Abstract
Objective
Due to a complex set of processes involved with the recording of health information in the Electronic Health Records (EHRs), the truthfulness of EHR diagnosis records is questionable. We present a computational approach to estimate the probability that a single diagnosis record in the EHR reflects the true disease.
Materials and Methods
Using EHR data on 18 diseases from the Mass General Brigham (MGB) Biobank, we develop generative classifiers on a small set of disease-agnostic features from EHRs that aim to represent Patients, pRoviders, and their Interactions within the healthcare SysteM (PRISM features).
Results
We demonstrate that PRISM features and the generative PRISM classifiers are potent for estimating disease probabilities and exhibit generalizable and transferable distributional characteristics across diseases and patient populations. The joint probabilities we learn about diseases through the PRISM features via PRISM generative models are transferable and generalizable to multiple diseases.
Discussion
The Generative Transfer Learning (GTL) approach with PRISM classifiers enables the scalable validation of computable phenotypes in EHRs without the need for domain-specific knowledge about specific disease processes.
Conclusion
Probabilities computed from the generative PRISM classifier can enhance and accelerate applied Machine Learning research and discoveries with EHR data.
Keywords: data quality, electronic health records, transfer learning, diagnosis records, generative models
INTRODUCTION
The continuing prevalence of Electronic Health Records (EHR) systems offers great promises for their secondary use in biomedical research. EHR data can accelerate real-time translation of evidence-based discoveries into everyday healthcare practice as we strive towards rapid-learning health care systems.1,2 However, because EHRs were not primarily designed for research and discovery, the utility of EHRs for research is hindered by legitimate and known data quality concerns.3–6 In its most recent publication, the Office of the National Coordinator (ONC) for Health IT has identified leveraging high-quality electronic health data for research as one of its two overarching goals.7
In general, the truthfulness (or plausibility) of clinical records stored in EHRs can be disputable to varying degrees.8 One of the more challenging aspect of assessing EHR data quality is quantifying the plausibility of diagnoses records. EHR records reflect a complex set of processes that may not be direct indicators of patients’ “true” health states at different time points, but rather reflect the clinical processes, the patients’ interactions with the system, and the recording processes.9–11 The existence of an International Classification of Diseases (ICD) code for a disease in a patient’s electronic records does not necessarily mean that the patient truly has the disease. This is an outstanding hurdle for a swift secondary use of EHR data to address pressing public health issues.
The central idea in this study is to obtain a generalizable model for scalable computation of probability of EHR disease records over time. We present a computational approach that leverages the concept of transfer learning to quantify the probability of diagnoses records in the EHRs for a wide range of diseases. The Generative Transfer Learning (GTL) approach involves developing generative classifiers using a small set of disease-agnostic features from EHRs that aim to represent Patients, pRoviders, and their Interactions within the healthcare SysteM—we call these PRISM features. Using EHR data on 18 diseases from the Mass General Brigham (MGB, formerly known as Partners Healthcare) Biobank,12,13 we demonstrate the competency, transferability, and generalizability of the PRISM features and exhibit use cases of PRISM classifiers for computing approximate disease labels and temporally updating disease probabilities. The GTL approach introduced in this study allows for validation of various EHR phenotypes without the need for domain-specific knowledge about individual disease processes.
BACKGROUND
The closest related work to computing disease probabilities is EHR phenotyping. To be able to make precise assumptions about the presence (or lack there) of a disease, given the presence of its diagnosis record in the EHRs, we would need to perform EHR phenotyping for all diseases. The key task in EHR phenotyping is to identify patient cohorts with (or without) certain phenotypes or clinical conditions of interest.14,15 Approaches to electronic phenotyping include rule-based methods, text processing, supervised/semi-supervised/unsupervised phenotyping using statistical learning techniques, and hybrid approaches.14,16 Computational phenotyping algorithms basically estimate, for each patient, the probability that they have the disease, given certain phenotypic characteristics that are inferred from a vector of medical records . Most of the statistical learning algorithms used in computational phenotyping apply discriminative models (such as logistic regression) that aim to learn the explicit hard/soft boundaries between different classes of in the data by directly modeling the conditional probability .
If we had computational or rule-based phenotyping algorithms for all diseases, plausibility of EHR diagnosis records would not be an issue. In most cases, however, phenotyping entails specialized computational or rule-based algorithms that are often costly, require expert involvement for curating phenotypic characteristics (vector of features), and do not scale over a wide range of diseases.
Further, most phenotyping algorithms need labeled data for training and testing. To soften this need, semi-supervised computational phenotyping relies on what is known as “silver-standard” labels that are curated based on the statistical computation of phenotype probabilities. Different data types and methods are utilized in computing phenotype probabilities from electronic medical records, for example, using disease-specific anchor features. Anchors are expected to signal positive when the phenotype is present, but are often uninformative when the phenotype is absent.17,18
Clinical notes and structured data are popular sources of information for mining disease-specific anchor features. For example, Halpern et al.18,19 and Agarwal et al.20 mined clinical notes for specific anchor features, such as phrases, that described the phenotype to compute probabilities. Yu et al.21–23 developed frameworks to automatically generate a list of anchor phrases (and clinical codes) from medical knowledge sources. Chiu and Hripcsak24 leveraged the International Classification of Diseases, the 9th revision (ICD-9) codes as surrogates to perform batch-phenotyping without annotated phenotype labels. Wagholikar et al.25 proposed the polar labeling (PL) that utilizes the distribution of disease-specific ICD codes to curate silver-standard labels, based on a threshold on the probability distribution function. Nevertheless, because anchors are disease-specific their selection/curation generally requires domain expertise (or prior knowledge) and thus does not generalize to other phenotypes. In this paper, we introduce a small set of disease-agnostic features that aim to reflect the Patients, pRoviders, and their Interactions within the healthcare SysteM (PRISM)—eg, the number of distinct dates in which a diagnosis code was recorded and the encounter type (inpatient, outpatient) for the diagnosis record. We argue and demonstrate that PRISM features can be used for training generative PRISM classifiers to estimate disease probabilities and are generalizable and transferable across disease and patient populations.
Our goal is to estimate the likelihood of disease in patient at time , given the diagnosis record for the disease at . Let us assume represents a vector of records and is an outcome variable (ie, means diagnosis record is true), obtained from electronic health records. We model both the PRISM features and the target variable as random (ie, stochastic) variables with a joint distribution . The list and description of PRISM features are provided in Table 1. Unlike discriminative models that are the prevalent modeling algorithms in computational phenotyping, generative models make structural assumptions on the data that preclude overfitting. They aim to learn the distribution of different classes of in the data, by learning the joint probability —ie, generative models care about both and .26 Generative models, therefore, can have a higher upside for the computing disease probabilities due to their ability to learn from small gold-standard labeled data.
Table 1.
An initial list of PRISM feature abbreviation and descriptions
| abbreviation | description |
|---|---|
| phenX | #diagnosis record(s) for the phenotype |
| enc_denom | #unique encounters for each patient |
| dx_denom | #unique diagnosis codes for each patient |
| enchphen_denom | #unique encounters between the first and last phenotype record |
| phenX_O | #OUTPATIENT diagnosis record(s) for the phenotype |
| rx_denom | #unique medication codes for each patient |
| different_dates | #unique dates in which the diagnosis codes for the phenotype recorded |
| distinc_providers | #unique provided who recorded the diagnosis codes for the phenotype |
| durate | #months between the first and the last phenotype record |
| sex_cd | patient gender |
| phenX_I | #INPATIENT diagnosis record(s) for the phenotype |
| age_mean | mean patient age at encounters when phenotype was recorded |
| age_min | youngest patient age at encounters when phenotype was recorded |
| age_max | oldest patient age at encounters when phenotype was recorded |
| phenx.rate | growth rate in phenotype record . The feature aims to represent the growth in the phenotype record over time |
| oldness | #months between the last record and the last phenotype record |
| phenX_E | #ED diagnosis record(s) for the phenotype |
We hypothesize that from PRISM features we can build generative PRISM classifiers that (1) can predict a target disease, and (2) are transferable across diseases and patient populations. The second hypothesis builds upon the transfer learning concept in Machine Learning (ML) theory (here, “transfer learning” refers to exploiting learning from one task to improve generalization on another task through transfer of knowledge27,28)
METHODS
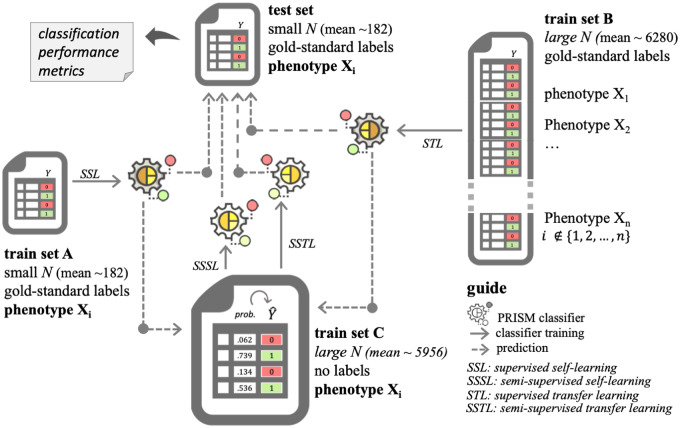
We test the study hypotheses through two approaches: supervised self-learning (SSL) and supervised transfer learning (STL) using PRISM features. We call the classifiers trained on PRISM features the PRISM classifiers. To further evaluate the second hypothesis, we expand the use case for the SSL and STL to curate silver standard labels on larger patient populations for semi-supervised learning (Figure 1).
Figure 1.
Study design for evaluating the feasibility, generalizability, and transferability of PRISM classifiers.
Setting
The experiment described in this paper was carried out on electronic health records data from the Mass General Brigham (MGB) Biobank in Boston. The use of data for this study was approved by the Mass General Brigham Institutional Review Board (2017P000282). To test the study hypotheses, we use EHR data on a diverse set of 18 diseases: alzheimer’s disease (AD), atrial fibrillation (AFIB), asthma, bipolar disorder (BD), breast cancer (BrCa), coronary artery disease (CAD), crohn's disease (CD), congestive heart failure (CHF), chronic obstructive pulmonary disease (COPD), epilepsy, gout, hypertension (HTN), rheumatoid arthritis (RA), schizophrenia (SCZ), stroke, type 1 diabetes mellitus (T1DM), type 2 diabetes mellitus (T2DM), and ulcerative colitis (UC). For each of the 18 diseases, a patient in the cohort has at least one record of the diagnosis code for the given disease and a gold-standard outcome label curated through expert review of clinical narratives. The gold-standard data includes labels for an average of 364 patients (ranging from 120 patients for schizophrenia to 1181 patients for hypertension). We demonstrate two use cases in this study. For the first use case, we also used unlabeled data from the MGB Biobank on all 18 diseases, which includes data from an average of 5732 patients—ranging from 1661 (AD) to 14 406 (CAD)—with at least one diagnosis code for the disease of interest. Supplementary Table S1 provides detailed information about data from each disease.
Self-learning
We conduct both supervised and semi-supervised self-learning (abbreviated as SSL and SSSL, respectively). In SSL, we start from a small (on average, ∼360 entries) data set with “gold-standard” labels (that were manually chart-reviewed) for a specific disease. For patients with gold-standard labels, we extract PRISM features from their medical records. We keep about half of these entries (on average, ∼182) as part of a held-out test set and call the other half the “train set A.” Train set C encompasses the rest (and vast majority) of the data set from the same disease, which is unlabeled. We compare two procedures:
(Supervised self-learning, SSL) Train a generative model on train set A and use it to label the test set.
(Semi-supervised self-learning, SSSL) Train a generative model on train set A and use it to compute probabilities and then curate labels on train set C. The resulting labels are called silver-standard labels. We then train a (possibly different) generative model on train set C with the silver-standard labels and use that to label the test set.
More specifically, the training works as follows. To minimize overfitting and redundancy between the features, we first run a filter-type feature extraction method using joint mutual information (JMI)29 as scoring criteria. The algorithm starts with a set containing the top feature according to mutual information, then iteratively adds to the feature maximizing the joint mutual information score
Here, denotes the mutual information between random variables and (a measure of the information shared by and —it can be expressed as the entropy of minus the entropy of given ). The random variable is simply the random variable corresponding to the joint distribution of and . In the end, we select the top features that were added to the set .
More precisely, suppose the features are ordered as , with corresponding JMI score (the scores are increasing because of the sum in the JMI). We retain features , where is least such that . Here, is a small positive number (we set it to ). In other words, we always keep the first four features, and after that keep a feature only if it increases the JMI score by a factor of at least .
The idea of using the joint mutual information score (as opposed to just the mutual information) is that it also takes into account the redundancy between the features: two features could each be highly relevant on their own, but also be strongly correlated. Once the features have been selected, we applied four generative models: Bayesian generalized linear regression (logistic link and student-T prior with one degree of freedom), linear and quadratic discriminant (LDA and QDA) analyses, and Naïve Bayes classifiers, to these data to model and hence . We chose the four generative models because they are well known, simple, and widely used.
Transfer learning
In the transfer learning approach, we again fix a single disease, , but use data for other diseases to train our model. Specifically, in addition to training set A (half the gold standard labels for disease ), we have at our disposal the labeled data from all the other diseases different from . We call this second data set training set . On average, the patient size for training set was above 6000, which provided a major boost to the training task, with a potential caveat that none of the labels were on the disease .
We first start as in the previous setup by using joint mutual information with training set to identify the top features on disease . Call this set of features .
We then identify the diseases for which these features also perform well. For this, we compute, for each disease and each feature the Kullback-Leibler (KL) divergence between and . Here, , refer to probability computed according to training set for and respectively. The Kullback-Leibler divergence is a well-known information-theoretic measure of similarity between probability distribution—smaller divergence means closer distributions, see for example Cover and Thomas (2012).30
Once has been computed for each and each , we let be the average of , for . Say we have . Then we let training set be training set without the last diseases in this list, where is a hyperparameter.
As before, we compare two approaches:
(Supervised transfer learning, STL) Train a generative model on train set C’ and use it to label a test set (labeled data for disease ).
(Semi-supervised transfer learning, SSTL) Train a generative model on train set C’ and use it to predict disease probabilities and curate silver-standard label on train set C (unlabeled data for disease ). We then train a (possibly different) generative model on train set C with the silver-standard labels and use that to label the test set.
Classifier evaluation
For evaluating the classifiers, we compute classification performance metrics from the held-out test sets. We use the area under the receiver operating characteristic curve (AUC ROC), as well as positive and negative predictive values (PPV and NPV) computed at the operating point 0.5. From the four classification algorithms, for each disease and learning approach (self- or transfer learning), we chose the best classifier using the AUC ROC and compare them with each other.
Use cases
For completing the assessment of the second hypothesis (transferability of the PRISM classifiers to a larger patient cohort), we evaluate a use case in which we utilize the supervised self- and transfer learning PRISM classifiers for curating silver-standard labels in a larger patient cohort. We use the supervised learning performance metrics as a benchmark to evaluate the performance of the semi-supervised learning with PRISM features. The idea here is that if a PRISM classifier trained on silver-standard labels (semi-supervised PRISM classifier) that were in turn curated from its corresponding supervised learning PRISM classifier provides comparable performance metrics on the test set, the silver-standard labels will have comparable properties to gold-standard labels.
As a second use case, we evaluate the feasibility of applying the self- and transfer learning PRISM classifiers to compute temporally updated record probabilities for a wide range of chronic diseases. For this use case, we update patient-level distribution of the PRISM features and use the trained PRISM classifiers to compute a record probability at each time stamp.
RESULTS
Overall, the classification performance metrics from the supervised self-learning (SSL) support the overall competency of the PRISM classifiers (ie, a classifier trained with PRISM features) to predict the target phenotype (Table 2). Compared with the state-of-the-art phenotyping results published in the literature, in eight of the 18 diseases, the PRISM classifiers outperformed computational phenotyping results, and in five disease the PRISM classifiers yielded slightly lower AUC ROCs. In seven diseases, we were not able to find computational phenotyping results in the literature. It is important to remember that computational phenotyping algorithms leverage a large number of features that encompass clinical notes, as well as diagnosis, medications, and laboratory records, often guided by domain experts or existing knowledge. Keep in mind, these are state-of-the-art, using many features, and clinical notes via NLP. As we will demonstrate in feature selection results, PRISM classifiers are trained on an average of seven general PRISM features.
Table 2.
Supervised learning performance comparison between self and transfer learning
| AUC ROC | PPVa | NPVa | AUC ROC | PPVa | NPVa | ||||
|---|---|---|---|---|---|---|---|---|---|
| − | lit. | 0.958 31 | lit. | ||||||
| AD | 0.877 | 0.727 | 0.880 | STL | Epilepsy | 0.955 | 0.938 | 0.756 | STL |
| 0.841 | 0.529 | 0.886 | SSL | 0.968 | 0.955 | 0.886 | SSL | ||
| 4% | 37% | −1% | Δ | −1% | −2% | −15% | Δ | ||
| – | lit. | – | lit. | ||||||
| AFIB | 0.927 | 0.931 | 0.529 | STL | Gout | 0.913 | 1.000 | 0.111 | STL |
| 0.931 | 0.808 | 0.909 | SSL | 0.981 | 1.000 | 0.222 | SSL | ||
| 0% | 15% | −42% | Δ | −7% | 0% | −50% | Δ | ||
| 0.942 31 | lit. | 0.952 31 | lit. | ||||||
| Asthma | 0.850 | 0.938 | 0.676 | STL | HTN | 0.903 | 0.943 | 0.534 | STL |
| 0.832 | 0.938 | 0.676 | SSL | 0.887 | 0.885 | 0.718 | SSL | ||
| 2% | 0% | 0% | Δ | 2% | 7% | −26% | Δ | ||
| 0.875 31 | lit. | 0.933–0.961 21 , 31 , 32 | lit. | ||||||
| BD | 0.852 | 0.727 | 0.732 | STL | RA | 0.973 | 0.704 | 0.981 | STL |
| 0.836 | 0.650 | 0.813 | SSL | 0.968 | 0.941 | 0.938 | SSL | ||
| 2% | 12% | −10% | Δ | 1% | −25% | 5% | Δ | ||
| 0.962 31 | lit. | 0.85–0.913 31 , 33 | lit. | ||||||
| BrCa | 0.965 | 1.000 | 0.611 | STL | SCZ | 0.873 | 0.333 | 0.950 | STL |
| 0.959 | 0.933 | 0.750 | SSL | 0.808 | 0.333 | 0.930 | SSL | ||
| 1% | 7% | −19% | Δ | 8% | 0% | 2% | Δ | ||
| 0.896–0.93 21 , 31 , 32 | lit. | – | lit. | ||||||
| CAD | 0.977 | 0.853 | 0.951 | STL | Stroke | 0.875 | 0.786 | 0.813 | STL |
| 0.968 | 0.903 | 0.938 | SSL | 0.844 | 0.750 | 0.881 | SSL | ||
| 1% | −6% | 1% | Δ | 4% | 5% | −8% | Δ | ||
| 0.94–0.963 22 , 31 , 32 | lit. | 0.981 31 | lit. | ||||||
| CD | 0.972 | 1.000 | 0.816 | STL | T1DM | 0.959 | 0.875 | 0.946 | STL |
| 0.966 | 0.935 | 0.879 | SSL | 0.994 | 1.000 | 0.931 | SSL | ||
| 1% | 7% | −7% | Δ | −4% | −13% | 2% | Δ | ||
| 0.72–0.87 33–35 | lit. | 0.9 31 , 33 | lit. | ||||||
| CHF | 0.868 | 0.500 | 0.897 | STL | T2DM | 0.947 | 0.902 | 0.884 | STL |
| 0.839 | 0.600 | 0.867 | SSL | 0.935 | 0.968 | 0.774 | SSL | ||
| 3% | −17% | 4% | Δ | 1% | −7% | 14% | Δ | ||
| – | lit. | 0.87–0.975 22 , 31–33 | lit. | ||||||
| COPD | 0.864 | 0.611 | 0.848 | STL | UC | 0.949 | 0.813 | 0.931 | STL |
| 0.872 | 0.647 | 0.851 | SSL | 0.955 | 0.885 | 0.857 | SSL | ||
| −1% | −6% | 0% | Δ | −1% | −8% | 9% | Δ | ||
| aPositive/negative predictive values at operating point 0.5 | mean Δ | 1% | 0% | −8% | |||||
|
STL: Supervised transfer learning SSL: Supervised self-learning |
std Δ | 3% | 14% | 17% | |||||
Δ is the performance delta between transfer learning and self-learning. Positive means transfer learning was better.
lit. represent AUC ROCs from the published state-of-the-art phenotyping research—to the best of our knowledge.
To test the transferability of the PRISM features, we compared the results between supervised self-learning (SSL) and supervised transfer learning (STL). As illustrated in Table 1, across the 18 diseases, supervised transfer learning (STL) improved the overall classification performance (AUC ROC) by about 1%. The delta in positive and negative predictive values (PPC and NPV) computed at operating point 0.5 was small with high divergence, meaning that reliable judgement cannot be achieved. Regardless, the results support that the PRISM features are transferable across diseases. That is, we can train a PRISM classifier from a group of diseases and use it to predict a different disease with negligible change in predictive power as compared with the self-learning.
Using the JMI feature selection also allowed us to draw a picture of the relative importance of PRISM features. As illustrated in Figure 2, the number unique encounters (enc_denom) and diagnosis (dx_denom), the number of times the disease code was recorded in an outpatient setting (phenX_O), the number of different dates (different_dates) and encounters (encphen_denom) the disease diagnosis code was recorded, the number of distinct providers, and the patient’s gender (sex_cd) were the most chosen PRISM features across the 18 diseases. Unlike the general perception that the overall diagnosis record (phenX) for a disease is predictive of the disease, we found that, when controlling for the outpatient diagnosis record (phenX_O), the overall count of all diagnosis records is unimportant for a true inference about the existence of the disease. Similarly, we also found that an in-patient diagnosis record is relatively unimportant for a true inference of underlying disease.
Figure 2.
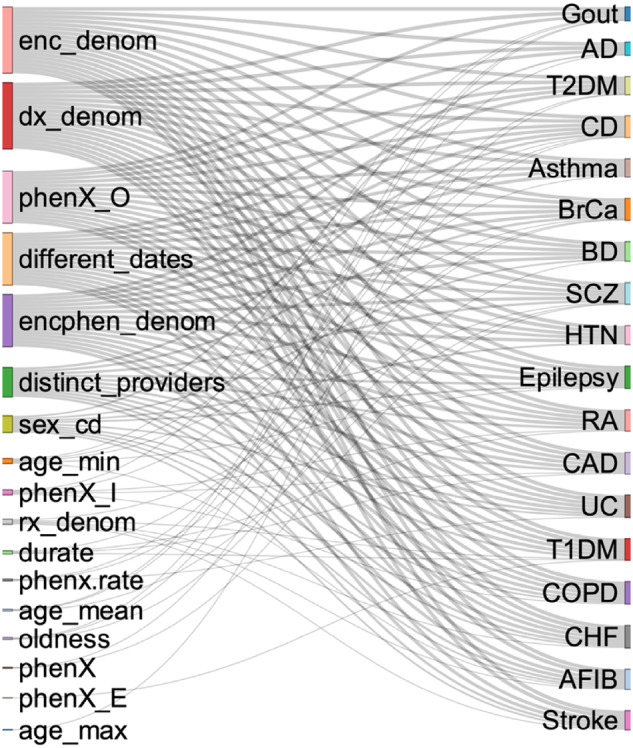
PRISM features and their use in predicting diseases. *PRISM features are listed on the left. The 18 diseases are listed on the right. The use of a PRISM feature to predict a disease is identified with connecting line.
To complement the feature importance findings from JMI, we also demonstrate the PRISM features’ regression coefficients obtained from the Bayesian generalized linear models (Figure 3). The coefficients allow us to understand how a PRISM feature associates with the true disease state. In a majority of the models, the number of times the disease code was recorded in an outpatient setting (phenX_O) was negatively associated with the respective disease outcome. The number of encounters (encphen_denom) in which the disease diagnosis code was recorded was positively associated with the diagnosis code being true. The higher the number of distinct providers who recorded the disease code, the higher are the chances that the disease code is true. In contrast, the higher the number of unique diagnosis and encounter records (which can be interpreted as how sick a patient might be), the lower the chances that any diagnosis record truly captures the existence of a disease in a patient.
Figure 3.
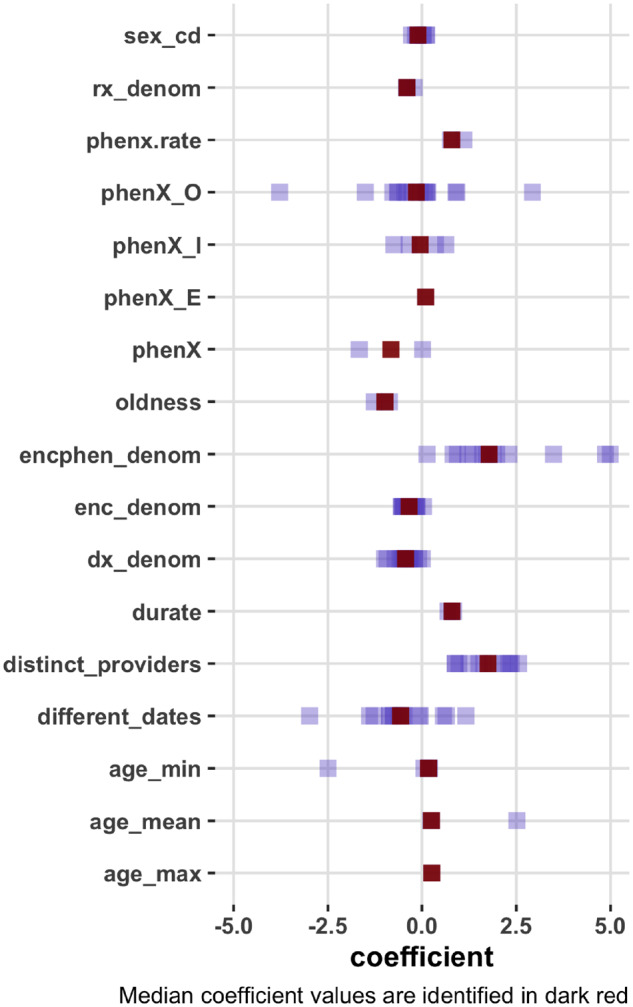
PRISM features’ regression coefficients for predicting different diseases. *Regression coefficients from Bayesian generalized linear models.
As a concrete example, we review the PRISM regression coefficients for Crohn’s Disease (CD), which is classified to ICD-9-CM category 555 (555.0, 555.1, 555.2, and 555.9). The significant PRISM features ranked by regression coefficients (from largest to smallest, in absolute values) for CD were: phenx.rate (1.11), distinct_providers (0.97), encphen_denom (0.94), dx_denom (-0.59), and enc_denom (-0.32). phenx.rate aims to represent the increase rate in the phenotype diagnosis record over time (). Using the PRISM features, we PRISM features, we were able to train supervised self-learning (SSL) and transfer learning (STL) classifiers that resulted in AUC ROC of 0.97, which can be compared to the specialized computational phenotyping performances between 0.94 and 0.96 in the literature.22,31,32
Using the selected features for each disease in the transfer learning step, we used the average Kullback-Leibler divergence to determine the transferability of learning with PRISM classifiers from different disease data sets. For example, if the self-learning models, identified certain PRISM features for predicting hypertension (HTN), we evaluated the transferability of labeled data from other diseases based on the pre-set HTN PRISM features. Based on this information, we found that data from alzheimer’s, schizophrenia, and bipolar disorder were transferable to learning the fewest diseases. In contrast, data from hypertension, chronic obstructive pulmonary disease (COPD), crohn's disease (CD), and asthma were transferable most (Figure 4).
Figure 4.
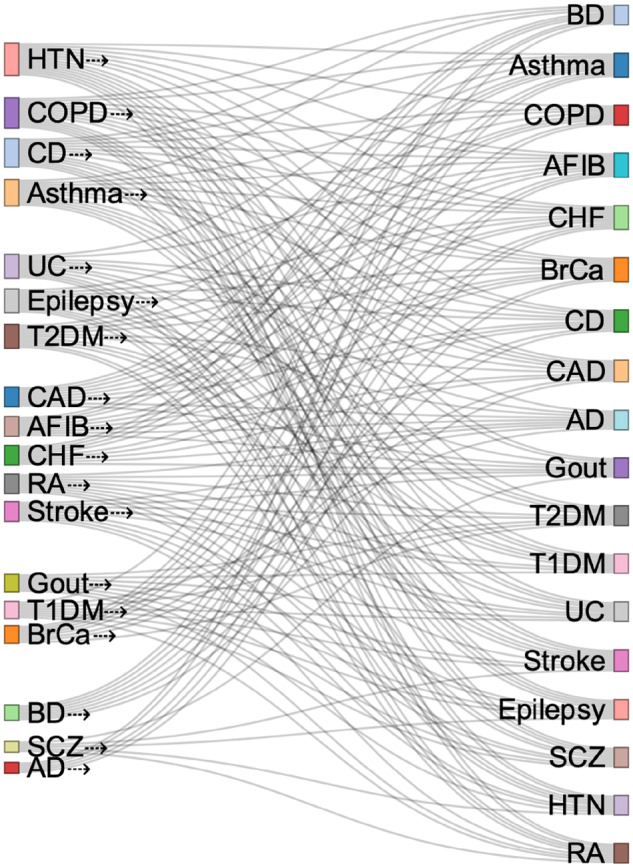
Transferability of labeled data for learning about other diseases using PRISM classifiers. *Training sets are identified on the left. Arrow means that data from a certain disease was used in the training set for learning about another disease (on the left).
Use case 1: curating silver standard labels
As the first use case, we evaluate the feasibility of using PRISM classifiers for curating silver-standard labels for various diseases. As described in the methods, we evaluate this by computing the performance delta obtained from the supervised learning (both self-learning and transfer learning) with the respective semi-supervised PRISM classier. The semi-supervised classifiers are trained based on silver-standard labels that are curated from the probabilities computed from the supervised learning classifiers—using 0.5 as the cutoff for determining positives and negatives. Table 3 shows the performance metrics for the semi-supervised learning as well as two deltas. Δ1 is the delta between semi-supervised and supervised learning performances in the transfer learning approach. Δ2 represents the delta between semi-supervised and supervised learning performances in the self-learning approach. The mean and standard deviation for each delta is also presented in Table 3.
Table 3.
Comparing supervised and semi-supervised learning performances
| AUC ROC | PPVa | NPVa | AUC ROC | PPVa | NPVa | ||||
|---|---|---|---|---|---|---|---|---|---|
| AD | 0.886 | 0.667 | 0.913 | SSTL | Epilepsy | 0.960 | 0.909 | 0.857 | SSTL |
| 1% | −8% | 4% | Δ1 | 1% | −3% | 13% | Δ1 | ||
| 0.836 | 0.533 | 0.870 | SSSL | 0.959 | 0.923 | 0.968 | SSSL | ||
| −1% | 1% | −2% | Δ2 | −1% | −3% | 9% | Δ2 | ||
| AFIB | 0.914 | 0.925 | 0.739 | SSTL | Gout | 0.925 | 1.000 | 0.286 | SSTL |
| −1% | −1% | 40% | Δ1 | 1% | 0% | 157% | Δ1 | ||
| 0.890 | 0.949 | 0.750 | SSSL | 0.975 | 0.930 | 1.000 | SSSL | ||
| −4% | 17% | −18% | Δ2 | −1% | −7% | 350% | Δ2 | ||
| Asthma | 0.806 | 0.789 | 0.827 | SSTL | HTN | 0.809 | 0.918 | 0.565 | SSTL |
| −5% | −16% | 22% | Δ1 | −10% | −3% | 6% | Δ1 | ||
| 0.748 | 0.733 | 0.717 | SSSL | 0.825 | 0.945 | 0.574 | SSSL | ||
| −10% | −22% | 6% | Δ2 | −7% | 7% | −20% | Δ2 | ||
| BD | 0.813 | 0.706 | 0.800 | SSTL | RA | 0.962 | 0.714 | 0.917 | SSTL |
| −4% | −3% | 9% | Δ1 | −1% | 2% | −7% | Δ1 | ||
| 0.805 | 0.609 | 0.828 | SSSL | 0.932 | 0.704 | 0.981 | SSSL | ||
| −4% | −6% | 2% | Δ2 | −4% | −25% | 5% | Δ2 | ||
| BrCa | 0.962 | 0.933 | 0.750 | SSTL | SCZ | 0.909 | 0.316 | 1.000 | SSTL |
| 0% | 0% | 0% | Δ1 | 4% | −5% | 7% | Δ1 | ||
| 0.919 | 1.000 | 0.688 | SSSL | 0.808 | 0.200 | 0.894 | SSSL | ||
| −4% | 17% | −28% | Δ2 | 0% | −75% | 10% | Δ2 | ||
| CAD | 0.926 | 0.879 | 0.952 | SSTL | Stroke | 0.905 | 0.708 | 0.921 | SSTL |
| −5% | 3% | 0% | Δ1 | 3% | −10% | 13% | Δ1 | ||
| 0.973 | 0.829 | 0.950 | SSSL | 0.851 | 0.750 | 0.826 | SSSL | ||
| 1% | −8% | 1% | Δ2 | 1% | 0% | −6% | Δ2 | ||
| CD | 0.968 | 1.000 | 0.816 | SSTL | T1DM | 0.961 | 0.833 | 0.914 | SSTL |
| 0% | 0% | 0% | Δ1 | 0% | −5% | −3% | Δ1 | ||
| 0.956 | 0.912 | 0.933 | SSSL | 0.988 | 0.833 | 1.000 | SSSL | ||
| −1% | −3% | 6% | Δ2 | −1% | −17% | 7% | Δ2 | ||
| CHF | 0.857 | 0.667 | 0.870 | SSTL | T2DM | 0.913 | 0.973 | 0.872 | SSTL |
| −1% | 33% | −3% | Δ1 | −4% | 8% | −1% | Δ1 | ||
| 0.777 | 0.450 | 0.914 | SSSL | 0.892 | 0.923 | 0.867 | SSSL | ||
| −7% | −25% | 5% | Δ2 | −5% | −5% | 12% | Δ2 | ||
| COPD | 0.849 | 0.583 | 0.900 | SSTL | UC | 0.926 | 0.926 | 0.912 | SSTL |
| −2% | −5% | 6% | Δ1 | −2% | 14% | −2% | Δ1 | ||
| 0.850 | 0.583 | 0.900 | SSSL | 0.964 | 0.917 | 0.838 | SSSL | ||
| −2% | −10% | 6% | Δ2 | 1% | 4% | −2% | Δ2 | ||
| Δ1 | Δ2 | ||||||||
| average | −1% | 0% | 14% | average | −3% | −9% | 19% | ||
| std | 3% | 11% | 37% | SSTL | std | 3% | 20% | 83% | SSSL |
Δ1 is the delta between semi-supervised and supervised learning performances in the transfer learning approach.
Δ2 is the delta between semi-supervised and supervised learning performances in the self-learning approach.
NPV and PPV values are computed at operating point 0.5.
Results show that, on average, the semi-supervised learning with PRISM features using the silver-standard labels from the supervised learning PRISM classifiers yield to classification performances that are only marginally inferior in AUC ROC. The delta in predictive and positive predictive values at the operating point 0.5 show that while there was a drop in positive predictive values (taking standard deviations into account), the negative predictive value is generally improved. Overall, our findings demonstrated that PRISM features can be scaled to transfer their phenotype prediction power to larger cohorts through using their estimated disease probabilities to curate silver-standard labels. We also compared the performance of semi-supervised learning between the self-learning and transfer learning classifiers (Supplementary Table S2). We found that, on average, the classification performance in semi-supervised transfer learning (SSTL) provides 2% (standard deviation of 5%) improvement over semi-supervised learning based on labels curated from self-learning classifiers (SSSL). The improvement was more notable in the positive predictive values at the operating point 0.5.
Use case 2: computing updated record probabilities
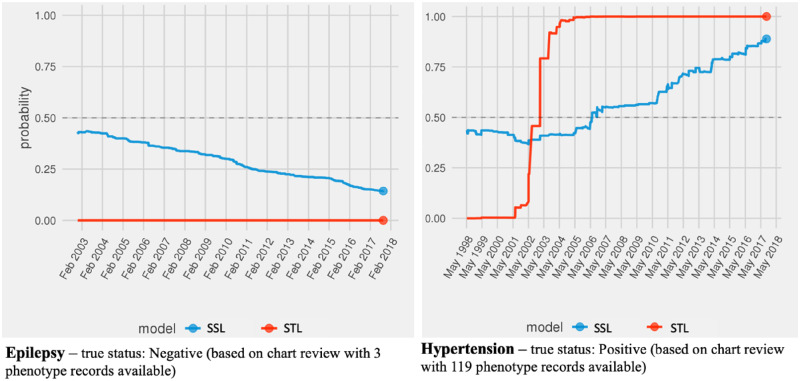
As the second use case, we demonstrate the feasibility of applying the self- and transfer learning PRISM classifiers to compute temporally updated record probabilities. Figure 5 presents examples of probability estimates for Epilepsy (plot on the left) and Hypertension (plot on the right) for two individual patients. The patient-level record probabilities are updated at each time stamp. In both examples, the patients had diagnosis records of the disease in their electronic health records. For the patient with a true hypertension diagnosis, the models can show different time points where disease probability passed over the 50% line.
Figure 5.
Updated probabilities of Epilepsy (left) and Hypertension (right) records over time.
This use case demonstrated that both self-learning and transfer learning PRISM classifiers can be used to compute disease probabilities at each time point operating on PRISM features that are updated over time. In these two examples, the SSL models appeared to show more gradual changes in the record probabilities, although this may not be true in all cases. The STL model in Figure 5 illustrated a steep increase (from 0 to 100) in the probability of the hypertension diagnosis record in a span of seven year (between 1998 and 2005) and eight encounters for the given patient. This can indicate that most of the phenotype records for hypertension for the given patient was recorded between 1998 and 2005. We found that both models demonstrated accurate predictions at the end point.
DISCUSSION
Over the past decade, billions have been spent to institute meaningful use of electronic health record (EHR) systems. For a multitude of reasons, however, EHR data are still complex and have ample quality issues, which make it difficult to leverage these data in order to address pressing health issues. Reliability of diagnosis records in the EHRs is questionable as they reflect healthcare processes and payer-provider policies. This issue is especially agonizing during pandemics such as COVID-19, when prompt responses are needed. Given the reliability issues with the diagnosis codes, rapid discovery of potential risk factors for any health outcome from the EHRs can be difficult, requiring significant resources to perform cohort identification on a long list of potential risk factors.
In this paper, we proposed a Generative Transfer Learning (GTL) approach for estimating the probability of diagnosis records in the EHRs using a small set of features we call PRISM features. We demonstrated that PRISM features and the generative PRISM classifiers are potent for estimating disease probabilities and exhibit generalizable and transferable distributional characteristics across diseases and patient populations. We characterize the GTL approach with PRISM features as a low-cost and disease-agnostic alternative to the computational phenotyping approach that is often expensive and disease-specific. The GTL approach allows for validation of various EHR phenotypes without the need for domain-specific knowledge about specific disease processes.
Comparing with the state-of-the-art computational phenotyping work published in the literature, we demonstrated that, with an average of seven PRISM features, the generative PRISM classifiers provided better classification performances in eight of the 18 diseases, while having slightly worse performance in five diseases. It is important to acknowledge that the AUCs used as points of comparison from the literature may be based on patient populations with different characteristics.
PRISM classifiers can be used for a variety of use cases. In the first use case, we demonstrated the utility of the generative transfer learning approach for curating silver-standard disease labels. Many healthcare institutions are actively involved in chart reviewing EHR data to facilitate development of Machine Learning algorithms. Yet, due to the high costs of performing manual chart reviews, the labeled data often encompass small numbers of patients, which provide issues with overfitting, but are available on a broad set of diseases. We showed that PRISM features from small set of labeled data on different diseases can be leveraged to train a generative PRISM classifier that can be used for curating silver standard labels on many diseases. Such a capability will have a notable impact on scaling up applied ML research using EHR data.
Because PRISM classifiers are computationally cheap to train and generalizable, they can also be applied to computing patient-level disease probabilities over time. In the second use case, we showed that PRISM classifiers can be used to estimate disease probabilities for all diagnosis codes with high accuracy. This will improve the capacity to advance rapid cohort identification from EHR data.
Overall, we demonstrated the generative PRISM classifiers are generalizable and transferable across diseases and patient populations. Potential portability of these classifier across multiple institutions remains to be evaluated in future research. Further, we anticipate that the list of PRISM features will keep growing and potentially can be tailored to medical ontologies in order to maximize the sensitivity or specificity of the classification tasks on various diseases. More research is also needed to study potential trend line differences in the probability estimations demonstrated in use case 2. Probabilities computed by the PRISM classifiers can be integrated into the Informatics for Integrating Biology & the Bedside (i2b2)36 as a probability dimension, representing the confidence level of a given condition. This would allow users to set a “confidence level” for a specific condition and adjust the sensitivity and specificity of a given i2b2 query depending on the specific use case.
CONCLUSION
We introduced a generative transfer learning approach using PRISM features for computing probabilities of disease records in electronic health records. We showed that the joint probabilities we learn from PRISM generative models about healthcare dynamics are transferable and generalizable to multiple diseases. Comparing the relative importance of PRISM features showed that, for example, when stratifying phenotype records by encounter types, an outpatient record is more important than an inpatient record or an unstratified record for a making inference about the underlying disease state. Because these models are inexpensive (ie, features are generalizable and uniform, and algorithms are not computationally intensive), this approach is scalable to compute disease probabilities for a wide range of diseases. Probabilities computed from the generative PRISM classifier can scale up applied Machine Learning research and knowledge discovery with EHR data.
FUNDING
This research was supported by the National Human Genome Research Institute grant number R01-HG009174.
AUTHOR CONTRIBUTIONS
Conceived study design: HE and SV. Contributed to data analysis and visualization: HE. Wrote the manuscript: HE and SV. Reviewed and edited manuscript: HE, SV, SNM. All authors approved the final manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
None declared.
Supplementary Material
REFERENCES
- 1.Institute of Medicine (US). Roundtable on Evidence-Based Medicine. In: Olsen L, Aisner D, McGinnis JM, eds. The Learning Healthcare System: Workshop Summary. Washingotn, DC: National Academies Press; 2007. [PubMed] [Google Scholar]
- 2. Stewart WF, Shah NR, Selna MJ, et al. Bridging the inferential gap: the electronic health record and clinical evidence. Health Aff 2007; 26: w181–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown JS, Kahn M, Toh S.. Data quality assessment for comparative effectiveness research in distributed data networks. Med Care 2013; 51 (8 Suppl 3): S22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kahn MG, Batson D, Schilling LM.. Data model considerations for clinical effectiveness researchers. Med Care 2012; 50 (Suppl): S60–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weiskopf NG, Hripcsak G, Swaminathan S, et al. Defining and measuring completeness of electronic health records for secondary use. J Biomed Inform 2013; 46 (5): 830–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gregori D, Berchialla P.. Quality of electronic medical records. In: Faltin FW, Kenett RS, Ruggeri F, eds. Statistical Methods in Healthcare. West Sussex, UK: Wiley; 2012: 456–76. [Google Scholar]
- 7.ONC. The National Health IT Priorities for Research: A Policy and Development Agenda. 2020. . https://www.healthit.gov/sites/default/files/page/2020-01/PolicyandDevelopmentAgenda.pdf Accessed February 20, 2020.
- 8. Kahn MG, Callahan TJ, Barnard J, et al. A harmonized data quality assessment terminology and framework for the secondary use of electronic health record data. eGEMs 2016; 4 (1): 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hripcsak G, Albers DJ, Perotte A.. Exploiting time in electronic health record correlations. J Am Med Informatics Assoc 2011; 18 (Suppl 1): i109–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hripcsak G, Albers DJ.. Next-generation phenotyping of electronic health records. J Am Med Informatics Assoc 2013; 20 (1): 117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Agniel D, Kohane IS, Weber GM.. Biases in electronic health record data due to processes within the healthcare system: Retrospective observational study. BMJ 2018; 361: k1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gainer VS, Cagan A, Castro VM, et al. The biobank portal for partners personalized medicine: A query tool for working with consented biobank samples, genotypes, and phenotypes using i2b2. J Pers Med 2016; 6 (1): 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karlson EW, Boutin NT, Hoffnagle AG, et al. Building the partners healthcare biobank at partners personalized medicine: Informed consent, return of research results, recruitment lessons and operational considerations. J Pers Med 2016; 6 (1): 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Banda JM, Seneviratne M, Hernandez-Boussard T, et al. Advances in electronic phenotyping: from rule-based definitions to machine learning models. Annu Rev Biomed Data Sci 2018; 1 (1): 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ding DY, Simpson C, Pfohl S, et al. The effectiveness of multitask learning for phenotyping with electronic health records data. Pac Symp Biocomput 2019; 24: 18–29. [PMC free article] [PubMed] [Google Scholar]
- 16. Shivade C, Raghavan P, Fosler-Lussier E, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc 2014; 21 (2): 221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Banda JM, Halpern Y, Sontag D, et al. Electronic phenotyping with APHRODITE and the Observational Health Sciences and Informatics (OHDSI) data network. AMIA Jt Summits Transl Sci Proc 2017; 2017: 48–57. [PMC free article] [PubMed] [Google Scholar]
- 18. Halpern Y, Choi Y, Horng S, et al. Using anchors to estimate clinical state without labeled data. AMIA Annu Symp Proc 2014; 2014: 606–15. [PMC free article] [PubMed] [Google Scholar]
- 19. Halpern Y, Horng S, Choi Y, et al. Electronic medical record phenotyping using the anchor and learn framework. J Am Med Inform Assoc 2016; 23 (4): 731–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Agarwal V, Podchiyska T, Banda JM, et al. Learning statistical models of phenotypes using noisy labeled training data. J Am Med Inform Assoc 2016; 23 (6): 1166–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu S, Liao KP, Shaw SY, et al. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. J Am Med Inform Assoc 2015; 22 (5): 993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu S, Ma Y, Gronsbell J, et al. Enabling phenotypic big data with PheNorm. J Am Med Informatics Assoc 2018; 25 (1): 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu S, Chakrabortty A, Liao KP, et al. Surrogate-assisted feature extraction for high-throughput phenotyping. J Am Med Inform Assoc 2017; 24: e143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chiu PH, Hripcsak G.. EHR-based phenotyping: bulk learning and evaluation. J Biomed Inform 2017; 70: 35–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wagholikar KB, Estiri H, Murphy M, et al. Polar labeling: silver standard algorithm for training disease classifiers. Bioinformatics 2020; 36 (10): 3200–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ng AY, Jordan MI.. On discriminative vs. generative classifiers: a comparison of logistic regression and naive Bayes. Adv Neural Inf Process Syst. 2002; 28: 169–87. [Google Scholar]
- 27. Goodfellow I, Bengio Y, Courville A.. Deep Learning (Adaptive Computation and Machine Learning. MIT Press; 2016. doi: 10.1016/B978-0-12-391420-0.09987-X.
- 28. Torrey L, Shavlik J. Transfer learning. n: Olivas ES, Guerrero JDM, Sober MM, et al., eds. Handbook of Research on Machine Learning Applications and Trends: Algorithms, Methods, and Techniques. Hershey, PA: IGI Global; .2010: 242–64.
- 29. Yang HH, Moody J. Feature selection based on joint mutual information. In: Proceedings of International ICSC Symposium on Advances in Intelligent Data Analysis; 1999; Rochester, New York.
- 30. Cover TM, Thomas JA.. Elements of Information Theory. 2nd ed. Hoboken, NJ: John Wiley & Sons; 2012. [Google Scholar]
- 31. Liao KP, Sun J, Cai TA, et al. High-throughput multimodal automated phenotyping (MAP) with application to PheWAS. bioRxiv 2019; doi: 10.1101/587436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ning W, Chan S, Beam A, et al. Feature extraction for phenotyping from semantic and knowledge resources. J Biomed Inform 2019; 91: 103122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miotto R, Li L, Kidd BA, et al. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep 2016. ; 6 (1): 26094. doi: 10.1038/srep26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu J, Roy J, Stewart WF.. Prediction modeling using EHR data: challenges, strategies, and a comparison of machine learning approaches. Med Care 2010; 48: S106–13. [DOI] [PubMed] [Google Scholar]
- 35. Liu C, Wang F, Hu J, et al. Temporal phenotyping from longitudinal electronic health records. In: Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining—KDD ’15. Sydney NSW, Australia: Association for Computing Machinery; 2015: 705–714. [Google Scholar]
- 36. Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 2010; 17 (2): 124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.