Significance
Recurrent histone H3K36M mutations are found in multiple cancer types yet their oncogenic mechanisms remain incompletely understood. Biochemically, H3K36M oncohistone dominantly inhibits several H3K36-specific methyltransferases such as NSD1/2 and SETD2, resulting in decreases in all methylation states of H3K36. We report here that genetic ablation of NSD1 and NSD2, methyltransferases specific for H3K36 dimethylation (H3K36me2), is sufficient to recapitulate H3K36M’s effects on enhancer activation, gene expression, differentiation blockade, and drug sensitivity. Our results suggest that depletion of H3K36me2 represents a key event downstream of the H3K36M mutation and also exposes potential therapeutic vulnerability of H3K36M-mutant tumor cells.
Keywords: cancer, chromatin and epigenomics, oncohistone, H3K36 methylation, NSD
Abstract
Hotspot histone H3 mutations have emerged as drivers of oncogenesis in cancers of multiple lineages. Specifically, H3 lysine 36 to methionine (H3K36M) mutations are recurrently identified in chondroblastomas, undifferentiated sarcomas, and head and neck cancers. While the mutation reduces global levels of both H3K36 dimethylation (H3K36me2) and trimethylation (H3K36me3) by dominantly inhibiting their respective specific methyltransferases, the relative contribution of these methylation states to the chromatin and phenotypic changes associated with H3K36M remains unclear. Here, we specifically deplete H3K36me2 or H3K36me3 in mesenchymal cells, using CRISPR-Cas9 to separately knock out the corresponding methyltransferases NSD1/2 or SETD2. By profiling and comparing the epigenomic and transcriptomic landscapes of these cells with cells expressing the H3.3K36M oncohistone, we find that the loss of H3K36me2 could largely recapitulate H3.3K36M’s effect on redistribution of H3K27 trimethylation (H3K27me3) and gene expression. Consistently, knockout of Nsd1/2, but not Setd2, phenocopies the differentiation blockade and hypersensitivity to the DNA-hypomethylating agent induced by H3K36M. Together, our results support a functional divergence between H3K36me2 and H3K36me3 and their nonredundant roles in H3K36M-driven oncogenesis.
A group of highly clustered missense mutations affecting histone H3 genes were first identified in pediatric glioblastomas and subsequently in tumors of other tissue types (1). Specifically, recurrent histone H3 lysine 36 to methionine (H3K36M) or isoleucine (H3K36I) mutations have been reported in cancers of mesenchymal origin, including 95% of chondroblastomas, a congenital soft-tissue neoplasm and a histiocytic tumor of the skull, as well as in head and neck squamous cell carcinomas (2–6). The mutant histone affects the binding and inhibits the catalytic activity of H3K36-directed methyltransferases by altering the hydrophobic pocket responsible for methylation (5, 7, 8). As a result, although the H3K36M/I mutation is heterozygous and only found in 1 of the 16 genes encoding canonical histone H3.1/2 or variant histone H3.3, it exerts a dominant effect on decreasing the global abundance of mono- (H3K36me1), di- (H3K36me2), and trimethyl (H3K36me3) marks (9, 10).
We and others have reported that the marked loss of H3K36 methylation in H3K36M/I mutant cells results in altered gene expression programs that are linked to cell differentiation and cancer (9, 10). Accordingly, the H3.3K36M mutation blocks the differentiation of mouse mesenchymal progenitor cells and promotes the development of undifferentiated sarcomas in vivo (10). Furthermore, tissue-specific transgenic mouse models of H3.3K36M expression show impaired hematopoiesis and adipogenesis (11, 12), pointing to a conserved and critical role of H3K36 methylation in tissue homeostasis and tumorigenesis. The reprogramming of the transcriptome by the H3K36M mutation appears to be coupled to the redistribution of another histone modification, H3K27 trimethylation (H3K27me3) (10, 11, 13). H3K36 methylation is known to antagonize the activity of polycomb repressive complex 2 (PRC2)—the methyltransferase for H3K27me3 (14, 15). In H3K36M-mutant cells, the reduction in H3K36 methylation causes a widespread increase of H3K27me3, which is accompanied by a relocation of the H3K27me3 “reader” polycomb repressive complex 1 (PRC1), thus leading to aberrant expression of genes that are repressed under normal conditions (10). However, a functional role of H3K27me3 in mediating the impact of the H3K36M oncohistone on gene expression remains to be fully established.
SET2, the homolog of human SETD2, is responsible for all H3K36 methylation states in yeast, where it serves a dual role of enabling transcriptional elongation through its association with RNA polymerase II while placing methyl marks that provide transcriptional memory and prevent cryptic transcription (16–19). In contrast, in mammalian cells, H3K36 methylation states are subject to more complex regulation. Whereas SETD2 retains the ability to catalyze all states of H3K36 methylation, its depletion in mouse and human cells only abolishes H3K36me3. Additional methyltransferases, including NSD1, NSD2, and ASH1L, have been shown to catalyze H3K36me1/2 (20, 21). In accordance with the divergence in their “writers,” the genomic distribution of H3K36me2 is distinct from that of H3K36me3. Within genic regions of actively transcribed genes, H3K36me2 covers areas downstream of the transcription start site through the first intron, followed by a marked switch to H3K36me3 after the first splice junction. In addition, H3K36me2 also demarcates intergenic regions where H3K36me3 is absent (17, 21). These observations suggest that in mammalian cells, H3K36me2 may have a distinct functional role from H3K36me3, which has been implicated in RNA splicing, prevention of intragenic transcription, and DNA damage repair (22, 23).
Our prior studies revealed that combined transient knockdown of Nsd1, Nsd2, and Setd2 led to defects in chondrocyte differentiation of the C3H10T1/2 (10T) mesenchymal stem cells mimicking the effects of H3.3K36M mutation (10). Here, we further dissected the specific contributions of reduction of H3K36me2 versus H3K36me3 to the epigenomic and phenotypic changes induced by H3K36M. Using CRISPR-Cas9–based gene editing, we generated Setd2 knockout (KO) and Nsd1 and Nsd2 double-knockout (Nsd1/2 DKO) cells that are specifically depleted of H3K36me3 and H3K36me2, respectively. We systematically assessed the chromatin landscape, gene expression, and phenotypic assays including differentiation and drug response in these cells and compared them with cells expressing H3.3K36M. Our studies revealed a major and nonredundant role of H3K36me2 loss in mediating the effect of H3.3K36M on epigenome remodeling, transcriptome, chondrogenesis, and drug sensitivity.
Results
Generation and Epigenome Profiling of Nsd1/2 DKO and Setd2 KO 10T Cells.
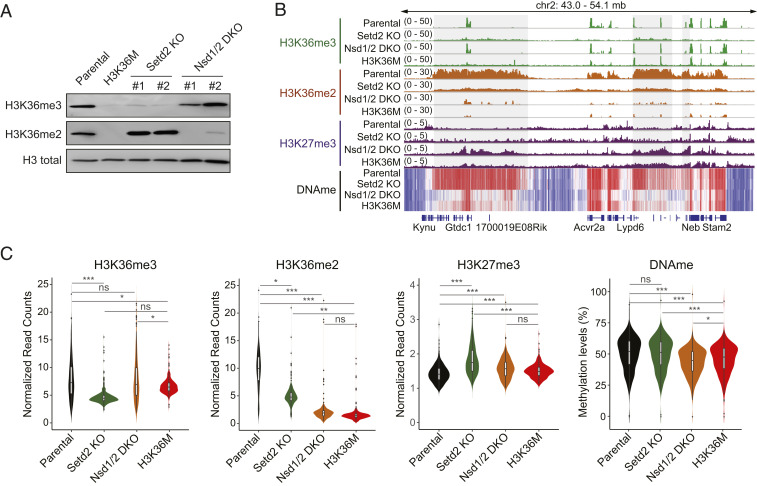
Using CRISPR-Cas9, we established clones of 10T cells that had single knockout of the Setd2 or double knockout of the Nsd1 and Nsd2 methyltransferases. Two independent clones of each genotype were derived. Immunoblotting confirmed that Setd2 KO cells and Nsd1/2 DKO cells exhibited specific depletion of H3K36me3 and H3K36me2, respectively, whereas expression of the H3.3K36M mutant histone resulted in loss of both methylation states (Fig. 1A). We performed semiquantitative chromatin immunoprecipitation-sequencing (ChIP-seq) with reference chromatin spike-in (ChIP-Rx) to profile genome-wide distributions of H3K36me2 and H3K36me3 in these cells. Compared with parental 10T cells, Setd2 KO cells showed a near-complete loss of H3K36me3 as shown at representative genomic regions (Fig. 1B and SI Appendix, Fig. S1A) and by genome-wide quantification (Fig. 1C). H3K36me3 levels were largely retained in Nsd1/2 DKO cells. Although H3K36M cells also displayed marked decreases in H3K36me3, peaks of H3K36me3 were retained at certain gene bodies, and the overall level was slightly but notably higher than that of Setd2 KO cells (Fig. 1 B and C, SI Appendix, Fig. S1, and Dataset S1). On the other hand, drastic decreases in H3K36me2, which are particularly pronounced at intergenic regions, were observed in Nsd1/2 DKO and H3K36M cells (Fig. 1 B and C and SI Appendix, Fig. S1A). Indeed, residual H3K36me2 peaks were mostly enriched at promoters and introns (SI Appendix, Fig. S1B), which may be catalyzed by additional K36M-insensitive H3K36 methyltransferases such as ASH1L (9). On the other hand, Setd2 KO led to a more modest, yet significant, depletion of H3K36me2 (Fig. 1 B and C).
Fig. 1.
Generation and epigenome profiling of Nsd1/2 DKO and Setd2 KO 10T cells. (A) Western blotting of H3K36me3 and H3K36me2 levels in parental, H3K36M, Nsd1/2 DKO, and Setd2 KO 10T cells. Total H3 is used as loading control. For Nsd1/2 DKO and Setd2 KO, clone 1 is used for all epigenome profiling analyses. (B) Integrative Genomics Viewer snapshot showing the chromatin landscape of H3K36me3, H3K36me2, H3K27me3, and DNA methylation at a representative genomic region in parental, H3K36M, Nsd1/2 DKO, and Setd2 KO 10T cells. (C) Violin plots showing the global levels of H3K36me3, H3K36me2, H3K27me3 (counts per million ChIP-seq reads normalized to reference chromatin spike-in control), and DNA methylation (average methylation level) in parental, H3K36M, Nsd1/2 DKO, and Setd2 KO cells. Bin size, 10 Mb. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant.
We recently demonstrated that H3K36me2 is required for the localization of the DNA methyltransferase DNMT3A to sustain levels of DNA methylation at intergenic regions (24). Therefore, as an independent validation, we performed whole-genome bisulfite sequencing (WGBS), which revealed that Nsd1/2 DKO and H3K36M cells exhibited similar genome-wide intergenic DNA hypomethylation compared with parental cells (Fig. 1 B and C). Despite the partial decrease in H3K36me2, patterns of DNA methylation in Setd2 KO are indistinguishable from those in parental cells. Taken together, these results indicate that the decreases in genic H3K36me3 and intergenic H3K36me2 can be largely recapitulated by genetic ablation of Setd2 and Nsd1/2, respectively.
Knockout of Nsd1/2 Mimics the Transcriptomic Changes Induced by H3.3K36M.
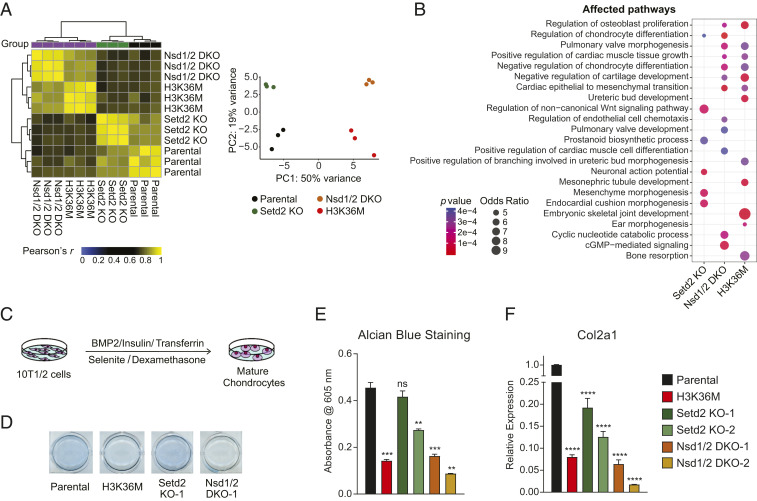
We next performed and analyzed RNA-sequencing (RNA-seq) data from our clones to determine the relative contribution of Nsd1/2 versus Setd2 to H3.3K36M-induced changes in gene expression. Unsupervised hierarchical clustering and principal-component analysis showed that Nsd1/2 DKO cells clustered closely with H3K36M cells, while the transcriptome profiles of Setd2 KO cells and parental cells were more similar (Fig. 2A). We also derived the lists of differentially expressed genes between knockout/mutant and parental 10T cells. Gene Ontology enrichment analysis of differentially expressed genes revealed that pathways related to mesenchymal cell differentiation and tissue development were commonly altered in Nsd1/2 DKO and H3K36M cells, whereas Setd2 KO affected expression of a distinct set of genes related to the Wnt signaling pathway and prostanoid biosynthetic process (Fig. 2B). While the gene expression profiles were relatively similar, a closer examination revealed that MAPK and ERK1/2 signaling cascades and extracellular matrix organization were among pathways that were differentially regulated between Nsd1/2 DKO and H3K36M cells (Dataset S2).
Fig. 2.
Depletion of H3K36me2 recapitulates transcriptomic and phenotypic changes induced by H3.3K36M. (A) Unsupervised hierarchical clustering based on correlation and principal component (PC) analysis of transcriptome profiles of parental, H3K36M, Nsd1/2 DKO (clone 1), and Setd2 KO (clone 1) 10T cells. n = 3. (B) Gene Ontology analysis of differentially expressed genes (fold change > 4, false discovery rate < 0.1) between parental cells and H3K36M, Nsd1/2 DKO, or Setd2 KO cells. n = 3. (C) Schematic of the 10T1/2 cell chondrocytic differentiation assay. (D) Images of 10T cells stained with Alcian blue after 11 d of induction of chondrocytic differentiation. (E) Bar graph showing quantification of Alcian blue staining as in D measured by absorbance. Error bars represent SD from n = 3. t test was performed to compare all samples with parental cells. *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001; ns, not significant. (F) Gene expression of Col2a1 in parental, H3K36M, Nsd1/2 DKO, and Setd2 KO 10T cells after 9 d of chondrocytic differentiation induction as measured by RT-PCR. Error bars represent SD from n = 3. t test was performed to compare all samples with parental cells. *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001; ns, not significant.
Knockout of Nsd1/2 Phenocopies H3.3K36M’s Effect on Differentiation Blockade.
As expression of genes involved in mesenchymal lineage specification was dysregulated in Nsd1/2 DKO and H3K36M cells, we determined and compared the differentiation potential of these cells with Setd2 KO cells in an in vitro assay of chondrogenesis. Following differentiation induction into mature chondrocytes using a specific mixture of growth factors (Fig. 2C), parental 10T cells readily accumulated proteoglycans as determined by Alcian blue staining (Fig. 2 D and E). Setd2 KO cells showed variable and modest delay in chondrocyte differentiation, whereas both H3K36M cells and Nsd1/2 DKO cells remained in an undifferentiated state (Fig. 2 D and E). These findings are corroborated by measurement of expression levels of Col2a1, a mature chondrocyte marker that encodes for a component of type II collagen (Fig. 2F). Therefore, it appears that the effects of H3.3K36M on the transcriptome and mesenchymal differentiation are largely mediated through depletion of H3K36me2.
Redistribution of H3K27me3 by H3.3K36M and Loss of Nsd1/2.
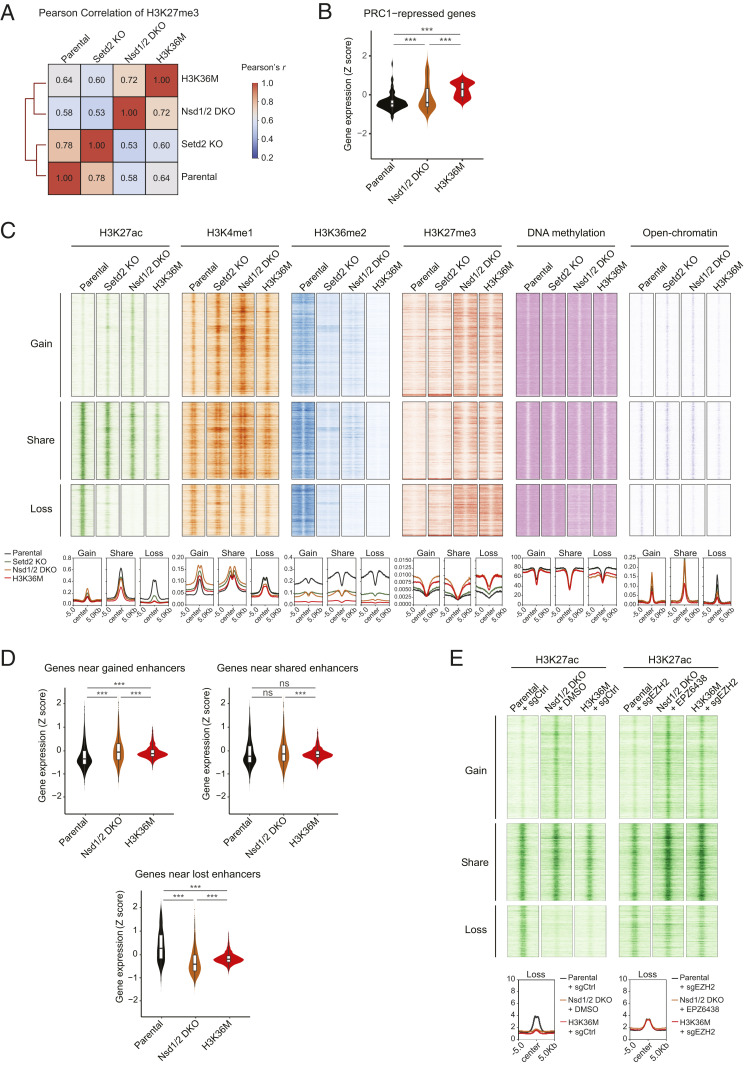
We next sought to understand the mechanisms by which H3K36me2 loss reprograms transcriptomes in Nsd1/2 DKO and H3K36M cells. H3K36me2 and DNA methylation have both been implicated in opposing the activity of PRC2 and H3K27me3 (14, 15, 25). Indeed, analysis of the enrichment patterns of H3K27me3, measured by ChIP-Rx, revealed a strong positive correlation between Nsd1/2 DKO and H3K36M cells, which is in sharp contrast to Setd2 KO and parental cells (Fig. 3A). This similarity can be readily appreciated at individual genomic loci (Fig. 1B and SI Appendix, Fig. S1), which demonstrated that the depletion of H3K36me2 in Nsd1/2 DKO and H3K36M cells was accompanied by both expansion of preexisting and accumulation of new H3K27me3 domains. We have previously suggested that such pervasive gain of H3K27me3 results in a “titration” of PRC1, the chromatin reader complex that binds to H3K27me3 to silence transcription, which in turn leads to derepression of PRC1-regulated genes in H3K36M cells (10, 26). We therefore analyzed genes that increased their expression upon RNA interference depletion of Ring1a/b in 10T cells (n = 130) (10). As expected, we observed a similar derepression of PRC1-silenced genes in Nsd1/2 DKO cells (Fig. 3B and SI Appendix, Fig. S2A).
Fig. 3.
Depletion of H3K36me2 reprograms gene expression via redistribution of H3K27me3. (A) Unsupervised hierarchical clustering based on correlation of the genomic distribution of H3K27me3. Bin size, 1 kb. (B) Violin plots comparing the expression levels of PRC1-repressed genes between parental, H3K36M, and Nsd1/2 DKO cells. n = 3. ***P < 0.001. (C) Landscape of chromatin modifications H3K27ac, H3K4me1, H3K36me2, and H3K27me3, DNA methylation, and chromatin accessibility near gained, shared, and lost enhancers in Nsd1/2 DKO cells, compared with the parental cells. ChIP-seq reads normalized to the reference chromatin spike-in control are shown as density plots relative to the center of H3K27ac peaks that are shared between parental and Nsd1/2 DKO cells and those that are gained/lost in Nsd1/2 DKO cells. (D) Violin plots comparing the expression levels of genes with gained, shared, and lost enhancers between parental, H3K36M, and Nsd1/2 DKO cells. n = 3. ***P < 0.001; ns, not significant. (E) Landscape of chromatin modification H3K27ac near gained, shared, and lost enhancers in Nsd1/2 DKO cells, with or without CRISPR-Cas9–mediated Ezh2 knockout (sgEZH2) or EZH2 inhibitor EPZ-6438. ChIP-seq reads normalized to the reference chromatin spike-in control are shown as density plots relative to the center of H3K27ac peaks that are lost in Nsd1/2 DKO cells.
To explore additional effects of the H3K27me3 increase on gene regulation, we profiled the landscape of active enhancers by performing ChIP-Rx of H3K27 acetylation (H3K27ac) and H3K4 monomethylation (H3K4me1). We also measured chromatin accessibility using ATAC-seq (assay for transposase-accessible chromatin with sequencing). We identified enhancers that are gained, shared, or lost in Nsd1/2 DKO cells based on quantification of nonpromoter H3K27ac peaks (Fig. 3C). Representative genomic regions containing gained or lost enhancers are shown in SI Appendix, Fig. S2B. We found that enhancers which became inactivated in Nsd1/2 DKO cells, as demonstrated by loss of H3K27ac, H3K4me1, and ATAC-seq signals, displayed the most profound decreases in H3K36me2 and DNA methylation and gain of H3K27me3 (Fig. 3C and SI Appendix, Fig. S2B). Transcriptome analysis suggested that whereas genes close to gained or shared enhancers displayed minimal expression changes, there was a marked decrease in expression for genes near enhancers that were lost in Nsd1/2 DKO cells (Fig. 3D). Importantly, this H3K27me3-associated aberrant silencing of enhancers was also found in H3K36M cells (Fig. 3 C and D). To determine the functional role of H3K27me3 in enhancer inactivation, we treated cells with EPZ-6438, a compound that inhibits the activity of the PRC2 catalytic subunit EZH2. Treatment of Nsd1/2 DKO cells with EPZ-6438 was sufficient to restore H3K27ac levels at those lost enhancers (Fig. 3E and SI Appendix, Fig. S2C).
Nsd1/2 DKO and H3K36M Cells Display Differential Drug Sensitivity from Setd2 KO Cells.
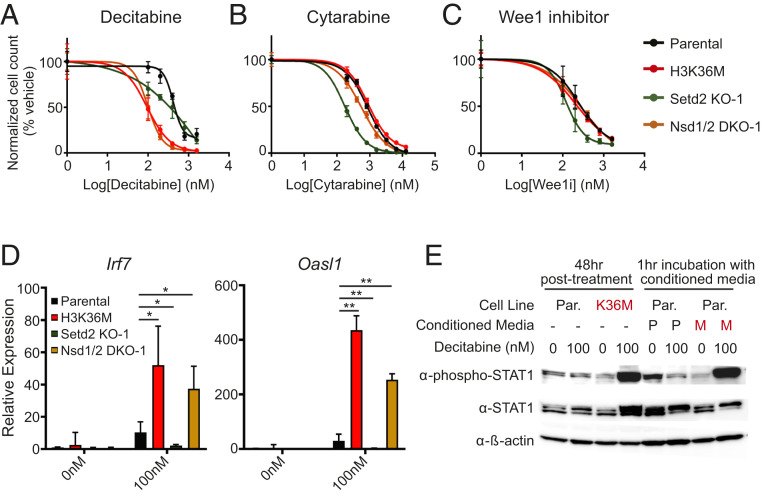
We reasoned that given the significant epigenomic, transcriptomic, and phenotypic overlap, Nsd1/2 DKO and H3K36M cells, but not Setd2 KO cells, may display coordinated sensitivity toward cancer therapeutics. Indeed, treatment of the DNA-hypomethylating agent decitabine produced selective toxicity toward Nsd1/2 DKO and H3.3K36M cells (Fig. 4A and SI Appendix, Table S1). In contrast, when given cytarabine, a chemotherapeutic antimetabolite that competitively inhibits DNA polymerase, Setd2 KO cells were more sensitive than Nsd1/2 DKO and H3K36M cells, which displayed similar sensitivity as parental cells (Fig. 4B and SI Appendix, Table S1) (27). Moreover, Setd2 KO cells, but not Nsd1/2 DKO, H3K36M, or parental cells, were hypersensitive to the treatment of a Wee1 kinase inhibitor, which has been reported to be synthetically lethal with loss of SETD2 (Fig. 4C and SI Appendix, Table S1) (28).
Fig. 4.
Differential drug sensitivity between H3K36me2-depleted vs. H3K36me3-depleted cells. (A–C) Normalized cell count after treatment with increasing concentrations of decitabine (A), cytarabine (B), or the Wee1 inhibitor AZD1775 (C). Error bars represent SD from n = 3. Two-way ANOVA was performed to compare all samples with parental cells and the P values are listed in SI Appendix, Table S1. (D) Gene expression of Irf7 and Oasl1 as measured by RT-PCR in parental, H3K36M, Nsd1/2 DKO, and Setd2 KO cells following decitabine treatment. Error bars represent SD from n = 4 for parental and n = 2 for others. t test was performed to compare all samples with parental cells. *P < 0.05, **P < 0.01. (E) Western blot of STAT1 phosphorylation in parental or H3K36M cells treated directly with decitabine or with conditioned media from decitabine-treated cells (M, H3K36M cells; P, parental cells). See SI Appendix, Fig. S3B for a schematic of the conditioned media treatment. Total STAT1 and β-actin were used as loading controls.
The anticancer effects of hypomethylating agents such as decitabine have been recently linked to the derepression of endogenous retroviruses that are normally silenced by DNA methylation, which in turn elicits an interferon response and host immune activation against the tumor cells (29–33). As we had established that H3K36M and Nsd1/2 DKO cells have decreased levels of DNA methylation (Fig. 1B), we hypothesized that the hypersensitivity of these cells to a DNA-hypomethylating agent (Fig. 4A and SI Appendix, Fig. S3A and Table S2) is linked to an enhanced activation of the interferon pathway. We examined the expression of Irf7 and Oasl1, two interferon-stimulated genes (ISGs) in cells that were treated with decitabine. Upon treatment of low-dose decitabine, H3K36M and Nsd1/2 DKO cells but not Setd2 KO cells showed augmented expression of Irf7 and Oasl1 (Fig. 4D). Furthermore, conditioned media from H3K36M but not parental cells treated with decitabine potently induced STAT1 phosphorylation, a marker of interferon pathway activity (34), suggesting that high levels of interferons are released by H3K36M cells into the media (Fig. 4E and SI Appendix, Fig. S3B). Treatment of ruxolitinib, a JAK inhibitor, rescued the selective toxicity of decitabine to Nsd1/2 DKO and H3K36M cells (SI Appendix, Fig. S3C and Table S3), indicating that the hypersensitivity of these cells to DNA hypomethylation involves the hyperactivation of the JAK/STAT pathway downstream of interferon stimulation.
Discussion
In this study, we sought to understand how alterations in H3K36me2 versus H3K36me3 contribute to the epigenome, transcriptome, and phenotype reprogramming by the H3.3K36M oncohistone. Our results indicated that Nsd1/2 DKO cells, compared with Setd2 KO, more closely resembled the chromatin and gene expression landscape of H3K36M cells. In agreement, in functional assays including chondrocytic differentiation and drug sensitivity, Nsd1/2 DKO cells phenocopied H3K36M cells. Taken together, our results suggest that the depletion of H3K36me2 represents a key event downstream of the H3K36M mutation and in the development of tumors driven by the H3K36M oncohistone.
Notably, we previously reported that both H3K36M mutations and NSD1 loss-of-function mutations are found in human head and neck squamous cell carcinomas (6). Furthermore, H3K36M and NSD1 mutations are mutually exclusive and together define a molecular subgroup with distinct signatures of DNA methylation and gene expression. These findings, together with the fact that H3K36M/I mutations are not found in cancer types harboring frequent SETD2 alterations such as renal cell carcinomas (35), offer strong human genetic evidence in support of our conclusion. Similarly, expression of H3K36M in preadipocytes blocks adipogenesis in vitro, and its effect can be recapitulated by knockdown of Nsd2 but not Setd2 (11). Despite the role of H3K36M as a driver for oncogenesis in certain contexts, in other cell types, such as a fibrosarcoma cell line, expression of H3K36M impaired cell proliferation; depletion of NSD2, but not SETD2, in these cells phenocopied H3K36M (36). A potential explanation for the distinction between H3K36M and SETD2 knockout/knockdown may be the incomplete inhibition of SETD2 activity by H3K36M. We noticed that residual levels of H3K36me3 persisted at some gene bodies in H3K36M cells (Fig. 1B). Similarly, a recent study found that in pancreatic cancer cells, expression of H3.3K36M had modest effects on H3K36me3 (37). In a histiocytic tumor of the skull where the H3K36I mutation was identified, the tumor cells had lost H3K36me2 while retaining H3K36me3 (4). Future studies are required to fully understand the variable and context-dependent inhibition of SETD2 by H3K36M, and to determine the functional importance of residual H3K36me3 in maintaining transcription elongation, fidelity, and alternative splicing.
Our data suggest a key role of the redistribution of H3K27me3 in connecting loss of H3K36me2 to transcriptome reprogramming. It appears that in the absence of the antagonism by H3K36me2, the pervasive increases in H3K27me3 serve to 1) titrate the dosage of PRC1 so that it can no longer efficiently suppress the canonical polycomb-regulated genes (10), and 2) reduce the accessibility and activity of a subset of active enhancers localized in H3K36me2 domains. The latter observation is consistent with several recent studies showing that H3K36me2 is required for maintaining active enhancers and the expression of their nearby genes in pancreatic ductal adenocarcinoma and multiple myeloma cells (37, 38). We further demonstrated that enhancers that became silenced in Nsd1/2 DKO and H3K36M cells could be reactivated by treatment with an EZH2 inhibitor (Fig. 3E). Thus, it will be interesting to determine the impact of PRC2 inhibition on the gene expression and growth of H3K36M-mutant tumors.
DNA methylation in intergenic regions is regulated by deposition of H3K36me2, and we found that in Nsd1/2 DKO and H3K36M cells, global DNA methylation was reduced (24, 39). Because prior studies have documented a vulnerability of cancer cells to DNA-hypomethylating agents that is linked to the activation of interferon response and tumor immunity, we reasoned that both of these cells would be selectively susceptible to further hypomethylation by decitabine treatment (29–33). Congruent with our hypothesis, decitabine treatment of H3K36M cells and Nsd1/2 DKO cells led to a selective toxicity that was accompanied by an increased expression of ISGs. These results mirror recent findings that the loss of PRC2 and subsequent reduction of H3K27me2/3 make malignant peripheral nerve sheath tumor cells sensitive to decitabine due to DNA hypomethylation, genomic instability, and up-regulation of interferon response (40). Similar observations have been made in H3K27M-mutant pediatric gliomas (41). Notably, the selective cytotoxicity of Nsd1/2 DKO and H3K36M cells to decitabine can be reversed by a JAK inhibitor. This finding is consistent with the critical role of the JAK/STAT pathway in interferon response. Alternatively, the JAK/STAT pathway has been implicated in the regulation of PRC2, either directly through EZH2 phosphorylation or indirectly through EZH2 transcription, which warrants future investigation (42, 43). Taken together, our studies imply that tumors with an imbalance between H3K27me3 and H3K36me2 may be therapeutically exploited with the treatment of demethylating agents, alone or in combination with immune checkpoint inhibitors.
Materials and Methods
Cell Culture and Generation of CRISPR-Cas9 Knockout Clones.
C3H10T1/2 cells (ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) with 10% fetal bovine serum (Sigma). CRISPR-Cas9 gene editing of C3H10T1/2 cells to create knockout clones has been previously described (24).
Chondrocyte Differentiation Assays.
Cells were plated on 6-well dishes in differentiation medium that was composed of DMEM with 1% serum and included 10 μg/mL insulin, 3 × 10−8 M sodium selenite, 10 μg/mL transferrin, 10−8 M dexamethasone, and 100 ng/mL rhBMP-2. Medium was replaced with fresh medium every 2 to 3 d, and cells were stained with Alcian blue after 11 d and absorbance at 605 nm was measured using a spectrophotometer. This signal was normalized to cell count. In parallel, RNA was extracted after 9 d from cells differentiated in the same manner and analyzed by RT-PCR for a genetic marker of chondrocyte differentiation.
ChIP-Rx and WGBS.
ChIP-Rx and WGBS were performed as described previously (10, 24). Antibodies used for the protocol included anti-H3K27ac: Active Motif (39133), lot 31814008, 10 micrograms per sample; anti-H3K4me1: Abcam (ab8895), lot GR3206285-1, 6 micrograms per sample; anti-H3K36me2: Cell Signaling Tech (2901), lot 5, 20 microliters per sample; and anti-H3K27me3: Cell Signaling Tech (9733). Libraries were prepared according to instructions from Illumina. Paired-end sequencing was performed on the Illumina HiSeq 2000/4000.
Western Blot.
Western blot was performed as previously described (10). Antibodies used for the protocol included anti-H3K36me3: Active Motif (61101); anti-H3K36me2: Cell Signaling Tech (2901); anti-H3: Abcam (ab1791); anti–phospho-STAT1 (Tyr701): Cell Signaling Tech (9167); anti-STAT1: Cell Signaling Tech (9172); and anti–beta-actin: Abcam (ab8224).
RNA Extraction and Quantitative RT-PCR.
RNA was harvested using TRIzol reagent. The Illumina TruSeq RNA Sample Prep Kit (FC-122-1001) was used with 1 μg of total RNA for the construction of sequencing libraries. RNA libraries were prepared with ribosomal RNA depletion according to instructions from the manufacturer (NEB). Paired-end sequencing was performed on the Illumina HiSeq 2500. Total RNA was reverse transcribed using the SuperScript IV First Strand System (Life Technologies). Quantitative RT-PCR was performed as described previously (10). The primer sets have been used previously (44). IRF7: forward (F) 5′-CAGCAGCAGTCTCGGCTTGTG-3′, reverse (R) 5′-TGACCCAGGTCCATGAGGAAGTG-3′; and Oasl1: F 5′-CCAGGAAGAAGCCAAGCACCATC-3′, R 5′-AGGTTACTGAGCCCAAGGTCCATC-3′.
Proliferation Assays.
Cell lines were plated in triplicate on 96-well plates at low density (∼10% confluency) for each condition. The next day, cells were treated using the indicated small molecules solubilized in dimethyl sulfoxide (DMSO) for 48 h with daily media changes. Following drug treatment, cells were trypsinized, harvested, and replated at 1:10 dilution onto new 96-well plates. Cell viability was determined 3 d later using the CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer’s guidelines.
Conditioned Media Assays.
Parental and H3K36M-expressing C3H10T1/2 cell lines were plated at low density (∼10% confluency) and treated the next day with DMSO or 100 nM decitabine for 48 h with daily media changes. Following drug treatment, cells were grown for another 48 h in fresh media in the absence of inhibitor. Conditioned media from both lines were harvested and incubated for 1 h with untreated parental C3H10T1/2 cells. Both drug-treated and conditioned media-treated cell lines were harvested for subsequent immunoblot analysis. See SI Appendix, Fig. S3B for a schematic.
ATAC-Seq.
For each cell line, 50,000 cells were harvested, washed in phosphate-buffered saline, and lysed in 50 μL of lysis buffer (10 mM Tris⋅HCl, 10 mM NaCl, 3 mM MgCl2, 0.1% [volume/volume] IGEPAL CA-630, pH 7.4) for 5 min on ice followed by centrifugation (500 × g at 4 °C for 10 min) and removal of supernatant. Transposition and library preparation were performed using the Nextera Kit (Illumina; F-121-1030) and Index Kit (Illumina; FC-121-1011). Cells were resuspended in 50 μL transposition reaction mix (2.5 μL TDE1 Tn5 transposase and 1 μL 0.5% digitonin in 1× Tagment DNA buffer) and incubated at 37 °C for 30 min. The reaction was quenched using 1 μL 10% sodium dodecyl sulfate and bead purification was performed using Agencourt AmpureXP beads (Beckman Coulter; A63881) according to the manufacturer’s instructions. Tagmented DNA was amplified using i7 and i5 indexing adapters using a five-cycle PCR program according to the manufacturer’s instructions, size-selected using AmpureXP beads to remove species >600 bp, and then normalized and pooled for sequencing. ATAC-seq libraries were sequenced using the Illumina HiSeq 2500.
ChIP-Seq Data Analysis.
ChIP-seq reads of C3H10T1/2 cell samples were mapped to mouse genome assembly mm10 using HISAT2 (v2.1.0). Peaks of H3K27ac ChIP-seq data are called using macs (v1.4) with immunoglobulin G input as control. The H3K27ac peaks are mapped to promoter regions by the function “map” of bedtools (v2.27.1) and are further identified as promoters and enhancers based on the mapping results. The distribution of ChIP-seq signals on promoter or enhancer regions is measured and visualized by the functions “computeMatrix” and “plotProfile” of deepTools (v3.3.2). The genomic distribution correlations of ChIP-seq signals are calculated and visualized by the function “plotCorrelation” of deepTools (v3.3.2).
RNA-Seq Data Analysis.
RNA-seq reads of C3H10T1/2 cell samples were mapped to mouse genome assembly mm10 using HISAT2 (v2.1.0). The mapped read count of each gene was measured by featureCounts (v1.6.1). The differential gene expression was calculated and visualized by the R packages DESeq2 (v1.28.0) and ggplot2 (v3.2.1). We performed hierarchical clustering on correlation among gene expression profiles of samples using the R package pheatmap (Pretty Heatmaps v1.0.10) with parameters clustering method = average, clustering_distance_cols = euclidean, and cutree_cols = 3. Distance (d) was assessed using d = 1 − r, where r is the Pearson product-moment coefficient. Gene set enrichment analysis based on Gene Ontology and MSigDB Hallmark annotation knowledgebases was performed by BiNGO (v3.0.3), which is integrated in Cytoscape (v3.4.0) and Enrichr (https://maayanlab.cloud/Enrichr).
Supplementary Material
Acknowledgments
We thank members of the C.L., J.M., and C.D.A. laboratories for critical reading of the manuscript. This research was supported by NIH grants (P01CA196539 to C.L., C.D.A., and J.M.; R35GM138181 to C.L.; T32GM007739 and F30CA224971 to D.N.W.; and T32HL105323 to K.N.R.) and the Rockefeller University (C.D.A.). This work was performed within the context of the I-CHANGE consortium and supported by funding from Genome Canada, Genome Quebec, and the Institute for Cancer Research of the Canadian Institutes of Health Research, McGill University, and the Montreal Children’s Hospital Foundation. C.L. is the Giannandrea Family Dale F. Frey Breakthrough Scientist of the Damon Runyon Foundation (DFS-28-18) and a Pew-Stewart Scholar for Cancer Research, and is supported by an American Association for Cancer Research Gertrude B. Elion Cancer Research Grant.
Footnotes
Competing interest statement: C.D.A. and B.D.S. are coauthors on the following article: M. J. Slaughter et al., HDAC inhibitors result in widespread alteration of the histone acetylation landscape and BRD4 targeting to gene bodies. Cell Rep. 34, 108638 (2021).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021795118/-/DCSupplemental.
Data Availability
The genomic data including RNA-seq, ChIP-seq, WGBS, and ATAC-seq reported in this article have been deposited in the Gene Expression Omnibus (accession nos. GSE160266 and GSE118785).
References
- 1.Lowe B. R., Maxham L. A., Hamey J. J., Wilkins M. R., Partridge J. F., Histone H3 mutations: An updated view of their role in chromatin deregulation and cancer. Cancers (Basel) 11, 660 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behjati S., et al., Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet. 45, 1479–1482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kernohan K. D.et al.; Care4Rare Canada Consortium , H3.1 K36M mutation in a congenital-onset soft tissue neoplasm. Pediatr. Blood Cancer 64, e26633 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Snuderl M., et al., Histone H3K36I mutation in a metastatic histiocytic tumor of the skull and response to sarcoma chemotherapy. Cold Spring Harb. Mol. Case Stud. 5, a004606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein B. J., et al., Recognition of cancer mutations in histone H3K36 by epigenetic writers and readers. Epigenetics 13, 683–692 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papillon-Cavanagh S., et al., Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat. Genet. 49, 180–185 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang S., et al., Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase. Genes Dev. 30, 1611–1616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y., et al., Molecular basis for the role of oncogenic histone mutations in modulating H3K36 methylation. Sci. Rep. 7, 43906 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang D., et al., The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science 352, 1344–1348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu C., et al., Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352, 844–849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhuang L., et al., Depletion of Nsd2-mediated histone H3K36 methylation impairs adipose tissue development and function. Nat. Commun. 9, 1796 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brumbaugh J., et al., Inducible histone K-to-M mutations are dynamic tools to probe the physiological role of site-specific histone methylation in vitro and in vivo. Nat. Cell Biol. 21, 1449–1461 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang D., Gan H., Wang H., Zhou H., Zhang Z., Probe the function of histone lysine 36 methylation using histone H3 lysine 36 to methionine mutant transgene in mammalian cells. Cell Cycle 16, 1781–1789 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan W., et al., H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 286, 7983–7989 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitges F. W., et al., Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42, 330–341 (2011). [DOI] [PubMed] [Google Scholar]
- 16.McDaniel S. L., Strahl B. D., Shaping the cellular landscape with Set2/SETD2 methylation. Cell. Mol. Life Sci. 74, 3317–3334 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao B., Shibata Y., Strahl B. D., Lieb J. D., Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol. Cell. Biol. 25, 9447–9459 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kizer K. O., et al., A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 25, 3305–3316 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrozza M. J., et al., Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123, 581–592 (2005). [DOI] [PubMed] [Google Scholar]
- 20.An S., Yeo K. J., Jeon Y. H., Song J. J., Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism. J. Biol. Chem. 286, 8369–8374 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo A. J., et al., NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol. Cell 44, 609–620 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikkelsen T. S., et al., Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J., Ahn J. H., Wang G. G., Understanding histone H3 lysine 36 methylation and its deregulation in disease. Cell. Mol. Life Sci. 76, 2899–2916 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinberg D. N., et al., The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 573, 281–286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddington J. P., et al., Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of polycomb target genes. Genome Biol. 14, R25 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinberg D. N., Allis C. D., Lu C., Oncogenic mechanisms of histone H3 mutations. Cold Spring Harb. Perspect. Med. 7, a026443 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reese N. D., Schiller G. J., High-dose cytarabine (HD araC) in the treatment of leukemias: A review. Curr. Hematol. Malig. Rep. 8, 141–148 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Pfister S. X., et al., Inhibiting WEE1 selectively kills histone H3K36me3-deficient cancers by dNTP starvation. Cancer Cell 28, 557–568 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiappinelli K. B., et al., Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu M., et al., Dual inhibition of DNA and histone methyltransferases increases viral mimicry in ovarian cancer cells. Cancer Res. 78, 5754–5766 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stone M. L., et al., Epigenetic therapy activates type I interferon signaling in murine ovarian cancer to reduce immunosuppression and tumor burden. Proc. Natl. Acad. Sci. U.S.A. 114, E10981–E10990 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roulois D., et al., DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bohl S. R., Bullinger L., Rücker F. G., Epigenetic therapy: Azacytidine and decitabine in acute myeloid leukemia. Expert Rev. Hematol. 11, 361–371 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Barrat F. J., Crow M. K., Ivashkiv L. B., Interferon target-gene expression and epigenomic signatures in health and disease. Nat. Immunol. 20, 1574–1583 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duns G., et al., Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 70, 4287–4291 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Sankaran S. M., Gozani O., Characterization of H3.3K36M as a tool to study H3K36 methylation in cancer cells. Epigenetics 12, 917–922 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan S., et al., Global regulation of the histone mark H3K36me2 underlies epithelial plasticity and metastatic progression. Cancer Discov. 10, 854–871 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lhoumaud P., et al., NSD2 overexpression drives clustered chromatin and transcriptional changes in a subset of insulated domains. Nat. Commun. 10, 4843 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu W., et al., DNMT3A reads and connects histone H3K36me2 to DNA methylation. Protein Cell 11, 150–154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wojcik J. B., et al., Epigenomic reordering induced by polycomb loss drives oncogenesis but leads to therapeutic vulnerabilities in malignant peripheral nerve sheath tumors. Cancer Res. 79, 3205–3219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krug B., et al., Pervasive H3K27 acetylation leads to ERV expression and a therapeutic vulnerability in H3K27M gliomas. Cancer Cell 36, 338–339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan J., et al., EZH2 phosphorylation by JAK3 mediates a switch to noncanonical function in natural killer/T-cell lymphoma. Blood 128, 948–958 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Pan Y. M., et al., STAT3 signaling drives EZH2 transcriptional activation and mediates poor prognosis in gastric cancer. Mol. Cancer 15, 79 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee M. S., Park C. H., Jeong Y. H., Kim Y. J., Ha S. J., Negative regulation of type I IFN expression by OASL1 permits chronic viral infection and CD8+ T-cell exhaustion. PLoS Pathog. 9, e1003478 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomic data including RNA-seq, ChIP-seq, WGBS, and ATAC-seq reported in this article have been deposited in the Gene Expression Omnibus (accession nos. GSE160266 and GSE118785).