Significance
We estimate the origin and spread of SARS-CoV-2 in Europe prior to spring 2020 border closures based on viral genome sequences using a phylodynamic model with geographic structure. We confirm that the predominant European outbreak most likely started in Italy and spread from there. This outbreak was probably seeded by a transmission event in either Hubei, China or Germany. In particular, we find that before the first border closures in Europe, the rate of new cases occurring from within-country transmission was within or exceeded the estimated bounds on the rate of new migration cases.
Keywords: SARS-CoV-2, transmission, disease import, phylogeography
Abstract
The investigation of migratory patterns during the SARS-CoV-2 pandemic before spring 2020 border closures in Europe is a crucial first step toward an in-depth evaluation of border closure policies. Here we analyze viral genome sequences using a phylodynamic model with geographic structure to estimate the origin and spread of SARS-CoV-2 in Europe prior to border closures. Based on SARS-CoV-2 genomes, we reconstruct a partial transmission tree of the early pandemic and coinfer the geographic location of ancestral lineages as well as the number of migration events into and between European regions. We find that the predominant lineage spreading in Europe during this time has a most recent common ancestor in Italy and was probably seeded by a transmission event in either Hubei, China or Germany. We do not find evidence for preferential migration paths from Hubei into different European regions or from each European region to the others. Sustained local transmission is first evident in Italy and then shortly thereafter in the other European regions considered. Before the first border closures in Europe, we estimate that the rate of occurrence of new cases from within-country transmission was within the bounds of the estimated rate of new cases from migration. In summary, our analysis offers a view on the early state of the epidemic in Europe and on migration patterns of the virus before border closures. This information will enable further study of the necessity and timeliness of border closures.
In response to the pandemic potential of the SARS-CoV-2 virus, many nations closed their borders in spring 2020 to curb the virus’ spread (1). These closures incurred high economic and social costs. To weigh the relative costs and benefits of border closures, it will be important to understand the efficacy of these policies. At the early stages of an outbreak, border closures can delay a pathogen’s arrival, thereby giving countries additional time to prepare (2). However, the success of this strategy depends on timely implementation and a good knowledge of where the pathogen is already circulating. To evaluate the efficacy of border closures in limiting the spread of SARS-CoV-2, it is important to reconstruct the timeline of the early international spread of the virus, before such policies were implemented.
In this analysis, we aim to estimate the early patterns of SARS-CoV-2 transmission into and across Europe. We also address the more specific question of where the predominant SARS-CoV-2 lineage circulating in Europe originated. We hope that by addressing these questions we can inform further analysis of the efficacy of border closures as a strategy to combat SARS-CoV-2.
The SARS-CoV-2 virus was identified as the cause of an epidemic in Wuhan, China in late 2019 (3). The epidemic in Wuhan was reported to the World Health Organization (WHO) on 31 December 2019 and within 1 mo, SARS-CoV-2 was confirmed to have spread to 19 additional countries (4). By the end of February 2020, the virus was detected in all WHO regions (https://covid19.who.int/). By late spring 2020, several lineages of the SARS-CoV-2 virus were circulating across the globe. The intermixing of these lineages in different countries and regions suggests that the virus was transmitted across borders many times (https://nextstrain.org/ncov/global).
Here we focus on estimating the early introductions of SARS-CoV-2 into Europe and the virus’ migration across European borders. Through national surveillance efforts, the first COVID-19 cases in Europe were detected in France on 24 January 2020 and in Germany on 28 January 2020 (5, 6). Of the 47 cases detected in Europe by 21 February 2020, 14 were infected in China, 14 were linked to the initial cases in Germany, 7 were linked to the initial cases in France, and 12 were of unknown origin (5). In addition to the unknown sources of transmission, some early introductions may not have been detected. This is especially probable given that a significant proportion of infected individuals are likely to be asymptomatic (7). In summary, it is difficult to draw firm conclusions about the source, number, and timing of SARS-CoV-2 introductions into Europe based on confirmed case data alone.
Viral genomes are an important secondary source of information on outbreak dynamics. If viruses acquire mutations on the same timescale as an outbreak, these mutations can provide information about past transmission events. Phylodynamic methods couple a model of viral evolution describing the mutational process to an epidemiological model describing the transmission process. By fitting the combined model to viral genomes sampled from a cohort of infected individuals, we can infer the evolutionary and epidemiological model parameters. Here we fit a phylodynamic model with geographic structure to SARS-CoV-2 genomes from Hubei, China and 19 European countries before the first borders were closed in these regions. We coinfer the transmission tree linking these sequences, the geographic location of ancestral lineages, migration rates of infected individuals between regions, the effective reproductive number, and the proportion of no-longer infectious cases sequenced in each region.
In addition to these inferences, we specifically focus on estimating the geographic origin of the predominant SARS-CoV-2 lineage in Europe. This lineage is defined by a characteristic amino acid substitution at position 314 in the ORF1b gene from proline to leucine and was provisionally named the A2a lineage by the Nextstrain team, later renamed to 20A. In the more dynamic, tree-based “pangolin” nomenclature suggested by Rambaut et al. (8), this lineage corresponds to the B.1 lineage described as “A large European lineage that corresponds to the Italian outbreak.” (9). As of 1 April 2020, two-thirds of the SARS-CoV-2 sequences collected in Europe belonged to this lineage and just 10% of sequences from the lineage were collected outside Europe [data from https://www.gisaid.org/; lineages assigned using Nextstrain (10)]. Here, we use the name A2a to refer to the group of SARS-CoV-2 viruses defined by the ORF1b:P314L mutation.
The origin of the A2a lineage was initially controversial, with conflicting reports in the academic and media press (11–14). Its characteristic ORF1b:P314L mutation was found in some of the earliest confirmed COVID-19 cases in Italy, Switzerland, Germany, Finland, Mexico, and Brazil in late February (11, 12). Intriguingly, a late-January sample from a cluster of infections in Bavaria, Germany linked to business travel from Shanghai, China (15, 16) shares a mutation at site 614 in the S gene with the A2a lineage, but does not have the A2a lineage-defining ORF1b:P314L mutation. This German sample is part of a smaller clade that is closely related to the larger clade of A2a sequences and which was originally named the A2 lineage but was later included in the larger 19A (Nextstrain nomenclature) or B (pangolin nomenclature) lineage (https://nextstrain.org/ncov/global). As a result, it was hypothesized that a German transmission cluster may have seeded the larger European outbreak (11–13). However, it was quickly pointed out that incomplete and biased sampling must be taken into account before this hypothesis can be rigorously addressed (12, 14, 17).
Phylodynamic models with geographic structure aim to account for such biases. First, parameter estimates are generated by integrating over a distribution of potential phylogenies, which acknowledges that we cannot reconstruct the true transmission tree with certainty. Second, sampling parameters are allowed to differ between regions, which acknowledges that testing and sequencing resources vary across regions. Here, we fit a phylodynamic model with geographic structure to full-length SARS-CoV-2 genomes collected before 8 March 2020 to: 1) Estimate the early patterns of SARS-CoV-2 spread into and across Europe, 2) weigh genomic evidence for competing hypotheses about the geographic origin of the predominant A2a lineage in Europe, 3) report on the epidemiological parameters, and 4) compare the rate of new cases arising from within-region transmission versus migration during the early epidemic.
Results
Testing Assumptions about Source and Sink Locations.
We assume that during the time span considered, the outbreak in Hubei, China and the different European outbreaks were only sources and not sinks for SARS-CoV-2 globally. The first assumption follows from the fact that Hubei is the location of the pandemic origin (see Materials and Methods for additional rationale). To test our second assumption that Europe was primarily a source and not a sink of infections before 8 March 2020, we analyzed A2a sequences collected from different global regions on or before that date. We aggregated sequences into five demes: Africa, Asia and Oceania, Europe, North America, and South and Central America (SI Appendix, Table S3), and then fit the multitype birth–death model described in the Materials and Methods to these data. The most recent common ancestor of the global set of A2a sequences was inferred to be in Europe with 95% posterior probability (SI Appendix, Fig. S5). The posterior distributions for the migration rates into Europe closely matched the prior, thus the data contain little information on these rates (SI Appendix, Fig. S6). However, in the analyzed dataset, 0 introduction events were inferred from other parts of the world into Europe, while in total 24 migration events were inferred from Europe to other parts of the world (SI Appendix, Table S5).
Inference Results.
SARS-CoV-2 transmission into and across Europe.
For our main analysis, we focused on estimating patterns of SARS-CoV-2 transmission into and across Europe. Based on the particular set of sequences analyzed, we infer that SARS-CoV-2 was introduced from Hubei into France, Germany, Italy, and other European countries approximately two to four times each before 8 March 2020 (Table 1). The largest number of estimated introductions was 18 from Italy to other European countries. Importantly, these estimates reflect only introductions occurring in the transmission history of the analyzed cases, not the full epidemic. In contrast, the inferred migration rate parameters should describe more general patterns of spread between regions. The sequence data were informative for inferring some, but not all, migration rates. We highlight here only the rates for which the data are the most informative (see SI Appendix, Fig. S1 for a full comparison of posterior and prior distributions). The highest migration rate was inferred to be from Italy into other European countries, with a median rate of 3.7/y. The lowest migration rate was from Italy to Germany, with a median rate of 0.43/y. We can translate these rates into the probability of an infected individual migrating using the fact that migration is modeled as a Poisson process. That is, we infer it is 10 times more likely that an infected individual traveled from Italy to a country in the “other European” region than to Germany. However, we note that the magnitude of the rates may be skewed by a bias toward genome sampling among recently returned travelers.
Table 1.
Median inferred number of introductions from each source region to each sink region along the transmission tree linking analyzed cases
| Source/sink | France | Germany | Italy | Other European |
| Hubei | 3 (0, 6) | 4 (1, 6) | 2 (0, 6) | 4 (0, 8) |
| France | — | 0 (0, 1) | 0 (0, 3) | 2 (0, 4) |
| Germany | 0 (0, 2) | — | 1 (0, 3) | 1 (0, 4) |
| Italy | 6 (1, 9) | 1 (0, 4) | — | 18 (6, 34) |
| Other European | 2 (0, 6) | 1 (0, 4) | 1 (0, 4) | — |
Hubei is assumed to be a source only. Values in parenthesis are the upper and lower bound of the 95% highest posterior density interval for these estimates.
A2a lineage origin.
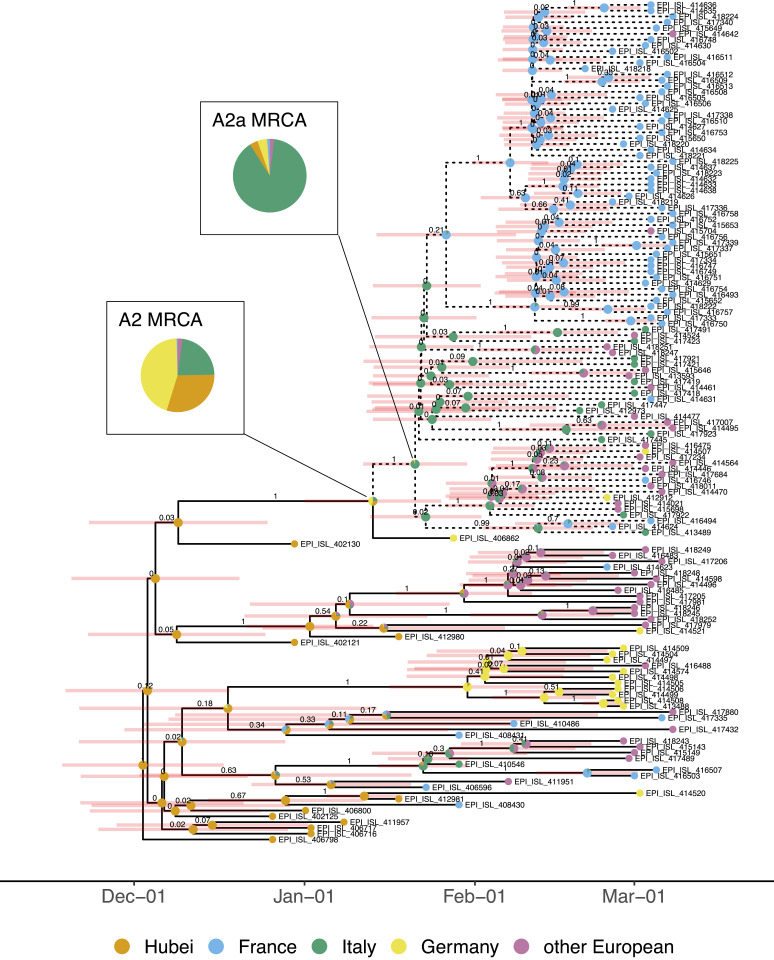
The maximum-clade credibility tree in Fig. 1 summarizes the posterior sample of transmission trees linking analyzed sequences. The A2a lineage sequences form a clear clade with posterior probability of 1. The most recent common ancestor of the analyzed A2a sequences is estimated to be in Italy with 89% posterior probability. In contrast, the location of the most recent common ancestor between this clade and the A2 Shanghai-linked German sequence is less certain. This ancestor is inferred to have been in either Germany (45% posterior probability), Hubei (30%), or Italy (23%). It is very improbable that this ancestor was in France or another European country (2% posterior probability). Using a lower prior for migration rates (results shown in SI Appendix, Fig. S8), Hubei is more likely to be the location of this ancestor than Germany (62% posterior probability for Hubei, 16% for Germany).
Fig. 1.
Maximum-clade credibility tree. The clade of A2a sequences analyzed is highlighted with dashed branches. The values above the branches are the posterior clade probabilities and the pale red bars show the 95% highest posterior density interval for node ages. The pie charts at nodes show posterior probability for the ancestor being located in each region (note that we assumed the root of the tree was in Hubei with probability 1). The region for each tip is the region in which the sequence was collected, irrespective of travel history. Tips are annotated with GISAID accession identifier.
Epidemiological parameters.
Several epidemiologically relevant parameters were coinferred along with the transmission tree. First, we report on the reproductive number in the different regions, which varied from 1.2 to 1.9 in Hubei to 2.5 to 3.5 in France (SI Appendix, Fig. S2A). Second, we report on the prevalence of no-longer infectious cases in each region as of the collection date of the last analyzed sequence. This quantity can be back-calculated from the inferred sampling proportion (prevalence = no. sequences analyzed/sampling proportion). We note that both the sampling proportion and prevalence estimates have large credible intervals (SI Appendix, Fig. S2 B and C). Of the European regions analyzed, the outbreak in Germany was estimated to be smaller in early March (150 to 485 cumulative cases) than the outbreaks in France (709 to 2,185 cases) and other European countries (719 to 1,782 cases), while the outbreak in Italy was the largest (2,600 to 4,923 cases).
Comparing Rates of Migration and within-Region Transmission.
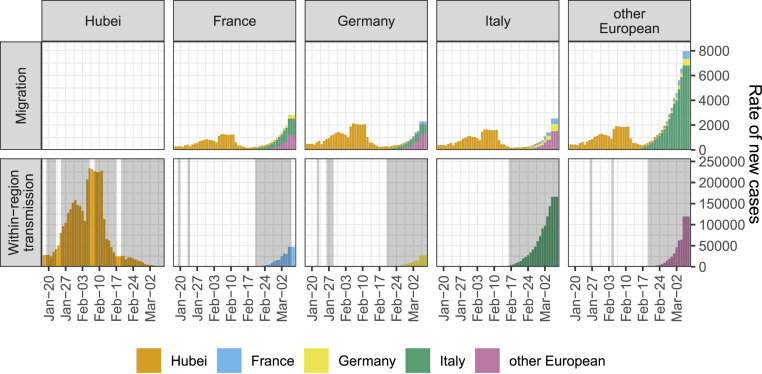
Fig. 2 compares the rate at which we estimate new cases to arise in each region from migration versus from within-region transmission. The estimated rates of new cases from migration and within-region transmission are represented here as point estimates 5 d before the date of case confirmation, which assumes a 5-d delay between infection and onward transmission or migration [the choice of 5 d is motivated by serial interval estimates for SARS-CoV-2 (18)]. We emphasize that we do not consider any non-European regions beyond Hubei; therefore, transmission from Hubei to a not-included location and then to Europe is considered to be migration directly from Hubei to Europe under our model.
Fig. 2.
Estimated rate of new cases arising from migration compared with the estimated rate of new cases arising from within-region transmission. For each day, we multiplied the (smoothed) number of newly confirmed cases in each source region by the posterior sample of migration rates from source to sink. The median of these rates is shown in the “Migration” row. We also multiplied the (smoothed) number of newly confirmed cases in each sink region by the posterior sample of transmission rates for the region. The median of these rates is shown in the “Within-region transmission” row. Gray shaded regions indicate dates on which new cases were reported in each region. Dates are lagged 5 d to account for a 5-d delay between infection and migration or onward transmission and daily case counts were smoothed by taking a rolling 7-d average. Case data comes from the Johns Hopkins Center for Systems Science and Engineering (https://github.com/CSSEGISandData/COVID-19).
Beginning with the first day on which we have case data from Hubei, we estimate a substantial risk of infected individuals migrating from Hubei into European regions. Throughout late January to mid-February 2020, cases were sporadically detected in each European region, each of which is associated with a risk of subsequent within-region transmission. Sustained within-region transmission is first evident in Italy in mid-February. Shortly thereafter, sustained within-region transmission occurred in other European countries, in France and in Germany. By 8 March 2020, the estimated rate of occurrence of new cases from within-region transmission is within or exceeds the estimated bounds on the rate of new cases from migration for each region considered (SI Appendix, Fig. S7A). We obtain the same qualitative result in our sensitivity analysis using a very different prior on the migration rate (SI Appendix, Fig. S7B). We note that the rates in Fig. 2 are underestimates of the rates of new cases arising due to migration or transmission due to the underreporting in the confirmed case data. However, assuming that the amount of underreporting is comparable across regions, we can indeed compare the rates.
Finally, we report support for a decrease in migration rates from Hubei into European regions at the date of the lockdown of Wuhan (SI Appendix, Fig. S1). We infer that migration decreased by 40% (95% highest posterior density interval 0–87%). Again, we note that the migration rate out of Hubei is not necessarily specific to Hubei, since we do not consider possible migration paths through other non-European locations.
Discussion
We inferred the early spread of the SARS-CoV-2 virus into and across Europe as well as the geographic origin of the predominant A2a lineage spreading in Europe. To do this, we applied a previously published phylodynamic model to analyze publicly available viral genome sequences from the epidemic origin in Hubei, China and from the earliest detected and largest European outbreaks before 8 March 2020. After performing Bayesian inference, we: 1) Report on inferred patterns of SARS-CoV-2 spread into and across Europe, 2) compare posterior probabilities for several hypotheses on the origin of the A2a lineage, 3) report on epidemiological parameters, and 4) compare the timeline of new cases resulting from migration versus within-region transmission in Europe before borders were closed.
Genome sequence data indicates that prior to 8 March 2020, SARS-CoV-2 was introduced from Hubei province into France, Germany, Italy, and other European countries at least two to four times each (Table 1). These estimates, which are based on genome sequence data and thus do not rely on having line list data for individual migration cases, provide a complementary account of introduction events compared to line list data (19) and phylogenetic inferences combining genome sequence and line list data (20–25). The introduction events we report here are inferred to have occurred along the transmission tree specific to the analyzed sequence set and are not attributable to individual cases. In comparison, line list data (5, 19) attributes introduction events to individual cases but cannot reconstruct previous, unobserved introductions. Since we analyze only a fraction of all cases, we expect our estimates to be a lower bound on the true number of introductions.
Ideally, we want to go beyond counting migration events among the analyzed sequences and investigate general dynamics. To do this, we would interpret inferred migration rates as representing more general patterns of SARS-CoV-2 spread. However, the sequence data were only informative for inferring some of these rates (SI Appendix, Fig. S1). In regions with few lineages circulating during the period considered, there is little signal for the amount of outward migration. We observe information about the per individual migration rate from Italy to other European countries (SI Appendix, Fig. S1). However, we do not find evidence for preferential migration paths from Hubei into different European regions or from each European region to the others, although we cannot exclude this possibility.
We estimate that the A2a viruses spreading in Europe by 8 March 2020 had a common ancestor in Italy sometime between mid-January and early February 2020 (Fig. 1). In contrast, at the time of this paper’s original submission, Nextstrain placed this ancestor in the United Kingdom with 100% confidence (https://nextstrain.org/ncov/europe). This Nextstrain result may have been an artifact of disproportionately high sequencing effort in the United Kingdom since biased sampling violates the assumptions of the “mugration” method employed (26). We additionally report that the A2a lineage was most likely carried from Hubei to Italy or from Hubei to Italy via Germany. Both transmission routes have substantial posterior probability under our main model assumptions (Fig. 1). Assuming a lower migration rate prior, transmission from Hubei to Italy instead of a route via Germany to Italy becomes the more likely scenario (SI Appendix, Fig. S9). Addressing the same question, recently developed phylodynamic methods accounting for undersampling and utilizing travel information from line list data have provided even stronger evidence for independent introductions from China into Germany and Italy instead of a route via Germany to Italy (27).
Although it is not the main focus of our analysis, we also report on epidemiological parameters of the early outbreaks considered. Estimates for the reproductive number fall roughly within the range of previous estimates (28), although we mention a particular caveat with respect to the reproductive number in Hubei below. Unsurprisingly, prevalence estimates in early March generally exceed confirmed case counts by a factor of 1 to 3 (SI Appendix, Fig. S4C). Our inferences of epidemiological parameters support the idea that the early reproductive number in different outbreaks is difficult to estimate precisely, but not hugely variable, and that there is substantial underreporting in line list data (29).
Finally, we estimated the rate of new cases arising from migration compared with the rate of new cases arising from within-region transmission in the regions analyzed. The magnitudes of these rates are quite uncertain due to uncertainty in the inferred migration and transmission rates (SI Appendix, Fig. S7) and underreporting in case counts, which we implicitly assume to be constant in time and between regions. However, the temporal trends suggested by these data are still compelling and robust toward different prior assumptions. We see that under sustained risk of case migration from abroad, isolated cases were confirmed throughout Europe beginning in late January 2020 but did not immediately cause large outbreaks. Shortly after the first evidence of sustained within-region transmission in Italy, outbreaks in the rest of Europe also took hold (Fig. 2).
Our results based on the multitype birth–death model take into account phylogenetic uncertainty and sampling biases between regions, which are two major concerns in genomic analyses of SARS-CoV-2 (14). Indeed, wide confidence intervals around internal nodes in the maximum-clade credibility tree and low clade support near the tips (Fig. 1) indicate a high degree of phylogenetic uncertainty. Therefore, it is important that the parameter estimates we report result from integrating over a distribution of potential phylogenies with different geographic locations assigned to ancestral lineages. In comparison, some initial studies that estimated international SARS-CoV-2 spread constructed a median-joining network instead of a phylogeny to account for this uncertainty (13, 30). In this approach, identical sequences are collapsed to single nodes and edges represent mutational differences. This disregards information from relative sampling times and means that ancestor-descendent relationships are highly dependent on the choice of the network root (31, 32). Unaccounted-for sampling biases in these analyses may also yield spurious results for the geographic origin of lineages (33, 34). Our analysis, which relies on a mechanistic model of migration and between-region sampling differences, should be robust to such biases.
Despite the advantages of the multitype birth–death model just mentioned, there are also several unique caveats to consider. The birth–death model assumes uniform-at-random sampling from the total infected population in each region. However, particularly in the early stages of outbreaks, infected individuals were identified by health ministries via contact tracing (5). Nonrandom sampling may be one possible explanation for why we infer markedly different transmission rates in China when analyzing cases from within Hubei (as in this analysis) as opposed to cases exposed in Hubei but sequenced elsewhere [as in our previous analysis (35)]. Furthermore, the multitype birth–death model assumes that parameters are constant through time and homogenous within regions. As a result, our inferences based on province-, country-, and continent-level regions are only coarse approximations of the true, heterogeneous epidemic dynamics occurring at a local level. Due to these limitations, we focus on estimating and interpreting particular events along the transmission tree of the analyzed sequences (e.g., Fig. 1 and Table 1) and advise caution when interpreting inferred migration rates (e.g., SI Appendix, Fig. S1).
We expect that our results will be useful in parameterizing more specialized models aimed at understanding the efficacy of border closures as a means to fight pandemic disease. So far, such analyses have primarily used line list data and information on travel networks to estimate SARS-CoV-2 migration patterns (36–38). Here we present independent estimates of migration patterns based on genome sequence data. By combining case count data and our estimates for migration and transmission rates, we provide a timeline of early SARS-CoV-2 introduction and spread before border closures were implemented. Despite migration risk from outside Europe being on the same order-of-magnitude as later migration risk from Italy, we only observe sustained outbreaks in other European regions after the onset of sustained within-region transmission in Italy. Finally, before the first border closures in Europe, we estimate the risk of new cases arising from within-region transmission to be within or exceeding the estimated range for the risk of new migration cases.
Materials and Methods
Model.
We fit a simplified version of the multitype birth–death model described in Scire et al. (39). Under this model, beginning with a single infected host in a single geographic region (deme), the virus can be transmitted from one host to another (a birth event), die out due to host recovery or death (a death event), be sequenced (a sampling event, assumed to correspond to a death event), or migrate from one deme to another (a migration event). The birth, death, and sampling processes are assumed to occur at deme-specific rates that are constant through time. Importantly, this model aims to capture heterogeneity in epidemiological parameters (birth and death rates) and sequencing effort (sampling proportion) among demes. Additionally, there is a unique migration rate from each deme to each other deme. All migration rates are assumed to be constant through time except for migration out of Hubei. In our main analysis, migration out of Hubei is assumed to be constant before and after the date of lockdown on 23 January 2020 and is assumed to decrease by a constant factor at the date of lockdown. This factor is a parameter of the model and is also inferred based on the genome sequence data. Finally, we used a version of the model parameterized in terms of the effective reproductive number, which allows us to additionally infer this epidemiologically relevant quantity for each deme.
Dataset.
We analyzed SARS-CoV-2 genome sequences from five different demes: Hubei province in China, France, Germany, Italy, and a composite deme of other European countries (“other European”). All sequences were accessed from GISAID (https://www.gisaid.org/). To represent the pandemic origin, we randomly chose 10 sequences from Hubei collected on or before the lockdown of Wuhan city on 23 January 2020. To investigate the earliest outbreaks in Europe, we considered all available sequences collected in France, Germany, and Italy on or before the lockdown of the Lombardy region of Italy on 8 March 2020. These countries had the first detected (France and Germany) and the largest (Italy) early outbreaks in Europe (4, 5). By limiting sampling to before regional lockdowns and border closures went into effect, we hope to 1) satisfy model assumptions that epidemiological and migration parameters are constant through time, and 2) get a picture of the early, unimpeded spread of SARS-CoV-2 within Europe. To represent the pool of SARS-CoV-2 circulating in other European countries during this time, we randomly down-sampled sequences from other countries to the cumulative number of confirmed COVID-19 deaths in each country by 8 March 2020 plus one (SI Appendix, Table S1). We used this quantity as a proxy value roughly proportional to the outbreak size in each country. Table 2 characterizes the sequences analyzed from each deme for the main analysis. As a sensitivity analysis, we repeated the analysis while down-sampling based on confirmed death data from 28 March 2020, considering that deaths occur with a delay after transmission. This yielded a slightly larger sequence set for analysis. For this analysis, we also did not consider a change in migration rates out of Hubei at 23 January 2020 (results in SI Appendix).
Table 2.
Analyzed sequence information
| Region | No. sequences | Locations represented | First sequence date (mo/d/y) | Last sequence date (mo/d/y) |
| Hubei | 10 | Hubei province, China | 26/12/2019 | 18/01/2020 |
| France | 66 | France | 23/01/2020 | 08/03/2020 |
| Germany | 15 | Germany | 28/01/2020 | 03/03/2020 |
| Italy | 13 | Italy | 29/01/2020 | 04/03/2020 |
| Other European | 41 | Spain (15), Netherlands (4), United Kingdom (4), Switzerland (3), Belgium (1), Czech Republic (1), Denmark (1), Finland (1), Iceland (1), Ireland (1), Luxembourg (1), Norway (1), Poland (1), Portugal (1), Slovakia (1), Sweden (1) | 07/02/2020 | 08/03/2020 |
Location is the location of sample collection and date is the date of sample collection.
Alignment Generation.
We prepared a sequence alignment from data publicly available on GISAID (https://www.gisaid.org/) on 1 April 2020 using the Nextstrain pipeline for SARS-CoV-2 (10). Short sequences (<25,000 bases), sequences without fully specified collection dates, and sequences in the Nextstrain exclude list (40) (duplicate sequences from the same case, or with suspicious amounts of nucleotide divergence) were excluded. We aligned selected sequences to reference genome GenBank accession no. MN908947. To eliminate sites identified by the Nextstrain team as prone to sequencing errors (41), we masked the first 130 and final 50 sites from the alignment, as well as sites 18,529, 29,849, 29,851, and 29,853.
Testing Assumptions about Source and Sink Locations.
We assume that during the time span considered that once a strain was in Europe, the strain could have been transmitted from Europe to other global regions, but subsequent reintroductions of this strain did not occur. Similarly, we assumed strains were not reintroduced into Hubei. These assumptions allow us to ignore sequences from outside of Hubei and Europe. To justify the second assumption, we argue there was not sufficient time between the pandemic origin in Hubei and 23 January 2020 for a significant amount SARS-CoV-2 export, transmission outside Hubei, and subsequent reintroduction into Hubei. Furthermore, confirmed case data shows that Hubei province was the epicenter of the SARS-CoV-2 pandemic until this time, with comparatively less transmission occurring outside of the province than within it (4). To justify the first assumption, we tested whether there was evidence for significant migration into European demes by running a separate analysis on A2a SARS-CoV-2 sampled from all global regions (results in SI Appendix).
Parameter Inference.
For inferences, we used the implementation of the multitype birth–death model in the bdmm package (39, 42) in the BEAST2 software (43). Since this is a parameter-rich model, we fixed some parameters to improve the identifiability of others. The values for fixed parameters, priors for estimated parameters, and the rationale behind these decisions are given in Table 3. We ran four Markov chain Monte Carlo chains to approximate the posterior distribution of the model parameters. The first 10% of samples from each chain were discarded as burn-in before samples from the chains were pooled. We used Tracer (44) to assess the convergence and confirm that the effective sample size (ESS) was >200 for all parameters.
Table 3.
Values and priors for the parameters of the multitype birth–death model
| Parameter | Value or prior | Rationale |
| Nucleotide substitution model | HKY + Γ | Unequal transition/transversion rates, unequal base frequencies, rate heterogeneity among sites |
| Clock rate | 0.0008 | Approximately 24 mutations per year (10) |
| Death rate | 36.5 y−1 | Period between infection and becoming uninfectious assumed exponentially distributed with a mean of 10 d |
| Sampling start time | 23 December 2019 | Just before date of first sample |
| Sampling end time (Hubei only) | 23 January 2019 | Only included sequences collected until lockdown |
| Location of origin | Hubei | Putative pandemic origin |
| Reproductive number | Lognormal (0.8, 0.5) | Median 2.2, 95% IQR 0.8 to 5.9 |
| Migration rates | Lognormal (0, 1) | Median time until travel is 1 y, 95% IQR 51 d to 7.1 y |
| Migration rate decrease from Hubei at lockdown | Uniform (0, 1) | Migration out of the Hubei deme is expected to decrease after lockdown |
| Time of origin | Lognormal (−1, 0.2) | Median 26 October, 95% IQR 22 August to 8 December 2019 |
| Sampling proportion | Upper bounds based on confirmed cases: | |
| Hubei | Uniform (0, 0.15) | 10 of 66 cases on 18 January 2020 |
| France | Uniform (0, 0.093) | 66 of 706 cases on 8 March 2020 |
| Germany | Uniform (0, 0.10) | 15 of 157 cases on 3 March 2020 |
| Italy | Uniform (0, 0.005) | 13 of 2,502 cases on 4 March 2020 |
| Other European | Uniform (0, 0.057) | 41 of 712 cases on 8 March 2020 |
Confirmed case data for Hubei came from Statistica (https://www.statista.com/statistics/1103040/cumulative-coronavirus-covid19-cases-number-worldwide-by-day/), for Germany, France, and Italy from the WHO (4), and for other European countries from the European Center for Disease Control (https://www.ecdc.europa.eu/en/publications-data/download-todays-data-geographic-distribution-covid-19-cases-worldwide). The number of analyzed sequences divided by the number of confirmed cases provides an upper bound to the sampling proportion since confirmed cases are only a fraction of total cases. IQR, interquartile range.
Comparing Rates of Migration and within-Region Transmission.
To weigh the significance of cases from migration versus within-region transmission during the early epidemic, we compare the rate at which new cases migrate into a region (= per individual migration rate × case count in source region) to the rate at which new cases arise from within-region transmission (= transmission rate × case count in sink region). When signal in the sequence data are low, for example, for some migration rates, our prior assumptions determine the magnitude of these rates. To assess the sensitivity of our main conclusions to the prior, we additionally analyzed the same sequences using a lower migration rate prior (SI Appendix, Fig. S7B). We note that the migration and transmission rates are assumed to be constant through time for this analysis, with the exception of the decrease in migration out of Hubei at 23 January 2020. Thus, the temporal trends depend largely on the confirmed case data, which we take from the Johns Hopkins Center for Systems Science and Engineering (https://github.com/CSSEGISandData/COVID-19).
Supplementary Material
Acknowledgments
We thank Louis du Plessis for helpful feedback on the original manuscript. S.A.N, T.G.V., J.S., J.S.H., and T.S. thank Eidgenössiche Technische Hochschule Zürich for funding. S.A.N. and T.S. are supported by the Swiss National Science Foundation (Grant 31CA30_196267).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2012008118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or supporting information. Code is available at https://github.com/SarahNadeau/cov-europe-bdmm.
References
- 1.Connor P., More than nine-in-ten people worldwide live in countries with travel restrictions amid COVID-19. Pew Research Center, https://www.pewresearch.org/fact-tank/2020/04/01/more-than-nine-in-ten-people-worldwide-live-in-countries-with-travel-restrictions-amid-covid-19/. Accessed 18 May 2020.
- 2.WHO , Updated WHO recommendations for international traffic in relation to COVID-19 outbreak, https://www.who.int/news-room/articles-detail/updated-who-recommendations-for-international-traffic-in-relation-to-covid-19-outbreak. Accessed 18 May 2020.
- 3.Wu F., et al., A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO , COVID-19 situation reports, https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. Accessed 20 April 2020.
- 5.Spiteri G., et al., First cases of coronavirus disease 2019 (COVID-19) in the WHO European region, 24 January to 21 February 2020. Euro Surveill. 25, 2000178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robert Koch Institute , Beschreibung des bisherigen Ausbruchsgeschehens mit dem neuartigen Coronavirus SARS-CoV-2 in Deutschland (Stand: 12 February 2020), https://edoc.rki.de/handle/176904/6487. Accessed 18 May 2020.
- 7.Mizumoto K., Kagaya K., Zarebski A., Chowell G., Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill. 25, 2000180 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rambaut A., et al., A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 5, 1403–1407 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Toole Á., PANGO lineages, https://cov-lineages.org/lineages.html. Accessed 28 January 2021.
- 10.Nextstrain, nextstrain/ncov: Nextstrain build for novel coronavirus (nCoV), https://github.com/nextstrain/ncov. Accessed 2 April 2020.
- 11.@trvb (Trevor Bedford). "Incredibly, it appears that this cluster containing Germany/BavPat1/2020 is the direct ancestor of these later viruses and thus led directly to some fraction of the widespread outbreak circulating in Europe today. 5/7" Twitter, 4 March 2020. https://twitter.com/trvrb/status/1235105849841872897. Accessed 20 April 2020.
- 12.Zehender G., et al., Genomic characterization and phylogenetic analysis of SARS-COV-2 in Italy. J. Med. Virol. 92, 1637–1640 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forster P., Forster L., Renfrew C., Forster M., Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. U.S.A. 117, 9241–9243 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mavian C., et al., Regaining perspective on SARS-CoV-2 molecular tracing and its implications. medRxiv: 10.1101/2020.03.16.20034470 (20 March 2020). [DOI]
- 15.Böhmer M. M., et al., Investigation of a COVID-19 outbreak in Germany resulting from a single travel-associated primary case: A case series. Lancet Infect. Dis. 20, 920–928 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothe C., et al., Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 382, 970–971 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.@trvb (Trevor Bedford). "A follow up to yesterday's thread on the possible connection between the Bavarian cluster and the Italian #COVID19 epidemic. https://twitter.com/trvrb/status/1235104921260675072… 1/5" Twitter, 5 March 2020. https://twitter.com/trvrb/status/1235382556863811584?lang=en. Accessed 20 April 2020.
- 18.Ali S. T., et al., Serial interval of SARS-CoV-2 was shortened over time by nonpharmaceutical interventions. Science 369, 1106–1109 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun K., Chen J., Viboud C., Early epidemiological analysis of the coronavirus disease 2019 outbreak based on crowdsourced data: A population-level observational study. Lancet Digit. Health 2, e201–e208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gámbaro F., et al., Introductions and early spread of SARS-CoV-2 in France, 24 january to 23 March 2020. Euro Surveill. 25, 2001200 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Díez-Fuertes F., et al., Phylodynamics of SARS-CoV-2 transmission in Spain. bioRxiv:2020.04.20.050039 (20 April 2020). [Google Scholar]
- 22.Du Plessis L., et al., Establishment & lineage dynamics of the SARS-CoV-2 epidemic in the UK. medRxiv:2020.10.23.20218446 (27 October 2020). [Google Scholar]
- 23.Oude Munnink B. B.et al.; Dutch-Covid-19 response team , Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in The Netherlands. Nat. Med. 26, 1405–1410 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Stefanelli P.et al.; On Behalf Of Iss Covid-Study Group , Whole genome and phylogenetic analysis of two SARS-CoV-2 strains isolated in Italy in January and February 2020: Additional clues on multiple introductions and further circulation in Europe. Euro Surveill. 25, 2000305 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker A., et al., Genetic structure of SARS-CoV-2 reflects clonal superspreading and multiple independent introduction events, North-Rhine Westphalia, Germany, February and March 2020. Euro Surveill. 25, 2000746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sagulenko P., Neher R., Inference of transition between discrete characters and ‘mugration’ models—TreeTime 0.7.6 documentation, https://treetime.readthedocs.io/en/latest/tutorials/mugration.html#sampling-biases. Accessed 21 May 2020.
- 27.Worobey M., et al., The emergence of SARS-CoV-2 in Europe and North America. Science 370, 564–570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y., Gayle A. A., Wilder-Smith A., Rocklöv J., The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 27, taaa021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stringhini S., et al., Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): A population-based study. Lancet 396, 313–319 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skums P., Kirpich A., Baykal P. I., Zelikovsky A., Chowell G., Global transmission network of SARS-CoV-2: From outbreak to pandemic. medRxiv:2020.03.22.20041145 (27 March 2020). [Google Scholar]
- 31.Sánchez-Pacheco S. J., Kong S., Pulido-Santacruz P., Murphy R. W., Kubatko L., Median-joining network analysis of SARS-CoV-2 genomes is neither phylogenetic nor evolutionary. Proc. Natl. Acad. Sci. U.S.A. 117, 12518–12519 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong S., Sánchez-Pacheco S. J., Murphy R. W., On the use of median-joining networks in evolutionary biology. Cladistics 32, 691–699 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Chookajorn T., Evolving COVID-19 conundrum and its impact. Proc. Natl. Acad. Sci. U.S.A. 117, 12520–12521 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mavian C., et al., Sampling bias and incorrect rooting make phylogenetic network tracing of SARS-COV-2 infections unreliable. Proc. Natl. Acad. Sci. U.S.A. 117, 12522–12523 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaughan T. G., Nadeau S., Scire J., Stadler T., Phylodynamic Analyses of outbreaks in China, Italy, Washington State (USA), and the Diamond Princess, https://virological.org/t/phylodynamic-analyses-of-outbreaks-in-china-italy-washington-state-usa-and-the-diamond-princess/439. Accessed 28 January 2021.
- 36.Linka K., Peirlinck M., Sahli Costabal F., Kuhl E., Outbreak dynamics of COVID-19 in Europe and the effect of travel restrictions. Comput. Methods Biomech. Biomed. Engin. 23, 710–717 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chinazzi M., et al., The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID-19) outbreak. Science 368, 395–400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wells C. R., et al., Impact of international travel and border control measures on the global spread of the novel 2019 coronavirus outbreak. Proc. Natl. Acad. Sci. U.S.A. 117, 7504–7509 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scire J., Barido-Sottani J., Kühnert D., Vaughan T. G., Stadler T., Improved multi-type birth-death phylodynamic inference in BEAST 2. bioRxiv: 10.1101/2020.01.06.895532 (6 January 2020). [DOI]
- 40.Nextstrain, ncov/exclude.txt at master nextstrain/ncov GitHub, https://github.com/nextstrain/ncov/blob/master/defaults/exclude.txt. Accessed 11 November 2020.
- 41.Nextstrain, ncov/parameters.yaml at master nextstrain/ncov GitHub, https://github.com/nextstrain/ncov/blob/master/defaults/parameters.yaml. Accessed 11 November 2020.
- 42.Kühnert D., Stadler T., Vaughan T. G., Drummond A. J., Phylodynamics with migration: A computational framework to quantify population structure from genomic data. Mol. Biol. Evol. 33, 2102–2116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouckaert R., et al., BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLOS Comput. Biol. 15, e1006650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A., Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. Code is available at https://github.com/SarahNadeau/cov-europe-bdmm.