Significance
Immune cells in the body encounter stimuli in complex temporal patterns, but our understanding of how the timing of stimulation determines immune responses is incomplete. We engineered T cells obtained from healthy human donors to respond to light as a stimulus in a laboratory setting. We discovered that T cells can filter out specific signals based only on signal frequency by stimulating engineered T cells with light-based temporal stimulation patterns designed to mimic the dynamic patterns a natural T cell might experience in the human body. This filtering mechanism could contribute to how immune cells distinguish between foreign/altered cells versus self-cells in vivo, since foreign/altered and self-cells exhibit highly distinct interaction dynamics with T cells within the body.
Keywords: synthetic biology, signal transduction, optogenetics, cell signaling
Abstract
T cells experience complex temporal patterns of stimulus via receptor–ligand-binding interactions with surrounding cells. From these temporal patterns, T cells are able to pick out antigenic signals while establishing self-tolerance. Although features such as duration of antigen binding have been examined, our understanding of how T cells interpret signals with different frequencies or temporal stimulation patterns is relatively unexplored. We engineered T cells to respond to light as a stimulus by building an optogenetically controlled chimeric antigen receptor (optoCAR). We discovered that T cells respond to minute-scale oscillations of activation signal by stimulating optoCAR T cells with tunable pulse trains of light. Systematically scanning signal oscillation period from 1 to 150 min revealed that expression of CD69, a T cell activation marker, reached a local minimum at a period of ∼25 min (corresponding to 5 to 15 min pulse widths). A combination of inhibitors and genetic knockouts suggest that this frequency filtering mechanism lies downstream of the Erk signaling branch of the T cell response network and may involve a negative feedback loop that diminishes Erk activity. The timescale of CD69 filtering corresponds with the duration of T cell encounters with self-peptide–presenting APCs observed via intravital imaging in mice, indicating a potential functional role for temporal filtering in vivo. This study illustrates that the T cell signaling machinery is tuned to temporally filter and interpret time-variant input signals in discriminatory ways.
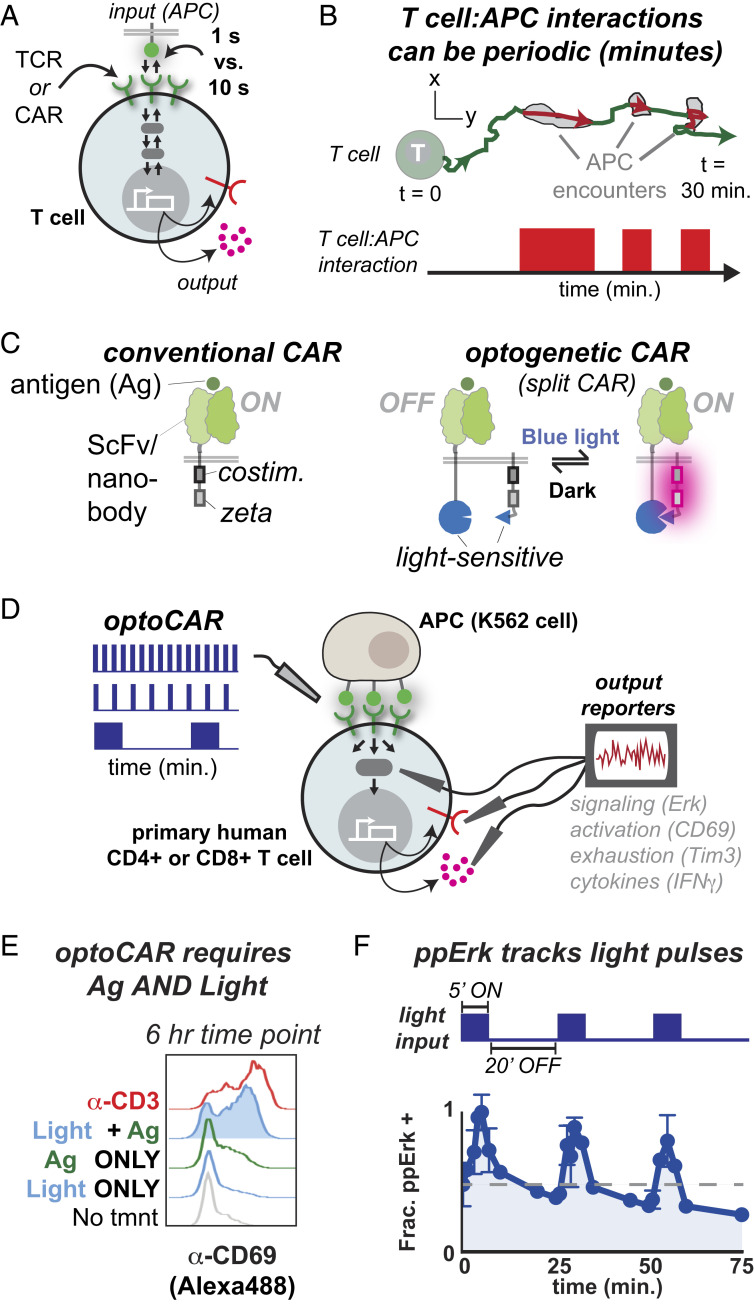
Receptor–ligand interactions on the surface of eukaryotic cells initiate reversible signaling and transcriptional cascades that ultimately culminate in long-lasting or irreversible changes in gene expression and functional activation (1, 2). In T cells, activating signals are initiated by either the native T cell receptor (TCR) or by engineered chimeric antigen receptors (CARs) that share the same downstream signaling pathways as a TCR but include user-defined ligand-recognition domains (3–5) (Fig. 1A). The intracellular signaling process initiated by TCRs or CARs is essential for the adaptive immune system to maintain homeostasis by first detecting and then responding to altered cells and tissues. The timing of a T cell signal may critically determine a T cell response to a potentially altered cell (2, 6–9). For instance, in the moth cytochrome c altered peptide ligand series, TCR–ligand interactions with molecular dwell times lasting only tens of seconds are sufficient to initiate T cell signaling and commit to changes in gene expression, whereas TCR–ligand interactions with molecular dwell times of just 1 s are not (10, 11) (Fig. 1A). These subtle differences can impact whether or not a T cell undergoes positive versus negative selection in the thymus (12) or whether a T cell is activated in a lymph node (13).
Fig. 1.
Systematically probing the transmission of periodic signals in T cells using optogenetics. (A) Dynamic receptor–ligand interactions regulate T cell gene expression by initiating reversible signaling and transcriptional cascades. Differences in receptor–ligand half-life as small as 1 s versus 10 s can result in dramatic differences in gene expression. (B) T cell–APC interactions can be highly periodic, introducing an element of T cell regulation on a longer, minutes timescale. (C, Left) A conventional CAR contains ITAM and costimulatory domains fused to a ligand-recognition domain, in this case an ScFv or nanobody. (C, Right) The optogenetic CAR is designed to split the ligand-recognition and signaling components of a CAR onto two separately expressed polypeptides, which are reversibly heterodimerized via light-sensitive domains. The system associates when blue light turns on and dissociates in the dark. (D) We used the optoCAR to stimulate primary human CD4+ T cells with temporally modulated waveforms and measured expression of cell surface receptors and cytokines. (E) The optoCAR induces CD69 expression at similar levels as the α-CD3 antibody OKT3 but only when light and antigen are present. Representative of over 10 independent experiments and seven donors. (F) Erk Thr202/Tyr204 phosphorylation turns on and off in response to several cycles of blue light illumination. The gray dashed line indicates initial ppErk levels. Representative of two independent experiments from one donor.
T cells are also highly motile compared to other cell types, which introduces an additional layer of temporal regulation to T cell activation (14, 15). As T cells traverse the body, they interact with antigen-presenting cells (APCs) displaying various self-peptides, with potential presentation of agonist or tumor-associated ligands (in the case of CARs). Intravital and in vitro imaging of both TCR and CAR T cells reveals that T cell interactions with APCs can be periodic, with irregular contact and rest intervals on the order of minutes to hours (16–22) (Fig. 1B). In analogy to the duration of a single molecule TCR–ligand interaction, the duration of a TCR T cell–APC contact interval is determined by the quality of the agonist ligand. APCs in the lymph node or skin displaying short dwell time self-ligand typically engage T cells for ∼3 to 11 min, whereas APCs displaying long dwell time agonist ligand form T cell–APC contacts for 30 min or more (16, 21, 22). Intravital imaging of α-CD19 CAR T cells indicates that CAR T cell–APC interactions are similarly periodic (19). Biophysical studies over several decades have quantitatively characterized the initial, seconds-scale receptor–ligand interactions and downstream signaling cascades driving T cell activation (6–11, 23) (Fig. 1A). However, our quantitative understanding of the impact of signal periodicity on T cell activation patterns remains relatively unexplored, despite the observed periodicity of TCR and CAR T cell–APC interactions (14–22) (Fig. 1B).
Our understanding of the impact of signal periodicity on cell signaling is more advanced in yeast and other mammalian cell types in which synthetically driven oscillatory signals with regular periodic intervals have uncovered mechanisms critical to cellular response and behavior. In yeast, synthetic control of oscillations in salt concentration was used to quantitatively dissect the role of negative feedback loops in the yeast osmotic shock response (24, 25). These negative feedback loops adapt the yeast signaling system to different salt environments and prepare the yeast for subsequent environmental stimuli. More recent work used a similar experimental system to reveal the existence of an “Achilles heel” in the same yeast osmotic shock response network—a specific resonant frequency at which the negative feedback loop regulating the shock response is defeated and the yeast consequently die (26). In mammalian cells, optogenetic approaches that enable reversible control of activation signals using light have revealed the existence of signal persistence detection via a paracrine cytokine circuit, a cancer mutation-dependent alteration in Erk signaling dynamics in lung cancer cell lines, and bandpass filtering of immediate early gene (IEG) expression (27–29). Prior applications of optogenetic tools to T cell signaling have focused on early signaling outcomes (Ca2+, diacylglycerol, nuclear factor of activated T cells [NFAT]) (30–32) or on in vivo applications (33, 34).
We therefore asked if mechanisms regulating primary human CD4+ T cell gene expression responses to periodic stimuli could be revealed using a synthetic biology approach. We designed an optogenetic CAR (optoCAR) regulated by blue light and demonstrated that this receptor is a reversible ON/OFF switch driving gene expression in primary human CD4+ T cells. By stimulating T cells with tunable pulse trains, we discovered that T cells respond to minute-scale signal oscillations with regular ON and OFF intervals between 1 and 40 min. Systematically scanning the signal oscillation period from 1 to 150 min revealed that expression of CD69, a T cell activation marker, reaches a minimum at a period of ∼25 min (corresponding to 5 to 15 min pulse widths). Regular oscillatory signals with a period of ∼25 min or pulse widths of 5 to 15 min are filtered out while signals with shorter or longer periods induce CD69 expression, even when signals with different oscillation periods have the same integrated input. These experiments reveal that T cells can temporally filter minute-scale signals, in addition to kinetically proofreading shorter, second-scale TCR–ligand dwell times. The duration of irregular periodic encounters between T cells and self-presenting APCs observed in vivo via intravital imaging in mice is approximately equal to the pulse widths of regular optogenetic signal oscillations leading to filtering of CD69 expression. Temporal filtering on this longer timescale could therefore lead to the selective attenuation of signals derived from self-peptide presented by APCs. These results also suggest mechanisms by which T cells can be engineered with more precise detection and signal-processing capabilities.
Results
Designing an Optogenetic CAR: A Light-Gated T Cell ON/OFF Switch.
To construct light-responsive T cells, we engineered an optogenetically controlled chimeric antigen receptor (optoCAR) using a split receptor design similar to that of endogenous antigenic receptors, that is, the TCR/CD3 and B cell receptor (BCR) complexes, whose ligand-recognition and signaling domains are expressed as separate polypeptides (3). We constructed the optoCAR in two parts, analogous to a previously reported rapalog-controlled ON-switch CAR (35), such that we could reversibly control the association and dissociation of the optoCAR with light. Part I contains a ligand-binding domain [either an α-CD19 ScFv or an α-green fluorescent protein (GFP) nanobody (36)] fused to one-half of a blue light–sensitive heterodimer pair (Fig. 1C). Part II features a key signaling element: the immunoreceptor tyrosine-based activation motifs (ITAMs). ITAMs are pairs of tyrosine residues that, when phosphorylated, serve as docking sites for the tandem Src homology 2 (SH2) domains of Zeta-chain associated protein kinase 70 (ZAP-70). Phosphorylation of ITAMs by the membrane-associated lymphocyte-specific protein tyrosine kinase (Lck) and subsequent recruitment of ZAP-70 is the first and critical step that initiates T cell activation (37). The optoCAR is designed with the ITAMs fused to the 41BB costimulatory domain and the second half of the blue light–sensitive heterodimer pair (38) (Fig. 1C).
We explored different arrangements of the signaling and heterodimerizing domains in order to minimize background activation in the absence of light while still allowing for reversible and strong T cell activation in the presence of light. To facilitate a reversible, light-sensitive ON/OFF switch for T cell activation, we used a set of structurally well-defined heterodimerizing components: the improved light inducible dimer (iLID) and its binding partner, the short peptide SspB (39, 40) (SI Appendix, Fig. S1A). The iLID–SspB pair bind in a 1:1 stoichiometry when illuminated with blue light (∼470 nm) and rapidly dissociate in the dark [<30 s half-life in vitro (39)] when blue light illumination is removed. We screened candidate receptors for light-dependent activation in primary human CD4+ T cells by coculturing optoCAR-expressing T cells with K562 APCs and assessed CD69 up-regulation via flow cytometry (Fig. 1D). Screening was performed in high throughput using black-walled 96-well plates illuminated by a custom-built optoPlate illumination device equipped with an array of blue light-emitting diodes (LEDs) (28).
The simplest optoCAR design consisted of a cytoplasmic ITAM fragment that could be recruited to an antigen-binding membrane receptor component upon illumination with light. This design exhibited extremely low basal activation but also failed to induce expression of the activation marker CD69 (SI Appendix, Fig. S1B). As with the rapalog-controlled ON-switch CAR, we found that localizing both components of the split receptor to the membrane led to a dramatic increase in T cell activation. Among all candidate designs, two (I.b + II.c and I.c + II.g) exhibited suitable activation properties under illumination with minimal basal activation in the dark (SI Appendix, Fig. S1B). In order to further minimize background activation, engineered T cells were sorted for high part II and low part I expression, and the lowest affinity SspB peptide was also cloned into part II (40) (SI Appendix, Fig. S1 C and D). The final I.b + II.c design, which was selected in order to completely separate the ligand-binding and signaling motifs on distinct polypeptides, is referred to as the optoCAR.
Characterization of a Dual Input-Gated optoCAR Construct.
The optoCAR (I.b + II.c) expressed CD69 only when stimulated with both light and cognate antigen. In addition, the fraction of CD69+ T cells and the up-regulated CD69 expression level were comparable to T cells stimulated with the α-CD3 antibody OKT3 (Fig. 1E). Stimulating conditions (6 h of ∼4 mW/cm2 470 nm light) led to only a modest increase in T cell death compared to T cells from the same culture kept in the dark (∼85% live cells compared to ∼90%) (SI Appendix, Fig. S1E), suggesting that toxicity due to blue light illumination was tolerable. We also measured the phosphorylation state of the mitogen-activated protein kinase (MAPK) Erk in response to repeated pulsatile stimulations with light. Erk phosphorylation at Thr202/Tyr204 increases in response to a 5-min pulse of light and rapidly decays during a 20-min off period (Fig. 1F). Successive 5- or 30-min pulses continue to restimulate Erk phosphorylation, although each subsequent pulse results in lower Erk phosphorylation than the previous pulse. Increasing the pulse duration accelerates the decline in Erk phosphorylation (SI Appendix, Fig. S1F). These results indicate that we successfully constructed a reversible, light-gated ON/OFF switch for T cell activation that responds rapidly to minute-scale oscillations of stimulus.
Fast, Minute-Scale Oscillations of T Cell Stimulus Are Sufficient for CD69 Expression.
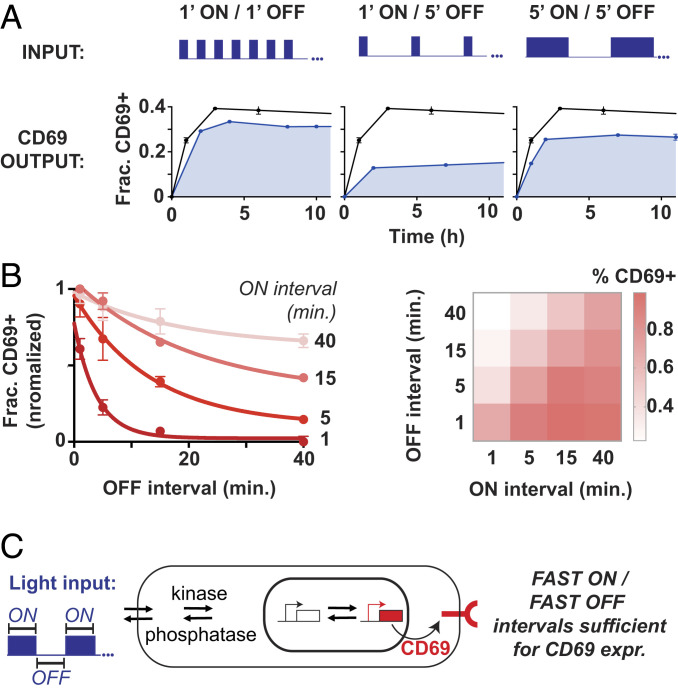
We next used the optoCAR to explore the timescales required to drive CD69 expression. We first programmed optoPlates with three types of pulse train—1′ ON/1′ OFF, 1′ ON/5′ OFF, and 5′ ON/5′ OFF, referred to as 1′/1′, 1′/5′, and 5′/5′ hereafter—and measured the fraction of T cells up-regulating CD69 in response to these pulse trains over a 10-h period. Within the first 2 h of stimulation, the fraction of CD69+ optoCAR T cells stimulated with a 1′/1′ pulse train reached a plateau with an amplitude ∼15% below the maximal amplitude reached with constant illumination (Fig. 2A). Increasing the OFF duration to 5 min—for a 1′/5′ pulse train—resulted in a similar plateau-like behavior but with a much lower response amplitude (∼10% versus ∼30%). This two-thirds decrease in the fraction of CD69+ T cells was almost completely rescued by increasing the ON interval to 5 min. The maximal amplitude in response to a 5′/5′ pulse train was persistently lower than the response to a 1′/1′ pulse train, despite the two waveforms having identical doses of light stimulus (Fig. 2A). Similar activation patterns were also observed in the expression of interferon-γ (IFN-γ), PD-1, and CD69 in CD8+ T cells (SI Appendix, Fig. S2).
Fig. 2.
CD69 expression is sensitive to fast, minute-scale oscillations in T cell stimulus. (A) optoPlates were programmed with 1′ ON/1′ OFF, 1′ ON/5′ OFF, and 5′ ON/5′ OFF waveforms, and the fraction of CD69+ T cells was assessed over a 10.5-h period. Black lines indicate the CD69 response to constant illumination, and shaded blue lines indicate the CD69 response to oscillating illumination. (B) The fraction of CD69+ T cells after 10 h of stimulation is plotted as a function of OFF interval for ON intervals of 1, 5, 15, and 40 min. The same matrix of ON/OFF intervals is represented graphically in the heatmap at right. Error bars are SEM, n = 2 technical replicates. (A and B) Representative of three independent experiments and two donors. (C) Oscillating optogenetic inputs may drive reversible signaling and transcriptional transitions that lead to CD69 expression. Fast ON/fast OFF periods are sufficient for CD69 expression.
We then systematically examined the timescales of ON and OFF pulse intervals necessary to stimulate CD69 expression by programming optoPlates with a matrix of square waveforms with pulse widths and off intervals ranging from 1 to 40 min. For all ON intervals tested, the fraction of CD69+ T cells decayed exponentially as a function of the OFF interval (Fig. 2B). Short, 1-min ON intervals required short OFF periods of at most 2 to 4 min in order to maintain half-maximal CD69 activation (Fig. 2B and SI Appendix, Fig. 2A). In contrast, longer ON intervals of 15 min maintained CD69 activation above half-maximal levels even when the OFF interval was extended as long as 15 min. In general, the OFF period required to maintain half-maximal CD69 activation increased nonlinearly as the ON interval was increased from 1 to 40 min (Fig. 2B and SI Appendix, Fig. S2B). This relationship might reveal the interplay between positive signaling regulators (such as an activating kinase) and negative signaling regulators (such as an inactivating phosphatase). For example, upon optoCAR stimulation, activating kinases and transcriptional activation likely dominated during ON intervals, while inactivating phosphatases and repression or deactivation of transcription dominated during OFF intervals (Fig. 2C).
Scanning Oscillation Period and Duty Cycle Reveals Attenuation of CD69 Expression at a Period of ∼25 Min.
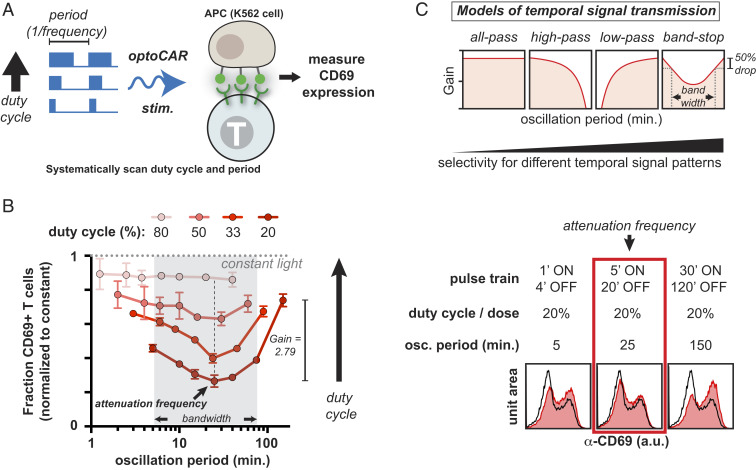
The observation that 5′/5′ pulse trains persistently stimulated a lower fraction of CD69+ T cells than 1′/1′ pulse trains raised the possibility that additional layers of signaling or transcriptional regulation may determine CD69 expression, since 1′/1′ and 5′/5′ pulse trains stimulate with identical doses of light (26). We therefore programmed optoPlates to emit pulse trains with oscillation periods ranging from 75 s to 150 min and duty cycles ranging from 20 to 80%. We then measured CD69 expression in response to these waveforms in optoCAR CD4+ T cells. Here, the duty cycle is simply the fraction of each period that the blue light is on (Fig. 3A). The pulse trains used were all regular—that is, for a 1′/4′ pulse train, the T cells in culture experienced 440 identical and consecutive stimulation periods composed of a 1′ illumination interval followed by a 4′ dark interval over the 22 h experiment duration. Although a T cell is likely to experience irregular periodic signals over a period of ∼24 h in vivo [e.g., in a lymph node or in the thymus (16, 21, 22)] (Fig. 1B), such a systematic scan with regular oscillation periods can reveal the timescales to which the T cell signaling network is tuned.
Fig. 3.
CD69 expression is a band-stop filter that is selectively attenuated at an oscillation period of ∼25 min. (A) optoPlates were programmed to systematically vary both the period (defined as 1/frequency) and duty cycle (the percentage of each period that the light is ON, from 1 to 100%) of blue light stimulation. optoCAR-expressing T cells cocultured with GFP+ K562 APCs were then stimulated with the programmed optoPlates for 22 h, and CD69 levels were assessed via flow cytometry. (B) The fraction of CD69+ T cells is plotted as a function of oscillation period and duty cycle, normalized to constant illumination. Trend lines indicate a single duty cycle. At constant duty cycles of 20, 33, and 50%, CD69 expression is attenuated at an oscillation period of ∼25 min, even though integrated input is constant. Bandwidth is calculated as the cutoff oscillation periods corresponding to an ∼50% drop in CD69 response. Gain is calculated as the ratio of CD69 response at the highest oscillation period versus at the attenuation frequency at a duty cycle of 20%. Error bars are SEM, n = 2 technical replicates. Representative of over seven independent experiments with CD4+ T cells derived from five healthy human donors. Histograms on the right represent raw data from which the plot at the left is derived. (C) Models of T cell signal transmission.
Higher duty cycle inputs increased CD69 up-regulation, as higher duty cycle inputs stimulated with a greater integrated signal. However, when duty cycle and therefore integrated input were kept constant, a high-resolution scan of oscillation period revealed that the fraction of CD4+ T cells expressing CD69 exhibited a minimum at a period of ∼25 min across several healthy human donors (Fig. 3 B and C and SI Appendix, Fig. S3 A and B). CD69 expression was attenuated at an oscillation period of ∼25 min in response to different duty cycle inputs (20, 33, and 50%) despite stimulation with variable ON and OFF intervals (equivalent to 5′/20′, 8′/16′, and 15′/15′ pulse trains, respectively) (Fig. 3B and SI Appendix, Fig. S3C). Differences in the fraction of CD69+ T cells in response to oscillation period appeared as early as 2 h after stimulation began and were maintained for at least 22 h (Fig. 3B and SI Appendix, Fig. S3D). As the duty cycle increased to 50% and above, the degree of attenuation of CD69 expression decreased, such that at an 80% duty cycle, the response to oscillation period was flat. The attenuation of CD69 expression occurred at a similar timescale as negative feedback acting on the Erk MAPK pathway (Fig. 1F and SI Appendix, Fig. S1F), indicating that the attenuation frequency and Erk negative feedback may be linked.
We also cultured CD4+ T cells in low (30 U/mL) and high (100 U/mL) interleukin-2 (IL-2) prior to optogenetic stimulation, since IL-2 is known to modulate T cell activation, and CD69 up-regulation could be influenced by cytokine signals. Increasing exogenous IL-2 concentration in culture increased the basal CD69 response but did not change the shape of the frequency response curve except for a slight decrease at an oscillation period of 150 min (SI Appendix, Fig. S3A).
Evolved biochemical systems can, in principle, filter frequency-modulated input signals with varying degrees of selectivity, in much the same way as engineered electronic or optical circuits (27, 41, 42) (Fig. 3C). At low duty cycles (thus, low integrated signals), the shape of the CD69 expression response to signal oscillations resembles a highly selective band-stop filter (Fig. 3 B and C) (42). This filter achieves an ∼50% drop in signal with a bandwidth of ∼70 min and a maximum gain of ∼2.79 with a mid or attenuation period of 25.8 ± 4.6 min (corresponding to 5 to 15 min pulse widths) (Fig. 3 B and C and SI Appendix, Fig. S3B). The error in the attenuation period represents the SD of the average of biological replicates from three healthy human donors (SI Appendix, Fig. S3B). At high duty cycles (thus, high integrated signals), the CD69 expression response resembles a promiscuous all-pass filter; in principle, gene expression could also exhibit low- or high-pass filtering behaviors, which have temporal selectivities intermediate between an all-pass and band-stop filters (Fig. 3C). These data represent a quantitative frequency response analysis of T cell gene expression and a reported observation of band-stop filtering in a living cell.
CD69 Expression Is a Highly Selective Temporal Filter Compared to Other Early T Cell Activation Responses.
We next tested the temporal filtering characteristics of other canonical early T cell activation expression responses. We stimulated optoCAR CD4+ T cells with the same duty cycles and oscillation periods as in Fig. 3 and assessed expression of eight genes (including CD69) chosen to encompass activating juxtracrine (CD69 and CD25), inhibitory juxtracrine (Tim3, PD-1, and Lag-3), and paracrine (IL-2, IFNγ, and TNFα) responses. We also measured the degree of GFP staining in optoCAR T cells. T cells appear as GFP+ presumably because of trogocytosis or endocytosis of GFP ligand (SI Appendix, Fig. S4A).
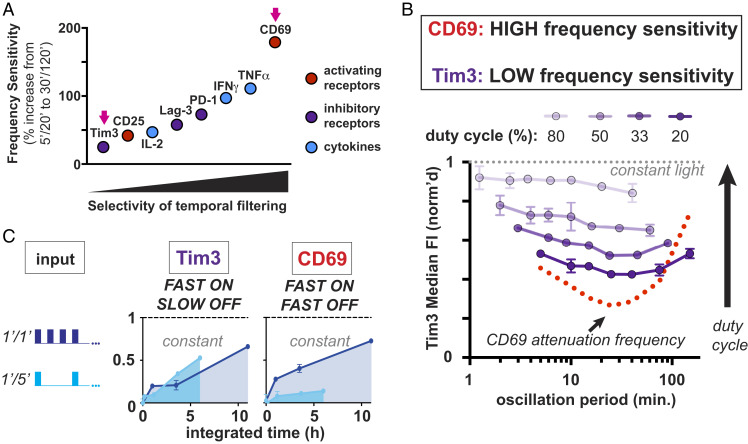
All responses increased as a function of duty cycle (i.e., integrated input) (Fig. 4 A and B and SI Appendix, Fig. S4A) but with varying sensitivity to signal oscillations. CD69 expression was by far the most sensitive to signal oscillations, with a gain of 2.79, whereas Tim3 achieved almost no gain as a function of oscillation period (gain = 1.25) (Fig. 4A). CD25, PD-1, Lag-3, IL-2, IFNγ, and TNFα expression had gain values intermediate between Tim3 and CD69. Although most responses exhibited clear sensitivity to signal oscillations, no clear trend emerged based on the type (activating juxtacrine, inhibitory juxtacrine, or paracrine) of response (Fig. 4A and SI Appendix, Fig. S4A).
Fig. 4.
CD69 expression is a more selective temporal filter than other early T cell activation responses. (A) Gain is plotted for Tim3, CD25, IL-2, Lag-3, PD-1, IFNγ, TNFα, and CD69. Gain is calculated as the normalized response at the highest oscillation period divided by the normalized response at the attenuation frequency at a duty cycle of 20%. (B) Tim3 expression (in purple), which has the lowest gain value, has a relatively flat response to oscillation period at all duty cycles tested compared to CD69 (in red), which has the highest gain value. (C) Tim3 and CD69 expression are plotted in response to the integrated input of 1′/1′ and 1′/5′ pulse trains. Tim3 expression remains high in response to 5′ OFF intervals that do not stimulate CD69 expression. Error bars are SEM, n = 2 technical replicates; representative of at least three independent experiments and two donors.
Tim3 expression was particularly insensitive to signal oscillations compared to other responses (Fig. 4B and SI Appendix, Fig. S4). One minute ON and 5 min OFF pulse intervals (1′/5′) that diminish CD69 expression do not have a similar impact on Tim3 expression (Fig. 4C). This effect is due to the fast, minute-scale oscillations of 1′/5′ versus 1′/1′ pulse trains and not to differences in integrated input, since impulses of constant stimulation with low versus high integrated inputs (3 h versus 8 h) resulted in similar expression levels of both CD69 and Tim3 (SI Appendix, Fig. S4C). Tim3 expression also reached maximal activation in response to shorter ON intervals and longer OFF intervals than CD69 regardless of the duration of ON/OFF interval (SI Appendix, Fig. S4D). Relatively high Tim3 expression in response to low duty cycle inputs is therefore due to relatively slow OFF kinetics (greater than ∼5 to 10 min) at one or more points in the signaling/transcriptional cascade downstream of the optoCAR. These kinetics result in Tim3 expression acting as a promiscuous all-pass filter even at the lowest duty cycles tested, in marked contrast to CD69 expression (Fig. 4B).
Signaling Downstream of Erk Is Responsible for the Shape of the CD69 Expression Response to Signal Oscillations.
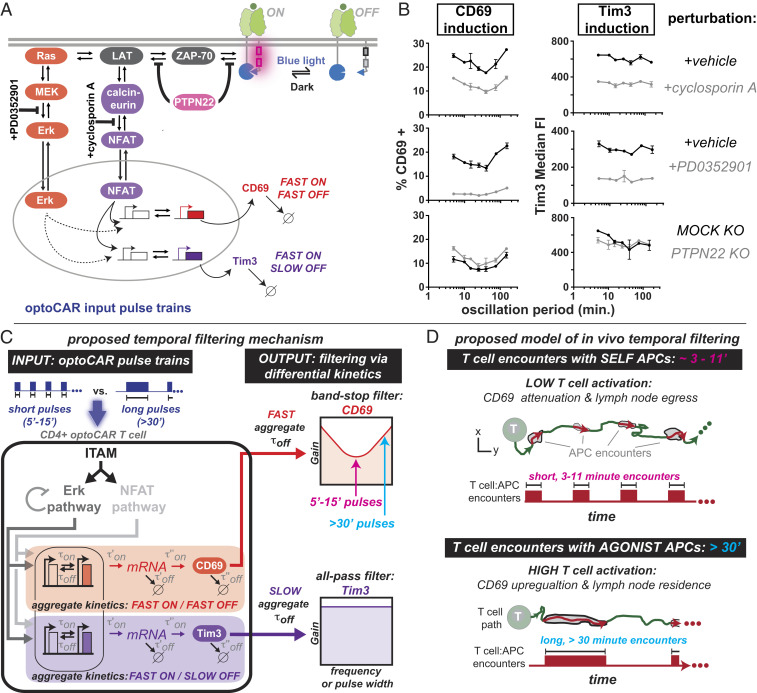
We next analyzed how selectively perturbing regulatory nodes in the T cell signaling network impacted CD69 expression in response to oscillatory signals. MEK in the Erk MAPK and calcineurin in the NFAT pathway, both key enzymes for the forward propagation of an input signal, were inhibited by small molecules PD03501 (MEKi) and cyclosporin A (CsA), respectively (Fig. 5A). The phosphatase PTPN22, which dampens input signals by dephosphorylating substrates immediately proximal to the signal-initiating receptor, was perturbed via gene knockout (KO) (Fig. 5A). We chose PTPN22 for KO because among the many phosphatases that regulate T cell signaling, PTPN22 correlates most strongly with the incidence of several autoimmune disorders, indicating that it plays a vital role in regulating T cell homeostasis (43–45). Tim3 expression was also measured simultaneously with CD69, since Tim3 and CD69 expression have dramatically different temporal filtering behaviors (Fig. 4).
Fig. 5.
Potential signaling mechanisms that sculpt selectively attenuated CD69 activation in response to specific oscillatory signals. (A) optoCAR stimulation was perturbed in three ways: 1) inhibition of MEK in the Erk MAPK pathway using 50 nM PD0352901, 2) inhibition of calcineurin in the NFAT pathway using 10 μM cyclosporin A, and 3) gene KO of the regulatory phosphatase PTPN22. (B) Inhibition of NFAT reduced CD69 and Tim3 expression evenly at all oscillation periods, whereas inhibition of Erk reduced CD69 expression selectively on either side of the attenuation frequency. PTPN22 KO had a relatively minor impact on the induction of CD69 and Tim3 expression. n = 2 technical replicates, representative of three independent experiments and two donors. (C) A proposed qualitative model of temporal filtering. Oscillatory inputs (e.g., 7.5-min versus 30-min pulses) stimulate an upstream signaling module, here represented by ITAMs, Erk, and NFAT. Differential filtering downstream of Erk—for example, via differences in one or more of the individual rate constants (τ’off, τ’’off, τ’’’off)—contributes to band-stop versus all-pass filtering profiles of CD69 and Tim3. The aggregate off-rate, τoff,, is a function of the individual rate constants (τoff = f(τ’off ; τ’’off ; τ’’’off)). Filtering may also be impacted by Erk negative feedback. (D) A proposed model of in vivo temporal filtering. The timescale of T cell–SELF-presenting APCs is strikingly similar to the timescale of CD69 expression attenuation.
Inhibition of the Erk MAPK pathway via treatment with MEKi selectively decreased CD69 expression at 5- and 150-min periods (1 and 30 min pulse widths) compared to the attenuation frequency (∼25 min period; 5 to 15 min pulse widths) (Fig. 5B and SI Appendix, Fig. S5A). In contrast, inhibition of NFAT signaling had an insignificant effect on how CD69 expression responded to signal oscillations, as the fraction of CD69+ T cells decreased by roughly 10% at all oscillation periods (Fig. 5B and SI Appendix, Fig. S5A). The concentration of both MEKi (50 nM) and CsA (10 μM) were chosen such that the degree of induction was roughly equal for each inhibitor. We therefore conclude that signaling downstream of the Erk pathway is responsible for the shape of the CD69 curve in response to oscillation period. The negative feedback loop acting on the Erk MAPK pathway (Fig. 1F and SI Appendix, Fig. S1F) may contribute to the selective attenuation of CD69 expression, given that Erk phosphorylation decreases over a similar timescale (∼30 min) as the attenuation period.
The PTPN22 gene was then excised using a recently developed CRISPR/Cas9 technique that achieves 80 to 90% PTPN22 KO efficiency in primary human CD4+ T cells (46). PTPN22 KO T cells exhibited significantly higher basal CD69 levels compared to mock KO T cells, but the effect on CD69 expression in response to signal oscillation was minimal. Upon KO of PTPN22, the degree of CD69 induction increased by less than 5% at all oscillation periods (Fig. 5B and SI Appendix, Fig. S5A).
Treatment with both CsA and MEKi decreased the induction of Tim3 expression evenly at all oscillation periods. Gene KO of PTPN22 had minimal impact on the degree of Tim3 induction and slightly increased basal Tim3 expression levels (Fig. 5B and SI Appendix, Fig. S5 A and B).
Discussion
It has long been appreciated that temporal filtering of seconds-scale TCR–ligand dwell times, or kinetic proofreading, is a critical mechanism determining whether or not a T cell responds to a stimulatory signal (6–11, 23). However, T cell–APC interactions can also be highly periodic on the timescale of minutes, raising the possibility that T cells also filter periodic signals with intervals and signaling durations on the order of minutes (14–22). By engineering a synthetic receptor capable of cycling T cell signaling on and off independently of ligand binding, we demonstrated that the T cell signaling machinery is capable of selectively filtering oscillatory signals on the timescale of minutes via a band-stop mechanism.
The attenuation period of the CD69 expression band-stop filter, corresponding to ON intervals of 5 to 15 min, strikingly coincides with the duration of T cell encounters with self-peptide–presenting APCs observed via intravital imaging in the lymph nodes and skin of mice [∼3 to 11 min (16, 21, 22)] (Fig. 5 C and D). There are caveats—most notably that the optoCAR likely induces T cell signaling differently than native TCRs—yet it is possible that a band-stop filtering mechanism could enhance TCR-mediated antigen discrimination by selectively attenuating expression of CD69, TNFα, IFNγ, and/or other genes at precisely the timescales corresponding to T cell–self APC interactions while allowing expression of those genes at timescales corresponding to T cell–agonist APC encounters. If CD69 is regulated in this way in vivo, then selective attenuation of CD69 up-regulation in T cells that have experienced many short encounters with self-peptide–presenting APCs could facilitate or even enhance egress of antigen-inexperienced T cells from the thymus and peripheral lymphoid organs (Fig. 5D) (47). Prior work has also demonstrated that integration of consecutive pulses of TCR signal in the thymus leads to positive selection, while sustained signaling leads to negative selection (48, 49). The gene expression patterns described here could selectively stabilize signaling intermediates required for positive selection only in T cells that have experienced brief, weak, and discontinuous signals, leaving T cells that have experienced continuous signals without these stabilized intermediates and committed to death (48).
Our data indicate that when a short stimulus (1 to 15 min) is removed, CD69 expression transitions to an “OFF” state if the subsequent OFF period duration lasts ∼4 to 20 min (Fig. 3). Tim3 expression remains high in response to these temporal stimulation patterns, indicating Tim3 has a slower transition to an “OFF” state compared to CD69 (Fig. 4). A plausible explanation is that the aggregate OFF kinetics of processes such as promoter transitions (50) and/or mRNA degradation is faster in the case of CD69 expression compared to Tim3 expression (Fig. 5C and SI Appendix, Fig. S5 C and D). Although the precise mechanism remains to be seen, loss of band-stop filtering upon inhibition of MEK but not NFAT (Fig. 5B) and observation of negative feedback in the Erk pathway (Fig. 1F and SI Appendix, Fig. S1F) indicate that the mechanism of band-stop filtering may lie downstream of the Erk pathway. Furthermore, above a threshold “ON” duration (between 30′ and 180′, SI Appendix, Fig. S4C), T cells commit to CD69 expression, which provides a mechanism for why the CD69 response to varying signal oscillations takes the form of a band-stop filter instead of a high-pass filter (Fig. 5C). Erk negative feedback may synergize with other band-stop mechanisms and would likely be mediated by dual specificity phosphatase (DUSP) 5/6 IEGs, since the timescale of Erk feedback is similar to the timescale of DUSP 5/6 expression and DUSP 5/6 dephosphorylate Erk on Thr202 and Tyr204 (51). Erk negative feedback may further compete with positive feedback mediated by the Ras GTPase SOS (52).
Band-stop and other temporal filtering mechanisms could also contribute to the function and efficacy of therapeutic T cells via TCR signaling, since most therapeutic T cells express a CAR in a T cell that also bears endogenous TCR. The filtering mechanisms described here could also be initiated directly by a CAR in vivo, since CAR T cells have been shown to engage in transient contacts with B cell lymphoma cells bearing the CAR’s cognate ligand (19). Finally, the optoCAR could be used to discover additional layers of temporal regulation in other areas of lymphocyte signaling, such as T cell differentiation (e.g., formation of Th1, Th2, Th17, and Treg subsets and the ratios between them) or signaling downstream of other ITAM-based receptors (e.g., FcR; BCR).
Materials and Methods
optoCAR Plasmid Construction.
All constructs were cloned via In-Fusion cloning (Clontech #ST0345) into a modified pHR’SIN:CSW transgene expression vector containing a spleen focus-forming virus promoter. Part one of the optoCAR receptor was built by fusing the α-CD19 scFv (53) or the Lag17 α-GFP nanobody (36) to the human CD8α hinge and transmembrane domains and the iLID light-sensitive domain (39) (SI Appendix, Fig. S1 A and B). Part one has an N-terminal human CD8α signal sequence for membrane targeting and an N-terminal Myc tag for assessment of surface expression. Part two of the optoCAR receptor was built by fusing the Dap10 extracellular domain to the CD8α transmembrane domain, the 41BB or the CD28 intracellular domain, the CD3ζ intracellular domain, and either the milli, micro, or nano SspB domain (39). Part two was preceded by the Dap10 signal sequence and an N-terminal FLAG tag for orthogonal assessment of surface expression. Full amino acid sequences for all optoCAR constructs are listed below.
optoCAR Amino Acid Sequences.
Amino acid sequences are labeled with the following abbreviations: TM = transmembrane, ECD = extracellular domain, and ICD = intracellular domain.
Sequence Number One, Anti-GFP Part One.
CD8α leader-myc tag-lag17-CD8α hinge-CD8αTM-GS linker-iLID.
MALPVTALLLPLALLLHAARPEQKLISEEDLMADVQLVESGGGLVQAGGSLRLSCAASGRTISMAAMSWFRQAPGKEREFVAGISRSAGSAVHADSVKGRFTISRDNTKNTLYLQMNSLKAEDTAVYYCAVRTSGFFGSIPRTGTAFDYWGQGTQVTVSTTTPAPRPPTPAPTIASQPLSLRPEACRPAAGGAVHTRGLDFACDIYIWAPLAGTCGVLLLSLVITLYCGSGSGSGSEFLATTLERIEKNFVITDPRLPDNPIIFASDSFLQLTEYSREEILGRNCRFLQGPETDRATVRKIRDAIDNQTEVTVQLINYTKSGKKFWNVFHLQPMRDYKGDVQYFIGVQLDGTERLHGAAEREAVCLIKKTAFQIAEAANDENYF*.
Sequence Number Two, Anti-CD19 Part One.
CD8α leader-myc tag-α-CD19 ScFv-CD8α hinge-CD8αTM-GS linker-iLID.
MALPVTALLLPLALLLHAARPEQKLISEEDLDIQMTQTTSSLSASLGDRVTISCRASQDISKYLNWYQQKPDGTVKLLIYHTSRLHSGVPSRFSGSGSGTDYSLTISNLEQEDIATYFCQQGNTLPYTFGGGTKLEITGGGGSGGGGSGGGGSEVKLQESGPGLVAPSQSLSVTCTVSGVSLPDYGVSWIRQPPRKGLEWLGVIWGSETTYYNSALKSRLTIIKDNSKSQVFLKMNSLQTDDTAIYYCAKHYYYGGSYAMDYWGQGTSVTVSSTTTPAPRPPTPAPTIASQPLSLRPEACRPAAGGAVHTRGLDFACDIYIWAPLAGTCGVLLLSLVITLYCGSGSGSGSEFLATTLERIEKNFVITDPRLPDNPIIFASDSFLQLTEYSREEILGRNCRFLQGPETDRATVRKIRDAIDNQTEVTVQLINYTKSGKKFWNVFHLQPMRDYKGDVQYFIGVQLDGTERLHGAAEREAVCLIKKTAFQIAEAANDENYF*.
Sequence Number Three, SspB Nano Part Two.
Dap10 leader–FLAG Tag-FLAG linker-Dap10 ECD-CD8αTM-41BB ICD-GS Linker-CD3zeta ICD-GS Linker-SspB nano.
MIHLGHILFLLLLPVAAADYKDDDDKGSQVQLQQQTTPGERSSLPAFYPGTSGSCSGCGSLSLPIYIWAPLAGTCGVLLLSLVITLYCSLKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCELGSGSGSGSRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPRGSGSGSGSMSGLRSRAQASNEFGIDLSGLTLQEFSSPKRPKLLREYYDWLVDNSFTPYLVVDATYLGVNVPVEYVKDGQIVLNLSASATGNLQLTNDFIQFNARFKGVSRELYIPMGAALAIYARENGDGVMFEPEEIYDELNIG*.
Sequence Number Four, SspB Micro Part Two.
Dap10 leader–FLAG Tag-FLAG linker-Dap10 ECD-CD8αTM-41BB ICD-GS Linker-CD3zeta ICD-GS Linker-SspB micro.
MIHLGHILFLLLLPVAAADYKDDDDKGSQVQLQQQTTPGERSSLPAFYPGTSGSCSGCGSLSLPIYIWAPLAGTCGVLLLSLVITLYCSLKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCELGSGSGSGSRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPRGSGSGSGSMSGLRSRAQASNEFGIDLSGLTLQEFSSPKRPKLLREYYDWLVDNSFTPYLVVDATYLGVNVPVEYVKDGQIVLNLSASATGNLQLTNDFIQFNAQFKGVSRELYIPMGAALAIYARENGDGVMFEPEEIYDELNIG*.
Sequence Number Five, SspB Milli Part Two.
Dap10 leader–FLAG Tag-FLAG linker-Dap10 ECD-CD8αTM-41BB ICD-GS Linker-CD3zeta ICD-GS Linker-SspB milli.
MIHLGHILFLLLLPVAAADYKDDDDKGSQVQLQQQTTPGERSSLPAFYPGTSGSCSGCGSLSLPIYIWAPLAGTCGVLLLSLVITLYCSLKRGRKKLLYIFKQPFMRPVQTTQEEDGCSCRFPEEEEGGCELGSGSGSGSRVKFSRSADAPAYQQGQNQLYNELNLGRREEYDVLDKRRGRDPEMGGKPRRKNPQEGLYNELQKDKMAEAYSEIGMKGERRRGKGHDGLYQGLSTATKDTYDALHMQALPPRGSGSGSGSMSGLRSRAQASNEFGIDLSGLTLQEFSSPKRPKLLREYYDWLVDNSFTPYLVVDATYLGVNVPVEYVKDGQIVLNLSASVTGNLQLTNDFIQFNAQFKGVSRELYIPMGAALAIYARENG DGVMFEPEEIYDELNIG*.
Primary Human T Cell Isolation and Culture.
Primary human CD4+ and CD8+ T cells were isolated from deidentified donor human peripheral blood obtained from STEMCELL Technologies. T cells were cryopreserved in Roswell Park Memorial Institute-1640 (University of California San Francisco [UCSF] cell culture core) with 20% human AB serum (Valley Biomedical, #HP1022) and 10% dimethyl sulfoxide. After thawing, T cells were maintained at 37 °C and 5% CO2 in human T cell medium (hTCM) consisting of X-Vivo 15 (Lonza #04-418Q), 5% human AB serum, and 10 mM neutralized N-acetyl l-Cysteine (Sigma-Aldrich #A9165) with 30 units/mL IL-2 or 100 units/mL IL-2 if indicated (National Cancer Institute Biological Resources Branch Preclinical Repository). T cells were resuspended in human T cell medium without IL-2 immediately prior to all optogenetic assays.
Lentiviral Transduction and Sorting of Human T Cells.
Pantropic vesicular stomatitis virus G pseudotyped lentivirus was produced via transfection of Lenti-X 293T cells (Clontech #11131D) with a pHR’SIN:CSW transgene expression vector and the viral packaging plasmids pCMVdR8.91 and pMD2.G using FuGENE HD (Promega, #E2312). Primary T cells were thawed the same day and after 24 h in culture, were stimulated with Dynabeads Human T-Activator CD3/CD28 (Life Technologies #11131D) at 25 μL per 1 × 106 T cells (defined as day 1). At 48 h (day 2), viral supernatant was harvested via centrifugation at 500 G for 5 min, and the primary T cells were exposed to the virus for 24 h in a 12-well tissue culture plate (0.5 mL of part one and 2 mL of part two). Dynabeads were removed at day 5 post-T cell stimulation. Part 1LO/Part 2HI T cells were sorted on day 6 post-T cell stimulation with a FACSAria II. Myc (part one) and FLAG (part two) tags were used for determination of surface expression for sorting with α-myc-Alexa647 (clone 9B11, Cell Signaling Technologies, #2233S) and α-DYKDDDDK-Alexa488 (clone 1042E, R&D Systems, #IC8529G-100) antibodies. Staining with α-Myc and α-FLAG antibodies was performed at a dilution of 1:50 in phosphate-buffered saline (PBS) without Ca2+ or Mg2+ salts for 30 min at 4 °C. Assays were performed starting on day 11 or 12 poststimulation until at most day 17 poststimulation with Dynabeads.
Antigen-Presenting Cell Lines.
K562 myelogenous leukemia antigen-presenting cell lines (ATCC #CCL-243) were lentivirally transduced to stably express human CD19 or surface GFP (GFP fused to the platelet-derived growth factor receptor transmembrane domain) and cytosolic mCherry. CD19 levels were determined by staining the cells with α-CD19 APC (Biolegend #302212). All cell lines were sorted for expression of the surface (CD19 or GFP) and cytosolic (mCherry) transgenes.
In Vitro Stimulation of optoCAR T Cells Using the optoPlate.
Black-walled, clear-bottom 96-well plates (Corning, #3603) were first plated with 100 μL/well of either 35,000 to 100,000 wild-type K562s, 35,000 to 100,000 CD19+ or GFP+ K562s, human T cell medium (negative control), and either 50 ng/mL phorbol 12-myristate 13-acetate (Millipore Sigma #P1585) and 1 μM ionomycin (Sigma-Aldrich, #I3909-1ML) or surface-bound OKT3 (Thermo Fisher Scientific, #MA1-10175) for positive controls. All wells were resuspended in a volume of 100 μL/well prior to addition of T cells; the outer two rows of each plate were filled with 200 μL/well Ca2+, Mg2+ free PBS (UCSF cell culture core), such that only the middle 64 wells contained samples. OKT3 was incubated at a concentration of 2 μg/mL for 2 h at 37 °C in Ca2+, Mg2+ free PBS (UCSF cell culture core), then washed twice with 200 μL/well PBS followed by two washes with 200 μL/well hTCM per well using a multichannel pipette. 35,000 to 100,000 CD4+ or CD8+ T cells were then added to the appropriate wells for a total volume of 200 μL/well. The 96-well plates containing T cell samples were then immediately transferred to the optoPlate in a cell culture incubator maintained at 37 °C and 5% CO2 for stimulation with light. Custom-made lids made of black plastic eliminated light contamination between wells.
optoPlate Intensity Calibration and Waveform Programming.
optoPlate illumination profiles were controlled using an Arduino Micro microcontroller programmed with a custom script through the Arduino Integrated Development Environment, as described previously (28). The Arduino Micro controlled the intensity of two blue LEDs (470 nm) underneath each well position of a 96-well plate. Intensity was controlled via 12-bit pulse wave modulation (4,096 intensity levels). Correct programming was confirmed by eye prior to initiation of experiments by speeding up the waveform frequency by a factor of ∼10.
Curve Fitting.
The oscillation period at which CD69 expression reaches a minimum was quantitatively estimated by fitting CD69 expression data collected at a duty cycle equal to 20% to a spline function using the BSplineFunction command in Mathematica (Wolfram) and calculating the minimum of the spline function.
Processing Samples for Flow Cytometry Analysis.
General methods.
Samples were transferred from a cell culture incubator to a biosafety cabinet with all the room and biosafety cabinet lights turned off. All subsequent sample processing was performed with the overhead light in the room and the light within the biosafety cabinet used for processing turned off and the lights in an immediately adjacent biosafety cabinet turned on. Samples were then transferred to a 96-well round-bottom tissue culture plate (Fisher Scientific, #0877217) prior to subsequent processing.
Cell surface receptor staining.
For analysis of cell surface receptor expression, samples were centrifuged at 500 × g for 5 min and resuspended in a 50 μL volume with the appropriate antibodies diluted 1:50 in Ca2+, Mg2+ PBS (UCSF cell culture core). After a 30-min incubation at 4 °C, samples were washed twice with Ca2+, Mg2+ free PBS prior to final resuspension in a solution of Ca2+, Mg2+ free PBS with 1% bovine serum albumin (BSA; Sigma-Aldrich #A9576). Samples were analyzed for protein expression on a BD LSRFortessa. Antibodies are as follows: APC Mouse anti-human CD69 clone FN50 (BioLegend #310910), BV421 Mouse anti-human Tim-3 clone 7D3 (BD Biosciences BDB565562), BUV737 Mouse anti-human PD-1 clone EH12.1 (BD Biosciences BDB612791), BUV395 Mouse anti-human Lag-3 clone T47-530 (BD Biosciences #745640), BV421 Mouse anti-human Lag-3 clone T47-530 (BD Biosciences #565720), and APC Mouse anti-human CD25 clone BC96 (BioLegend #102012).
Assessment of intracellular cytokine expression.
For analysis of intracellular cytokine expression, samples were incubated for the final 6 h of stimulation with brefeldin A and monensin protein transport inhibitors diluted 1:1,000 in hTCM (BD #555028 and BD #554715). After stimulation, cells were transferred to a 96-well round-bottom plate and washed twice with 200 μL/well PBS without Ca2+ or Mg2+ salts, centrifuged for 5 min at 500 × g, then resuspended in 100 μL/well eBioscience Fixable Viability Dye eFluor 780 (Thermo Fisher Scientific #65-0865-14). Samples were incubated for 20 to 30 min at 4 °C, then washed twice with 150 to 250 μL/well PBS without Ca2+ or Mg2+ salts. The tissue culture plate was vortexed vigorously immediately prior to addition of 100 μL/well Fixation/Permeabalization buffer (BD #555028). Cells were incubated for 30 min at room temperature with the Fix/Perm buffer, and then washed twice with 250 μL/well Perm/Wash buffer (BD #555028), centrifuging 5 min now at 800 × g for each wash. At this stage, cells could be stored overnight at 4 °C. The sample was then centrifuged once more for 5 min at 800 × g and resuspended in 35 μL/well of the following antibodies diluted 1:20 in Perm/Wash buffer: APC Mouse α-human IFN-γ clone 4S.B3 (BioLegend #502512), BV421 Mouse α-human TNF-α clone MAb11 (Fisher Scientific #BDB562783), and BUV737 Rat α-human IL-2 clone MQ1-17H12 (Fisher Scientific # BDB612836). After incubation, samples were washed twice with 250 μL/well Perm/Wash buffer (centrifuged for 5 min at 800 × g) prior to final resuspension in 80 to 100 μL/well PBS without Ca2+ or Mg2+ salts supplemented with 1% BSA. Analysis via flow cytometry was performed as described above on a BD Fortessa.
Assessment of Erk Thr202/Tyr204 phosphorylation.
For analysis of Erk Thr202/Tyr204 phosphorylation, T cell samples were prepared as described above, but the final volume per well was 150 μL instead of 200 μL. After stimulation, an equal 150 μL volume of prewarmed CytoFix Fixation Buffer (BD #554655) was added simultaneously to all sample wells using a multichannel pipette. (Light stimulation patterns included delays for early time points, such that all time points were synchronized to end at the same time.) Cells were incubated for 10 to 12 min at 37 °C, then transferred to round-bottom 96-well plates prior to centrifugation for 6 min at 600 × g. Supernatant was removed with a multichannel pipette, taking care not to disturb the pellet, prior to vigorous vortexing to disrupt the pellets prior to permeabalization. Cells were permeabalized by adding 100 μL of chilled Perm Buffer III (BD #558050), mixed with a multichannel pipette, and incubated 30 min at 4 °C. After incubation, cells were washed three times with 250 μL/well of PBS supplemented with 5% BSA. Samples were then resuspended in 35 μL/well of α-ppErk Thr202/Tyr204-Alexa647 antibody diluted 1:50 in PBS supplemented with 5% BSA and incubated for 30 min at 4 °C. After incubation, cells were washed three times with 250 μL/well PBS supplemented with 1% BSA before final resuspension in 100 μL/well PBS supplemented with 1% BSA. Cells were then analyzed via flow cytometry as described above.
Inhibition of Erk and NFAT Signaling Pathways.
To inhibit Erk and NFAT signaling downstream of optoCAR stimulation, T cells were incubated with either 50 nM MEKi or 10 μM cyclosporin A in hTCM without IL-2 for 15 min at 37 °C prior to mixing in a 1:1 volume ratio with GFP+ K562s in hTCM without IL-2. T cells were then stimulated and analyzed via flow cytometry as described above.
CRISPR/Cas9 KO of PTPN22.
The following is adapted from Anderson, et al. (46). For CRISPR/Cas9 KO of PTPN22, T cells were expanded with Dynabeads and transfected with lentivirus according to the same protocols described above with the following modifications. Virus was removed on day 3 poststimulation instead of day 2, and α-CD3/α-CD28 Dynabeads were removed on day 2 poststimulation instead of on day 5. Dynabeads were removed using an Easy 50 EasySep Magnet (Stemcell Technologies #18002). On day 3, poststimulation T cells were electroporated with 2.5 μg complexed guide RNA (gRNA; Synthego)/Cas9 (Integrated DNA Technologies #260682666) and 20 pmol short, single-stranded oligodeoxynucleotide (ssODN; Integrated DNA Technologies) per 3 × 105 cells. Sequences for the gRNA and ssODN are given below. Samples were electroporated in 20 μL volumes using the P3 Primary Cell 96-well Nucleofector Kit (Lonza #V4SP-3096) and a Nucleofector II + 96-well Nucleofector Shuttle System (Lonza) using program EO-115. Immediately after electroporation, samples were supplemented with 80 μL/well prewarmed hTCM + 100 U/mL IL-2 and incubated 15 min at 37 °C. After incubation, samples were diluted 1:3 in a new 96-well round-bottom plate for a final volume of 250 μL/well hTCM + 100 U/mL. Electroporated samples were maintained in hTCM + 100 U/mL until assays were performed starting on day 11 or day 12.
gRNA, PTPN22.
5′-AAGGCAAUCUACCAAGUACA-3′.
ssODN, PTPN22 Exon 2 (* Represents Phosphorothioate Linkages).
5′-T*T*TAGAGATAGCTTCTTACCTCATTTCTCTAGGAAGATTAATTTTTCTTGGAATCTTATTACAGAAGCTGAAAAGGCAATCTACCAAGTTAAGTGACTAGAATAAAACAAGGCAGACAAAACCTATCCTACAACTGTGGCTGAGAAGCCCAAGAATATCAAGAAAAACAGATATAAGGATATTTTGCCCTGTAAG*T*T-3′.
Supplementary Material
Acknowledgments
We thank Art Weiss for his generous advice and Judith Ashouri for the gift of MEKi and CsA inhibitors. We thank the Lim and Weiss laboratories for their feedback and support. Research reported in this publication was supported by the NIH under award number F32GM116489 (to G.P.O.) and grant number R01 CA196277 (to W.A.L.) and by HHMI (to W.A.L.).
Footnotes
Competing interest statement: W.A.L. is a shareholder of Gilead Sciences and a scientific advisor for Allogene.
This article is a PNAS Direct Submission. M.L.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019285118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Lemmon M. A., Schlessinger J., Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chakraborty A. K., Weiss A., Insights into the initiation of TCR signaling. Nat. Immunol. 15, 798–807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss A., Littman D. R., Signal transduction by lymphocyte antigen receptors. Cell 76, 263–274 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Gross G., Waks T., Eshhar Z., Expression of immunoglobulin-T-cell receptor chimeric molecules as functional receptors with antibody-type specificity. Proc. Natl. Acad. Sci. U.S.A. 86, 10024–10028 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irving B. A., Weiss A., The cytoplasmic domain of the T cell receptor ζ chain is sufficient to couple to receptor-associated signal transduction pathways. Cell 64, 891–901 (1991). [DOI] [PubMed] [Google Scholar]
- 6.Matsui K., Boniface J. J., Steffner P., Reay P. A., Davis M. M., Kinetics of T-cell receptor binding to peptide/I-Ek complexes: Correlation of the dissociation rate with T-cell responsiveness. Proc. Natl. Acad. Sci. U.S.A. 91, 12862–12866 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huse M., et al., Spatial and temporal dynamics of T cell receptor signaling with a photoactivatable agonist. Immunity 27, 76–88 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Iezzi G., Karjalainen K., Lanzavecchia A., The duration of antigenic stimulation determines the fate of naive and effector T cells. Immunity 8, 89–95 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Huppa J. B., Gleimer M., Sumen C., Davis M. M., Continuous T cell receptor signaling required for synapse maintenance and full effector potential. Nat. Immunol. 4, 749–755 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Corse E., Gottschalk R. A., Krogsgaard M., Allison J. P., Attenuated T cell responses to a high-potency ligand in vivo. PLoS Biol. 8, 1–12 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pielak R. M., et al., Early T cell receptor signals globally modulate ligand:receptor affinities during antigen discrimination. Proc. Natl. Acad. Sci. U.S.A. 114, 12190–12195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniels M. A., et al., Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature 444, 724–729 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Skokos D., et al., Peptide-MHC potency governs dynamic interactions between T cells and dendritic cells in lymph nodes. Nat. Immunol. 8, 835–844 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Germain R. N., Robey E. A., Cahalan M. D., A decade of imaging cellular motility and interaction dynamics in the immune system. Science 336, 1676–1681 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dustin M. L., Stop and go traffic to tune T cell responses. Immunity 21, 305–314 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Bousso P., T-cell activation by dendritic cells in the lymph node: Lessons from the movies. Nat. Rev. Immunol. 8, 675–684 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Mempel T. R., Henrickson S. E., Von Andrian U. H., T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature 427, 154–159 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Faroudi M., Zaru R., Paulet P., Müller S., Valitutti S., Cutting edge: T lymphocyte activation by repeated immunological synapse formation and intermittent signaling. J. Immunol. 171, 1128–1132 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Cazaux M., et al., Single-cell imaging of CAR T cell activity in vivo reveals extensive functional and anatomical heterogeneity. J. Exp. Med. 216, 1038–1049 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunzer M., et al., Antigen presentation in extracellular matrix: Interactions of T cells with dendritic cells are dynamic, short lived, and sequential. Immunity 13, 323–332 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Natsuaki Y., et al., Perivascular leukocyte clusters are essential for efficient activation of effector T cells in the skin. Nat. Immunol. 15, 1064–1069 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Gaylo-Moynihan A., et al., Programming of distinct chemokine-dependent and -independent search strategies for Th1 and Th2 cells optimizes function at inflamed sites. Immunity 51, 298–309.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Donoghue G. P., Pielak R. M., Smoligovets A. A., Lin J. J., Groves J. T., Direct single molecule measurement of TCR triggering by agonist pMHC in living primary T cells. eLife 2, e00778 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mettetal J. T., Muzzey D., Gómez-Uribe C., Van Oudenaarden A., The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science 319, 482–484 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muzzey D., Gómez-Uribe C. A., Mettetal J. T., van Oudenaarden A., A systems-level analysis of perfect adaptation in yeast osmoregulation. Cell 138, 160–171 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell A., Wei P., Lim W. A., Oscillatory stress stimulation uncovers an Achilles’ heel of the yeast MAPK signaling network. Science 350, 1379–1383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toettcher J. E., Weiner O. D., Lim W. A., Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–1434 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bugaj L. J., et al., Cancer mutations and targeted drugs can disrupt dynamic signal encoding by the Ras-Erk pathway. Science 361, eaao3048 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson M. Z., Ravindran P. T., Lim W. A., Toettcher J. E., Tracing information flow from Erk to target gene induction reveals mechanisms of dynamic and combinatorial control. Mol. Cell 67, 757–769.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hannanta-Anan P., Chow B. Y., Optogenetic control of calcium oscillation waveform defines NFAT as an integrator of calcium load. Cell Syst. 2, 283–288 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tischer D. K., Weiner O. D., Light-based tuning of ligand half-life supports kinetic proofreading model of T cell signaling. eLife 8, 1–25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yousefi O. S., et al., Optogenetic control shows that kinetic proofreading regulates the activity of the T cell receptor. eLife 8, 1–33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bohineust A., Garcia Z., Corre B., Lemaître F., Bousso P., Optogenetic manipulation of calcium signals in single T cells in vivo. Nat. Commun. 11, 1143 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Z., et al., Engineering light-controllable CAR T cells for cancer immunotherapy. Sci. Adv. 6, eaay9209 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu C.-Y., Roybal K. T., Puchner E. M., Onuffer J., Lim W. A., Remote control of therapeutic T cells through a small molecule-gated chimeric receptor. Science 350, aab4077 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fridy P. C., et al., A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 11, 1253–1260 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H., et al., ZAP-70: An essential kinase in T-cell signaling. Cold Spring Harb. Perspect. Biol. 2, a002279 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milone M. C., et al., Chimeric receptors containing CD137 signal transduction domains mediate enhanced survival of T cells and increased antileukemic efficacy in vivo. Mol. Ther. 17, 1453–1464 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guntas G., et al., Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. U.S.A. 112, 112–117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimmerman S. P., et al., Tuning the binding affinities and reversion kinetics of a light inducible dimer allows control of transmembrane protein localization. Biochemistry 55, 5264–5271 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim W. A., Designing customized cell signalling circuits. Nat. Rev. Mol. Cell Biol. 11, 393–403 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zverev A. I., Handbook of Filter Synthesis (Wiley-Interscience, 1967). [Google Scholar]
- 43.Begovich A. B., et al., A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am. J. Hum. Genet. 75, 330–337 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bottini N., et al., A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat. Genet. 36, 337–338 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Kyogoku C., et al., Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am. J. Hum. Genet. 75, 504–507 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson W., Thorpe J., Long S. A., Rawlings D. J., Efficient CRISPR/Cas9 disruption of autoimmune-associated genes reveals key signaling programs in primary human T cells. J. Immunol. 203, 3166–3178 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shiow L. R., et al., CD69 acts downstream of interferon-α/β to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature 440, 540–544 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Au-Yeung B. B., et al., Quantitative and temporal requirements revealed for Zap70 catalytic activity during T cell development. Nat. Immunol. 15, 687–694 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melichar H. J., Ross J. O., Herzmark P., Hogquist K. A., Robey E. A., Distinct temporal patterns of T cell receptor signaling during positive versus negative selection in situ. Sci. Signal. 6, ra92 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hao N., O’Shea E. K., Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat. Struct. Mol. Biol. 19, 31–39 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaud G., Lesourne R., Love P. E., Regulatory mechanisms in T cell receptor signalling. Nat. Rev. Immunol. 18, 485–497 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Das J., et al., Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell 136, 337–351 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grupp S. A., et al., Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N. Engl. J. Med. 368, 1509–1518 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.