Significance
Immunotherapy benefits some aggressive breast cancers, but many breast tumors do not respond to checkpoint blockade. Novel strategies to increase breast cancer immunogenicity are needed to improve immunotherapy. Here, we used epithelial cell adhesion molecule (EpCAM) aptamer-linked small-interfering RNA chimeras (AsiC) to selectively knock down genes in mouse breast cancers to induce tumor neoantigens or overcome immune evasion. Individual gene knockdown markedly delayed tumor growth and enhanced antitumor immunity. Cd47 and Parp1 AsiCs outperformed anti-CD47 antibody and the PARP1 inhibitor Olaparib, respectively. Combining EpCAM-AsiCs targeting multiple pathways worked better than single agents and enhanced tumor inhibition by a checkpoint inhibitor. EpCAM-AsiCs have the potential to boost immunity to tumors that are poorly responsive to checkpoint blockade.
Keywords: EpCAM aptamer, gene knockdown, immunotherapy, checkpoint blockade, CD47
Abstract
New strategies for cancer immunotherapy are needed since most solid tumors do not respond to current approaches. Here we used epithelial cell adhesion molecule EpCAM (a tumor-associated antigen highly expressed on common epithelial cancers and their tumor-initiating cells) aptamer-linked small-interfering RNA chimeras (AsiCs) to knock down genes selectively in EpCAM+ tumors with the goal of making cancers more visible to the immune system. Knockdown of genes that function in multiple steps of cancer immunity was evaluated in aggressive triple-negative and HER2+ orthotopic, metastatic, and genetically engineered mouse breast cancer models. Gene targets were chosen whose knockdown was predicted to promote tumor neoantigen expression (Upf2, Parp1, Apex1), phagocytosis, and antigen presentation (Cd47), reduce checkpoint inhibition (Cd274), or cause tumor cell death (Mcl1). Four of the six AsiC (Upf2, Parp1, Cd47, and Mcl1) potently inhibited tumor growth and boosted tumor-infiltrating immune cell functions. AsiC mixtures were more effective than individual AsiC and could synergize with anti–PD-1 checkpoint inhibition.
The impressive successes of current immune therapy in some cancers, but lack of effectiveness in most, suggest additional strategies to promote antitumor immunity are needed. The most successful current immune therapies use checkpoint blockade to restore functionality to exhausted tumor-infiltrating lymphocytes (TIL) or adoptively transferred, cytotoxic T cells expressing chimeric receptors (CAR T cells) that kill tumor cells (1). However, no CAR T cell therapy is approved for solid tumors, and only a minority of solid tumors respond to checkpoint blockade. Some tumors do not respond to checkpoint inhibition because they are not recognized by immune cells as foreign. Tumors that respond have high somatic mutation rates (i.e., ∼100/Mb for melanoma and non-small cell lung cancer), which contribute to their immunogenicity by causing the tumor to express nonself tumor antigens (2). Much of the ongoing work to improve responses focuses on combining checkpoint inhibitors, with some, mostly incremental, successes, but with the risk of increased autoimmune toxicity, because most checkpoint inhibitors remove the brakes on all T cells, not just antitumor T cells (3). As tumors grow in the face of innate and adaptive immune surveillance, their survival depends on tumor cell editing that enables them to avoid recognition and resist killing by cytotoxic lymphocytes. Some of the strategies tumors use to stymie immune control include loss of expression of antigenic or antigen-presenting and processing proteins or natural killer (NK) receptor ligands; induction of inhibitory receptor ligands (i.e., programmed death-ligand 1 [PD-L1], PD-L2), immunosuppressive cytokines (i.e., transforming growth factor [TGF]-β, interleukin [IL]-10), antiapoptotic proteins or the “don’t-eat-me signal” CD47 that interferes with phagocytosis and tumor antigen cross-presentation; and development of insensitivity to interferon [IFN]-γ (4). This study is based on the hypothesis that increasing the response rate for immunotherapies will require new strategies that directly address the lack of tumor recognition by killer cells, that could be used with or without conventional chemotherapy, targeted therapies, or checkpoint inhibitors.
To evaluate new ways to make tumors susceptible to immune control, we took advantage of a method for tumor-directed in vivo gene knockdown. This method uses RNA aptamers (which can be thought of as RNA “antibodies”)—structured RNAs that bind with high affinity to a cell receptor—to deliver covalently linked small-interfering RNAs (siRNAs) selectively into tumor cells (5). In a previous study, we showed that uptake and knockdown occur selectively in receptor-bearing tumor xenografts when epithelial cell adhesion molecule EpCAM (a tumor-associated antigen highly expressed on common epithelial cancers and their tumor-initiating cells)-targeting aptamer-linked siRNA chimeras (AsiCs) (Fig. 1A) are injected subcutaneously (6). EpCAM, the first described tumor antigen, is much more highly expressed in epithelial cancers (97% of breast cancer, 100% of lung, pancreas, colon, ovarian, prostate cancers) and their “cancer stem cells” than normal epithelia, making it an attractive target for selective tumor targeting (7). A 19-nt EpCAM aptamer (8) binds with low nanomolar affinity to both mouse and human EpCAM, making it ideal for studying immunotherapy in immunocompetent mice and potential translation to humans. EpCAM-AsiC are chemically synthesized with 2′-fluoropyrimidine substitutions and 3′-dTdT overhangs to enhance RNase resistance and in vivo stability and block activating innate immune sensors of foreign RNA. This configuration is stable for >36 h in serum in vitro, does not activate IFN or inflammatory cytokines and is cleaved in cells by Dicer to release an active siRNA (6). After subcutaneous injection, EpCAM-AsiC targeting PLK1 were selectively taken up by human HER2+ and basal-A triple negative breast cancer (TNBC) xenografts, knocked down PLK1, and blocked tumor growth without any apparent toxicity. Moreover, PLK1 EpCAM-AsiC were taken up by “cancer stem cells” within EpCAM+ breast cancers ex vivo and blocked tumor initiation.
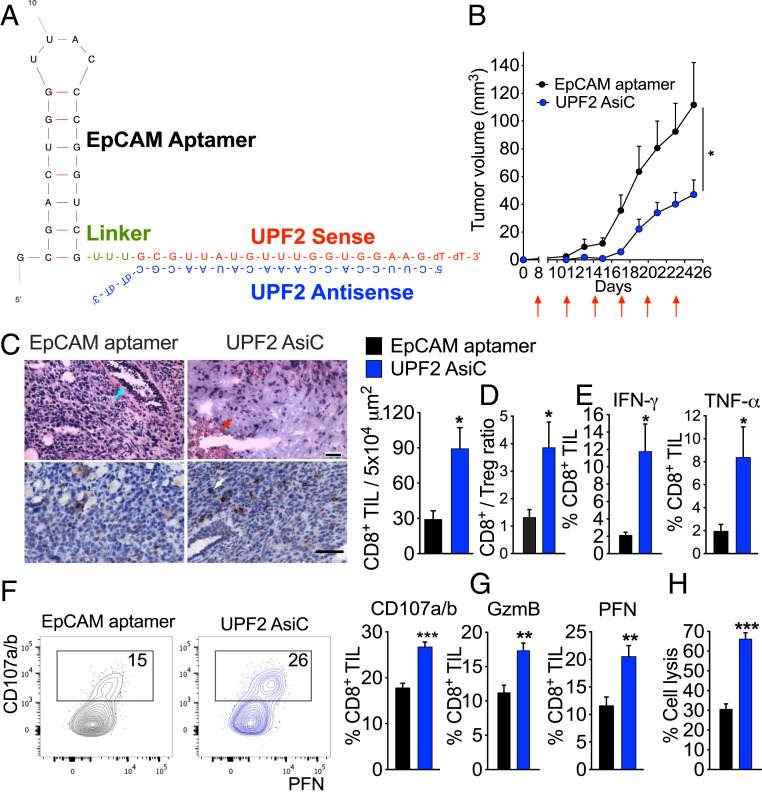
Fig. 1.
Tumor inhibition and immune modulation by UPF2 AsiC. (A) Design of UPF2 EpCAM AsiC. (B) Orthotopic 4T1E tumor growth after subcutaneous injection (red arrows) of EpCAM aptamer or UPF2 AsiC (5 mg/kg every third day after tumors became palpable); EpCAM aptamer (n = 7), UPF2 AsiC (n = 8). (C to H) Mice bearing orthotopic 4T1E tumors, treated as in B, were killed on day 15 and tumors were analyzed by IHC (n = 2) (C), flow cytometry (n = 5) (D–G), and 51Cr release assay (n = 3) (H). (C) Representative sections (Left) of H&E (Upper) and CD8 (Lower) stained tumors and quantification of CD8+ TIL counts (Right). On left, blue arrow indicates mammary gland; red arrow, necrotic area; white arrow, CD8+ TIL. (Scale bars, 500 μm [Upper] and 50 μm [Lower].) (Right) CD8+ TIL counts per selected area. For each mouse, five fields per section were counted. (D) CD8+/CD4+Foxp3+ Treg ratio. (E) Percentage of CD8+ TIL producing IFN-γ and TNF-α after PMA and ionomycin stimulation. (F) Percentage of degranulating CD8+ TIL measured by CD107a/CD107b surface expression. (Left) representative flow plots. (G) Percentage of CD8+ TIL expressing GzmB and PFN after 6 h incubation with 4T1E knocked down for Upf2. (H) Specific cytotoxicity of CD8+ TIL against 4T1E knocked down for Upf2. (effector:target ratio, 5:1). (F to H) were performed with pooled TIL. Data show mean + SEM and are representative of at least two experiments. Statistical tests: (B) tumor growth curves were compared by calculating the area under the curve values for each sample followed by Student’s t test. (C, E–H) Student’s t test. (D) Mann–Whitney test. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
To study potential targets, we designed EpCAM-AsiC to knock down genes that might increase immune recognition of aggressive HER2+ and TNBC mouse cancers. TNBC and HER2+ breast cancers are the worst prognosis breast cancers (9, 10). There is not much targeted therapy for TNBC (except for PARP-1 inhibition for a subset of BRCA1-mutated TNBC tumors, which only extends mean survival by a few months), and a large fraction of patients relapse and develop metastases after chemotherapy (11). Although HER2-targeted therapies have radically improved HER2+ breast cancer outcome, >20% of patients develop recurrent disease within 5 y (12). Thus, novel strategies are needed. Many breast cancers are immunologically quiescent, which has been attributed to, at least in part, their low nonsynonymous mutational burden (about 1/Mb) (13). However, abundant evidence suggests that breast cancers are under immune surveillance, and anti–PD-L1 was approved last year in conjunction with nanoparticle-delivered chemotherapy for TNBC patients whose tumors express PD-L1 (14). Moreover, increases in TIL are associated with better overall and disease-free survival in TNBC and HER2+ breast cancer, with each 10% increase in TIL linked to a 15 to 25% decrease in risk of relapse and death (15). The long-term effectiveness of some conventional chemotherapy drugs, targeted therapy, and radiotherapy depends on their ability to trigger antitumor T cells (16). These findings highlight the opportunity to develop more effective immunotherapies to improve aggressive breast cancer outcome.
In this study we designed EpCAM-AsiC to knock down a TNBC-dependency gene (Mcl1), a nonsense-mediated decay (NMD) enzyme involved in RNA quality control (Upf2), and DNA damage repair genes (Parp1, Apex1) to induce tumor neoantigens, Cd274 encoding PD-L1 for checkpoint inhibition, and a “don’t eat me” signal that inhibits tumor phagocytosis (Cd47). Four of these six AsiC markedly suppressed tumor growth and enhanced TIL functions in an orthotopic 4T1 TNBC model. One therapeutic advantage of AsiC is that they can easily be combined to generate AsiC mixtures to target multiple immune evasion pathways. AsiC combinations were more effective than individual AsiC and inhibited metastatic disease and aggressive spontaneous breast cancers in transgenic mice expressing an inducible oncogenic HER2 (ErbB2ΔEx16) in the mammary epithelium.
Results
EpCAM Aptamer-siRNAs Cause Selective Knockdown in EpCAM+ Mouse Breast Cancer Lines.
To investigate using EpCAM-AsiC for cell-specific gene knockdown for breast cancer, we first verified that fluorescently labeled EpCAM aptamer was taken up by mouse EpCAM+ breast cancer cell lines (4T1, 4T1E [4T1 sorted for E-cadherinhi cells], N202.1A), but not EpCAM− mouse cell lines (L929, P815, B16-F10), as previously shown for human breast cancer lines. To test whether knocking down Upf2, Parp1, Apex1, Mcl1, Cd47, or Cd274 in mouse breast cancer cell lines could enhance antitumor immunity, EpCAM-AsiC were designed using siRNAs that each caused ∼90% knockdown after transfection of 4T1E TNBC with 100 nM siRNA (SI Appendix, Fig. S1A). None of these siRNAs affected cell viability or proliferation, except for the Mcl1 siRNA (SI Appendix, Fig. S1 B–D), which was expected since Mcl1 was previously identified as a TNBC-dependency gene (17, 18). To construct EpCAM-AsiC, the sense (passenger or inactive) strand of each selected siRNA was linked to the 3′ end of the 19-nt EpCAM aptamer via a U-U-U linker (Fig. 1A and SI Appendix, Table S1). This RNA was chemically synthesized and then annealed to the antisense (guide or active) strand of each siRNA. The EpCAM-AsiC were synthesized with 2′-fluoropyrimidine substitutions and 3′-dTdT overhangs to enhance their resistance to RNases and in vivo stability. Each of these EpCAM-AsiC knocked down target gene expression in EpCAM+ 4T1E tumor cells in vitro by 50 to 90% when measured 72 h later (SI Appendix, Fig. S1E). As expected, EpCAM-AsiC did not affect target gene expression in EpCAM-L929. Subcutaneous injection of 100 μg (5 mg/kg) AsiC in mice knocked down target gene expression by 50 to 70% in 4T1E tumors implanted orthotopically (SI Appendix, Fig. S1 F and G). Knockdown was specific since injection of the EpCAM aptamer on its own or an EpCAM-AsiC directed against eGFP did not knock down endogenous genes. Moreover, knockdown did not occur in CD45−EpCAM− cells within the tumor.
UPF2 EpCAM-AsiC Inhibit Tumor Growth and Enhance Antitumor T Cell Immunity.
Knocking down Upf2, which encodes a protein that binds to prematurely terminated mRNAs to activate NMD, has been postulated to induce tumor cell expression of neoantigens to promote T cell recognition. In fact, Upf2 knockdown using prostate-specific membrane antigen (PSMA)-AsiC to target mouse colorectal cancer and melanoma cells ectopically expressing PSMA reduced tumor growth (19). To verify that subcutaneously injected EpCAM-AsiC decreased NMD activity in orthotopic 4T1E tumor cells, we compared the ratio of fully spliced mRNA to its precursor pre-mRNA for four known NMD-targeted transcripts (Gadd45a, Gadd45b, Cdkn1a, Nat9). An increased ratio indicates diminished NMD activity (20). The mRNA/pre-mRNA ratio for all four genes was significantly higher in the tumors of UPF2 EpCAM-AsiC–treated mice than in control mice treated with just the aptamer (SI Appendix, Fig. S1H), indicating impaired NMD activity in vivo. To determine whether UPF2 EpCAM-AsiC have antitumor activity, mice bearing palpable orthotopic 4T1E tumors were treated with 5 mg/kg of EpCAM aptamer or UPF2 EpCAM-AsiC subcutaneously every 3 d. The 4T1E tumor growth was significantly inhibited in UPF2 EpCAM-AsiC–treated mice (Fig. 1B). The effect of tumor-targeted Upf2 knockdown on TIL was assessed by immunohistochemistry (IHC) and flow cytometry. UPF2 EpCAM-AsiC strongly increased the density of CD8+ TIL measured by IHC by threefold (Fig. 1C). The ratio of CD8+ T cells to CD4+Foxp3+ Treg, a parameter strongly associated with antitumor immunity and response to immunotherapy for aggressive breast cancer (21, 22), also increased threefold in UPF2 AsiC-treated tumors (Fig. 1D). CD8+ TIL from UPF2 AsiC-treated tumors also produced more IFN-γ and tumor necrosis factor (TNF)-α after ex vivo stimulation with phorbol 12-myristate 13-acetate (PMA) and ionomycin (Fig. 1E). After coincubation with UPF2 siRNA-treated 4T1E ex vivo for 6 h, these CD8+ TIL also degranulated more as measured by CD107a/b surface expression (Fig. 1F) and stained more for the cytotoxic effectors, granzyme B (GzmB) and perforin (PFN) (Fig. 1G). Indeed, CD8+ TIL from UPF2 AsiC-treated tumors were twice as effective at killing Upf2-knocked down 4T1E cells as aptamer-treated tumors (Fig. 1H). Thus, UPF2 EpCAM-AsiC significantly enhanced antitumor CD8+ T cell immunity and delayed 4T1E tumor growth.
UPF2 Knockdown Induces Novel mRNA Transcripts.
To investigate whether UPF2 knockdown in breast cancer generates novel mRNA isoforms, bulk RNA sequencing (RNA-seq) compared an EpCAMhi MDA-MB-231 human breast cancer cell line transfected with noncoding control or UPF2 siRNA for 72 h. We identified 222 examples of differential exon usage (DEU) within 281 genes (Dataset S1). For example, UPF2 knockdown significantly reduced usage of exon 8 in RINL mRNA (transcript ID ENSG00000187994) (log2 fold-change −15.2, adjusted P = 0.03) and significantly enhanced usage of exon 6 (log2 fold-change of 14.5, adjusted P = 0.02) in ATP11B mRNA transcript (ENSG00000058063), which was almost not detected in control cells. These DEU events could lead to expression of novel polypeptides and novel T cell epitopes. The number and diversity of DEUs suggest that UPF2 knockdown could have caused alternative splicing. To test this idea, UPF2 knockdown-related transcriptional diversity was deconvoluted to identify and estimate the abundance of transcript isoforms. Forty-two genes with potential differential isoform usage (DIU) were identified (Dataset S2). These included seven genes identified as having DEU (CENPH, PFKFB4, UCN2, SNHG8, CDKAL1, TRIM4, TMEM242). These DIU events included examples of novel mRNA isoforms that may encode new polypeptides (e.g., in DNAJC2, LAT2, and TMPRSS5) (SI Appendix, Fig. S2A). In addition, some genes with DIU—such as CENPH, SNRPA1, and EBPL—increased mRNA isoforms known to be sensitive to NMD. For example, UPF2 knockdown increased a CENPH isoform with exon-skipping predicted to have premature termination codons sensitive to NMD (SI Appendix, Fig. S2B). Collectively, our data suggest that UPF2 knockdown may induce expression of tumor neoantigens.
NMD inhibition could inhibit tumor growth and promote antitumor immunity by other mechanisms besides generating neoantigens. NMD inhibition has been reported to regulate transcripts involved in cellular stress responses and nutrient homeostasis (23, 24). Amino acid starvation and endoplasmic reticulum (ER) stress in the tumor inhibit NMD activity, which may be a tumor strategy to up-regulate stress-responsive transcripts to adapt to environmental challenges (25). Both DEU and DIU changes after UPF2 knockdown were noted in PFKFB4, UCN2, CDKAL1, and TRIM4, genes involved in oxidative stress or ER stress responses. Gene ontology (GO) analyses of pathways enriched in DEU genes using DAVID or Metascape showed significant enrichment for pathways involved in RNA processing, nucleotide biosynthesis, responses to glucose and oxidative stress, and cell proliferation and others that might influence tumor proliferation independently of immune recognition (SI Appendix, Fig. S3).
Parp1 Knockdown Reduces Tumor Growth and Enhances Antitumor Immunity.
Inhibiting tumor cell DNA repair might be another way to promote tumor immunity. Poly(ADP-ribose) polymerase 1 (PARP1) senses DNA damage and recruits and activates the DNA repair machinery at break sites. PARP1 inhibition, which is an approved therapy of TNBC and ovarian cancers with BRCA1/2 mutation or other defects in homologous recombination, leads to chromosomal abnormalities and genome instability that can trigger an innate protective IFN response (26). Knocking down PARP1 in tumor cells might potentially lead to more DNA damage-related mutations, thereby introducing tumor-specific neoantigens that could be recognized by T cells. To test whether Parp1 knockdown activates antitumor immunity, mice bearing palpable orthotopic 4T1E tumors were treated with the EpCAM aptamer, PARP1 EpCAM-AsiC, or the PARP1 inhibitor, olaparib. The PARP1 AsiC more effectively inhibited 4T1E tumor growth than olaparib, which showed a trend toward inhibition that did not reach significance (Fig. 2A).
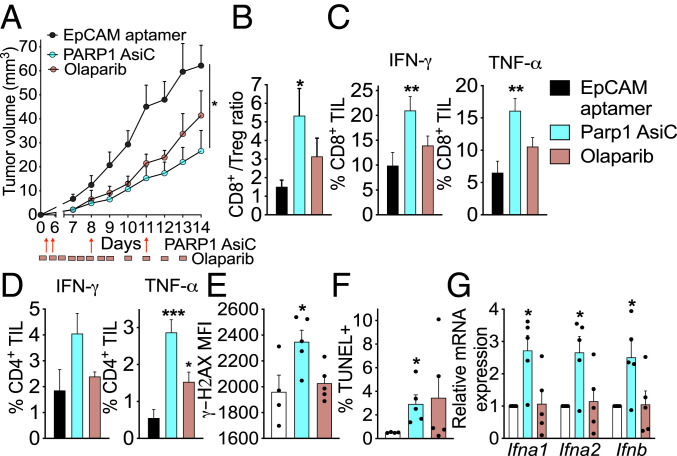
Fig. 2.
Tumor inhibition and immune modulation by PARP1 AsiC and PARP1 inhibitor olaparib. (A) Growth of 4T1E orthotopic tumors in mice treated beginning on day 3 with EpCAM aptamer or PARP1 EpCAM-AsiC (5 mg/kg, every third day) or olaparib (50 mg/kg daily). (B to D) Flow cytometry analysis of TIL, harvested on day 14 after implantation from mice treated as in A, for (B) CD8+/CD4+Foxp3+ Treg and percentage of CD8+ (C) and CD4+ (D) TIL producing IFN-γ and TNF-α after PMA and ionomycin stimulation. (n = 4 in A–D). (E and F) Flow cytometry analysis of unrepaired DNA damage in CD45−EpCAM+ tumor cells as measured by mean fluorescence intensity (MFI) of γ-H2AX (E) and percentage of TUNEL+ tumor cells (F) in each group of day 14 tumors. (G) Relative mRNA expression by qRT-PCR of type I interferon genes in each group of day 14 tumors. (E–G) EpCAM aptamer: n = 4, PARP1 AsiC and olaparib: n = 5. Data shown are mean + SEM and are representative of two experiments. Statistical tests: (A) tumor growth curves were compared by calculating the area under the curve values for each sample followed by one-way ANOVA with Holm–Sidak’s multiple comparisons. (B–E) One-way ANOVA with Holm–Sidak’s multiple comparisons. (F) Kruskal–Wallis test with Dunn’s multiple comparisons. (G) Two-way ANOVA with Holm–Sidak’s multiple comparisons. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
The PARP1 AsiC also enhanced TIL antitumor properties more than olaparib. It potently and significantly increased the CD8+/CD4+ Treg ratio in the tumor (Fig. 2B), activation-stimulated IFN-γ and TNF-α production by CD8+ TIL (Fig. 2C), and TNF-α production by CD4+ TIL (Fig. 2D) compared to control aptamer-treated tumors. Olaparib had a more subtle effect on antitumor immunity that did not reach significance except for an increase in TNF-α production by CD4+ TIL. Why Parp1 knockdown was more effective than PARP1 enzymatic inhibition is unclear, but removing PARP1 protein would interfere with the recognition and assembly of repair proteins at sites of DNA damage, whereas inhibiting the poly(ADP)ribosylation (PARylation) activity of PARP1 would only act more downstream to inhibit repair. As a consequence, unrepaired DNA damage and genomic instability after Parp1 knockdown might be more extensive than after inhibiting PARP1 enzymatic activity. Supporting this hypothesis, unrepaired DNA damage, measured by tumor cell γ-H2AX and TUNEL positivity, was significantly enhanced in the tumors of mice that received PARP1 AsiC, but not in olaparib-treated tumors (Fig. 2 E and F). Tumor cell IFN-I mRNA (IFN-α1, IFN-α2, IFN-β), which also promotes antitumor immune cell functionality (27), was also significantly increased in PARP1 AsiC-treated, but not olaparib-treated, mice, presumably in response to increased unresolved DNA damage (Fig. 2G).
Knocking Down Apex1 or Cd274 Modestly Affects Tumor Growth.
APEX1 is a key endonuclease in base excision repair, which repairs the most common DNA damage in cells, abasic sites formed by oxidative DNA damage (28). Apex1 genetic deficiency leads to early embryonic lethality (embryonic days 4 to 6.5) and cell lines deficient in Apex1 do not grow (29). Of note, tumors do not mutate this essential gene. We therefore thought Apex1 knockdown should be investigated since it might be directly cytotoxic and also induce mutations that could activate T cell immunity. Perhaps because it is such as essential gene, in vitro Apex1 knockdown by EpCAM-AsiC was only 50%, less effective than for other EpCAM-AsiC (SI Appendix, Fig. S1E). When administered in the same dose and schedule as other EpCAM-AsiC, tumor-targeted Apex1 knockdown reduced 4T1E tumor growth, but the difference compared to mice treated with just the aptamer did not reach significance (SI Appendix, Fig. S4A).
Checkpoint inhibition induces protective immunity with dramatic and durable responses in some cancers; 4T1E strongly and uniformly express PD-L1 (SI Appendix, Fig. S4B). We therefore assessed the antitumor activity of CD274 EpCAM-AsiC targeting PD-L1 (SI Appendix, Fig. S1E). CD274 EpCAM-AsiC inhibited tumor growth, but the effect was not statistically significant (SI Appendix, Fig. S4C). CD274 AsiC treatment alone also did not affect the number and function of CD8+ TIL (SI Appendix, Fig. S4 D and E), suggesting that combining CD274 EpCAM-AsiC with other therapies might be necessary to improve antitumor immunity.
CD47 EpCAM-AsiC Promotes EpCAM+ Breast Cancer Cell Phagocytosis by Macrophages and Enhances Antitumor T Cell Immunity.
Tumor cells change expression of many genes to avoid immune elimination during tumor editing. One strategy is up-regulation of the surface glycoprotein CD47, which binds to signal-regulatory protein SIRPα on macrophages and dendritic cells (DCs) and acts as a potent “don’t eat me” signal (30). CD47-blocking antibodies, which are being evaluated for clinical use, induce macrophages to phagocytose tumor cells, suppress tumor growth, and synergize with chemo- and radiotherapy, by promoting tumor antigen presentation (31, 32). To evaluate the antitumor effect of Cd47 knockdown, orthotopic 4T1E tumor-bearing mice were treated with EpCAM aptamer or CD47 EpCAM-AsiC. CD47 EpCAM-AsiC inhibited tumor growth (Fig. 3A) and promoted antitumor immunity, as indicated by an increased CD8+/CD4+ Treg TIL ratio (Fig. 3B), reduced coinhibitor PD-1 expression on CD44+CD8+ TIL (Fig. 3C), and increased CD8+ and CD4+ TIL production of IFN-γ (Fig. 3 D and E) and CD8+ TIL expression of GzmB (Fig. 3F), compared to mice treated with EpCAM aptamer.
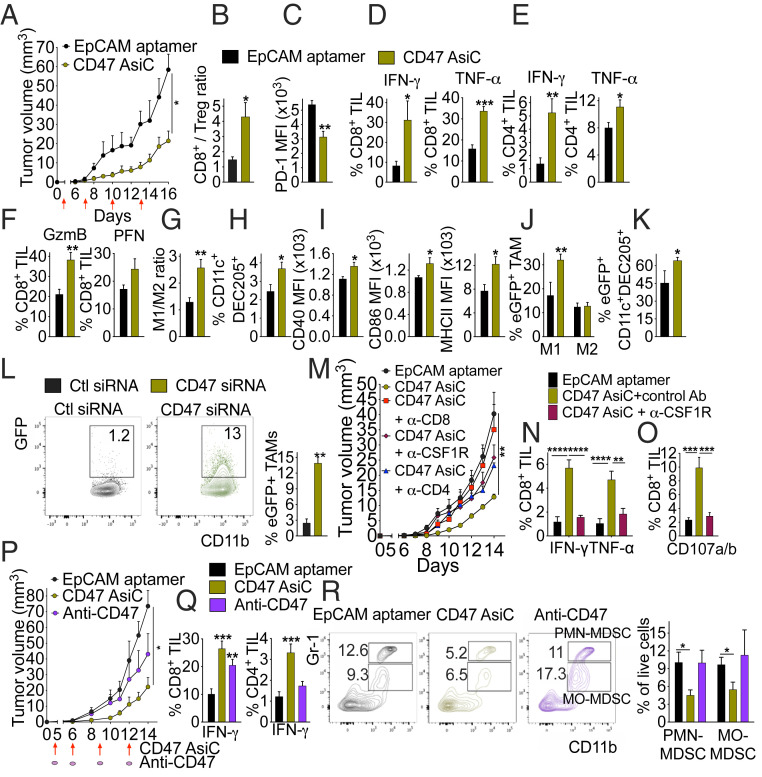
Fig. 3.
CD47 AsiC inhibit tumor growth by enhancing antitumor immunity. (A) Growth of orthotopic 4T1E tumors treated with EpCAM aptamer or CD47 EpCAM-AsiC. Red arrows indicate treatment. (B–I) 4T1E tumor-bearing mice, treated as in A, were killed on day 16 and single-cell tumor suspensions were analyzed by flow cytometry for (B) CD8+/CD4+Foxp3+ Treg ratio, (C) PD-1 expression on CD44+CD8+ TIL, percentage of CD8+ (D) and CD4+ (E) TIL producing IFN-γ and TNF-α after PMA and ionomycin stimulation, (F) percentage of CD8+ TIL staining for GzmB and PFN, (G) M1/M2 TAM ratio, (H) percentage of CD11c+DEC205+ DC in CD45+ cells, and (I) CD40, CD86, and MHC II MFI on CD11c+DEC205+ DC. (J and K) Mice bearing orthotopic 4T1E-eGFP tumors were treated as in A and analyzed for (J) percentage of M1 TAM and M2 TAM that are eGFP+ and (K) percentage of CD11c+DEC205+ DC that are eGFP+. (A–K, n = 5). (L) Ex vivo phagocytosis by CD11b+F4/80+ TAM, isolated on day 14 from untreated 4T1E-bearing mice, of 4T1E-eGFP tumor cells pretreated with nontargeting control or CD47 siRNA. (Left) Representative flow plots and (Right) mean of (n = 5) samples. TAM were pooled from n = 3 mice. (M) Comparison of 4T1E tumor growth in tumor-bearing mice treated with EpCAM aptamer or CD47 AsiC and injected with isotype control Ab or depleted of CD8+ T cells, CD4+ T cells, or macrophages using anti-CSF1R. (N) Cytokine production (Left) and degranulation (Right) of CD8+ TIL, isolated on day 14 from tumors of mice treated with EpCAM aptamer or CD47 AsiC with or without macrophage depletion, after incubation with 4T1E for 6 h. (M and N, n = 5). (O) 4T1E tumor growth in mice treated with EpCAM aptamer, CD47 AsiC, or anti-CD47. (P and Q) Percentage of PMA and ionomycin-stimulated CD8+ TIL (P) and CD4+ TIL (Q) producing IFN-γ. (R) Percentage of PMN-MDSC and MO-MDSC among tumor-infiltrating live cells. (Left) Representative flow plots and (Right) mean. (O, R, n = 5). Data shown are mean + SEM and are representative of two experiments. Statistical tests: (A, M, and O) tumor growth curves were compared by calculating the area under the curve values for each sample followed by Student’s t test (A) or one-way ANOVA with Holm–Sidak’s multiple comparisons (M and O). (B–I and L) Student’s t test. (K) Mann–Whitney test. (J and R) Multiple t tests with Holm–Sidak correction. (N, Left) Two-way ANOVA with Holm–Sidak correction. (N, Right, P and Q) One-way ANOVA with Holm–Sidak’s multiple comparisons. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Next, we analyzed the impact of Cd47 knockdown on tumor-associated macrophages (TAM) and DCs. TAM can polarize into either proinflammatory, classically activated M1-like macrophages with antitumor properties or immunosuppressive, alternatively activated M2-like macrophages that correlate with tumor progression, metastasis, and poor prognosis (33). Although CD47 EpCAM-AsiC did not significantly change TAM numbers, the ratio of M1/M2 TAM significantly increased in CD47 EpCAM-AsiC–treated tumors (Fig. 3G and SI Appendix, Fig. S5 A and B). In addition, the percentage of CD11c+DC205+ DCs that are specialized in taking up extracellular antigens within the CD45+ cells in the tumor was significantly higher after CD47 EpCAM-AsiC, compared to aptamer treatment (Fig. 3H). DCs in CD47 EpCAM-AsiC–treated tumors expressed more costimulatory molecules CD40, CD86, and surface MHC-II, suggesting they were more effective antigen-presenting cells (Fig. 3I). To determine whether TAM phagocytosis of tumor cells increased in vivo after aptamer or CD47 EpCAM-AsiC treatment, we used 4T1E stably expressing eGFP (4T1E-eGFP) and examined TAM GFP fluorescence. Significantly more M1, but not M2, TAM were GFP+ in CD47 EpCAM-AsiC–treated tumors, indicating increased in vivo phagocytosis (Fig. 3J). More CD11c+DC205+ DCs were also GFP+ after CD47 AsiC treatment (Fig. 3K), implying their increased potential to present tumor antigens. To confirm that enhanced TAM phagocytosis was due to reduced CD47 expression on tumor cells, TAM enriched from 4T1E tumors were cocultured with 4T1E-eGFP cells that were pretreated with nontargeting or Cd47 siRNA. TAM phagocytosis of Cd47 knocked down 4T1E-eGFP was increased fourfold compared to control tumors (Fig. 3L).
To determine whether the tumor-suppressive effect of CD47 EpCAM-AsiC was mediated by TIL or TAM, we depleted CD8+ or CD4+ T cells or macrophages in orthotopic 4T1E tumor-bearing mice before treatment with CD47 EpCAM-AsiC using CD4, CD8, or CSF1R antibodies, respectively (SI Appendix, Fig. S5 C–E). Depletion of CD8+ T cells completely abrogated the antitumor effect of CD47 EpCAM-AsiC, but CD4+ T cell or macrophage depletion had less of an effect (Fig. 3M). However, macrophage depletion was less complete than T cell depletion since ∼30% of MHCII+TAM persisted after depletion. Increased CD8+ TIL functionality in CD47 EpCAM-AsiC–treated tumors, assessed by IFN-γ and TNF-α production and degranulation in response to 4T1E, was abrogated in mice depleted of macrophages, indicating the importance of TAM in promoting CD8+ TIL antitumor immunity in CD47 AsiC-treated tumors (Fig. 3 N and O).
Next, the antitumor effect of CD47 AsiC and anti-CD47 was compared. Although both reduced tumor size, the difference was only significant for CD47 AsiC (Fig. 3P). CD8+ TIL from CD47 EpCAM-AsiC and anti-CD47–treated mice both produced more IFN-γ after PMA and ionomycin stimulation than those in control tumors, but only CD47 AsiC significantly increased stimulated IFN-γ production of CD4+ TIL (Fig. 3Q). In addition, CD47 EpCAM-AsiC, but not anti-CD47, significantly reduced the numbers of tumor-infiltrating immunosuppressive polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs) and mononuclear (MO)-MDSCs compared to control tumors (Fig. 3R and SI Appendix, Fig. S4A). Thus, CD47 EpCAM-AsiC more effectively controlled tumor growth and enhanced antitumor immunity than anti-CD47.
MCL1 EpCAM-AsiC Induce Antitumor Immunity.
Because TNBC are heterogeneous cancers, genome-wide siRNA screens to identify shared dependencies of human basal-A TNBC cell lines identified few shared dependency genes (17, 18). One of the strongest hits was the antiapoptotic BCL-2 family gene MCL1, which is commonly amplified in TNBC and whose overexpression correlates with poor prognosis (34). To determine whether tumor cell death induced by Mcl1 knockdown (SI Appendix, Fig. S1B) promotes cross-presentation of tumor antigens to CD8+ T cells, thereby improving antitumor immunity, we first verified that MCL1 EpCAM-AsiC reduced 4T1E viability in vitro (SI Appendix, Fig. S6A). MCL1 EpCAM-AsiC, injected subcutaneously every 3 d after orthotopic 4T1E tumors became palpable, slowed tumor growth significantly (SI Appendix, Fig. S6B). MCL1 EpCAM-AsiC markedly improved the CD8+/CD4+ Treg ratio and antitumor CD8+ and CD4+ TIL functions (SI Appendix, Fig. S6 C–F). A similar improvement in antitumor T cell immunity was also observed using another cytotoxic EpCAM-AsiC targeting the essential gene Plk1, encoding a kinase required for mitosis. Thus, some EpCAM-AsiC that are cytotoxic also promote effective tumor-protective immunity.
Enhanced Antitumor Activity of EpCAM-AsiC Combinations.
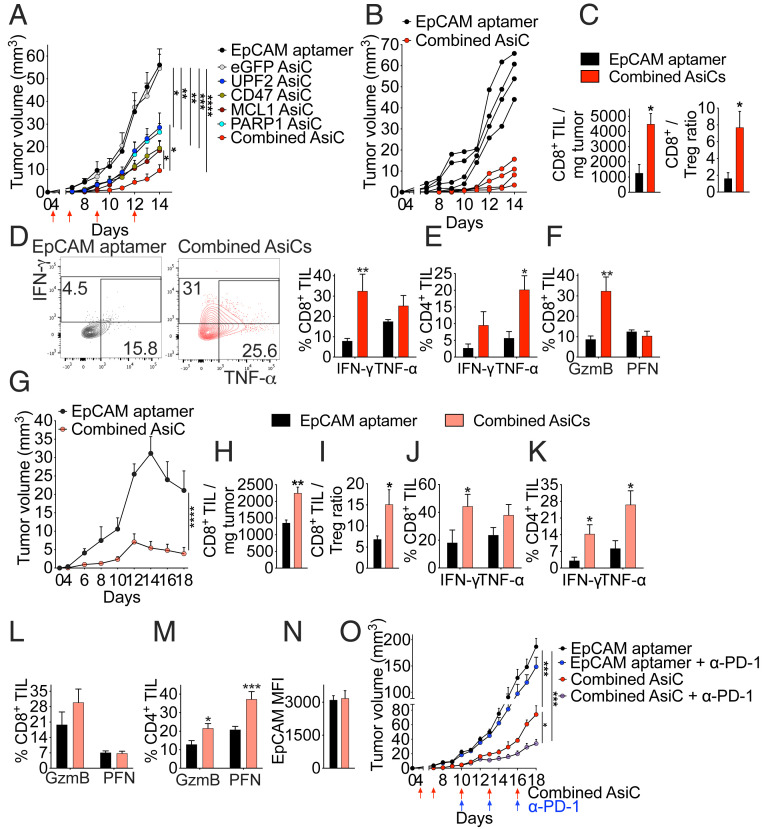
One advantage of AsiC for cancer treatment is that it is relatively easy to combine AsiC to produce drug mixtures that could have additive or synergistic effects by knocking down genes that promote tumor immunity by different mechanisms. To investigate EpCAM-AsiC combinations, 4T1E orthotopic tumor-bearing mice were treated with the four most effective EpCAM-AsiC, targeting Upf2, Parp1, Cd47, or Mcl1, individually or in combination, using EpCAM aptamer or eGFP EpCAM-AsiC as controls (Fig. 4 A and B). Each EpCAM-AsiC on its own markedly delayed tumor progression, but the mixture was significantly better. The mixture increased the number of CD8+ TIL by approximately fourfold (Fig. 4C), improved the CD8+/CD4+ Treg TIL ratio by approximately fivefold (Fig. 4C), and increased stimulated production of cytokines and cytotoxic molecules by CD8+ and CD4+ TIL (Fig. 4 D–F). The combined EpCAM-AsiC were also evaluated in mice bearing 4T1E-eGFP, whose expression of the immunogenic foreign protein causes tumor regression (Fig. 4G). Tumors treated with the AsiC mixture grew much more slowly and started to regress earlier. The combination also potently boosted T cell immunity in 4T1E-eGFP tumors (Fig. 4 H–M). Importantly, five injections of EpCAM-AsiC combinations did not change EpCAM expression on 4T1E-eGFP tumors, ruling out down-regulation of the aptamer target as a possible source of drug resistance (Fig. 4N).
Fig. 4.
Combination of immune-modulating EpCAM-AsiC enhances tumor suppression. (A) Orthotopic 4T1E tumor growth in mice treated as indicated (red arrows) with EpCAM aptamer or EpCAM-AsiC targeting Upf2, Cd47, Mcl1, or Parp1, individually or in combination, or eGFP as control. (B) Growth of 4T1E tumors in individual mice treated with either EpCAM aptamer or the four EpCAM-AsiC combination. (C–F) Mice treated as in A were killed on day 14 and tumors were analyzed by flow cytometry. (C) Numbers of CD8+ TIL per milligram of tumor (Left) and CD8+/CD4+Foxp3+ Treg ratio (Right). (D) Percentage of CD8+ TIL producing IFN-γ and TNF-α after PMA and ionomycin stimulation. Shown at Left are representative flow plots. (E) Percentage of CD4+ TIL producing IFN-γ and TNF-α induced by PMA and ionomycin. (F) Percentage of CD8+ TIL staining for GzmB and PFN. (A–F, n = 4). (G) Growth of 4T1E-eGFP tumors in mice treated with EpCAM aptamer or the four EpCAM-AsiC combination. (H–N) Mice treated as in G were killed on day 18 and tumors were analyzed by flow cytometry. (H) Numbers of CD8+ TIL per mg of tumor. (I) CD8+/CD4+Foxp3+ Treg ratio. (J and K) Percentage of CD8+ (J) and CD4+ (K) TIL producing IFN-γ and TNF-α after PMA and ionomycin stimulation. (L and M) Percentage of CD8+ (L) and CD4+ (M) TIL staining for GzmB and PFN. (N) EpCAM MFI of eGFP+ tumor cells. (G–N, n = 5). (O) Growth of 4T1E tumors in mice treated with EpCAM aptamer or the combined AsiC (red arrows) in combination with anti-PD-1 or isotype control antibody (blue arrows) (n = 4). Data shown are mean + SEM. Statistical tests: (A, G, and O) tumor growth curves were compared by calculating the area under the curve values for each sample followed by one-way ANOVA with Holm–Sidak’s multiple comparisons or Student’s t test. (C, H, and N) Student’s t test. (D–F and J–M) Multiple t tests with Holm–Sidak corrections. (I) Mann–Whitney test. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
We next investigated whether tumor inhibition by the EpCAM-AsiC mixture could be improved by anti–PD-1 (Fig. 4O). Treating control mice receiving EpCAM aptamer with anti–PD-1 did not affect 4T1E tumor growth. However, combining anti–PD-1 and the EpCAM-AsiC mixture significantly reduced tumor growth more than the AsiC mixture on its own. The EpCAM-AsiC mixture markedly reduced PD-1 levels on CD44+CD8+ TIL (SI Appendix, Fig. S7A). The addition of anti–PD-1 further reduced PD-1 staining when the same antibody clone (29F.1A12) was used for detection, presumably because the bound therapeutic antibody blocked staining. Although anti–PD-1 or the combined AsiC did not significantly affect CD44+CD8+ TIL expression of other coinhibitory receptors (CD244, CTLA-4, TIM-3, LAG-3), combining anti–PD-1 with the AsiC significantly reduced expression of CD244 and CTLA-4 (SI Appendix, Fig. S7B). Furthermore, adding anti–PD-1 to the AsiC mixture strongly increased the number of CD8+ (SI Appendix, Fig. S7C) and NK (SI Appendix, Fig. S7D) TIL and stimulated cytokine production by CD8+ TIL (SI Appendix, Fig. S7E) compared to mice treated with just the AsiC mixture. Thus, an AsiC mixture targeting tumor cell immune evasion could potentially synergize with T cell inhibitory receptor blockade.
EpCAM-AsiC Mixture Broadly Augments Tumor-Infiltrating T Cell and Macrophage Functionality.
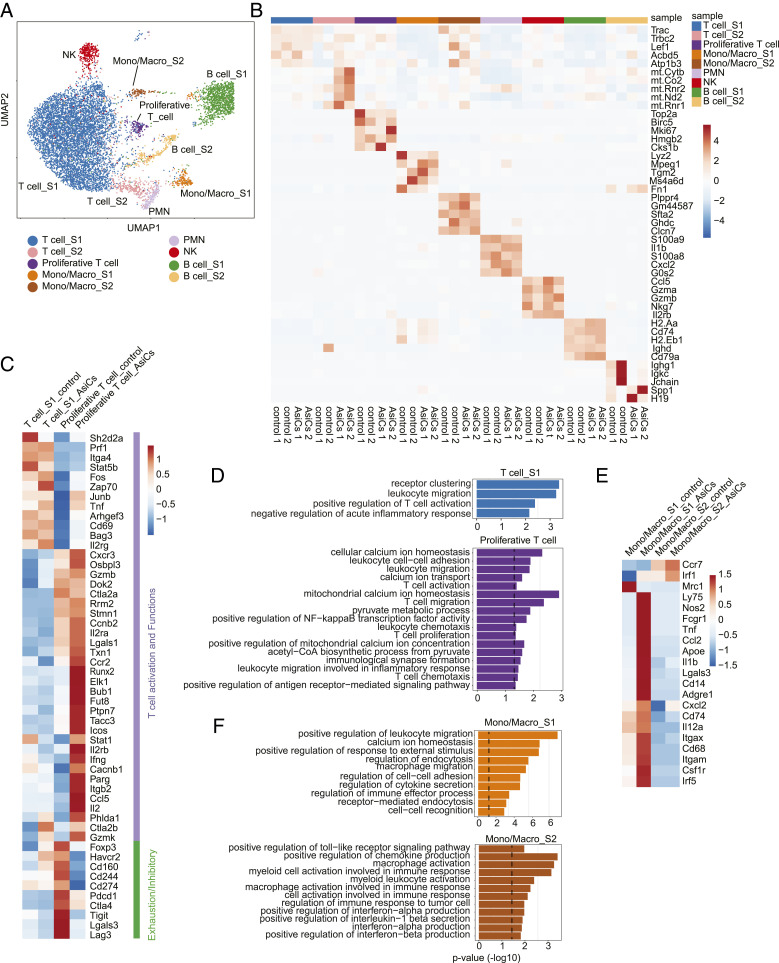
To assess without bias changes in tumor-infiltrating immune cells induced by the four EpCAM-AsiC, single-cell RNA-seq (scRNA-seq) compared CD45+ tumor-infiltrating cells from mice bearing 4T1E orthotopic tumors treated with EpCAM aptamer or the AsiC mixture (Fig. 5 A and B). The abundance of immune cell subsets, defined by expression of key subtype-defining genes, did not significantly change. Because cell numbers were limiting even with samples pooled from multiple mice, our analysis focused on tumor-infiltrating T cells and monocyte/macrophages, which showed the greatest gene-expression changes after EpCAM-AsiC treatment. Three distinct subpopulations of T cells were identified (T cell_s1, T cell_s2, proliferative T cells). However, T cell_s2 highly expressed mitochondrial genes, suggesting this subset contained a high proportion of dying cells (Fig. 5B). Therefore, our analysis focused on T cell_s1 and proliferative T cells. Compared to proliferative T cells, T cell_s1 were less activated with lower expression of proliferative and functional genes (e.g., Mki67, CD44, Gzmb), suggesting this cell subpopulation might be mostly bystander T cells (Fig. 5 B and C), as described in previous studies of human tumors (35). On the other hand, many of the proliferative T cells could be tumor-specific. Analysis of differentially expressed genes (DEG) in AsiC-treated versus control cells revealed higher expression of genes related to leukocyte migration and early stages of T cell activation in AsiC-treated T cell_s1 cells, while AsiC-treated proliferative T cells significantly increased expression of genes associated with T cell activation, proliferation and metabolism, migration/chemotaxis, immunological synapse formation, and effector functions (Fig. 5 C and D). The T cell_s1 subpopulation more highly expressed genes involved in the early signaling events of T cell activation (e.g., Fos, Zap70, Junb, and Cd69), which were further up-regulated by EpCAM-AsiC treatment (Fig. 5C). Proliferative T cells in AsiC-treated tumors up-regulated effector and memory and functional T cell genes (e.g., transcripts encoding effector molecules Ifng, Tnf, Il2, Gzmb, Gzmk); the costimulatory gene Icos; IL-2 receptor complex genes IL2ra, IL2rb, and IL2rg; and Runx2, which promotes the long-term persistence of CD8+ memory T cells. T cell functional genes (e.g., Gzmb, Gzmk, Prf1, and Tnf) were also increased in the T cell_s1 subset after EpCAM-AsiC treatment. In contrast, genes encoding coinhibitory molecules( e.g., Pdcd1, Ctla4, Tigit, Lag3, and Havcr2), and the Treg signature gene Foxp3 were mostly down-regulated in T cells from AsiC-treated tumors, especially in the proliferative T cell cluster (Fig. 5C), suggesting that EpCAM-AsiC could ameliorate T cell exhaustion.
Fig. 5.
Functional improvement of tumor-infiltrating immune cells after treatment with immune-modulating EpCAM-AsiC as analyzed by scRNA-seq. CD45+ cells from 4T1E tumors in mice treated with EpCAM aptamer or EpCAM-AsiC targeting Upf2, Cd47, Mcl1, and Parp1 were pooled (n = 2) for scRNA-seq analysis. (A) Uniform manifold approximation and projection (UMAP) plot of CD45+ immune cells in all four samples. Each dot represents a cell that was colored by inferred cluster identity. (B) Heatmap showing the z-score–normalized expression of canonical cell-type DEGs across different clusters. Each column represents a biological sample. (C and D) Heatmap showing expression of genes involved in T cell activation, effector function, memory and exhaustion in T cell clusters, averaged per cluster and z-score standardized across clusters (C) and GO enrichment for DEGs up-regulated in EpCAM-AsiC-treated T cells (D). (E and F) Heatmap showing expression of genes involved in monocyte/macrophage phenotype and function in monocyte/macrophage clusters, averaged per cluster and z-score standardized across clusters (E) and GO enrichment for DEGs up-regulated in EpCAM-AsiC–treated monocytes/macrophages (F). Dashed lines in D and F indicate P = 0.05 (one-sided Fisher’s exact test).
Two monocyte/macrophage clusters (mono/macro_s1, mono/macros2) were identified in the tumor. Gene expression in the mono/macro_s1 subpopulation was more affected by AsiC treatment. Genes related to monocyte/macrophage migration, activation, and endocytosis were markedly up-regulated in the EpCAM-AsiC–treated mono/macro_s1 subpopulation, while genes involved in inflammation and IFN-I and chemokine production, including genes that regulate the immune response to tumors, were up-regulated in the mono/macros2 subpopulation (Fig. 5 E and F). Furthermore, monocyte/macrophages in the AsiC-treated tumors up-regulated expression of genes associated with myeloid cell maturation (e.g., Cd74); M1 TAM functionality (e.g., Nos2, Fcgr1, Cd68, Il12a, and Ccr7), phagocytosis and antigen processing (e.g., Lgals3, Il1b, Apoe, Cd14, and Ly75); and inflammatory cytokine/chemokine production (e.g., Tnf, Ccl2, Cxcl2, and Il12a); while genes associated with M2-like TAMs (e.g., Mrc1) were markedly down-regulated, suggesting improved antitumor functionality.
EpCAM-AsiC Reduce Metastatic Tumor Growth.
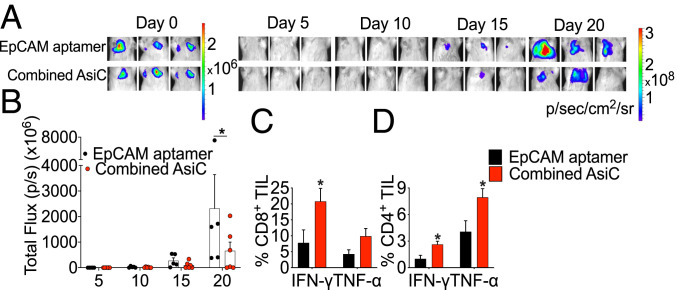
All the experiments described so far treated mice when orthotopic tumors became palpable. However, breast cancer patients often present with more advanced local or metastatic disease, which is more difficult to treat. Moreover, metastases are what usually kills patients. Many breast cancer patients also have microscopic metastases before metastatic disease becomes clinically apparent. Therefore, the ability to target metastatic tumor cells is critical for effective therapy. To determine whether EpCAM-AsiC are active against metastatic TNBC, we generated a 4T1E cell line stably expressing firefly luciferase (4T1E-Luc), detectable by bioluminescence imaging of live animals. After intravenous injection of 4T1E-Luc, lung tumor cells could be detected 7 to 10 d later. Mice bearing 7-d-old metastatic 4T1E-Luc lung tumors were treated with EpCAM aptamer or an EpCAM-AsiC mixture targeting Upf2, Cd47, Parp1, and Mcl1. EpCAM-AsiC significantly inhibited lung metastases (Fig. 6 A and B). Twenty days after tumor challenge, CD8+ and CD4+ T cells isolated from the lungs were analyzed for PMA and ionomycin-stimulated production of IFN-γ and TNF-α (Fig. 6 C and D). Significantly more lung CD8+ and CD4+ T cells from mice treated with EpCAM-AsiC produced these cytokines. Thus EpCAM-AsiC inhibited the growth of metastatic tumors and augmented tumor immunogenicity in the lung.
Fig. 6.
Effect of immune-modulating EpCAM-AsiC on metastatic 4T1E-Luc lung tumors. (A) Representative luminescent images of mouse lung areas at indicated days after tail-vein injection of 4T1E-Luc tumor cells. Mice were treated with EpCAM aptamer (Upper) or EpCAM-AsiC targeting Upf2, Cd47, Mcl1, and Parp1 (Lower). (B) Total luminescent photon flux of lung metastases in each mouse on indicated days after tumor cell injection. (C and D) Percentage of CD8+ (C) and CD4+ (D) T cells from lungs isolated on day 21 producing IFN-γ and TNF-α after PMA and ionomycin stimulation. (EpCAM aptamer-treated group, n = 5; AsiC group, n = 6). Data show mean ± SEM. Statistical tests: (B–D) multiple t tests with Holm–Sidak’s multiple comparisons. *P ≤ 0.05.
EpCAM-AsiC Inhibit Aggressive Breast Cancers in Erb2ΔEx16 Transgenic Mice.
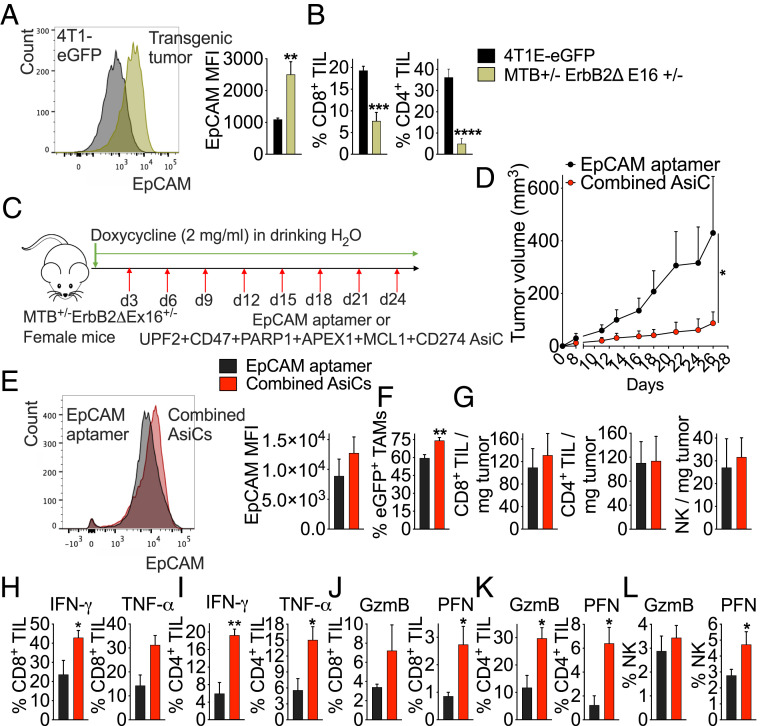
Genetically engineered mouse (GEM) models of cancer are notoriously difficult to control with immunotherapy, in part because genetically engineered tumors contain few mutations that can be recognized as foreign. To evaluate EpCAM-AsiC, we chose an aggressive HER2+ breast cancer model in which eGFP and a truncated HER2 gene (ErbB2ΔEx16, encoding a constitutively active HER2 receptor) are doxycycline-inducible under the control of the mouse mammary tumor virus promoter (36). At least 80% of these mice develop multifocal rapidly growing and metastatic HER2+ breast tumors that are uniformly EpCAM+ (Fig. 7A) within ∼10 to 28 d of adding doxycycline. Untreated tumors had few infiltrating CD4 or CD8 T cells (Fig. 7B). Erb2ΔEx16 mice were treated subcutaneously beginning 3 d after starting doxycycline and every third day thereafter with either EpCAM aptamer as a control or a combination of six EpCAM-AsiC targeting genes (Upf2, Parp1, Apex1, Cd47, Mcl1, and Cd274) (Fig. 7C). Although treatment with the EpCAM-AsiC mixture did not alter the number of mice with detectable tumors (four of six mice in each group developed tumors in 10 d), it greatly reduced tumor size after 4 wk of treatment (Fig. 7D). After eight treatments, EpCAM expression by the GFP+ tumor did not change, suggesting that down-regulation of EpCAM is not a mechanism of tumor resistance (Fig. 7E). Moreover, antitumor immune function was significantly enhanced in mice receiving the EpCAM-AsiC mixture. More TAM were GFP+, indicating increased tumor cell phagocytosis (Fig. 7F). Although the EpCAM-AsiC mixture did not change the numbers of TIL within spontaneously arising ErbB2ΔEx16 tumors (Fig. 7G), stimulated production of IFN-γ and TNF-α by CD8+ and CD4+ TIL was enhanced (Fig. 7 H and I) and GzmB and PFN expression were increased in CD8+, CD4+, and NK TIL (Fig. 7 J–L). Thus, an EpCAM-AsiC mixture suppressed tumor growth and mobilized antitumor immunity in an immunologically “cold” highly aggressive spontaneous GEM breast tumor model.
Fig. 7.
Immune-modulating EpCAM-AsiC suppress spontaneous tumors in ErbB2ΔEx16 transgenic mice. (A) EpCAM MFI of ErbB2ΔEx16 transgenic tumor cells compared to 4T1E-eGFP tumor cells. Shown at Left are representative histograms of EpCAM expression in GFP+ tumor cells and at Right, mean MFI of mice in each group. (B) Percentage of CD8+ (Left) and CD4+ (Right) TIL in ErbB2ΔEx16 transgenic and 4T1E-eGFP tumors. (A and B) 4T1E-eGFP, n = 5; ErbB2ΔEx16, n = 7. (C) Experimental scheme of tumor induction and treatment (red arrows) in ErbB2ΔEx16 transgenic mice. (D) Tumor growth in doxycycline-fed ErbB2ΔEx16 transgenic mice treated with EpCAM aptamer or EpCAM-AsiC mixture. (E–L) Flow cytometry characterization of single cell suspensions of tumors from treated mice isolated on day 26 after starting doxycycline. (E) Representative flow histograms (Left) and mean EpCAM MFI (Right) of tumor cells from ErbB2ΔEx16 transgenic mice treated with EpCAM aptamer or the AsiC mixture. (F) TAM phagocytosis measured by percentage of GFP+ TAMs. (G) Number of CD8+ (Left), CD4+ (Center), and NK (Right) TIL per milligram tumor. (H, I) Percentage of CD8+ (H) and CD4+ (I) TIL producing IFN-γ and TNF-α after stimulation with PMA and ionomycin. (J–L) GzmB and PFN staining of CD8+ (J), CD4+ (K), and NK (L) TIL. (D–L, n = 6). Data shown are mean + SEM. Statistical tests: (A, H, and l) Mann–Whitney test. (B, E–G and I–K) Student’s t test. (D) Multiple t tests with Holm–Sidak’s multiple comparisons. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Discussion
In this study we show that tumor cell-targeted gene knockdown with immune-modulating EpCAM-AsiC improves tumor immunogenicity and inhibits tumor growth in aggressive mouse TNBC and HER2+ breast cancer models. We used EpCAM-AsiC to knockdown six genes participating in different pathways and stages of the cancer-immunity cycle (Upf2, Parp1, Apex1, Mcl1, CD47, CD274) focusing on making breast cancers more visible to the immune system. Two of these gene products are targets of Food and Drug Administraiton-approved drugs (PARP1, PD-L1) and an antibody targeting a third (CD47) is in clinical development. Four of the AsiC (UPF2, PARP1, MCL1, CD47) significantly improved immune responses to the tumor and suppressed orthotopic tumor growth, while the remaining two AsiC (APEX1, CD274) were less active but trended toward reduced tumor growth. PD-L1 expression on some tumor-infiltrating immune cells also suppresses the antitumor functions of PD-1+ TIL, which may explain why knocking down PD-L1 selectively in tumor cells did not boost protective immunity. In two cases where compounds are in clinical use or in development to inhibit the knocked down gene product (olaparib for PARP1, anti-CD47), AsiC performed better than the inhibitors. Pharmacokinetic/pharmacodynamic characterization of systemically administered AsiC will be important both for future preclinical development and to help understand why AsiC were more active than a PARP1 inhibitor or anti-CD47 antibody.
We took advantage of the ease of putting together AsiC mixtures to show that AsiC combinations could be much more effective than single agents at activating antitumor immunity and suppressing tumor growth and could synergize with checkpoint inhibition. Mixtures could provide strategies to compensate for the shortcomings of knocking down an individual gene. For example, getting the tumor to express neoantigens might not be effective if the tumor microenvironment (TME) in the established tumor is immunosuppressive, but an AsiC combination that both induces new antigens and increases tumor phagocytosis or reduces checkpoint inhibition could be more effective. Future experiments will be needed to identify which combinations are synergistic and which AsiC are most important in a mixture. A limitation of our study was that we initiated therapy when the tumors were still small and followed tumor growth for only a few weeks. Longer-term studies will be needed to evaluate whether AsiC suppress larger tumors, lead to tumor regression or drug resistance, and induce durable immune memory.
Importantly, we showed that AsiC combinations potently inhibited metastatic disease and a very aggressive GEM cancer. To our knowledge, it has not been previously possible to inhibit any GEM model using immunotherapy because the genetically engineered oncogenes so potently promote tumorigenesis that the mutational burden in spontaneous GEM tumors is low, precluding effective immune recognition. The AsiC combination strongly enhanced CD8+ and CD4+ TIL cytokine production and cytotoxic functions and increased TAM phagocytosis of tumor cells. These increased functional properties in flow cytometry experiments were confirmed by scRNA-seq data comparing gene expression of tumor-infiltrating T cells and macrophages, which also showed strong down-regulation of Foxp3 and inhibitory receptors associated with immunosuppressive Treg and T cell exhaustion, respectively.
Tumor targeted gene knockdown could be used both to identify immune-modulating drug targets for conventional small-molecule or biologics drug development and as the basis for candidate drugs. Four RNA interference (RNAi) drugs that knock down gene expression in the liver (patisiran, givosiran, lumasiran, inclisiran) were recently approved for clinical use for diseases mediated by genes expressed in the liver (37, 38). Gene knockdown in the liver is now being developed for many indications using GalNAc-conjugated siRNAs targeted to the abundant asialoglycoprotein receptor on hepatocytes, but achieving targeted siRNA gene knockdown in other tissues and disseminated cancer cells has been challenging (39). EpCAM-AsiC are an attractive platform for selective gene knockdown in the common epithelial cancers, which highly express EpCAM on the plasma membrane. Here we showed effective antitumor effects after subcutaneous AsiC injection not only in distal orthotopic tumors, but also in lung metastases. Normal epithelial cells express much lower levels of EpCAM than epithelial cancers and expression is restricted to gap junctions, which may be largely inaccessible to AsiC. In fact, in a previous study tumor targeting was so selective that knockdown of an essential gene in dividing cells (PLK1) did not cause any apparent uptake or toxicity to normal epithelia (6). In this study, tumors from mice that received seven AsiC injections did not become resistant to EpCAM-AsiC by down-modulating EpCAM, perhaps because EpCAM acts as an oncogenic signaling protein that promotes breast cancer cell proliferation and migration and its down-modulation might suppress tumor growth (40). Future studies should investigate whether other mechanisms of resistance develop, such as by tumor cell mutation of the targeted gene sequence that the siRNA sense strand recognizes or by altering other genes that function in the same pathway (41).
AsiC are a flexible platform for targeted gene knockdown (41) that offers the ability to target proteins that are “undruggable” by small-molecule drugs or intracellular proteins that antibodies can’t reach. By changing the aptamer, the platform can target different types of tumors or normal tissues. By changing the siRNA sequence, the same overall design can be used to knock down any gene. Moreover, as illustrated in this study, AsiC targeting multiple genes can be easily identified and combined to make drug mixtures that attack multiple targets with synergy and reduce the risk of tumor escape. This study offers further proof that AsiC could be potent agents of immune modulation, as already shown by the Gilboa, Rossi, and Kortylewski laboratories (19, 41–43), but we have just scratched the surface by studying a small number of plausible candidate genes. Future work could screen AsiC libraries to identify novel immune-enhancing gene candidate targets and the combinations that are most synergistic at inducing tumor immunity. Compared to conventional antibody-mediated targeting, RNA aptamers provide many additional advantages, such as their smaller size, low manufacturing cost, ability to be chemically synthesized, lack of biological variability, higher in vivo stability, low immunogenicity, lack of need for refrigeration, and ability to be lyophilized (42). We previously showed that EpCAM-AsiC do not stimulate immune responses, thereby allowing repeat administration without affecting their in vivo pharmacokinetics (6). EpCAM-AsiC also are unlikely to activate their receptor presumably because they do not cross-link it (44). AsiC could be optimized by chemical modifications to improve their pharmacological properties and the potency and durability of gene knockdown, as has been done for GalNAc-conjugated siRNA drugs (45). Chemically synthesized drugs, which are feasible for AsiC that use a small aptamer like the EpCAM aptamer, are simpler to develop and manufacture and cheaper than biological drugs, such as antibodies (42).
E. Gilboa pioneered using tumor-targeted AsiC to get tumors to express neoantigens. He used PSMA-AsiC to knock down NMD genes (Upf2, Smg1) to enhance tumor vaccination (19) and nucleolin AsiC to knockdown TAP, the gene encoding the transporter for antigen processing (46). TAP knockdown caused presentation of neoantigens. However, new antigen expression as the mechanism responsible for improved immune protection by NMD gene knockdown was not experimentally verified and may not be its mechanism of action. Cells deficient in NMD up-regulate aberrant mRNA splicing variants. NMD inhibition by Upf1 knockdown in N2A neuroblastoma was previously shown to alter expression of more than 200 exons (47). Here we showed that knocking down UPF2 reduced NMD activity in breast cancer cells grown in vitro and in vivo, induced DEU events in 281 genes, and generated a number of novel mRNA isoforms and NMD-sensitive transcripts that could encode tumor neoantigens. Upf2 knockdown in mouse 4T1 tumors led to increased numbers and function of CD8+ TIL and potently inhibited orthotopic tumor growth. Although some neoantigens induced by NMD inhibition may vary from cell to cell in the tumor, other changes in exon usage might lead to common expression of novel isoforms that could contain novel antigens (48). Determining whether this is the case would be challenging and require identifying the novel peptides presented by MHC on NMD-inhibited tumor cells, generating peptide-specific T cells that recognize them, and determining whether these T cells respond to a large fraction of cells within the tumor. Immunopeptidomics study of MHC class I-loaded peptides after UPF2 knockdown in breast cancer cells would help determine whether neoepitopes are generated that stimulate antitumor immunity.
However, the enhanced immune response generated after Upf2 knockdown slowed down tumor growth but did not cause tumor regression, whereas expressing a foreign antigen, such as GFP, in tumor cells often causes regression. One reason for this difference in antitumor efficacy could be that tumor neoantigen-specific T cells elicited by Upf2 knockdown would be primed and activated in an immunosuppressive TME, where their antitumor activities could be impaired compared to tumor-specific T cells that recognize a constitutively expressed tumor antigen primed in the early stages of tumorigenesis before immunosuppression sets in. Combining Upf2 knockdown with knockdown of other genes that reverse immune suppression in the TME or with checkpoint inhibitors might address this problem. NMD inhibition could also promote antitumor immunity by other mechanisms. NMD inhibition regulates transcripts involved in cellular stress responses and nutrient homeostasis, and environmental stresses also inhibit NMD (24). Our RNA-seq analysis identified changes in both DEU and DIU after UPF2 knockdown that might affect tumor growth independently of immune regulation. Future studies to analyze changes in tumor growth after treatment with UPF2 AsiC in immunodeficient mice could be used to determine whether some of the antitumor effect of Upf2 knockdown is independent of immune recognition.
A subset of TNBC have defective homologous recombination, a high-fidelity DNA double-strand break repair mechanism, which makes them susceptible to PARP1 inhibition. PARP1 recognizes sites of DNA damage of all sorts and recruits and activates DNA repair factors. Knocking down Parp1 should interfere with repair of ongoing DNA damage due to DNA oxidation, as well as DNA damage induced by radiation or cytotoxic chemotherapy, increasing the tumor DNA mutational burden, and causing genomic instability, which could be immunogenic by triggering cytosolic innate immune DNA sensors, like cGAS and IFI16, resulting in increased IFN-I production and cell death (49). Parp1 knockdown might also induce neoantigens, although any individual antigen would most likely be “private,” because it would be due to random sites of DNA damage and hence might be presented by a minority of cells in the tumor (although it could be transmitted to a single tumor cell’s progeny during cell proliferation). Future studies, such as T cell receptor cloning of TIL from control and PARP1 AsiC-treated tumors, could help determine whether neoepitopes are generated after Parp1 knockdown. PARP1 AsiC led to significantly more unrepaired DNA damage as measured by γ-H2AX staining, more TUNEL+ apoptotic tumor cells, and enhanced IFN-I production, which was associated with improved CD8+/CD4+ Treg ratio and cytokine production by both CD8+ and CD4+ TIL. On the other hand, the Food and Drug Administration-approved PARP1 inhibitor olaparib did not affect DNA damage repair or boost IFN-I expression and failed to significantly inhibit 4T1E tumor growth or boost antitumor T cell immunity. The reason for the difference between Parp1 knockdown and olaparib needs to be studied in more detail, but knockdown should interfere more strongly with DNA damage repair than inhibiting PARylation since it would block the initial recruitment of DNA repair factors to DNA damage sites.
Phagocytosis of dying cancer cells and tumor antigen cross-presentation are critical for initiating effective antitumor T cells. Tumor cells often foil T cell recognition by up-regulating CD47 to inhibit phagocytosis (32). The antitumor effect of CD47 AsiC was abrogated by anti-CD8 and significantly suppressed by anti-CD4 or anti-CSF1R, which can deplete not only macrophages but also tissue DCs (50). Indeed, CD47 AsiC enhanced the M1/M2 macrophage ratio and increased the percentage and maturation of tumor CD11c+DEC205+ DCs, which are specialized in taking up extracellular antigens and could cross-present tumor antigens. CD47 AsiC also promoted M1 TAM and CD205+ DC tumor phagocytosis. The antigen cross-presentation capacity of these DCs likely was also improved, which can be examined in future studies. CD47 AsiC outperformed anti-CD47 in suppressing 4T1E tumor growth for unclear reasons. Better tumor penetration of EpCAM-AsiC compared to antibodies might contribute to their superior antitumor activity. Although both therapies increased the function of CD8+ TIL, only CD47 AsiC improved CD4+ TIL function and reduced MDSCs in the tumor. Anti-CD47 therapy in mice was previously reported not to improve CD4+ T cell function (32). However, CD4+ T cell depletion markedly impaired the therapeutic efficacy of CD47 AsiC, clearly indicating that CD4+ T cells are important in CD47 AsiC effectiveness.
Materials and Methods
EpCAM-AsiCs designed to knock down Upf2, Parp1, Apex1, CD47, CD274, and Mcl1 were injected subcutaneously into mice bearing breast tumors and evaluated for tumor growth. Mice injected with the EpCAM aptamer were used as controls. At killing, single-cell suspensions of tumors were analyzed for infiltrating immune cell numbers, phenotypes, and functions. The cell lines, mouse strains, and all animal experiments are described in detail in the SI Appendix, Materials and Methods. Mouse studies were conducted in compliance with all ethical regulations and were approved by the Harvard Medical School Institutional Animal Care and Use Committee. All relevant materials and methods, including design of RNA constructs and methods for analyzing gene knockdown, histology, and IHC, isolation of immune cells from mice, antibody staining and flow cytometry, CD8+ TIL degranulation and cytotoxicity, ex vivo phagocytosis, scRNA-seq, bulk RNA-seq, and statistical analysis, are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank past and present members of the J.L. laboratory who contributed to the development of this work and provided helpful discussions; Dr. W. J. Muller for providing the genetically engineered mouse model of breast cancer; the Harvard University Single Cell Core for providing inDrop single-cell encapsulation service; and the Specialized Histopathology and Tissue Microarray Imaging Cores of the Dana-Farber/Harvard Cancer Center for histology analysis. The Dana-Farber/Harvard Cancer Center is supported in part by National Cancer Institute Cancer Center Support Grant NIH 5 P30 CA06516. This work was supported by NIH Grant R01CA184718 (to J.L.) and a Department of Defense Breast Cancer Breakthrough Fellowship Award W81XWH-19-1-0039 (to Y.Z.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022830118/-/DCSupplemental.
Data Availability
The scRNA-seq and RNA-seq data have been deposited in the Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo [accession nos. GSE156214 (51) and GSE156185 (52)]. All other study data are included in the article and supporting information.
References
- 1.Yang Y., Cancer immunotherapy: Harnessing the immune system to battle cancer. J. Clin. Invest. 125, 3335–3337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence M. S., et al., Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinmann S. C., Pisetsky D. S., Mechanisms of immune-related adverse events during the treatment of cancer with immune checkpoint inhibitors. Rheumatology (Oxford) 58 (suppl. 7), vii59–vii67 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vinay D. S., et al., Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin. Cancer Biol. 35 (suppl.), S185–S198 (2015). [DOI] [PubMed] [Google Scholar]
- 5.J. O. McNamara, 2nd, et al., Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 24, 1005–1015 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Gilboa-Geffen A., et al., Gene knockdown by EpCAM aptamer-siRNA chimeras suppresses epithelial breast cancers and their tumor-initiating cells. Mol. Cancer Ther. 14, 2279–2291 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Imrich S., Hachmeister M., Gires O., EpCAM and its potential role in tumor-initiating cells. Cell Adhes. Migr. 6, 30–38 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shigdar S., et al., RNA aptamer against a cancer stem cell marker epithelial cell adhesion molecule. Cancer Sci. 102, 991–998 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Kumar P., Aggarwal R., An overview of triple-negative breast cancer. Arch. Gynecol. Obstet. 293, 247–269 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Padmanabhan R., Kheraldine H. S., Meskin N., Vranic S., Al Moustafa A. E., Crosstalk between HER2 and PD-1/PD-L1 in breast cancer: From clinical applications to mathematical models. Cancers (Basel) 12, 636 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Mahmood S., Sapiezynski J., Garbuzenko O. B., Minko T., Metastatic and triple-negative breast cancer: Challenges and treatment options. Drug Deliv. Transl. Res. 8, 1483–1507 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cameron D.et al., Herceptin Adjuvant (HERA) Trial Study Team , 11 years’ follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive early breast cancer: Final analysis of the HERceptin adjuvant (HERA) trial. Lancet 389, 1195–1205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerji S., et al., Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 486, 405–409 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams S., et al., Current landscape of immunotherapy in breast cancer: A review. JAMA Oncol. 5, 1205–1214 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao G., Wang Z., Qu X., Zhang Z., Prognostic value of tumor-infiltrating lymphocytes in patients with triple-negative breast cancer: A systematic review and meta-analysis. BMC Cancer 20, 179–15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kroemer G., Senovilla L., Galluzzi L., André F., Zitvogel L., Natural and therapy-induced immunosurveillance in breast cancer. Nat. Med. 21, 1128–1138 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Chan S., et al., Basal-A triple-negative breast cancer cells selectively rely on RNA splicing for survival. Mol. Cancer Ther. 16, 2849–2861 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrocca F., et al., A genome-wide siRNA screen identifies proteasome addiction as a vulnerability of basal-like triple-negative breast cancer cells. Cancer Cell 24, 182–196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pastor F., Kolonias D., Giangrande P. H., Gilboa E., Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature 465, 227–230 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popp M. W., Maquat L. E., Attenuation of nonsense-mediated mRNA decay facilitates the response to chemotherapeutics. Nat. Commun. 6, 6632 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takada K., et al., Use of the tumor-infiltrating CD8 to FOXP3 lymphocyte ratio in predicting treatment responses to combination therapy with pertuzumab, trastuzumab, and docetaxel for advanced HER2-positive breast cancer. J. Transl. Med. 16, 86–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng G.-L., et al., CD8+ cytotoxic and FoxP3+ regulatory T lymphocytes serve as prognostic factors in breast cancer. Am. J. Transl. Res. 11, 5039–5053 (2019). [PMC free article] [PubMed] [Google Scholar]
- 23.Gardner L. B., Nonsense-mediated RNA decay regulation by cellular stress: Implications for tumorigenesis. Mol. Cancer Res. 8, 295–308 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardner L. B., Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol. Cell. Biol. 28, 3729–3741 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendell J. T., Sharifi N. A., Meyers J. L., Martinez-Murillo F., Dietz H. C., Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36, 1073–1078 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Pantelidou C., et al., PARP inhibitor efficacy depends on CD8+ T-cell recruitment via intratumoral STING pathway activation in BRCA-deficient models of triple-negative breast cancer. Cancer Discov. 9, 722–737 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zitvogel L., Galluzzi L., Kepp O., Smyth M. J., Kroemer G., Type I interferons in anticancer immunity. Nat. Rev. Immunol. 15, 405–414 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Shah F., et al., Exploiting the Ref-1-APE1 node in cancer signaling and other diseases: From bench to clinic. npj Precision Oncology 1, 19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xanthoudakis S., Smeyne R. J., Wallace J. D., Curran T., The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl. Acad. Sci. U.S.A. 93, 8919–8923 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrissey M. A., Kern N., Vale R. D., CD47 ligation repositions the inhibitory receptor SIRPA to suppress integrin activation and phagocytosis. Immunity 53, 290–302.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X., et al., CD47 blockade triggers T cell-mediated destruction of immunogenic tumors. Nat. Med. 21, 1209–1215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tseng D., et al., Anti-CD47 antibody-mediated phagocytosis of cancer by macrophages primes an effective antitumor T-cell response. Proc. Natl. Acad. Sci. U.S.A. 110, 11103–11108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genard G., Lucas S., Michiels C., Reprogramming of tumor-associated macrophages with anticancer therapies: Radiotherapy versus chemo- and immunotherapies. Front. Immunol. 8, 828 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell K. J., et al., MCL-1 is a prognostic indicator and drug target in breast cancer. Cell Death Dis. 9, 19–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simoni Y., et al., Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557, 575–579 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Turpin J., et al., The ErbB2ΔEx16 splice variant is a major oncogenic driver in breast cancer that promotes a pro-metastatic tumor microenvironment. Oncogene 35, 6053–6064 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Setten R. L., Rossi J. J., Han S.-P., The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 18, 421–446 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Scott L. J., Givosiran: First approval. Drugs 80, 335–339 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Lieberman J., Tapping the RNA world for therapeutics. Nat. Struct. Mol. Biol. 25, 357–364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baeuerle P. A., Gires O., EpCAM (CD326) finding its role in cancer. Br. J. Cancer 96, 417–423 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lieberman J., Manipulating the in vivo immune response by targeted gene knockdown. Curr. Opin. Immunol. 35, 63–72 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J., Rossi J., Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 16, 440 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Sakib Hossain D. M., Duttagupta P., Kortylewski M., The aptamer-siRNA conjugates: Reprogramming T cells for cancer therapy. Ther. Deliv., 10.4155/tde.14.92 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wheeler L. A., et al., TREX1 knockdown induces an interferon response to HIV that delays viral infection in humanized mice. Cell Rep. 15, 1715–1727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nair J. K., et al., Impact of enhanced metabolic stability on pharmacokinetics and pharmacodynamics of GalNAc-siRNA conjugates. Nucleic Acids Res. 45, 10969–10977 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garrido G., et al., Tumor-targeted silencing of the peptide transporter TAP induces potent antitumor immunity. Nat. Commun. 10, 3773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ni J. Z., et al., Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 21, 708–718 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tani H., et al., Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol. 9, 1370–1379 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dziadkowiec K. N., Gąsiorowska E., Nowak-Markwitz E., Jankowska A., PARP inhibitors: Review of mechanisms of action and BRCA1/2 mutation targeting. Przegl. Menopauz. 15, 215–219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacDonald K. P. A., et al., The colony-stimulating factor 1 receptor is expressed on dendritic cells during differentiation and regulates their expansion. J. Immunol. 175, 1399–1405 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y., Naderi Yeganeh P., Hide W., Lieberman J., Immunotherapy for breast cancer using EpCAM aptamer tumor-targeted gene knockdown efficacy_UPF2 knockdown in EpCAMhi MDA-MB-231 breast cancer cell lines. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE156185. Deposited 13 August 2020.
- 52.Zhang Y., Xie X., Lieberman J., Immunotherapy for breast cancer using EpCAM aptamer tumor-targeted gene knockdown efficacy. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE156214. Deposited 13 August 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq and RNA-seq data have been deposited in the Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo [accession nos. GSE156214 (51) and GSE156185 (52)]. All other study data are included in the article and supporting information.