Abstract
Objective
The study sought to describe the prevalence and nature of clinical expert involvement in the development, evaluation, and implementation of clinical decision support systems (CDSSs) that utilize machine learning to analyze electronic health record data to assist nurses and physicians in prognostic and treatment decision making (ie, predictive CDSSs) in the hospital.
Materials and Methods
A systematic search of PubMed, CINAHL, and IEEE Xplore and hand-searching of relevant conference proceedings were conducted to identify eligible articles. Empirical studies of predictive CDSSs using electronic health record data for nurses or physicians in the hospital setting published in the last 5 years in peer-reviewed journals or conference proceedings were eligible for synthesis. Data from eligible studies regarding clinician involvement, stage in system design, predictive CDSS intention, and target clinician were charted and summarized.
Results
Eighty studies met eligibility criteria. Clinical expert involvement was most prevalent at the beginning and late stages of system design. Most articles (95%) described developing and evaluating machine learning models, 28% of which described involving clinical experts, with nearly half functioning to verify the clinical correctness or relevance of the model (47%).
Discussion
Involvement of clinical experts in predictive CDSS design should be explicitly reported in publications and evaluated for the potential to overcome predictive CDSS adoption challenges.
Conclusions
If present, clinical expert involvement is most prevalent when predictive CDSS specifications are made or when system implementations are evaluated. However, clinical experts are less prevalent in developmental stages to verify clinical correctness, select model features, preprocess data, or serve as a gold standard.
Keywords: clinical decision support, machine learning, electronic health records, nurses, physicians, hospitals
INTRODUCTION
Machine learning, a type of artificial intelligence that involves computers initiating and executing learning from data without human intervention,1 is being increasingly applied in the health care domain for prognostication and treatment.2,3 Examples span specialties and settings from predicting risk for poor glycemic control among patients with diabetes,4 to forecasting likely interventions in the intensive care unit (ICU).5 The pace of machine learning research in health care, for any purpose, is especially increased by the ubiquity of electronic health records (EHRs), which store large volumes of patient data.
Many machine learning models that make prognosis or treatment predictions are intended to be used in clinical decision support systems (CDSSs) to assist clinicians in making informed decisions about patient care. Historically, CDSSs have used known relationships between variables in patient data to provide clinicians with evidence-based recommendations, alerts, or patient summaries to support their decision making.6,7 Alternatively, machine-learning-based CDSSs, which use relationships between patient variables and target outputs learned by the machine learning model, face challenges to clinician adoption.8–10 Shortliffe and Sepúlveda10 recently outlined 6 challenges to CDSS in “the era of artificial intelligence”: (1) complicated models lack transparency, which prohibits clinicians’ ability to understand and accept predictions or recommendations; (2) clinician time is scarce; (3) systems must be usable and easily learnable; (4) recommendations must be relevant to the clinicians in the targeted domain; (5) delivery must respect clinician expertise; and (6) recommendations must be based on rigorous science. Many of these challenges apply to CDSSs that do not use machine learning (ie, expert systems). For example, all CDSSs must be usable. However, machine learning offers unique technical capabilities that exacerbate these challenges. For example, investigators can engineer new features not present in the original dataset, creating a model likely more difficult to quickly understand as an end user than a system using Boolean logic on original data. Additionally, while all listed challenges apply to machine-learning-based CDSS for any type of clinical decision (eg, diagnosis, prognosis, treatment) in any setting (eg, outpatient, inpatient), they are especially difficult when systems are designed (1) to assist with prognosis or treatment decision making (hereafter referred to as, predictive CDSSs) and (2) for the hospital setting. This is because, unlike many diagnostic decisions, prognosis and treatment decisions often cannot be linked to a gold standard, such as a biopsy. Even expert clinicians may disagree.10 Additionally, in the hospital setting, clinicians are under increased time pressure and making decisions that impact the patient in the immediate or near terms.11,12
Addressing each of the challenges outlined previously requires interdisciplinary collaboration between experts in informatics, data science, human factors, and the clinical domain that the system aims to target. For example, to mitigate the issues of model transparency and understandability, researchers have suggested using domain knowledge to assess model complexity3 or to identify features likely to prove reasonable to an end user.13 However, as machine learning is increasingly researched (eg, PubMed articles tagged with the “machine learning” MeSH [Medical Subject Heading] Major Topic increased by nearly 10-fold from 2014 to 2019)14 and applied to clinical decision making, it is unclear the frequency with and capacity in which clinical experts are involved in predictive CDSS development, evaluation, and implementation.
The process of building and testing a predictive CDSS for use in a hospital setting can be situated within Stead et al’s15 framework describing medical informatics system design. The framework has 5 sequential stages: (1) specification, (2) component development, (3) combination of components into system, (4) integration of system into environment, and (5) routine use. The specification stage involves eliciting system needs and technical functionalities from end users. The component development stage involves development of an “isolatable subset of a system”15 with clear inputs and outputs. In predictive CDSS design, these components might include the machine learning model, the CDSS interface, the database, etc. Combination of components into a system involves integrating previously developed components. Integration of system into environment involves incorporating the system into the technical and cultural ecosystem in the intended setting. Finally, routine use is achieved when the system is a normal function of work in the environment.15 Each successive stage depends on rigorous evaluation of the previous.15,16
Though the routine use stage will inherently involve clinicians, it is possible for developers and investigators of predictive CDSSs to move through the other stages with or without engaging clinical experts, potentially missing an opportunity to mitigate 1 or more known challenges to adoption. The objective of this review is to describe the literature on predictive CDSS research for in-hospital decision making as it pertains to clinician involvement in Stead et al’s 5 stages of system design. A scoping review is best suited to this objective because the intent is both (1) to examine the range and nature of research on this topic and (2) to inform future research and development.17–19 Our ultimate goal is to inform the broader discourse on rigorous methods for overcoming challenges to predictive CDSS adoption.
MATERIALS AND METHODS
Information sources and search terms
This scoping review was conducted following the PRISMA-ScR (Preferred Reporting Items for Systematic reviews and Meta-Analyses extension for Scoping Reviews) guidelines (Supplementary Table 1).20 Three scholarly databases were searched in October 2019: PubMed, CINAHL, and IEEE Xplore; proceedings from the Machine Learning for Healthcare conference and CHI: Conference on Human Factors in Computing Systems were hand searched in May 2020. Our search strategy combined terms representing 3 elements of the topic of interest: machine learning, clinical decision support, and clinicians. Keywords were identified to represent each element as comprehensively as possible, searched using truncation and standardized subject headings when appropriate, and combined using Boolean operators (Supplementary Table 2).
Eligibility criteria
Inclusion and exclusion criteria are presented in Table 1. Publications describing predictive CDSS targeting nurses, physicians, or advanced practice providers for prognostic or treatment decision making in the hospital using EHR data were included.
Table 1.
Study eligibility criteria
| Inclusion criteria | Exclusion criteria |
|---|---|
|
|
Note: CDSS: clinical decision support system; ECG: electrocardiogram; ED: emergency department; EHR: electronic health record.
We focused on systems using EHR data because EHRs are the primary health information system clinicians use in patient care and thus, are rich with potential signals for predicting patient outcomes. EHR data also present unique complexity (eg, varying temporal granularity) and opportunity for clinical expertise. Publication year was limited to the last 5 years given the rapid evolution of machine learning science.21,22 For example, deep learning research in health care has exponentially increased,22,23 reflecting a broader evolution in technical methods and thus evolving opportunities for clinical expert involvement.
Imaging and pathology interpretation systems were excluded because they are not strictly hospital-based and represent computer-assisted diagnosis. We determined if described models were for a CDSS according to the article’s stated goal and its alignment with patient care workflows. For example, modeling research database searches were excluded.
To operationalize our criteria, we defined machine learning a priori by referencing Beam and Kohane,24 who defined machine learning as a spectrum from minimal to maximal machine involvement. Using their spectrum, we included machine learning models with more machine involvement than linear and logistic regression.1,24 Though standard regression models are frequently used for machine learning, because they are at the bottom of the machine involvement spectrum, they are less computationally complex and standardly require human involvement. Thus, clinical expert involvement in regression model design is expected and challenges to adoption (eg, model transparency) are less likely to apply. More computationally complex models offer novel opportunities for modeling complex relationships25 and for clinician involvement.
Data screening and charting
Two authors (J.M.S. and K.D.C.) screened titles and abstracts of identified articles for eligibility criteria. A calibration exercise was conducted between the 2 screening authors with approximately 50 titles or abstracts. A third author (A.J.M.) resolved any screening disagreements and any disagreements between the 3 authors were discussed until a consensus was reached. Two authors (J.M.S. and K.D.C.) reviewed the full texts of remaining articles. Reference lists of included articles were reviewed to identify additional articles meeting criteria. Covidence software (Covidence, Melbourne, Australia) was used to assist with screening.26
Data charting was completed by one author (J.M.S.) using Microsoft Excel and Word (Microsoft Corporation, Redmond, WA)27 and verified by a second author (K.D.C.). Data items extracted include study characteristics such as country of origin, clinical specialty, study design, method of machine learning model(s) described, and size of cohort used to train and test the model or number of clinicians who evaluated a system. Charted items related to the review objective include the study objective, model outcomes, target decision maker (eg, nurses or physicians), stage(s) in system design,15 and clinician involvement. Author affiliation or licensure was charted subsequently to provide perspective on the possibility that authors may have served as clinical experts themselves.
Charting of particular data items involved our interpretation. If the authors did not specify if the system was intended for nurses or providers, the term “clinician” is used. Similarly, when the profession of clinicians involved was not explicitly stated, we used the term “clinical experts.” General indicative language such as “based on expert medical knowledge and opinion”28 was charted as clinical expert involvement. Study objectives are not direct quotes, but one author’s (J.M.S.) summary. The stage in system design was not explicitly stated in any of the articles. Stead et al’s framework15 and its subsequent elaboration by Kaufman et al16 were used to identify the aligned stage. Additionally, the charted study location is the country where the majority of investigators are affiliated and does not necessarily reflect the location of the data source. We have also described the machine learning model method according to a larger class in many cases. For example, “neural networks” is used to describe studies that involved any type of neural network (eg, back-propagation or convolutional). This serves 2 purposes: (1) synthesis of model methods and (2) clarity for an audience not necessarily expert in machine learning.
RESULTS
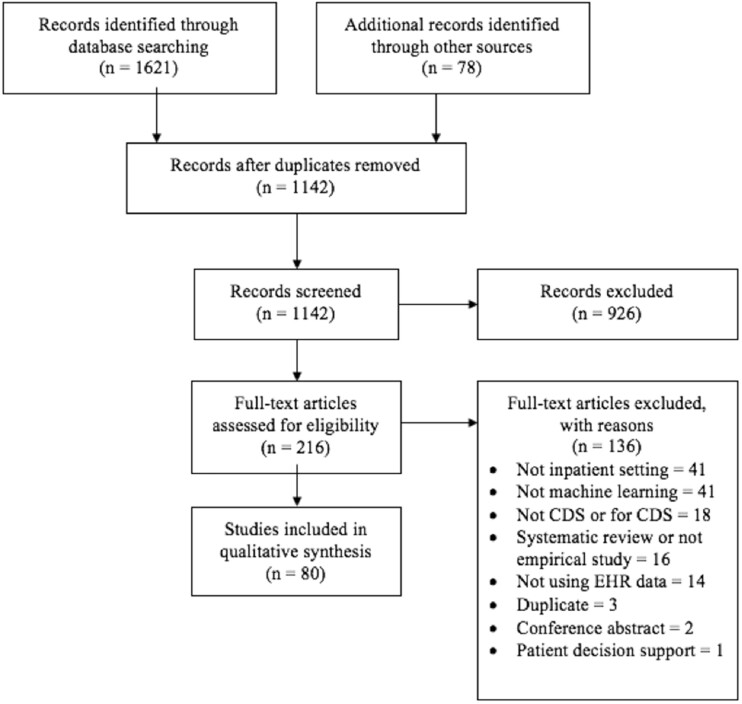
The initial database search yielded 1621 articles (Figure 1). Seventy-eight additional articles were identified from hand-searching conference proceedings and reference lists. After duplicates were removed, 1142 article titles or abstracts were screened. A total of 926 articles were excluded in title or abstract screening, leaving 216 full-text articles assessed for eligibility. A total of 136 articles were excluded after full-text screening, leaving 80 articles eligible for synthesis.
Figure 1.
PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) flow diagram of study eligibility screening. CDS: clinical decision support; EHR: electronic health record.
Summary of study characteristics
Table 2 summarizes the characteristics of the 80 included studies (further detailed in Supplementary Table 3). Studies were primarily of retrospective cohort design (n = 66), were conducted in the United States (n = 55), were published in 2019 (n = 23), were designed for intensive care (n = 37), and used neural networks (n = 34).
Table 2.
Summary of study characteristics
| Characteristic | Variable | Results |
|---|---|---|
| Study Design | Retrospective cohort | 66 (82.5)5,28–92 |
| Prospective cohort | 5 (6.3)93–97 | |
| Case-control | 3 (3.8)98–100 | |
| Qualitative | 3 (3.8)12,101,102 | |
| Retrospective + prospective cohort | 3 (3.8)103–105 | |
| Locationa | United States | 55 (68.8)5,28,29,31,32,34,36,40–44,47–50,52,54–60,62,63,65–70,73–80,82,83,85,90–93,95,97–100,102,104,105 |
| United Kingdom | 7 (8.8)39,53,55,61,63,67,77 | |
| Taiwan | 5 (6.3)38,39,45,67,88 | |
| Canada | 4 (5)12,33,81,87 | |
| China | 4 (5)37,39,71,103 | |
| Australia | 3 (3.8)86,94,96 | |
| Germany | 2 (2.5)51,101 | |
| India | 2 (2.5)64,89 | |
| Switzerland | 2 (2.5)35,72 | |
| South Korea | 1 (1.3)30 | |
| Saudi Arabia | 1 (1.3)46 | |
| Spain | 1 (1.3)84 | |
| France | 1 (1.3)76 | |
| Portugal | 1 (1.3)75 | |
| Year | 2019 | 23 (28.8)12,42,43,60,64,65,67,68,70,72,79,82,83,92,95–99,101–103,105 |
| 2018 | 18 (22.5)30,36,39,41,46,61,63,66,69,71,74,78,81,88,89,91,94,104 | |
| 2017 | 15 (18.8)5,29,31,44,51–53,57,58,75,77,80,85,87,100 | |
| 2016 | 12 (15)28,32,37,40,47,49,50,62,73,84,86,93 | |
| 2015 | 12 (15)33–35,38,45,48,54–56,59,76,90 | |
| Machine learning methodsa | Neural networks | 34 (42.5)28–33,35,39,41,43,45,47–51,54,60,66,68,70,72,76,79–84,89,94,96,99,100 |
| Random forests | 32 (40)30,35–38,40,42–44,49–51,54,57,63–66,71,76,79,83,88,90,91,94,95,97,98,103–105 | |
| Regressionb | 27 (33.8)30,32,35,39–41,43,47,48,50,54,56,61,62,65,70,76,79,83,84,87,88,90–92,94,95 | |
| Support vector machines | 24 (30)32,34,35,37–39,44,46–48,50,51,55–57,59,61,62,66,75,79,94,98,103 | |
| Boosting | 17 (21.3)32,47,49,50,52,53,65–67,69,76,78,90,91,94,98,103 | |
| Decision trees | 15 (18.8)32,35,38–40,47,48,50,52,53,56,62,76,78,88 | |
| Penalized regressionc | 13 (16.3)5,35,37,44,55,57,65,66,91,95,98,100,103 | |
| Bayesian | 13 (16.3)5,35,37,39,44,57,62,66,76,79,86,93,98 | |
| Topic modeling | 9 (11.3)55,57,59,66,79,80,85,91,99 | |
| Ensemble | 7 (8.8)52,53,62,76,78,81,95 | |
| Nearest neighbors | 6 (7.5)35,50,54,72,98,103 | |
| Gaussian process | 5 (6.3)5,29,31,55,98 | |
| Clustering | 5 (6.3)51,61,87,89,92 | |
| Reinforcement learning | 4 (5)34,63,73,77 | |
| Generalized additive models | 3 (3.8)76,90,95 | |
| Bagging | 3 (3.8)35,50,76 | |
| Discriminant analysis | 2 (2.5)35,61 | |
| Word vectorization | 2 (2.5)64,89 | |
| Other methodsd | 10 (12.5)32,35,37,44,54,58,72,74–76 | |
| Sample size | Qualitative | 10-19 clinicians |
| Model development | 127 patients to 296,724 hospital admissions | |
| Clinical specialtya | Intensive care (adult) | 37 (46.3)5,12,28,30,34,43,44,51–53,55,56,58–61,63,64,66–69,71–77,79,80,83,86,87,89,91,101 |
| In-hospital, acute care/not further specified | 14 (17.5)30,31,34,46,50,54,69,82,85,90,97,103–105 | |
| Emergency medicine | 14 (17.5 )12,35,36,40,42,57,69,84,86,88,90,93,95,99 | |
| Cardiology | 7 (8.8)37,71,81,86,88,92,102 | |
| Pediatric (acute and intensive care) | 6 (7.5)32,58,70,78,81,100 | |
| Nephrology | 5 (6.3)62,66,75,79,83 | |
| Neonatal intensive care | 4 (5)33,94,96,98 | |
| Stroke | 4 (5)47–49,86 | |
| Surgery | 3 (3.8)38,41,86 | |
| Diabetes | 2 (2.5)39,65 | |
| Pulmonology/respiratory | 2 (2.5)42,90 | |
| Nursing | 1 (1.3)45 | |
| Trauma | 1 (1.3)95 | |
| In-hospital, specifically: emergency medicine, intensive care, cardiothoracic surgery/transplant, neurology/vascular/stroke, gastrointestinal, oncology/hematology/immunology/pharmacology | 1 (1.3)86 |
Values are n (%) or range.
Studies may fall into more than 1 category.
Does not meet our definition of machine learning but included for purposes of reporting all methods authors compared.
Including LASSO, Ridge, and Elastic Net.
Generalized linear models, conditional inference recursive partition, conditional random fields, Weibull-Cox proportional hazards, lazy learner, piecewise-constant conditional intensity model, analysis of covariance (see footnote b), analysis of variance (see footnote b), fuzzy modeling (see footnote b), switching-state autoregressive model, mimic learning, nearest shrunken centroids, J48 algorithm, PART rule.
Predictive CDSSs and clinician involvement
Clinician involvement according to system design stage is summarized in Table 3 and further detailed along with decision-support objectives, machine learning model outcomes, target clinicians, and author affiliation in Supplementary Table 4. Most study authors described decision support for clinicians nonspecifically (n = 50). Fewer targeted physicians only (n = 18), nurses only (n = 5), or either nurses or physicians or providers (n = 4), with 1 including respiratory therapists.101 Three studies specifically identified hospital interprofessional rapid response teams as target end users.29–31
Table 3.
Summary of findings of clinician involvement by stage of system design
| Clinician Involvement Category | Clinician Involvement Details | Results |
|---|---|---|
| Specification | 3 (4) | |
| Identified system needs and design | Clinicians interviewed regarding: | 3 (100) |
| Component development | 76 (95) | |
| Clinical relevance/correctness |
|
10 (13) |
| Feature selection | 8 (11) | |
| Data preprocessing | 4 (5) | |
| Gold standard | 4 (5) | |
| Combination of components into system | 5 (6) | |
| Integration of system into environment | 5 (6) | |
| Evaluation | 2 (40) | |
| Routine use | 0 (0) | |
Values are n (%). Some studies described more than 1 stage of design and/or method of clinician involvement. Stages based on framework outlined by Stead et al.15
ICU: intensive care unit.
Studies investigated predicting a variety of target outcomes (Supplementary Table 4). The most common were mortality (both after discharge and in-hospital; n = 31), sepsis and septic shock (n = 16), transfer to the ICU (n = 6), ICU or hospital readmission (n = 6), and length of stay (n = 5).
Specification
Three studies investigated the specification stage of predictive CDSS, each seeking to illuminate the needs of clinicians using predictive CDSS at varying levels of specificity—all of which involved clinical experts. ICU clinicians were interviewed to specify their predictive CDSS needs for patient monitoring.101 Clinicians in the emergency department and ICU were interviewed to understand when and how explanations are needed to understand predictive CDSSs.12 Finally, clinicians in cardiology co-designed a predictive CDSS for ventricular assist device decision making and advised on fit with clinical workflow.102
Component development
Of the 76 studies describing component development, 21 (28%) involved clinical experts in this stage. All component development studies described developing the machine learning model. Clinical expert involvement in component development can be organized into 5 categories: clinical relevance or correctness,28,31–38,93 feature selection,35,38–41,93,94,98 data preprocessing,5,42–44 gold standard,35,37,45,99 and no clinician involvement described.29, 30, 46-92, 95, 96, 100, 103-105,25–28,30–34,36,50–88,93,96,100,103–105
Clinical relevance or correctness
Of the 21 studies involving clinical experts in the component development stage, nearly half (n = 10) involved clinical experts advising on the clinical relevance or correctness of the model(s). Clinicians set performance requirements such as sensitivity, specificity, and false positive rates,33,93 and established alert trigger thresholds.93 Alternatively, clinicians advised on model outcomes – defining outcomes31,34 or identifying their presence in the data.34–36 Clinicians also helped to derive meaning from features.28,32,36,37 For example, Jalali et al28 consulted clinical experts to group features by body organ to improve their mortality model and its intuitiveness in the ICU. More broadly, clinical experts advised on implications for clinical practice34,36 and research design.38
Feature selection
Eight studies included clinical experts in determining which features should be used as inputs in the machine learning model—something that can be done according to domain knowledge or purely using computational methods. Clinical experts functioned to both narrow down the set of all possible features available from their datasets38–40,94 and to identify candidate features from which to start working.35,41,93,98 Half of these studies additionally used computational feature selection methods.39,40,93,98
Data preprocessing
Four studies involved clinical experts in data preprocessing. Clinical experts advised on the periodicity of interventions in practice, from which investigators established intervention gap thresholds for interpolation in their forecasting model.5 In 2 studies, clinicians identified invalid or outlier values in feature distributions.42,43 Finally, clinical experts advised on correctly constructing features from ICU data.44
Gold standard
In 4 studies, clinicians established the gold standard against which model performance was judged; models either learned from clinician judgement data or were assessed based on clinician-determined ground truth. In one example, physicians documented impressions of patient severity of illness.35 Clinicians also annotated progress notes for the presence of outcomes.37,99 In Liao et al45 experienced nurses compared documented nursing diagnoses to an established ontology used in model development.
No clinician involvement described
Fifty-five (72%) of the studies on component development did not mention any involvement from clinical experts.25–28,30–34,36,50–88,100,103–105,9396 However, 4 of these studies discuss this as a future improvement.56,65,74,82 For example, J.Y. Kwon et al.65 positioned their paper as an argument for using nursing knowledge to improve predictive CDSSs, describing how nurse experts may have improved the performance and clinical relevance of their model.65
Additionally, in multiple articles, authors did not explicitly describe clinician involvement but indirectly imply using clinical expertise. For example, Kaji et al60 mentioned identifying model features based on “known clinical relevance to [their] target end points.” Each author was affiliated with a medical school.
Combination of components into system
All 5 studies that reported on combination of components into system also reported on component development. These studies either described or presented a predictive CDSS prototype or interface; none of them described involving clinical experts in this stage.29,33,34,84,93
Integration into environment
Five studies described integrating the system into its environment,45,93,97,104,105 with 4 also describing component development.45,93,104,105 Two studies involved clinical experts in evaluating the predictive CDSS after integration.45,97 In both studies, clinicians responded to surveys evaluating their impressions of the predictive CDSS and clinicians helped develop the surveys.45,97 For example, in Ginestra et al,97 attending physicians, residents, and nurses helped develop an evaluation survey that was completed by 252 clinicians.
Author affiliation or licensure
It is possible that clinically trained authors made clinically relevant decisions in their predictive CDSS design process but did not describe it in their article. Of the 55 articles that did not mention any clinician involvement, 39 (71%) were authored by at least 1 person with a health-related affiliation (eg, MD in title, located at school of nursing),29,30,46,50–54,56–58,60,62,63,65–71,74,76,78,79,81,82,84–86,90–92,95,96,100,103–105 14 (25%) were not authored by investigators with health-related affiliations,47–49,55,61,64,72,73,75,77,80,87–89 and 2 studies did not report affiliation.59,83
DISCUSSION
The results of this scoping review indicate that clinical expert involvement is most prevalent in the specification and integration into environment stages of predictive CDSS design for nurses and providers in the hospital. Clinical expert involvement was less prevalent in the intermediary stages of predictive CDSS design (Table 3, Figure 2). This is not entirely surprising, especially for articles describing machine learning model development—as clinician involvement in machine learning is likely not considered customary; clinicians are not standardly trained in machine learning. However, recent literature on improving the understandability of machine learning models describe involving domain experts in development, particularly for verifying and increasing clinical relevance or correctness and for feature selection.13,65,106,107 However, with only 21% (n = 16) of studies on component development involving clinical experts for verifying clinical relevance or correctness or for feature selection, our findings indicate that this is not widespread practice. We note that using clinical expertise for feature selection is not necessarily state of the art. However, recent literature illustrates creative ways that expert knowledge can be integrated with computational feature selection.106,108 For example, Boulet et al106 used a power prior to incorporate numerical clinical relevance weights assigned by clinical experts with stochastic search variable selection.
Figure 2.
Prevalence of clinician involvement per stage of system design.
Notably, 17 studies described in this review used the publicly available MIMIC (Medical Information Mart for Intensive Care) datasets (Supplementary Table 3),109,110 which has implications for clinical expert involvement, as the experts may be advising on EHR data derived outside of the institution and workflows with which they are most familiar.111 Alternatively, clinical experts may serve to advise on public data generalizability.
By including studies across the stages of system design, our findings highlight where in the process predictive CDSS research is published. It is clear that evidence on implementations of predictive CDSS for nurses and providers in the hospital are lacking, as 90% of included studies are of, at most, the component development stage and no studies are of the routine use stage. This dearth may be a reflection of known adoption challenges,10 the extensive resource and time investment required to implement predictive CDSS, or lack of evaluation research being conducted or published. The low prevalence of studies from the specification stage may indicate that most investigators evaluate the need for a predictive CDSS through literature review, rather than qualitative research, which is a valid approach.15,16 However, if investigators are conducting research on component development in the absence of rigorous specification evaluations, they may struggle overcoming the known challenge of ensuring relevance to the clinical domain.10 The 3 studies of the specification stage were published in the most recent year (2019), which may indicate that investigators are considering adoption challenges by working to thoroughly understanding the needs and desires of clinician end users from the outset, as has recently been recommended.112
Beyond universal CDSS adoption challenges outlined in foundational literature (eg, importance of human-computer interface)113 and those described by Shortliffe and Sepúlveda,10 Lenert et al114 recently detailed another specific challenge that predictive CDSSs may face after implementation—model degradation if behavior change occurs. The authors suggest modeling the intervention space (eg, modeling antibiotic administration for sepsis CDSS) in development so that likely changes to the outcome distribution are learned. This is a unique challenge to predictive CDSS and certainly a potential area for clinician involvement not described in our review findings.
Predictive CDSS adoption also has implications for clinician reasoning. Clinicians must consider the merit and consequence of a data-driven prediction though they are accustomed to looking to EHR data for documented observations.115,116 While we have reviewed one approach to easing adoption—involving clinicians in system research and development—new training programs are likely needed to equip clinicians with the skills needed to understand the strengths and limitations of predictive CDSS.111
Work done outside the hospital setting demonstrates the potential promise of clinical expert involvement.106,108,117 Simon et al117 described and advocated for engaging clinical experts across the stages of designing and implementing a predictive CDSS for oncology treatment, attributing the success and veracity of their predictive CDSS to this collaboration. Others have described clinician involvement in the development of a machine learning–based diagnostic decision support system, which clinicians co-designed by thinking aloud as they interacted with a model explanation interface.118 Such efforts have the potential to overcome known challenges namely, increasing usability, clinical relevance, understandability, and delivering CDSS in a respectful manner.
Future directions
The structure and timing of this scoping review is optimal for informing future predictive CDSS research and development. First, the multitude of strategies for involving clinical experts in predictive CDSS research found in this review should be empirically evaluated. One approach may be to measure clinician end-user adoption of or trust in implemented systems. Alternatively, researchers may compare using clinical expertise in any of the ways described (eg, feature selection, verifying clinical relevance or correctness) with taking a purely computational approach or relying on nonclinical developers. Outcomes may be simple comparisons of model accuracy or more long-term evaluations of later stages of design. Aptly, informaticians are calling for a shift in focus away from the mechanics of predictive modeling toward the sustained benefit of predictive systems in practice.21 As such, clinician involvement in predictive CDSS research should be evaluated according to the value added to patient care and clinician workflows.
Second, the lack of studies reporting on implemented systems indicates these findings may inform future implementations and that this is an area of needed research. Those planning a predictive CDSS implementation should review and consider the merit of using the methods for clinician involvement described in this review. We also suggest investigations of evidence-based implementation strategies, such as Expert Recommendations for Implementing Change119 or evaluating implementation success according to a validated framework such as RE-AIM.120
Third, we advise that future publications of predictive CDSS research standardly report clinician involvement explicitly. Interpretation of our findings should consider that this is not yet standard practice and authors may have omitted detail on clinician involvement. However, because machine learning is burgeoning in the healthcare domain, there is an opportunity to institute standard reporting guidelines, including description of clinical expert involvement. There is a recent call to expand the TRIPOD (Transparent Reporting of Multivariable Prediction Model for Individual Prognosis or Diagnosis) checklist (criteria for reporting findings from development or validation of medical prediction models) to better suit machine learning, as the original was created primarily for regression models.121 The panel convened to expand the TRIPOD checklist may use these findings to consider the relevance of incorporating guidelines on reporting clinician involvement.
Finally, these findings show that predictive CDSS for nurses and for clinical specialties outside of intensive care are underrepresented. Many of the included studies modeled outcomes that nursing care certainly impacts (eg, sepsis, in-hospital mortality) but do not all name nurses as target end users. Additionally, 46% of reviewed systems were developed for adult ICUs, pointing to opportunities for research in less represented (eg, pediatrics) and unrepresented (eg, orthopedics) specialties. We suggest a follow-up review in a few years to illuminate the progression of research on predictive CDSS with regard to methods, implementation, and variety of target users and specialties.
LIMITATIONS
In consideration of feasibility, relevant journals were not hand-searched and additional databases were not queried, potentially limiting the number of eligible studies located. We attempted to mitigate this by strategically selecting popular databases and including conference proceedings. Additionally, our analysis was restricted to the scope of Stead et al’s15 framework of system design; thus, we could not comment on clinician involvement in work outside of the stages, for example, curation of datasets. Finally, our review search did identify CDSSs that are further along in stage of development but were excluded because they exclusively used logistic regression.122,123 While not covered here, lessons from their implementations should be considered in the discourse of mitigating challenges to predictive CDSS adoption.
CONCLUSION
This scoping review found clinical expert involvement in predictive CDSS research for the hospital most prevalent at the specification and integration into environment stages. However, most published research is of the component development stage, where clinician involvement is less prevalent but has been proposed as a method for mitigating challenges to adoption. Further empirical research is needed to understand the impact of involving clinical experts throughout the predictive CDSS design process.
FUNDING
This work was supported by National Institute for Nursing Research grant 5T32NR007969 and National Library of Medicine grant 5T15LM007079.
AUTHOR CONTRIBUTIONS
JMS and KDC conceptualized the review and NE and SCR advised on the scope. JMS, KDC, and AJM conducted the title/abstract screening. JMS and KDC conducted full text screening. JMS conducted data extraction. KDC verified extracted data. JMS drafted the manuscript with revisions and feedback from AJM, SCR, NE, and KDC.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online
DATA AVAILABILITY
There are no new data associated with this article.
CONFLICT OF INTEREST STATEMENT
The authors declare no competing interests with respect to this publication.
Supplementary Material
REFERENCES
- 1.U.S. National Library of Medicine. Machine learning - MeSH. https://www.ncbi.nlm.nih.gov/mesh/2010029 Accessed December 9, 2019.
- 2.Topol EJ.High-performance medicine: the convergence of human and artificial intelligence. Nat Med 2019; 25 (1): 44–56. [DOI] [PubMed] [Google Scholar]
- 3.Rajkomar A, Dean J, Kohane I.. Machine learning in medicine. N Engl J Med 2019; 380 (14): 1347–58. doi: 10.1056/NEJMra1814259 [DOI] [PubMed] [Google Scholar]
- 4.Romero-Brufau S, Wyatt KD, Boyum P, et al. A lesson in implementation: a pre-post study of providers’ experience with artificial intelligence-based clinical decision support. Int J Med Inform 2020; 137: 104072. [DOI] [PubMed] [Google Scholar]
- 5.Ghassemi M, Wu M, Hughes MC, et al. Predicting intervention onset in the ICU with switching state space models. AMIA Jt Summits Transl Sci Proc 2017; 2017: 82–91. [PMC free article] [PubMed] [Google Scholar]
- 6.Middleton B, Sittig DF, Wright A.. Clinical decision support: a 25 year retrospective and a 25 year vision. Yearb Med Inform 2016; 25 (Suppl 1): S103–S116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.U.S. National Library of Medicine. Decision support systems, clinical - MeSH. https://www.ncbi.nlm.nih.gov/mesh/? term=clinical+decision+support+systems Accessed December 9, 2019.
- 8.Stead WW.Clinical implications and challenges of artificial intelligence and deep learning. JAMA 2018; 320 (11): 1107–8. [DOI] [PubMed] [Google Scholar]
- 9.Mehta N, Devarakonda MV.. Machine learning, natural language programming, and electronic health records: The next step in the artificial intelligence journey? J Allergy Clin Immunol 2018; 141 (6): 2019–40. [DOI] [PubMed] [Google Scholar]
- 10.Shortliffe EH, Sepúlveda MJ.. Clinical decision support in the era of artificial intelligence. JAMA 2018; 320 (21): 2199–200. [DOI] [PubMed] [Google Scholar]
- 11.Lynn LA.Artificial intelligence systems for complex decision-making in acute care medicine: a review. Patient Saf Surg 2019; 13 (1): 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tonekaboni S, Joshi S, McCradden MD, Goldenberg A.. et al. What clinicians want: contextualizing explainable machine learning for clinical end use. Proc Mach Learn Res 2019; 106: 359–80. [Google Scholar]
- 13.Murdoch WJ, Singh C, Kumbier K, et al. Definitions, methods, and applications in interpretable machine learning. Proc Natl Acad Sci U S A 2019; 116 (44): 22071–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.U.S. National Library of Medicine. “Machine learning” [MeSH major topic]. https://pubmed.ncbi.nlm.nih.gov/? term=%22machine+l Accessed September 23, 2020.
- 15.Stead WW, Haynes BR, Fuller S, et al. Designing medical informatics research and library - resource projects to increase what is learned. J Am Med Inform Assoc 1994; 1 (1): 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaufman D, Roberts D, Merrill J, Lai T-Y, et al. Applying an evaluation framework for health information system design, development, and implementation. Nurs Res 2006; 55 (2 Suppl): S37–S42. [DOI] [PubMed] [Google Scholar]
- 17.Arksey H, O'Malley L.. Scoping studies: towards a methodological framework. Int J Soc Res Methodol Theory Pract 2005; 8 (1): 19–32. [Google Scholar]
- 18.Tricco AC, Lillie E, Zarin W, et al. A scoping review on the conduct and reporting of scoping reviews. BMC Med Res Methodol 2016; 16 (1): 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cochrane Training. Scoping reviews: what they are and how you can do them. https://training.cochrane.org/resource/scoping-reviews-what-they-are-and-how-you-can-do-them Accessed September 21, 2020.
- 20.Tricco AC, Lillie E, Zarin W, et al. PRISMA extension for scoping reviews (PRISMA-ScR): Checklist and explanation. Ann Intern Med 2018; 169 (7): 467–73. [DOI] [PubMed] [Google Scholar]
- 21.Lenert L.The science of informatics and predictive analytics. J Am Med Inform Assoc 2019; 26 (12): 1425–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kocheturov A, Pardalos PM, Karakitsiou A.. Massive datasets and machine learning for computational biomedicine: trends and challenges. Ann Oper Res 2019; 276 (1–2): 5–34. [Google Scholar]
- 23.U.S. National Library of Medicine. Deep neural network [MeSH]. https://pubmed.ncbi.nlm.nih.gov/?term=deep+neural+network+%5BMeSH%5D&timeline=expanded&sort=date Accessed September 23, 2020.
- 24.Beam AL, Kohane IS.. Big data and machine learning in health care. JAMA 2018; 319 (13): 1317–8. [DOI] [PubMed] [Google Scholar]
- 25.Orfanoudaki A, Chesley E, Cadisch C, et al. Machine learning provides evidence that stroke risk is not linear: The non-linear Framingham stroke risk score. PLoS One 2020; 15 (5): e0232414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veritas Health Innovation. Covidence systematic review software. 2019. https://www.covidence.org Accessed September 23, 2020.
- 27.Microsoft Office. 2019. https://products.office.com/en-us/home Accessed December 20, 2019.
- 28.Jalali A, Bender D, Rehman M, et al. Advanced analytics for outcome prediction in intensive care units. Annu Int Conf IEEE Eng Med Biol Sci 2016; 2016: 2520–4. Orlando, FL. doi : 10.1109/EMBC.2016.7591243. [DOI] [PubMed] [Google Scholar]
- 29.Futoma J, Hariharan S, Sendak M, et al. An improved multi-output gaussian process RNN with real-time validation for early sepsis detection. arXiv: 1708.05894; 2017.
- 30.Kwon JM, Lee Y, Lee Y, et al. An algorithm based on deep learning for predicting in-hospital cardiac arrest. J Am Heart Assoc 2018; 7 (13): e0086789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Futoma J, Hariharan S, Heller K. Learning to detect sepsis with a multitask Gaussian process RNN classifier. In: ICML ’17: Proceedings of the 34th International Conference on Machine Learning – Volume 70; 2017; 1174–82; Sydney, Australia.
- 32.Che Z, Purushotham S, Khemani R, et al. Interpretable deep models for ICU outcome prediction. AMIA Annu Symp Proc. 2017; 2016: 371–80. [PMC free article] [PubMed] [Google Scholar]
- 33.Frize M, Gilchrist J, Martirosyan H, et al. Integration of outcome estimations with a clinical decision support system: application in the neonatal intensive care unit (NICU). In: 2015 IEEE International Symposium on Medical Measurements and Applications (MeMeA) Proceedings; 2015: 175–9. doi : 10.1109/MeMeA.2015.7145194
- 34.Tsoukalas A, Albertson T, Tagkopoulos I.. From data to optimal decision making: a data-driven, probabilistic machine learning approach to decision support for patients with sepsis. JMIR Med Inform 2015; 3 (1): e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenny MA, Hertwig R, Ackermann S, et al. Are mortality and acute morbidity in patients presenting with nonspecific complaints predictable using routine variables? Acad Emerg Med 2015; 22 (10): 1155–63. [DOI] [PubMed] [Google Scholar]
- 36.Levin S, Toerper M, Hamrock E, et al. Machine-learning-based electronic triage more accurately differentiates patients with respect to clinical outcomes compared with the emergency severity index. Ann Emerg Med 2018; 71 (5): 565–74.e2. [DOI] [PubMed] [Google Scholar]
- 37.Hu D, Huang Z, Chan T-M, et al. Utilizing Chinese admission records for MACE prediction of acute coronary syndrome. Int J Environ Res Public Health 2016; 13 (9): 912. doi : 10.3390/ijerph13090912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chuang M, Hu Y, Tsai C, et al. The identification of prolonged length of stay for surgery patients. In: proceedings of the 2015 IEEE International Conference on Systems, Man, and Cybernetics; 2015: 3000–3. doi : 10.1109/SMC.2015.522
- 39.Cui S, Wang D, Wang Y, et al. An improved support vector machine-based diabetic readmission prediction. Comput Methods Programs Biomed 2018; 166: 123–35. [DOI] [PubMed] [Google Scholar]
- 40.Taylor RA, Pare JR, Venkatesh AK, et al. Prediction of in-hospital mortality in emergency department patients with sepsis: a local big data-driven, machine learning approach. Acad Emerg Med 2016; 23 (3): 269–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee CK, Hofer I, Eilon G, et al. Development and validation of a deep neural network model for prediction of postoperative in-hospital mortality. Anesthesiology 2018; 129 (4): 649–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo G, Stone BL, Nkoy FL, et al. Predicting appropriate hospital admission of emergency department patients with bronchiolitis: secondary analysis. JMIR Med Inform 2019; 7 (1): e12591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shickel B, Loftus TJ, Adhikari L, et al. DeepSOFA: a continuous acuity score for critically ill patients using clinically interpretable deep learning. Sci Rep 2019; 9 (1): 1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu M, Ghassemi M, Feng M, et al. Understanding vasopressor intervention and weaning: Risk prediction in a public heterogeneous clinical time series database. J Am Med Inform Assoc 2017; 24 (3): 488–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao P-H, Hsu P-T, Chu W, et al. Applying artificial intelligence technology to support decision-making in nursing: a case study in Taiwan. Health Informatics J 2015; 21 (2): 137–48. [DOI] [PubMed] [Google Scholar]
- 46.Alhassan Z, Budgen D, Alshammari R, et al. Stacked denoising autoencoders for mortality risk prediction using imbalanced clinical data. In: proceedings of the 2018 17th IEEE International Conference on Machine Learning and Applications (ICMLA); 2018: 541–6. doi : 10.1109/ICMLA.2018.00087
- 47.Alotaibi NN, Sasi S. Stroke in-patients’ transfer to the ICU using ensemble based model. In: proceedings of the 2016 International Conference on Electrical, Electronics, and Optimization Techniques (ICEEOT); 2016: 2004–10. doi : 10.1109/ICEEOT.2016.7755040
- 48.Alotaibi NN, Sasi S. Predictive model for transferring stroke in-patients to intensive care unit. In: proceedings of the 2015 International Conference on Computing and Network Communications (CoCoNet); 2015: 848–53. doi : 10.1109/CoCoNet.2015.7411288
- 49.Alotaibi NN, Sasi S. Tree-based ensemble models for predicting the ICU transfer of stroke in-patients. In: proceedings of the 2016 International Conference on Data Science and Engineering (ICDSE); 2016: 1–6. doi : 10.1109/ICDSE.2016.7823951
- 50.Churpek MM, Yuen TC, Winslow C, et al. M;ulticenter comparison of machine learning methods and conventional regression for predicting clinical deterioration on the wards. Crit Care Med 2016; 44 (2): 368–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dervishi A.Fuzzy risk stratification and risk assessment model for clinical monitoring in the ICU. Comput Biol Med 2017; 87: 169–78. [DOI] [PubMed] [Google Scholar]
- 52.Desautels T, Calvert J, Hoffman J, et al. Using transfer learning for improved mortality prediction in a data-scarce hospital setting. Biomed Inform Insights 2017; 9: 11782226177122994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desautels T, Das R, Calvert J, et al. Prediction of early unplanned intensive care unit readmission in a UK tertiary care hospital: A cross-sectional machine learning approach. BMJ Open 2017; 7 (9): e017199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Escobar GJ, Ragins A, Scheirer P, et al. Nonelective rehospitalizations and postdischarge mortality predictive models suitable for use in real time. Med Care 2015; 53 (11): 916–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghassemi M, Pimentel MAF, Brennan T, et al. A multivariate timeseries modeling approach to severity of illness assessment and forecasting in ICU with sparse, heterogeneous clinical data. Proc Natl Conf Artif Intell 2015; 1: 446–53. [PMC free article] [PubMed] [Google Scholar]
- 56.Guillén J, Liu J, Furr M, et al. Predictive models for severe sepsis in adult ICU patients. In: proceedings of the 2015 Systems and Information Engineering Design Symposium; 2015: 182–7. doi : 10.1109/SIEDS.2015.7116970
- 57.Horng S, Sontag DA, Halpern Y, et al. Creating an automated trigger for sepsis clinical decision support at emergency department triage using machine learning. PLoS One 2017; 12 (4): e0174708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Islam KT, Shelton CR, Casse JI, et al. Marked point process for severity of illness assessment. Proc Mach Learn Res 2017; 68: 255–70. [Google Scholar]
- 59.Jo Y, Loghmanpour N, Rosé CP. Time series analysis of nursing notes for mortality prediction via a state transition topic model. In: CIKM ’15: Proceedings of the International Conference on Information and Knowledge Management; 2015: 1171–80. doi : 10.1145/2806416.2806541
- 60.Kaji DA, Zech JR, Kim JS, et al. An attention based deep learning model of clinical events in the intensive care unit. PLoS One 2019; 14 (2): e0211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karunarathna KMDM. Predicting ICU death with summarized patient data. In: proceedings of the 2018 IEEE 8th Annual Computing and Communication Workshop and Conference (CCWC); 2018: 238–47. doi : 10.1109/CCWC.2018.8301645
- 62.Kate RJ, Perez RM, Mazumdar D, et al. Prediction and detection models for acute kidney injury in hospitalized older adults. BMC Med Inform Decis Mak 2016; 16 (1): 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Komorowski M, Celi LA, Badawi O, et al. The artificial intelligence clinician learns optimal treatment strategies for sepsis in intensive care. Nat Med 2018; 24 (11): 1716–20. [DOI] [PubMed] [Google Scholar]
- 64.Krishnan GS, Sowmya Kamath S. Evaluating the quality of word representation models for unstructured clinical text based ICU mortality prediction. In: ICDCN ’19: Proceedings of the 20th International Conference on Distributed Computing and Networking; 2019: 480–5.
- 65.Kwon JY, Karim ME, Topaz M, et al. Nurses “seeing forest for the trees” in the age of machine learning: using nursing knowledge to improve relevance and performance. Comput Inform Nurs 2019; 37 (4): 203–12. [DOI] [PubMed] [Google Scholar]
- 66.Li Y, Yao L, Mao C, et al. Early prediction of acute kidney injury in critical care setting using clinical notes. In: proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); 2018: 683–6. doi : 10.1109/BIBM.2018.8621574 [DOI] [PMC free article] [PubMed]
- 67.Lin PC, Huang HC, Komorowski M, et al. A machine learning approach for predicting urine output after fluid administration. Comput Methods Programs Biomed 2019; 177: 155–9. [DOI] [PubMed] [Google Scholar]
- 68.Lin Y-W, Zhou Y, Faghri F, et al. Analysis and prediction of unplanned intensive care unit readmission using recurrent neural networks with long short-term memory. PLoS One 2019; 14 (7): e0218942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mao Q, Jay M, Hoffman JL, et al. Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU. BMJ Open 2018; 8 (1): e017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Messinger AI, Bui N, Wagner BD, et al. Novel pediatric‐automated respiratory score using physiologic data and machine learning in asthma. Pediatr Pulmonol 2019; 54 (8): 1149–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miao F, Cai Y-P, Zhang Y-X, et al. Predictive modeling of hospital mortality for patients with heart failure by using an improved random survival forest. IEEE Access 2018; 6: 7244–53. [Google Scholar]
- 72.Moor M, Horn M, Rieck B, et al. Early recognition of sepsis with Gaussian process temporal convolutional networks and dynamic time warping. arXiv: 1902.01659; 2019.
- 73.Nemati S, Ghassemi MM, Clifford GD. Optimal medication dosing from suboptimal clinical examples: A deep reinforcement learning approach. In: proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); 2016: 2978–81. doi : 10.1109/EMBC.2016.7591355 [DOI] [PubMed]
- 74.Nemati S, Holder A, Razmi F, et al. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit Care Med 2018; 46 (4): 547–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pacheco R, Salgado CM, Deliberato R, et al. Short-term prediction of low kidney function in ICU patients. In: proceedings of the 2017 IEEE International Conference on Fuzzy Systems (FUZZ-IEEE); 2017: 1–5.
- 76.Pirracchio R, Petersen ML, Carone M, et al. Mortality prediction in intensive care units with the Super ICU Learner Algorithm (SICULA): A population-based study. Lancet Respir Med 2015; 3 (1): 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Raghu A, Komorowski M, Celi LA, et al. Continuous state-space models for optimal sepsis treatment - a deep reinforcement learning approach. arXiv: 1705.08422; 2017.
- 78.Rubin J, Potes C, Xu-Wilson M, et al. An ensemble boosting model for predicting transfer to the pediatric intensive care unit. Int J Med Inform 2018; 112: 15–20. [DOI] [PubMed] [Google Scholar]
- 79.Sun M, Baron J, Dighe A, et al. Early prediction of acute kidney injury in critical care setting using clinical notes and structured multivariate physiological measurements. Stud Health Technol Inform 2019; 264: 368–72. [DOI] [PubMed] [Google Scholar]
- 80.Suresh H, Hunt N, Johnson A, et al. Clinical intervention prediction and understanding using deep networks. Proc Mach Learn Res 2017; 68: 322–37. [Google Scholar]
- 81.Tonekaboni S, Mazwi M, Laussen P, et al. Prediction of cardiac arrest from physiological signals in the pediatric ICU. Proc Mach Learn Res 2018; 85: 534–50. [Google Scholar]
- 82.Wang JX, Sullivan DK, Wells AJ, et al. Neural networks for clinical order decision support. AMIA Jt Summits Transl Sci Proc 2019; 2019: 315–24. [PMC free article] [PubMed] [Google Scholar]
- 83.Zimmerman LP, Reyfman PA, Smith ADR, et al. Early prediction of acute kidney injury following ICU admission using a multivariate panel of physiological measurements. BMC Med Inform Decis Mak 2019; 19 (S1): 16. doi : 10.1186/s12911-019-0733-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zlotnik A, Alfaro MC, Pérez MCP, et al. Building a decision support system for inpatient admission prediction with the manchester triage system and administrative check-in variables. Comput Inform Nurs 2016; 34 (5): 224–30. [DOI] [PubMed] [Google Scholar]
- 85.Chen JH, Goldstein MK, Asch SM, et al. Predicting inpatient clinical order patterns with probabilistic topic models vs conventional order sets. J Am Med Inform Assoc 2017; 24 (3): 472–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cai X, Perez-Concha O, Coiera E, et al. Real-time prediction of mortality, readmission, and length of stay using electronic health record data. J Am Med Inform Assoc 2016; 23 (3): 553–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rouzbahman M, Jovicic A, Chignell M.. Can cluster-boosted regression improve prediction of death and length of stay in the ICU? IEEE J Biomed Health Inform 2017; 21 (3): 851–8. [DOI] [PubMed] [Google Scholar]
- 88.Chang H, Wu C, Liu J, et al. Using machine learning algorithms in medication for cardiac arrest early warning system construction and forecasting. In: proceedings of the 2018 Conference on Technologies and Applications of Artificial Intelligence (TAAI); 2018: 1–4. doi: 10.1109/TAAI.2018.00010
- 89.Krishnan GS, Kamath SS. A Supervised learning approach for ICU mortality prediction based on unstructured electrocardiogram text reports. In: Silberztein M, Atigui F, Kornyshova E, Métais E, Meziane F, eds. NLDB 2018: Natural Language Processing and Information Systems. New York, NY: Springer; 2018: 126–34.
- 90.Caruana R, Lou Y, Gehrke J, et al. Intelligible models for healthcare: predicting pneumonia risk and hospital 30-day readmission. In: KDD ’15: Proceedings of the 21st ACM SIGKDD International Conference on Knowledge Discovery and Data Mining;2015: 1721–30. doi : 10.1145/2783258.2788613
- 91.Weissman GE, Hubbard RA, Ungar LH, et al. Inclusion of unstructured clinical text improves early prediction of death or prolonged ICU stay. Crit Care Med 2018; 46: 1125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jiang W, Siddiqui S, Barnes S, et al. Readmission risk trajectories for patients with heart failure using a dynamic prediction approach: retrospective study. JMIR Med Inform 2019; 7 (4): e14756.doi : 10.2196/14756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brown SM, Jones J, Kuttler KG, et al. Prospective evaluation of an automated method to identify patients with severe sepsis or septic shock in the emergency department. BMC Emerg Med 2016; 16 (1): 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hu Y, Lee VCS, Tan K. Prediction of clinicians’ treatment in preterm infants with suspected late-onset sepsis—An ML approach. In: 2018 13th IEEE Conference on Industrial Electronics and Applications (ICIEA); 2018:1177–1182.
- 95.Christie SA, Conroy AS, Callcut RA, et al. Dynamic multi-outcome prediction after injury: Applying adaptive machine learning for precision medicine in trauma. PLoS One 2019; 14 (4): e0213836.doi : 10.1371/journal.pone.0213836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hu Y, Lee VCS, Tan K. An application of convolutional neural networks for the early detection of late-onset neonatal sepsis. In: 2019 International Joint Conference on Neural Networks (IJCNN); 2019:1–8. doi : 10.1109/IJCNN.2019.8851683
- 97.Ginestra JC, Giannini HM, Schweickert WD, et al. Clinician perception of a machine learning-based early warning system designed to predict severe sepsis and septic shock. Crit Care Med 2019; 47 (11): 1477–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Masino AJ, Harris MC, Forsyth D, et al. Machine learning models for early sepsis recognition in the neonatal intensive care unit using readily available electronic health record data. PLoS One 2019; 14 (2): e0212665–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Obeid JS, Weeda ER, Matuskowitz AJ, et al. Automated detection of altered mental status in emergency department clinical notes: a deep learning approach. BMC Med Inform Decis Mak 2019; 19 (1): 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wellner B, Grand J, Canzone E, et al. Predicting unplanned transfers to the intensive care unit: a machine learning approach leveraging diverse clinical elements. JMIR Med Inform 2017; 5 (4): e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Poncette A-S, Spies C, Mosch L, et al. Clinical requirements of future patient monitoring in the intensive care unit: qualitative study. JMIR Med Inform 2019; 7 (2): e13064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang Q, Steinfeld A, Zimmerman J. Unremarkable AI: fitting intelligent decision support into critical, clinical decision-making processes. In: CHI ’19: Proceedings of the 2019 CHI Conference on Human Factors in Computing Systems; 2019; 1–11. doi : 10.1145/3290605.3300468
- 103.Ye C, Wang O, Liu M, et al. A real-time early warning system for monitoring inpatient mortality risk: prospective study using electronic medical record data. J Med Internet Res 2019; 21 (7): e13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dziadzko MA, Novotny PJ, Sloan J, et al. Multicenter derivation and validation of an early warning score for acute respiratory failure or death in the hospital. Crit Care 2018; 22 (1): 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Giannini HM, Ginestra JC, Chivers C, et al. A machine learning algorithm to predict severe sepsis and septic shock: development, implementation, and impact on clinical practice. Crit Care Med 2019; 47 (11): 1485–92. doi : 10.1097/CCM.0000000000003891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Boulet S, Ursino M, Thall P, et al. Integration of elicited expert information via a power prior in Bayesian variable selection: application to colon cancer data. Stat Methods Med Res 2020; 29 (2): 541–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Holzinger A, Biemann C, Pattichis CS, et al. What do we need to build explainable AI systems for the medical domain? arXiv: 1712.09923; 2017.
- 108.Suleiman M, Demirhan H, Boyd L, et al. Incorporation of expert knowledge in the statistical detection of diagnosis related group misclassification. Int J Med Inform 2020; 136: 104086. [DOI] [PubMed] [Google Scholar]
- 109.Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3 (1): 160035. doi : 10.1038/sdata.2016.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Saeed M, Villarroel M, Reisner AT, et al. Multiparameter intelligent monitoring in intensive care II: A public-access intensive care unit database. Crit Care Med 2011; 39 (5): 952–60. doi : 10.1097/CCM.0b013e31820a92c6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Amarasingham R, Patzer RE, Huesch M, et al. Implementing electronic health care predictive analytics: Considerations and challenges. Health Aff (Millwood) 2014; 33 (7): 1148–54. [DOI] [PubMed] [Google Scholar]
- 112.Levy-Fix G, Kuperman GJ, Elhadad N. Machine learning and visualization in clinical decision support: current state and future directions. arXiv: 1906.02664; 2019.
- 113.Sittig DF, Wright A, Osheroff JA, et al. Grand challenges in clinical decision support. J Biomed Inform 2008; 41 (2): 387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lenert MC, Matheny ME, Walsh CG.. Prognostic models will be victims of their own success, unless. J Am Med Inform Assoc 2019; 26 (12): 1645–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Berndt M, Fischer MR.. The role of electronic health records in clinical reasoning. Ann N Y Acad Sci 2018; 1434 (1): 109–14. [DOI] [PubMed] [Google Scholar]
- 116.Häyrinen K, Lammintakanen J, Saranto K.. Evaluation of electronic nursing documentation-Nursing process model and standardized terminologies as keys to visible and transparent nursing. Int J Med Inform 2010; 79 (8): 554–64. [DOI] [PubMed] [Google Scholar]
- 117.Simon G, DiNardo CD, Takahashi K, et al. Applying artificial intelligence to address the knowledge gaps in cancer care. Oncologist 2019; 24 (6): 772–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang D, Yang Q, Abdul A, et al. Designing theory-driven user-centric explainable AI. In: CHI ’19: Proceedings of the 2019 CHI Conference on Human Factors in Computing Systems; 2019: 1–15.
- 119.Powell BJ, Waltz TJ, Chinman MJ, et al. A refined compilation of implementation strategies: Results from the Expert Recommendations forImplementing Change (ERIC) project. Implement Sci 2015; 10 (1): 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bakken S, Ruland CM.. Translating clinical informatics interventions into routine clinical care: how can the RE-AIM Framework help? J Am Med Inform Assoc 2009; 16 (6): 889–97. doi : 10.1197/jamia.M3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Collins GS, Moons KGM.. Reporting of artificial intelligence prediction models. Lancet 2019; 393 (10181): 1577–9. doi : 10.1016/S0140-6736(19)30235-1 [DOI] [PubMed] [Google Scholar]
- 122.Shimabukuro DW, Barton CW, Feldman MD, et al. Effect of a machine learning-based severe sepsis prediction algorithm on patient survival and hospital length of stay: a randomised clinical trial. BMJ Open Resp Res 2017; 4 (1): e000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Calvert J, Hoffman J, Barton C, et al. Cost and mortality impact of an algorithm-driven sepsis prediction system. J Med Econ 2017; 20 (6): 646–51. doi : 10.1080/13696998.2017.1307203 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
There are no new data associated with this article.