA results (Fig. 3E). HCMV genomic DNA was also measured at each time point and, while CHR-4487 treatment led to viral genome replication, GSK726 did not (Fig. 3D). Notably, GSK726 was also able to restrict HCMV genome replication and infectious virion production in lytically infected HFFFs (SI Appendix, Fig. S9). Hence, we tested whether cotreatment with both compounds could have cooperative effects in inducing HCMV IE gene expression while inhibiting virus DNA replication. Indeed, cotreatment of latently infected CD14+ monocytes showed an additive effect in the induction of IE gene expression at 3 nM (SI Appendix, Fig. S10A). However, restriction of full HCMV reactivation did not occur, with equivalent numbers of plaques seen after HFFF overlay (SI Appendix, Fig. S10B).
Fig. 3.
BET inhibitors dysregulate HCMV transcription from latency and restrict viral DNA replication. (A–E) CD14+ monocytes isolated from apheresis cones infected with HCMV TB40e wild type for 5 d were treated with DMSO (black lines), HDACi (CHR-4487 30 nM, red lines), I-BET (GSK726 30 nM, blue lines) and PMA (20 nM, green lines). RNA, DNA, and protein were isolated from cells every 24 h for 4 d. RT-qPCR was then performed for virus (A) IE72, (B) early UL44, and (C) late UL83/pp65 transcripts relative to host GAPDH with (D) HCMV genome copy number determined by qPCR of UL44 promoter relative to host GAPDH promoter and (E) immunoblotting carried out for virus targets relative to β-actin (images representative of n = 3). *P < 0.05, **P < 0.01, ***P < 0.001; mean ± SEM, n = 3. (F–H) RNA-seq of flow cytometry isolated YFP-positive HCMV-TB40e-IE86-eYFP–infected CD14+ monocytes after 72 h treatment with compounds (F). Data are presented as a heat map of means of HCMV transcripts identified from biological duplicates, with data ranked by I-BET (GSK726) fold-change relative to DMSO control. HCMV gene groups indicated by associated asterisk color (red, structural protein; green, long noncoding RNA; orange, immunomodulatory protein; blue, DNA replication protein). Venn diagrams show HCMV transcript number from RNA-seq data either (G) up-regulated (>1.5-fold) or (H) down-regulated (>50%) relative to DMSO control.
To better understand the effects of these compounds on the full transcriptional profile of latent HCMV, we next performed RNA-sequencing (RNA-seq) analysis. After 72 h of drug or control treatment of CD14+ monocytes, which had been latently infected with HCMV-TB40e-IE86-eYFP so that IE gene induction could be identified, we isolated YFP-positive cells via fluorescence-activated cell sorting (FACS) from CHR-4487, GSK726, and control-treated (DMSO and PMA) samples. RNA-seq of these bulk samples was then carried out essentially as per the massively parallel single-cell RNA-seq (MARS-seq) protocol used previously on latent cells (32) (Fig. 3F). Using a stringent threshold of a minimum mean of 10 HCMV reads per million total reads (from at least one of the conditions), of the 93 HCMV transcripts found across all conditions (SI Appendix, Table S2), over 40% were up-regulated greater than 1.5-fold in comparison with the DMSO control (Fig. 3G), including many surface glycoproteins (e.g., gN, gM) and tegument proteins (e.g., pp150, pp28) that are immunogenic T cell targets (33). Interestingly, I-BET GSK726 caused the greatest number of HCMV transcripts to decrease by greater than 50% in comparison with DMSO (Fig. 3H), including four viral DNA replication machinery proteins (TRS1, UL36, UL105, UL112), providing a rationale for why the viral genome is not replicated and infectious virus is not induced by this I-BET. I-BET–mediated regulation of specific viral gene expression in latently infected cells, be it up-regulated or down-regulated, appeared to show no correlation to the canonical expression class of the viral gene (e.g., IE, early or late) (SI Appendix, Table S2) and could not be attributed to specific regions of viral genes/open reading frames in the genome (SI Appendix, Fig. S11).
BRD4 Inhibition Modulates P-TEFb Association with HCMV Promoters.
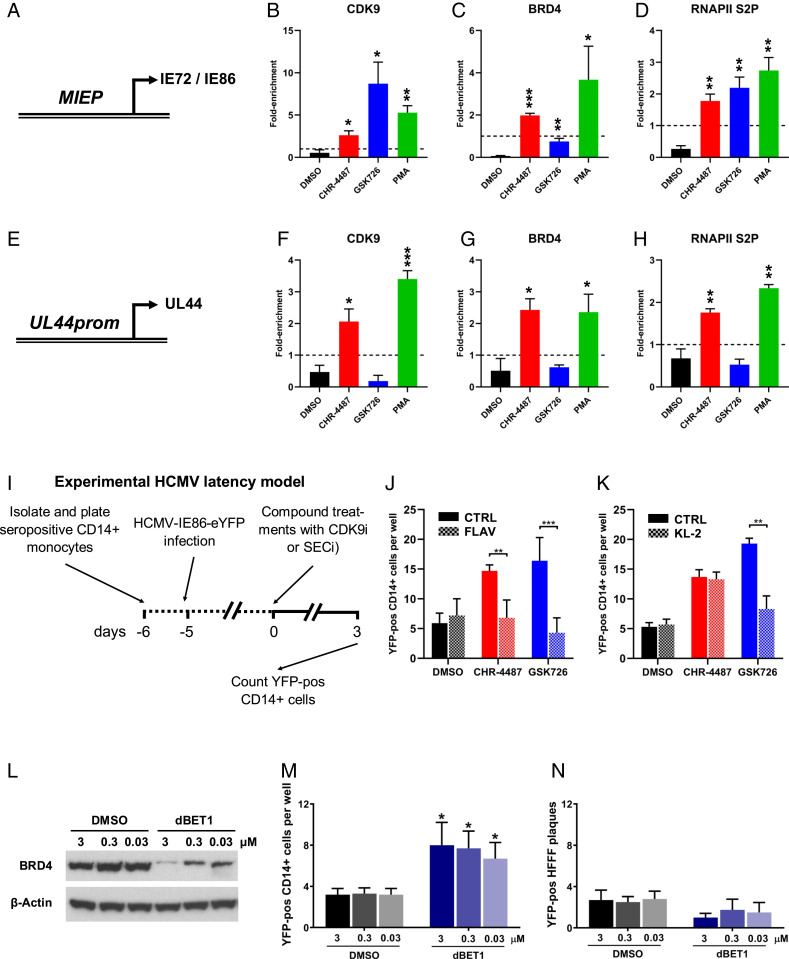
I-BET treatment resulted in an HCMV transcription profile from latently infected cells quite different from the usual temporal cascade of viral gene expression associated with virus reactivation via PMA-driven differentiation, with a correlation coefficient only lower between GSK726 and DMSO (SI Appendix, Fig. S12). I-BETs are known to target BRD4 in a manner that causes release of cyclin-dependent kinase-9 (CDK9), a component of the P-TEFb complex and activator of RNA polymerase-II (RNAPII), from cytosolic repressive 7SK-snRNP complexes into the nucleus and induce reactivation of other herpesviruses and HIV (34, 35). Additionally, CDK9 inhibition has been shown to interfere with the initiation of HCMV IE gene expression during lytic infection (36). Therefore, we aimed to use chromatin immunoprecipitation (ChIP) analysis to investigate the association of known transcriptional activator CDK9 with HCMV genomes during drug treatment of latently infected cells. Due to technical constraints using primary CD14+ monocytes, including the number of cells required for ChIP analysis, these experiments were carried out in latently infected THP-1 monocytic cells, which have routinely been used as a model of HCMV latency, at 48 h posttreatment of compound (Fig. 4). CDK9 was found enriched at the HCMV MIEP (Fig. 4A) with all IE activating treatments, although highest with GSK726 (Fig. 4B), possibly due in part to a cytosolic-to-nuclear shift in protein levels as confirmed by immunoblot and immunofluorescence analysis (SI Appendix, Fig. S13). Interestingly, this was in contrast to BRD4 association with the MIEP where GSK726 treatment resulted in a below-background level (Fig. 4C), consistent with inhibition of BRD4 binding to acetylated histones with I-BET treatment, which was also confirmed at control host promoters (SI Appendix, Fig. S14). However, the association of elongating phosphorylated RNAPII serine-2 (RNAPII S2P) at the MIEP reflected transcriptional activity with all treatments (Fig. 4D). Additionally, histone acetylation at all promoters analyzed paralleled transcriptional activity, while CHR-4487 treatment caused pronounced hyper-acetylation (SI Appendix, Fig. S15). In contrast, association of all immunoprecipitated targets at the UL44-promoter with GSK726 treatment was below that of background (Fig. 4 E–H), consistent with the absence of UL44 transcript (Fig. 3B). In order to confirm that, not only the presence, but also the activity of CDK9 were essential for activation of HCMV IE gene expression from latently infected CD14+ monocytes, we applied the CDK9 inhibitor (CDK9i) Flavopiridol synchronously with CHR-4487 or GSK726 (Fig. 4I). The results show a decrease in the number of YFP-positive cells with CDK9i to the level of DMSO with both CHR-4487 and GSK726 at 30 nM (Fig. 4J). Conversely, cotreatment of a SEC inhibitor (KL-2) only decreased the number of YFP-positive cells induced by GSK726, whereas CHR-4487 MIEP activation was unaffected, confirming the association of SEC:P-TEFb at the MIEP after I-BET treatment (Fig. 4K). Use of a BRD4-degrader (dBET1) resulted in BRD4 protein loss, as expected, but also in induction of IE gene expression in latently infected CD14+ monocytes (Fig. 4 L and M). This BRD4 ablation also shows some evidence of a decrease in virus replication below that of background, although this is not statistically significant due to the already low negative control level of virus plaques (Fig. 4N).
Fig. 4.
BRD4 inhibition modulates SEC:P-TEFb association with HCMV promoters. (A–H) Monocytic THP-1 cells infected with HCMV TB40e IE86-eYFP for 3 d were treated with DMSO (black bars), HDACi (CHR-4487 30 nM, red bars), I-BET (GSK726 30 nM, blue bars) or PMA (100 nM, green bars). After 48 h, ChIP qPCR analysis was employed to determine the enrichment of (A–D) MIEP and (E–H) UL44 promoter DNA using antibodies specific for (B and F) CDK9, (C and G) BRD4, and (D and H) RNAPII S2P relative to isotype controls (dashed line, set to 1) (mean + SEM, n = 3). (I) Treatment regime of cells in J–K. CD14+ monocytes isolated from apheresis cones infected with HCMV TB40e IE86-eYFP for 5 d were treated for 72 h with DMSO (black bars), HDACi (CHR-4487, red bars), or I-BET (GSK726, blue bars) (all 30 nM) with concurrent treatment of half of wells with either (J) CDK9 inhibitor (CDK9i), Flavopiridol (FLAV 40 nM), or (K) SECi KL-2 (10 μM) before the number of YFP-positive cells per well were then counted. (L–N) CD14+ monocytes isolated from apheresis cones infected with HCMV TB40e IE86-eYFP for 5 d were treated with DMSO (black bars) or BRD4-degrader dBET1 (blue bars) (3 to 0.03 μM) with immunoblot data confirming BRD4 degradation at 24 h (L) (images representative of n = 3). After 72 h, the number of YFP-positive cells per well then counted (M) before monocytes were overlaid with HFFFs and resulting plaques counted after 7 d (N) (mean + SEM, n = 5; *P < 0.05, **P < 0.01, ***P < 0.001).
BET Inhibitors Allow Efficient Killing of Latent HCMV-Infected Cells and Reduce Peripheral Blood Virus Carriage.
Although I-BET treatment caused HCMV IE gene induction and increases in transcription of genes that would ordinarily be limited by the virus due to their immunogenicity for T cell recognition (Fig. 3F), it remained to be tested whether treatment of latently infected cells with I-BET would allow recognition and killing of these cells by CTLs. Interestingly, GSK726 treatment caused decreases in transcript levels of a number of HCMV immunoevasion genes (Fig. 3F: UL21.5, US8, UL18, US10, UL6-11) (SI Appendix, Fig. S16: US2, US3, US6, US11) that would normally interfere with antigen processing, presentation, and CTL recognition. However, use of inhibitors such as HDACis and, to some extent, I-BETs has previously been shown to inhibit the cytotoxic effector function of T cells (16, 37). Hence, we determined if these inhibitors had any effect on T cell functionality (interferon-γ [IFN-γ] secretion) using total peripheral blood mononuclear cells (PBMCs) in quantitative anti–IFN-γ Fluorospot assays. PBMCs from one HCMV seronegative and three seropositive donors were tested with a pool of overlapping HCMV peptides covering the IE72/IE86 and UL83/pp65 proteins, as well as a positive-control polyclonal CD3/CD28 cross-linking for T cell receptor and costimulator activation (SI Appendix, Fig. S17). The results showed that the presence of CHR-4487 inhibited IFN-γ production at the higher concentration of 30 nM, whereas GSK726 had little to no effect. Additionally, a phenotypic analysis of cell-surface markers of inhibitor-treated CD14+ monocytes showed little change in surface expression of MHC class I and II molecules (HLA-A, -B, -C and HLA-DR, respectively) with GSK726, whereas CHR-4487 caused decreased surface expression, especially at the higher compound concentration (SI Appendix, Fig. S18). Cell-surface marker phenotyping also confirmed that induction of HCMV IE gene expression in treated monocytes was not simply being driven by differentiation through treatment with GSK726, whereas CHR-4487 did affect surface expression of differentiation markers of monocytes, macrophage, and DCs.
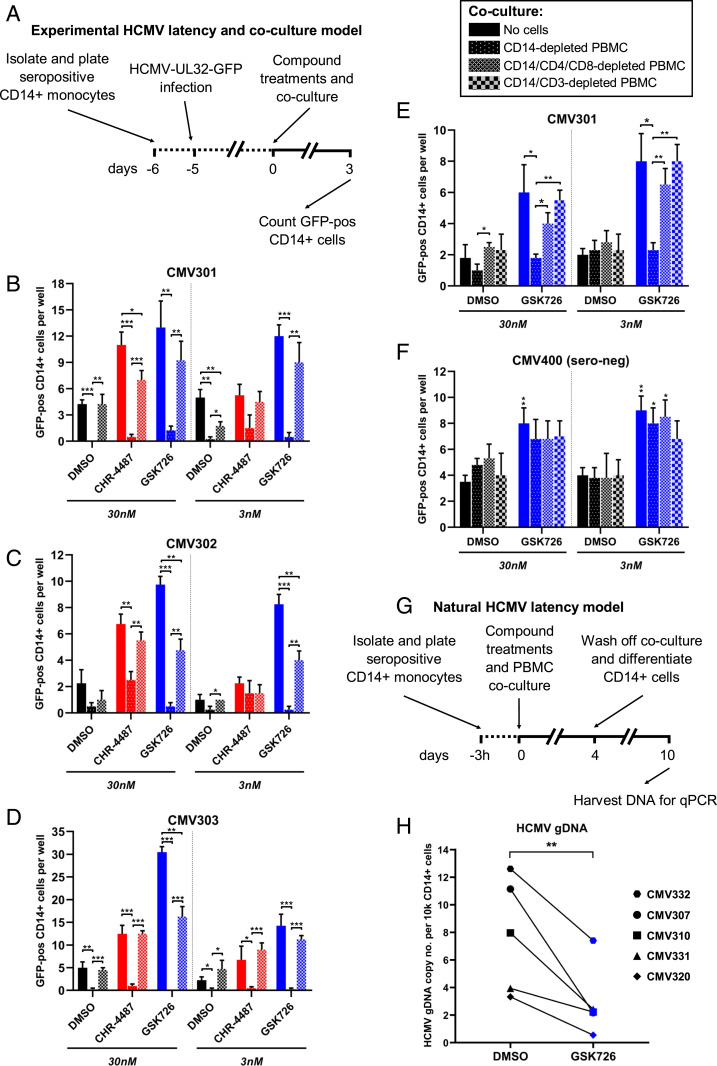
As I-BET treatment of CD14+ monocytes and T lymphocytes did not show major changes in either phenotypic or T cell effector function, we assessed whether I-BET would allow T cell recognition and killing of HCMV latently infected cells in our ex vivo models (Fig. 5). CD14+ monocytes were isolated from peripheral blood of three HCMV seropositive donors, while the remaining PBMCs were retained for later use. After 24 h, monocytes were infected with HCMV-TB40e-UL32-GFP, a virus expressing a pp150-GFP fusion protein that is highly expressed after both CHR-4487 and GSK726 treatment (Fig. 3F). After another 5 d, latently infected CD14+ monocytes were then cocultured with “no cells,” “CD14-depleted PBMCs” (containing CD4+ and CD8+ T cells), or “CD14/CD4/CD8-depleted PBMCs” from the original donor, and the number of resulting GFP-positive cells were counted over 3 d (Fig. 5A). Across the three donors, while GSK726 caused increased levels of GFP-positive cells above DMSO at both concentrations (3 and 30 nM), CHR-4487 significantly raised only GFP-positive cell numbers at the higher concentration (Fig. 5 B–D). However, in almost all cases, including DMSO treatment, coculture with CD14-depleted PBMCs led to eradication of GFP-positive cells, apart from CHR-4487 treatment of donor CMV302 where GFP-positive cell numbers decreased down to background levels (Fig. 5C). As expected, depleting CD4+ and CD8+ T cells from the coculture prevented the removal of GFP-positive cells, which were similar in number to the control wells (Fig. 5 B–D), whereas depletion of all CD3-expressing cells from PBMCs restored the number of GFP-positive cells back to control level (Fig. 5E); the difference in GFP-positive cell number is likely due to residual CD4‒CD8‒CD3+ γ/δ T cells in the CD14/CD4/CD8-depleted population (SI Appendix, Fig. S19). A HCMV seronegative donor was also tested, demonstrating that removal of activated latently infected cells was specific (Fig. 5F), as HCMV seronegative donors do not have HCMC-specific CTLs.
Fig. 5.
BET inhibitors allow efficient killing of latent HCMV-infected cells and reduce peripheral blood virus carriage. (A–D) CD14+ monocytes isolated from seropositive peripheral blood were infected with HCMV TB40e UL32-GFP for 5 d before cocultures were established (no cells, full bars; CD14-depleted PBMCs, dotted bars; CD14/CD4/CD8-depleted PBMCs, hatched bars) and treated with either DMSO (black bars), HDACi (CHR-4487, red bars), or I-BET (GSK726, blue bars) (30 and 3 nM) for 72 h (A). The number of GFP-positive cells per well was then counted for seropositive donor (B) CMV301, (C) CMV302, and (D) CMV303 (mean + SEM, n = 3). (E and F) Experiments were repeated with additional coculture of CD14/CD3-depleted PBMCs (checkered bars), and GFP-positive cells were counted for (E) seropositive donor CMV301 and (F) seronegative donor CMV400 (mean + SEM, n = 4; *P < 0.05, **P < 0.01, ***P < 0.001). (G) Treatment regime of cells in H. CD14+ monocytes isolated from peripheral blood of five seropositive donors were plated for 3 h before cocultures were established using total remaining PBMCs and either DMSO (black symbol) or I-BET (GSK726, 30 nM; blue symbol). After 4 d, coculture was withdrawn and CD14+ monocytes were differentiated (GM-CSF/IL-1β 5-d, LPS 5-d) before cells were harvested, DNA isolated and (H) HCMV genome copy number determined by qPCR of glycoprotein B DNA relative to host GAPDH promoter (mean, n = 3). **P value = 0.0092 (Student’s t test).
Finally, to determine if GSK726 would allow targeting of naturally latent HCMV-infected cells, PBMCs were derived from five HCMV seropositive donors, CD14+ monocytes were isolated and adhered to plastic and, after 3 h, the total remaining PBMCs were added back to the cultures and treated with either DMSO or GSK726 (Fig. 5G). After 4 d of treatment and coculture, the nonadherent CD14‒ PBMCs were removed and the remaining CD14+ monocytes were differentiated to DCs in order to reactivate remaining naturally latent HCMV. After another 10 d, DNA was harvested from these cultures, and HCMV genomic DNA (gDNA) copy number analysis was carried out by qPCR (Fig. 5H). Comparison of treatment with GSK726 to DMSO clearly showed a statistically significant (**P = 0.0092) depletion of HCMV gDNA levels ranging from 41 to 83% (mean: 64%). Plaque assays show that the ability of remaining latent HCMV to reactivate with cellular differentiation, even after prior GSK726 treatment, was not impeded (SI Appendix, Fig. S20). Overall, the results demonstrate that treatment of CD14+ monocytes containing naturally HCMV latently infected cells with GSK726 allows these cells to be targeted by host T cells and killed.
Discussion
In the context of immunosuppression following HSCT and SOT, HCMV reactivation continues to be a significant contributing factor to both morbidity and mortality within this patient group. In the present study, we assessed a wide-ranging panel of epigenetic inhibitors for their ability to induce lytic gene expression from HCMV latently infected cells and, thus, allow for targeting by endogenous HCMV-specific T cells. Treatment of HCMV latently infected CD14+ monocytes with HDACis in our in vitro model showed their ability to induce IE gene expression such that cells could then be targeted by T cells. However, the continuous presence of the inhibitor (mimicking a clinical treatment regime), rather than a single pulse as previously used (15), resulted in full reactivation of HCMV with production of infectious particles. Transcription from the HCMV genome, which would rapidly be chromatinized by host nucleosome deposition machinery upon entry to the nucleus (38), is, at least in part, controlled by dynamic association of histone posttranslational modifications during latency and reactivation to lytic infection (39–43) and companion epigenetic modifying enzymes (44). Histone acetylation is known to be present in all distinct classes of temporally controlled viral promoters, and HDACis have also been shown by our group and others to increase lytic infection of HCMV (42, 45, 46). Hence, it is possible that overarching inhibition of HDAC activity could therefore lead to full HCMV reactivation from latency as seen here. Although VPA treatment of epileptic patients showed a trend toward reductions in HCMV genome latent loads, patients would be expected to have fully functional immune systems. However, this is unlikely to be the case for transplant recipients, especially HSCT patients. Therefore, a treatment of this nature may itself be harmful to the patient.
Intriguingly, we found that I-BET compounds (i.e., JQ1, iBET151, GSK726), which more specifically target the BRD2/3/4 proteins, while inducing IE gene expression did not result in production of infectious virus. This was in contrast to BRDis targeting the other host proteins ATAD2 and BAZ2A/B (i.e., GSK8814, GSK388, GSK2801). In fact I-BET GSK726, a tetrahydroquinoline compound (GSK1324726A) first designed for the oncology and inflammatory setting at 30 nM concentration (BET protein dissociation selectivity being found previously at Kd 20 to 25 nM) (28) far surpassed any other compound and experimental positive controls in inducing the highest level of IE transcript level and IE-positive cells while restricting virus DNA replication. Interestingly, induction of IE transcripts was phenocopied with the BD2-selective I-BET RVX-208, but not with the BD1-selective I-BET ZL0580, suggesting that IE induction by GSK726 may be linked to a high sensitivity of BRD4 BD2 to GSK726 inhibition in this setting. This is also consistent with BRD4 BD2 having a higher recognition specificity for an acetylated lysine motif in CycT1 (47) and hence a greater ability of GSK726 to release the transcription-activating P-TEFb complex from BRD4 interaction either in the cytoplasm or in the nucleus itself for potential recruitment to the HCMV MIEP. In parallel, the failure of GSK726 to induce IE86 expression at higher concentrations than 30 nM may be due to complete pan-BET (BRD2, 3, 4; BD1/BD2) inhibition, associated with a global cellular transcriptional modulation, as described elsewhere (48), which could lead to a block in HCMV MIEP activation.
RNA-seq analysis determined that GSK726 was able to dysregulate HCMV transcription such that the usual temporal cascade of HCMV gene expression, consistent with full reactivation and seen with HDACi treatment (CHR-4487) and differentiation (PMA), did not occur. GSK726 caused a substantial down-regulation of viral DNA replication machinery genes, including UL44 (DNA-binding protein and polymerase processivity factor) (49), UL105 (helicase-primase subunit) (50), and UL112 (DNA replication complex recruitment protein) (51) in comparison to control treated cells. This alone would be likely to cause restriction of viral DNA replication and is consistent with our qPCR results analyzing HCMV gDNA copy number during treatment of both latent and lytic cell models. Another study found a decrease in GFP-tagged pp28 tegument protein level during HCMV lytic infection following treatment with another I-BET, OTX015 (52). In this work, the Moorman laboratory surmised that possible interference of BRD2 activity during HCMV DNA replication could be playing a role in restricting late gene expression. This view is comparable with observations using Epstein-Barr virus (EBV), which showed that treatment with the I-BET JQ1 during lytic EBV infection limited the virus life cycle in two ways: preventing expression of the EBV IE protein BZLF1 but also inhibiting interaction of BRD4 with the origin of lytic replication (oriLyt) (53), consistent with our use of the BRD4-degrader dBET1. Thus, despite possible different mechanisms, the restriction of HCMV reactivation by I-BETs might allow safer treatment of already immunocompromised patients (24). Cotreatment with GSK726 and CHR-4487 still induced reactivation and virus replication, likely due to HDACis aiding virus DNA replication. Therefore, cotreatment regimens, which have previously been used on HIV models to good effect (54), might not be advisable in the context of HCMV treatment.
ChIP-qPCR analysis confirmed the association of RNAPII-activating CDK9 at the MIEP with all IE-activating treatments, although the greatest enrichment was mediated by GSK726 treatment, in line with CDK9 relocation to or within the nucleus. This was despite an absence of BRD4 association at the MIEP (consistent with I-BET activity), compatible with P-TEFb recruitment via the SEC (55) as shown here with use of SEC inhibitor (SECi) KL-2 and work elsewhere on HSV-1 (34, 56). Significantly, others have previously shown the redundancy of BRD4 to recruit CDK9 to the MIEP of HCMV but also the necessity for active transcription to be occurring (57). This may explain, at least in part, why transcription of three of the four HCMV long noncoding RNAs is so highly up-regulated with GSK726 treatment. As shown by previous single-cell RNA-seq analysis, RNA2.7 (also known as β2.7) and RNA4.9 are two of the top three transcripts present in latently infected cells (58). Therefore, it is not surprising that SEC:P-TEFb recruitment might be highest in these active promoters and drive amplification of an already present transcription profile. Furthermore, the 2.3-fold up-regulation of expression of UL69, an HCMV transactivator protein (59), with GSK726 treatment may further aid recruitment of CDK9 to certain promoters including the MIEP, as shown previously elsewhere (36). This rapid overall increase in RNA synthesis, caused here by GSK726, could also have a fortuitous knock-on effect on HCMV replication, as this phenomenon induces DNA synthesis stress through transcription-replication conflicts (60); this would be even more likely to occur on the HCMV genome, as oriLyt is located within the up-regulated RNA4.9 locus.
In contrast to the MIEP, the restriction of transcription from other HCMV promoters associated with GSK726 treatment appears to be mediated by inhibition of BRD4-dependent recruitment of P-TEFb, consistent with dBET1-driven ablation of BRD4 resulting in HCMV replication below that of background control levels. The UL44-promoter is unique to HCMV in having three separate transcription start sites (TSSs) (two early and one late) (61), with RNAPII regulation as well as genomic sequence likely controlling TSS selection. BRD4 has been shown to be an important factor in the regulation of alternative RNA splicing (62), and, more recently, over 100 possible alternative splice sites have been described across the HCMV genome (63, 64). Whereas redistribution of BRD4 described in a study of herpes simplex virus increased recruitment of P-TEFb to virus genomes (65), here inhibition of BRD4 interaction directly with the HCMV genome may have effects on transcription and translation. Moreover, global inhibition of BRD4 interaction with acetylated histones, and other proteins, could also have indirect effects on HCMV and the host. I-BETs have routinely been shown to cause myriad effects on cellular transcriptional expression, most commonly suppressing superenhancer driven transcription (66). Additionally, HCMV DNA replication could also be impacted by BRD4 inhibition through interruption of virus-mediated host expression modulation, such as that of interferon via virus-driven increases in histone acetylation (67), or, indeed, more indirect processes such as de-compaction of chromatin structure that can induce the DNA damage and innate immune responses that would hinder virus DNA replication, as seen elsewhere with pseudorabies virus (68).
The finding that GSK726 induces production of viral proteins such as IE72 (IE1), pp65, and pp150 (UL32) (33), which are also immunodominant for T cell recognition, but restricts virus DNA replication (e.g., UL44), could be considered highly advantageous. Additionally, in contrast to HDACi CHR-4487, I-BET GSK726 restricts expression of a number of virus-encoded immunoevasins (e.g., US2, 3, 6, 8, and 11), while allowing full effector function of T cells. This combination of properties provides the ideal conditions for killing of latent cells following GSK726 treatment, as seen in both our experimental and naturally infected HCMV cell models. This ultimately led to a clear and significant depletion of the latent HCMV gDNA load by up to 83% as compared to controls. These data show that short-term use, which is less likely to result in any adverse effects, of I-BETs within a clinical setting should be able to reduce the HCMV latent peripheral reservoir of both donors and recipients prior to transplantation. Previous studies have shown efficacy of I-BETs in various models of human myeloid leukemia, and many inhibitors are now in clinical development (phases I, II, and III) as single treatments of both oncology and nononcology diseases (66). Our work also shows that I-BETs can initiate IE gene expression from HCMV latently infected CD34+ cells, which are good models for HCMV latent infection (69), whereas HDACis such as CHR-4487 (a myeloid-selective HDACi) can have differential effects on CD34+ models with little response in Kasumi-3 cells, consistent with others’ previous work (70). Hence, with good compound penetrance and known immune surveillance in bone marrow, treatment with I-BETs like GSK726 may also allow targeting of latently infected bone-marrow resident pluripotent CD34+ progenitor cells.
Overall, our study shows that bromodomain proteins have a regulatory role in HCMV gene expression control during virus latency and reactivation and that their inhibition or degradation can also restrict viral DNA replication. Moreover, our data show that pre-emptive treatment of all HCMV seropositive immunocompetent transplant donors and/or recipients with I-BETs, such as GSK726, could reduce an individual’s latent reservoir. This should lead to HCMV reactivation events occurring less often after transplantation in immunocompromised patients and, therefore, reduce the rate of associated patient morbidity and mortality. Ultimately, the findings with GSK726 warrant further investigation in patient populations.
Materials and Methods
All human biological samples were obtained ethically and after approval of protocols from the Cambridgeshire 2 Research Ethics Committee (REC reference 97/092) conducted in accordance with the Declaration of Helsinki. Informed written consent was obtained from all of the volunteers included in this study under an Institutional Review Board/Ethics Committee (IRB/EC)-approved protocol before providing blood samples, and all experiments were carried out in accordance with the approved guidelines.
Cells, Viruses, and HCMV Infection.
All details regarding cell origin, culture, HCMV infection thereof, and reactivation analyses are included in SI Appendix, Supplementary Methods. All HCMV used were the TB40e strain, and each stock was titrated on the appropriate cell type for experimental use.
Epigenetic Inhibitors.
Cells were treated with epigenetic inhibitors provided by GlaxoSmithKline (Epigenetics toolbox) and commercially available inhibitors (SI Appendix, Table S1) at a range of concentrations across various time courses dependent on experimental set up. Full details, including cytotoxicity testing, are included in SI Appendix, Supplementary Methods.
RNA, DNA, and Protein Analyses.
Full details of all methods are included in SI Appendix, Supplementary Methods.
ChIP Analysis.
Full details of experimental approaches are included in SI Appendix, Supplementary Methods.
YFP-Positive Cell Sorting and RNA Library Construction.
Briefly, CD14+ monocytes latently infected with HCMV-TB40e-IE86-eYFP were treated with compounds for 72 h before being FACS-sorted and snap-frozen on dry ice. Library preparation for RNA-seq was then carried out essentially as per the MARS-seq protocol (71), producing libraries equating to the 3′ end of HCMV and host mRNA transcripts. Full details are included in SI Appendix, Supplementary Methods.
Sequencing and Data Analysis.
In short, RNA-seq libraries were produced in duplicate and sequenced using NextSEq. 500 (Illumina). Alignment and analysis were done as described previously (58). Full details are included in SI Appendix, Supplementary Methods.
Statistical Analysis.
All statistical analyses (Student’s t test) were carried out using GraphPad Prism v8.4 (*P < 0.05, **P < 0.01, and ***P < 0.001 throughout).
Supplementary Material
Acknowledgments
A.N., B.R., and M.S. are members of Noam Stern-Ginossar’s laboratory, whom we thank for RNA-seq technical assistance. R.K.P. is Vice-President, Head of Adaptive Immunity and Immuno-epigenetics Research Unit, and D.F.T. is a Senior Fellow, Senior Scientific Director of the Adaptive Immunity Research Unit, GlaxoSmithKline. This research was supported by the Cambridge National Institute for Health Research Biomedical Research Centre (NIHR BRC) Cell Phenotyping Hub. In particular, we thank Nika Romashova for advice and support. M.R.W. and J.H.S. are funded by the British Medical Research Council (Grant MR/S00081X/1) and a GSK-Varsity Initiative award (to M.R.W. and J.H.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023025118/-/DCSupplemental.
Data Availability
The data discussed in this publication have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus and are accessible through GEO Series accession number GSE156169 (72).
References
- 1.Sinclair J., Sissons P., Latency and reactivation of human cytomegalovirus. J. Gen. Virol. 87, 1763–1779 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Griffiths P., Baraniak I., Reeves M., The pathogenesis of human cytomegalovirus. J. Pathol. 235, 288–297 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Haidar G., Boeckh M., Singh N., Cytomegalovirus infection in solid organ and hematopoietic cell transplantation: State of the evidence. J. Infect. Dis. 221 (suppl. 1), S23–S31 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Styczynski J., Who is the patient at risk of CMV recurrence: A review of the current scientific evidence with a focus on hematopoietic cell transplantation. Infect. Dis. Ther. 7, 1–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plotkin S. A., Preventing infection by human cytomegalovirus. J. Infect. Dis. 221 (suppl. 1), S123–S127 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Field H. J., Vere Hodge R. A., Recent developments in anti-herpesvirus drugs. Br. Med. Bull. 106, 213–249 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Douglas C. M., et al., Letermovir resistance analysis in a clinical trial of cytomegalovirus prophylaxis for hematopoietic stem cell transplant recipients. J. Infect. Dis. 221, 1117–1126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wills M. R., Poole E., Lau B., Krishna B., Sinclair J. H., The immunology of human cytomegalovirus latency: Could latent infection be cleared by novel immunotherapeutic strategies? Cell. Mol. Immunol. 12, 128–138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reeves M., Sissons P., Sinclair J., Reactivation of human cytomegalovirus in dendritic cells. Discov. Med. 5, 170–174 (2005). [PubMed] [Google Scholar]
- 10.Weekes M. P., et al., Latency-associated degradation of the MRP1 drug transporter during latent human cytomegalovirus infection. Science 340, 199–202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishna B. A., et al., Targeting the latent cytomegalovirus reservoir with an antiviral fusion toxin protein. Nat. Commun. 8, 14321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nehme Z., Pasquereau S., Herbein G., Control of viral infections by epigenetic-targeted therapy. Clin. Epigenetics 11, 55 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinclair J., Chromatin structure regulates human cytomegalovirus gene expression during latency, reactivation and lytic infection. Biochim. Biophys. Acta 1799, 286–295 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Reeves M. B., MacAry P. A., Lehner P. J., Sissons J. G., Sinclair J. H., Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. U.S.A. 102, 4140–4145 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishna B. A., et al., Transient activation of human cytomegalovirus lytic gene expression during latency allows cytotoxic T cell killing of latently infected cells. Sci. Rep. 6, 24674 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones R. B., et al., Histone deacetylase inhibitors impair the elimination of HIV-infected cells by cytotoxic T-lymphocytes. PLoS Pathog. 10, e1004287 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tough D. F., Lewis H. D., Rioja I., Lindon M. J., Prinjha R. K., Epigenetic pathway targets for the treatment of disease: Accelerating progress in the development of pharmacological tools: IUPHAR review 11. Br. J. Pharmacol. 171, 4981–5010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reeves M., Sinclair J., Regulation of human cytomegalovirus transcription in latency: Beyond the major immediate-early promoter. Viruses 5, 1395–1413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gan X., et al., Epigenetically repressing human cytomegalovirus lytic infection and reactivation from latency in THP-1 model by targeting H3K9 and H3K27 histone demethylases. PLoS One 12, e0175390 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang Y., et al., Targeting the JMJD2 histone demethylases to epigenetically control herpesvirus infection and reactivation from latency. Sci. Transl. Med. 5, 167ra5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang Y., Vogel J. L., Narayanan A., Peng H., Kristie T. M., Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat. Med. 15, 1312–1317 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abraham C. G., Kulesza C. A., Polycomb repressive complex 2 silences human cytomegalovirus transcription in quiescent infection models. J. Virol. 87, 13193–13205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arbuckle J. H., et al., Inhibitors of the histone methyltransferases EZH2/1 induce a potent antiviral state and suppress infection by diverse viral pathogens. MBio 8, e01141-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groves I. J., Sinclair J. H., Wills M. R., Bromodomain inhibitors as therapeutics for herpesvirus-related disease: All BETs are off? Front. Cell. Infect. Microbiol. 10, 329 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaware N., Zhou M. M., Bromodomain biology and drug discovery. Nat. Struct. Mol. Biol. 26, 870–879 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tough D. F., Tak P. P., Tarakhovsky A., Prinjha R. K., Epigenetic drug discovery: Breaking through the immune barrier. Nat. Rev. Drug Discov. 15, 835–853 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Van Damme E., et al., Glucocorticosteroids trigger reactivation of human cytomegalovirus from latently infected myeloid cells and increase the risk for HCMV infection in D+R+ liver transplant patients. J. Gen. Virol. 96, 131–143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gosmini R., et al., The discovery of I-BET726 (GSK1324726A), a potent tetrahydroquinoline ApoA1 up-regulator and selective BET bromodomain inhibitor. J. Med. Chem. 57, 8111–8131 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Picaud S., et al., RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. Proc. Natl. Acad. Sci. U.S.A. 110, 19754–19759 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niu Q., et al., Structure-guided drug design identifies a BRD4-selective small molecule that suppresses HIV. J. Clin. Invest. 129, 3361–3373 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dawson M. A., et al., Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaitin D. A., et al., Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sylwester A. W., et al., Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 202, 673–685 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alfonso-Dunn R., et al., Transcriptional elongation of HSV immediate early genes by the super elongation complex drives lytic infection and reactivation from latency. Cell Host Microbe 21, 507–517.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartholomeeusen K., Xiang Y., Fujinaga K., Peterlin B. M., Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. J. Biol. Chem. 287, 36609–36616 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kapasi A. J., Spector D. H., Inhibition of the cyclin-dependent kinases at the beginning of human cytomegalovirus infection specifically alters the levels and localization of the RNA polymerase II carboxyl-terminal domain kinases cdk9 and cdk7 at the viral transcriptosome. J. Virol. 82, 394–407 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Georgiev P., et al., BET bromodomain inhibition suppresses human T cell function. Immunohorizons 3, 294–305 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Nitzsche A., Paulus C., Nevels M., Temporal dynamics of cytomegalovirus chromatin assembly in productively infected human cells. J. Virol. 82, 11167–11180 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nitzsche A., Steinhäusser C., Mücke K., Paulus C., Nevels M., Histone H3 lysine 4 methylation marks postreplicative human cytomegalovirus chromatin. J. Virol. 86, 9817–9827 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy J. C., Fischle W., Verdin E., Sinclair J. H., Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 21, 1112–1120 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuevas-Bennett C., Shenk T., Dynamic histone H3 acetylation and methylation at human cytomegalovirus promoters during replication in fibroblasts. J. Virol. 82, 9525–9536 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Groves I. J., Reeves M. B., Sinclair J. H., Lytic infection of permissive cells with human cytomegalovirus is regulated by an intrinsic ‘pre-immediate-early’ repression of viral gene expression mediated by histone post-translational modification. J. Gen. Virol. 90, 2364–2374 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Zalckvar E., et al., Nucleosome maps of the human cytomegalovirus genome reveal a temporal switch in chromatin organization linked to a major IE protein. Proc. Natl. Acad. Sci. U.S.A. 110, 13126–13131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dupont L., et al., Src family kinase activity drives cytomegalovirus reactivation by recruiting MOZ histone acetyltransferase activity to the viral promoter. J. Biol. Chem. 294, 12901–12910 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michaelis M., et al., Increased human cytomegalovirus replication in fibroblasts after treatment with therapeutical plasma concentrations of valproic acid. Biochem. Pharmacol. 68, 531–538 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Nevels M., Paulus C., Shenk T., Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc. Natl. Acad. Sci. U.S.A. 101, 17234–17239 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vollmuth F., Blankenfeldt W., Geyer M., Structures of the dual bromodomains of the P-TEFb-activating protein Brd4 at atomic resolution. J. Biol. Chem. 284, 36547–36556 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilan O., et al., Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation. Science 368, 387–394 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pari G. S., Kacica M. A., Anders D. G., Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J. Virol. 67, 2575–2582 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith J. A., Jairath S., Crute J. J., Pari G. S., Characterization of the human cytomegalovirus UL105 gene and identification of the putative helicase protein. Virology 220, 251–255 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto T., Suzuki S., Radsak K., Hirai K., The UL112/113 gene products of human cytomegalovirus which colocalize with viral DNA in infected cell nuclei are related to efficient viral DNA replication. Virus Res. 56, 107–114 (1998). [DOI] [PubMed] [Google Scholar]
- 52.Arend K. C., et al., Kinome profiling identifies druggable targets for novel human cytomegalovirus (HCMV) antivirals. Mol. Cell. Proteomics 16 (4, suppl. 1), S263–S276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keck K. M., et al., Bromodomain and extraterminal inhibitors block the Epstein-Barr virus lytic cycle at two distinct steps. J. Biol. Chem. 292, 13284–13295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Halper-Stromberg A., et al., Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell 158, 989–999 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen F. X., Smith E. R., Shilatifard A., Born to run: Control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 19, 464–478 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Alfonso-Dunn R., Arbuckle J. H., Vogel J. L., Kristie T. M., Inhibition of the super elongation complex suppresses herpes simplex virus immediate early gene expression, lytic infection, and reactivation from latency. MBio 11, e01216-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kapasi A. J., Clark C. L., Tran K., Spector D. H., Recruitment of cdk9 to the immediate-early viral transcriptosomes during human cytomegalovirus infection requires efficient binding to cyclin T1, a threshold level of IE2 86, and active transcription. J. Virol. 83, 5904–5917 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shnayder M., et al., Defining the transcriptional landscape during cytomegalovirus latency with single-cell RNA sequencing. MBio 9, e00013-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winkler M., Rice S. A., Stamminger T., UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J. Virol. 68, 3943–3954 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bowry A., Piberger A. L., Rojas P., Saponaro M., Petermann E., BET inhibition induces HEXIM1- and RAD51-dependent conflicts between transcription and replication. Cell Rep. 25, 2061–2069.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leach F. S., Mocarski E. S., Regulation of cytomegalovirus late-gene expression: Differential use of three start sites in the transcriptional activation of ICP36 gene expression. J. Virol. 63, 1783–1791 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uppal S., et al., The bromodomain protein 4 contributes to the regulation of alternative splicing. Cell Rep. 29, 2450–2460.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stern-Ginossar N., et al., Decoding human cytomegalovirus. Science 338, 1088–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gatherer D., et al., High-resolution human cytomegalovirus transcriptome. Proc. Natl. Acad. Sci. U.S.A. 108, 19755–19760 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ren K., et al., An epigenetic compound library screen identifies BET inhibitors that promote HSV-1 and -2 replication by bridging P-TEFb to viral gene promoters through BRD4. PLoS Pathog. 12, e1005950 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cochran A. G., Conery A. R., Sims R. J. III, Bromodomains: A new target class for drug development. Nat. Rev. Drug Discov. 18, 609–628 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Parekh B. S., Maniatis T., Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol. Cell 3, 125–129 (1999). [DOI] [PubMed] [Google Scholar]
- 68.Wang J., et al., BRD4 inhibition exerts anti-viral activity through DNA damage-dependent innate immune responses. PLoS Pathog. 16, e1008429 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poole E., et al., An iPSC-derived myeloid lineage model of herpes virus latency and reactivation. Front. Microbiol. 10, 2233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Albright E. R., Kalejta R. F., Myeloblastic cell lines mimic some but not all aspects of human cytomegalovirus experimental latency defined in primary CD34+ cell populations. J. Virol. 87, 9802–9812 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Keren-Shaul H., et al., MARS-seq2.0: An experimental and analytical pipeline for indexed sorting combined with single-cell RNA sequencing. Nat. Protoc. 14, 1841–1862 (2019). [DOI] [PubMed] [Google Scholar]
- 72.Edgar R., Domrachev M., Lash A. E., Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30 (1), 207–10, 10.1093/nar/30.1.207 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this publication have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus and are accessible through GEO Series accession number GSE156169 (72).