Abstract
Background
Growing evidence suggests that the pluripotent state of mesenchymal stem cells (MSCs) relies on specific local microenvironmental cues such as adhesion molecules and growth factors. Fibronectin (FN), fibroblast growth factor 2 (FGF2), and bone morphogenetic protein 4 (BMP4) are the key players in the regulation of stemness and lineage commitment of MSCs. Therefore, this study was designed to investigate the pluripotency and multilineage differentiation of bone marrow-derived MSCs (BMSCs) with the introduction of FN, FGF-2, and BMP4 and to identify the metabolic and proteomic cues involved in stemness maintenance.
Methods
To elucidate the stemness of BMSCs when treated with FN, FGF-2, and BMP4, the pluripotency markers of OCT4, SOX2, and c-MYC in BMSCs were monitored by real-time PCR and/or western blot. The nuclear translocation of OCT4, SOX2, and c-MYC was investigated by immunofluorescence staining. Multilineage differentiation of the treated BMSCs was determined by relevant differentiation markers. To identify the molecular signatures of BMSC stemness, gas chromatography-mass spectrometry (GC-MS), liquid chromatography-tandem mass spectrometry (LC-MS/MS), and bioinformatics analysis were utilized to determine the metabolite and protein profiles associated with stem cell maintenance.
Results
Our results demonstrated that the expression of stemness markers decreased with BMSC passaging, and the manipulation of the microenvironment with fibronectin and growth factors (FGF2 and BMP4) can significantly improve BMSC stemness. Of note, we revealed 7 differentially expressed metabolites, the target genes of these metabolites may have important implications in the maintenance of BMSCs through their effects on metabolic activity, energy production, and potentially protein production. We also identified 21 differentially abundant proteins, which involved in multiple pathways, including metabolic, autophagy-related, and signaling pathways regulating the pluripotency of stem cells. Additionally, bioinformatics analysis comfirned the correlation between metabolic and proteomic profiling, suggesting that the importance of metabolism and proteome networks and their reciprocal communication in the preservation of stemness.
Conclusions
These results indicate that the culture environment supplemented with the culture cocktail (FN, FGF2, and BMP4) plays an essential role in shaping the pluripotent state of BMSCs. Both the metabolism and proteome networks are involved in this process and the modulation of cell-fate decision making. All these findings may contribute to the application of MSCs for regenerative medicine.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13287-021-02227-7.
Keywords: Mesenchymal stem cells (MSCs), Growth factor, Stemness maintenance, Metabolite, Proteome, Regenerative medicine
Introduction
Mesenchymal stem cells (MSCs) are being exploited as an experimental therapy for many diseases in view of their self-renewable capacity and the potential to differentiate into various mesodermal lineages [1–3]. Under certain pathological conditions, the homing, recruitment, and transplantation of MSCs could help tissue repair and regeneration, leading to the recovery of injuries and degeneration disorders, making MSCs promising candidates for cell-based therapies [3–5]. However, the study of MSC populations leads to the observation that in vitro expansion of these cells is coupled with a limited cell population doubling. That is, MSCs tend to senesce and lose their multi-potentiality and self-renewal capability with time in the current cell culture method [6–8]. Therefore, creating a favorable microenvironment for maintaining the pluripotency of MSCs is of significant importance.
Growing evidence suggests that the cell fate decision of MSCs relies on genes orchestrating cell stemness or commitment to specific lineages [7, 9–11]. Recent progress in our understanding of the transcriptional regulatory circuitry in regulating the pluripotency and self-renewal of stem cells underlines the importance of transcription factors such as OCT4, SOX2, and c-MYC, which are considered to be responsible for pluripotency and self-renewal of stem cells [11–15]. Multiple levels of cell-autonomous and extrinsic signals tightly control fate determination of MSCs [11, 16–19]. It is possible that extrinsic factors derived from the local niche and systemic environment shape the epigenetic landscape of stem cells, which influences the gene expressions of stemness markers to dictate cell fate [20–26]. Previous studies revealed that designing artificial niches through adding extracellular matrix (ECM) proteins or growth factors in the ex vivo culture condition proved to be an effective way to maintain the potential capability of BMSCs [7, 26–28]. Fibronectin (FN) is a promising extracellular matrix factor for the maintenance of stemness characteristics due to its Arg-Gly-Asp containing central cell-binding domain that is essential to mesenchymal cell survival and growth [29]. Fibroblast growth factor 2 (FGF2) is a prerequisite for stem cell multi-potency since its absence triggers lineage differentiation [26, 27]. Bone morphogenetic protein-4 (BMP4) has been recognized as an extracellular pluripotency cue, and it has distinct effects on the fate determination of stem cells by shielding them from differentiation through interaction with a specific niche [30]. Interestingly, the effect of FN, FGF2, or BMP4 on self-renewal capabilities is frequently associated with transcriptional factors (OCT4, SOX2, and c-MYC) [31–34].
Therefore, based on the above, this study aimed at investigating the expression pattern of OCT4, SOX2, and c-MYC in MSCs with successive passaging and then examined these stemness markers (OCT4, SOX2, and c-MYC) and the multilineage differentiation capability of MSCs when treated with FN, FGF2, and BMP4. Furthermore, we focused on the identification of molecular signatures for stemness maintenance of MSCs via metabolic and proteomic cues in this process. The findings from this study will be of significance in facilitating the application of MSCs and extending the field of clinical applications for cell-based therapy in regenerative medicine.
Materials and methods
Cell culture and treatments
Bone marrow-derived MSCs (BMSCs) were isolated and cultured as described previously [1]. Briefly, the bone marrow was obtained from patients (50–60 years old) undergoing elective knee replacement surgery at the Prince Charles Hospital after informed consent was given (n = 5). The procedure was approved by the Office of Research Ethics and Integrity, Queensland University of Technology (ethics approval number 1400001024). Lymphoprep (Axis-Shield PoC AS, Oslo, Norway) was added to isolate the mononuclear cells from the bone marrow by density gradient centrifugation. The obtained cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco®, Life Technologies Pty Ltd., Australia) supplemented with 10% fetal bovine serum (FBS; In Vitro Technologies, Australia) and 1% (v/v) penicillin/streptomycin (P/S; Gibco®, Life Technologies Pty Ltd., Australia) at 37 °C in a humidified CO2 incubator containing 5% CO2. The culture medium was changed every 3 days, and unattached hematopoietic cells were removed by the media changes. The confluent cells were expanded and only early passages (passage 1–5) of cells were used in this study.
To determine the effect of FN (R&D Systems, Minneapolis, MN, USA), FGF2 (R&D Systems, Minneapolis, MN, USA) and/or BMP4 (R&D Systems, Minneapolis, MN, USA) on the expression of OCT4, SOX2, and c-MYC, six different experimental groups were designed in this study as follows: (1) a control group where BMSCs were cultured with DMEM supplemented with 10% FBS, (2) BMSCs cultured on 5 μg/mL FN-coated plates with DMEM supplemented with 10% FBS, (3) BMSCs cultured with DMEM supplemented with 2 ng/mL FGF2 and 10% FBS, (4) BMSCs cultured on FN-coated plates and DMEM supplemented with 2 ng/mL FGF2 and 10% FBS, (5) BMSCs cultured on FN-coated plates and DMEM supplemented with 10 ng/mL BMP4 and 10% FBS, and (6) BMSCs cultured on FN-coated plates and DMEM supplemented with 2 ng/mL FGF2, 10 ng/mL BMP4, and 10% FBS.
Quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR)
Total RNA of cultured cells at passage 5 from six different experimental groups was isolated with TRIzol® reagent (Life Technologies Pty Ltd., Australia) as per manufacturer’s instructions. Reverse transcription was performed using DyNAmo™ cDNA Synthesis Kit (Genesearch Pty Ltd., Australia). SYBR Green qPCR Master Mix (Life Technologies Pty Ltd., Australia) was used for detection, and OCT4, SOX2, and c-MYC mRNA expression were assayed on the ABI PRISM® 7500 FAST Sequence Detection System (Applied Biosystems, California, USA). The relative gene expression was obtained by normalizing the mean cycle threshold (Ct) value of each target gene with the Ct value of the house-keeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The corresponding primer sequences of the reference gene and the target genes were designed based on cDNA sequences from the NCBI Sequence Database (Table 1).
Table 1.
Primer sequences used in a quantitative real-time polymerase chain reaction
| GENE | Primer |
|---|---|
| OCT4 | Forward: 5′-GCTCGAGAAGGATGTGGTC-3′ |
| Reverse: 5′-ATCCTCTCGTTGTGCATAGTCG-3′ | |
| SOX2 | Forward: 5′- CACTGCCCCTCTCACACATG-3′ |
| Reverse: 5′- CCCATTTCCCTCGTTTTTCTT-3′ | |
| c-MYC | Forward: 5′- GGCTCCTGGCAAAAGGTCA-3′ |
| Reverse: 5′- AGTTGTGCTGATGTGTGGAGA-3′ | |
| ACAN | Forward: 5′- AGACTTGGTGGGGTCAG-3′ |
| Reverse: 5′- GATGTTTCCCACTAGTG-3′ | |
| OPN | Forward: 5′- CTGAGGCTGAGAATACCACACTT-3′ |
| Reverse: 5′- GGTGATGTCCTCGTCTGTA-3′ | |
| PPAγ2 | Forward: 5′-CTGTTGACTTCTCCAGCA-3′ |
| Reverse: 5′-GTCAGCGGACTCTGGA-3′ | |
| GAPDH | Forward: 5′-TCAGCAATGCCTCCTGCAC-3′ |
| Reverse: 5′-TCTGGGTGGCAGTGATGGC-3′ |
Western blot analysis
The BMSCs at passage 5 were cultured on FN-coated plates and DMEM supplemented with 2 ng/mL FGF2, 10 ng/mL BMP4, and 10% FBS. The whole-cell lysates of stimulated BMSCs were collected after 3 days of culture. Equivalent amounts of diluted protein samples (15 μg) were resolved by 10% SDS-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes. Bands were detected with polyclonal antibodies (1:1000) against OCT4 (Abcam, USA), SOX2 (Abcam, USA), and c-MYC (Abcam, USA). a-Tubulin (1:5000; Abcam, USA) was used as a loading control. The protein bands were visualized using the Odyssey Infrared Imaging System (LI-COR Biotechnology, Nebraska, USA). The relative intensity of protein bands compared with α-Tubulin was quantified using Image Studio™ Software.
Immunofluorescence staining
BMSCs in the FN + FGF2 + BMP4 treatment group and untreated control group at passages 3 and 5 were cultured in Nunc™ chamber slides (Thermo Fisher Scientific, USA). Briefly, the cells were fixed with 4% (w/v) paraformaldehyde (PFA) for 15 min and then permeabilized with 0.1% Triton X-100 in PBS for 10 min, incubated in 3% bovine serum albumin in PBS for 30 min and with the primary monoclonal anti-OCT4 antibody (1:100), anti-SOX2 antibody (1:100), and anti-c-MYC antibody (1:100) at 4 °C overnight. Finally, all samples were rinsed and then incubated with fluorochrome labeled (Alexa Fluor 488) secondary antibody (1:150, Invitrogen, USA). Images were visualized and captured using a fluorescence microscope (Carl Zeiss Microimaging Gmbh, Gottingen, Germany).
In vitro differentiation of BMSC populations
Differentiation capabilities of the FN + FGF2 + BMP4 treatment group and untreated control group at passage 5 were determined by stimulating cells in selective differentiation media for osteogenic, chondrogenic, and adipogenic lineage.
Chondrogenic differentiation
Cells at passage 5 from both groups were differentiated into chondrocytes according to a previously described micro-mass pellet culture [35]. The cell pellets were cultured in a chondrogenic medium. After 21 days, the cell pellets were stained with Alcian blue for evidence of proteoglycan deposition, and the mRNA expression of aggrecan (ACAN) was measured.
Osteogenic differentiation
Osteogenic differentiation was induced on a monolayer of confluent populations with complete media supplemented with 50 mM ascorbic acid, 10 mM β-glycerol phosphate, and 100 nM dexamethasone (Sigma-Aldrich, Castle Hill, NSW, Australia). After 21 days, the cells were stained with Alizarin Red S to observe calcium depositions and the mRNA expression of osteopontin (OPN) was measured.
Adipogenic differentiation
Cells at passage 5 for both groups were differentiated into adipocytes as previously described [36]. Lipid droplets were stained with Oil Red O and the mRNA expression of peroxisome proliferator-activated receptor γ2 (PPAγ2) was measured.
Metabolomic and proteomic characterization
Details of the procedure were provided in the Supplementary Materials and Methods.
Genomic biological prediction of target genes regulated by OCT4, SOX2, and c-MYC
The Homo sapiens genes regulated by three transcription factors, POU5F1/OCT4, SOX2, and MYC/c-MYC, were predicted by the Gene Transcription Regulation Database (GTRD, http://gtrd.biouml.org/). Ensembl Database (https://rest.ensembl.org) was used for the annotation process of the data from GTRD. Venn diagrams in the R package were utilized to compare 3 datasets from the transcription factors, among which the co-regulated coding-protein genes were selected. Based on these genes, functional enrichment analysis in terms of pathway analysis was performed by the cluster profiler R package, and Venn analysis was performed with the known metabolic pathways identified from the metabolomic characterization. In addition, the co-regulated genes and 21 differential genes based on proteomic analysis were further analyzed using Venn analysis. Furthermore, we explored the potential regulatory feedback between the metabolic and proteomic data through analyzing their common KEGG pathways and subsequently correlated metabolites and differential proteins in the common pathways.
Statistical analysis
All experiments were performed in triplicate. The results obtained from this study were expressed as mean ± SD (standard deviations). Statistical analysis was performed using GraphPad Prism 7 (Version 7.02) for Windows (GraphPad Software Inc., USA). Statistical differences between groups were determined by one-way ANOVA with Bonferroni’s multiple comparison tests. p < 0.05 was considered statistically significant.
Results
Expression pattern of stemness marker in BMSCs with successive passaging
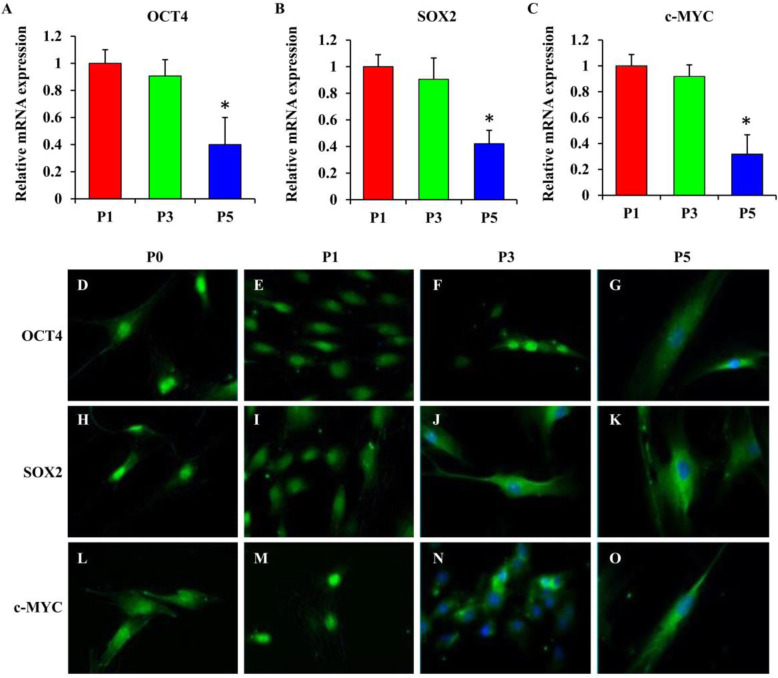
The mRNA expressions of OCT4, SOX2, and c-MYC were downregulated in culture from passage 1 to passage 5 (Fig. 1a–c). Among all groups, the cells at passage 5 had the most significant reduction in stemness marker expressions compared to other groups (p < 0.05). Immunofluorescence staining revealed that OCT4 (Fig. 1d–g), SOX2 (Fig. 1h–k), and c-MYC (Fig. 1l–o) while detected in the nucleus as early as the primary culture exhibited cytoplasmic expression with subsequent passaging.
Fig. 1.
Expression of OCT4, SOX2, and c-MYC in BMSCs at various passages. a–c The mRNA expressions of OCT4, SOX2, and c-MYC were downregulated in BMSC cultures at passage 5 compared to those at passages 1 and 3. The primary antibody of each target protein OCT4 (d–g), SOX2 (h–k), and c-MYC (l–o) was detected with Alexa Fluor 488 (green stain) at various passages. Statistical significance was accepted at *p < 0.05
Manipulation of microenvironment counteracted the change of stemness marker expression patterns
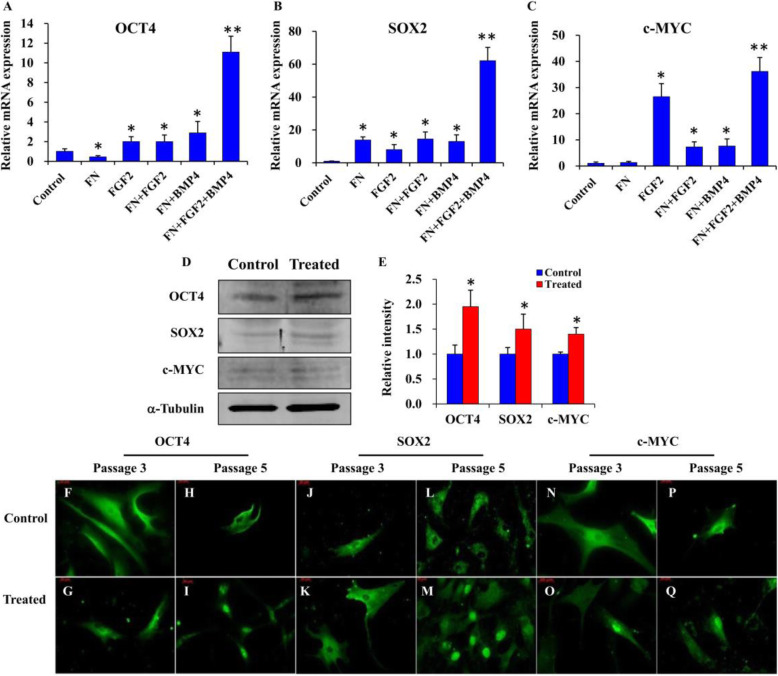
The qRT-PCR analysis of BMSCs at passage 5 showed that the mRNA expressions of OCT4 (Fig. 2a), SOX2 (Fig. 2b), and c-MYC (Fig. 2c) were all significantly upregulated relative to the control in the experimental groups, FGF2, FN+ FGF2, and FN + BMP4-treated groups, respectively (p < 0.05). Importantly, the most significant upregulation occurred in BMSC cultures stimulated with FN + FGF2 + BMP4 (p < 0.001) (Fig. 2a–c). Western blot analysis showed similar expression patterns to the qRT-PCR results. Expression of all the stemness markers (OCT4, SOX2, and c-MYC) was enhanced in the FN + FGF2 + BMP4 group (Fig. 2d, e). Immunofluorescence staining revealed that OCT4 (Fig. 2f–i), SOX2 (Fig. 2j–m), and c-MYC (Fig. 2n–q) while detected in the cytoplasm at passages 3 and 5 in the control group exhibited nuclear expression at passage 5 in the FN + FGF2 + BMP4-treated group, indicating cytoplasmic to nuclear translocation.
Fig. 2.
The expression and translocation pattern of OCT4, SOX2, and c-MYC with the stimulation of FN + FGF2 + BMP4. a–c All treatments transiently induced the expressions of OCT4, SOX2, and c-MYC, and the induction effect of FN + FGF2 + BMP4 was most obvious. d, e Treated cells with FN + FGF2 + BMP4 enhanced the protein expression of OCT4, SOX2, and c-MYC. The primary antibody of each target protein OCT4 (f–i), SOX2 (j–m), and c-MYC c (n–q) was detected with Alexa Fluor 488 (green stain). Nuclear translocation of the target transcription factors was observed in all treated samples (FN + FGF2 + BMP4) at passage 5. Statistical significance was accepted at *p < 0.05 or **p < 0.001
Manipulation of microenvironment increased differentiation potential of BMSCs
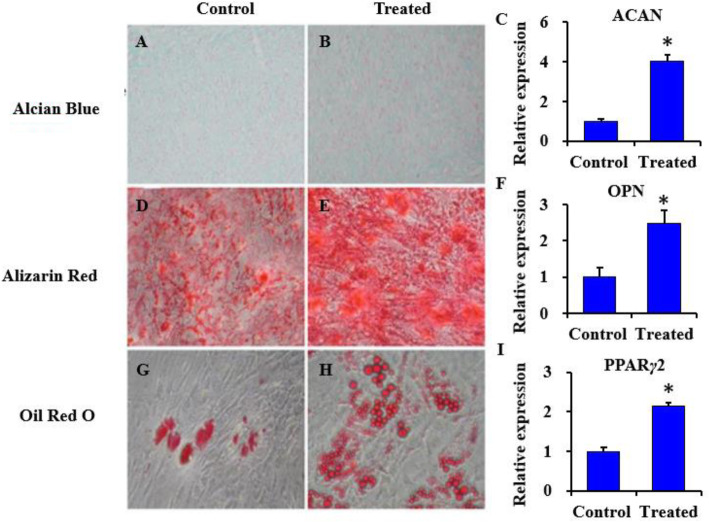
After 21 days of multi-lineage differentiation induction, the treated BMSCs at passage 5 appeared to be more multi-potent as revealed by the staining pattern and differentiation transcript expression. Alcian blue staining was more prominent in the treated samples compared with the untreated control group (Fig. 3a, b). The ACAN gene transcript expression was significantly higher in the treated samples (p < 0.05) (Fig. 3c). When cultured in osteogenic media, both groups exhibited mineralization and calcium deposition (Fig. 3d, e). The treated samples were almost completely covered with calcium deposits as revealed by Alizarin Red S staining (Fig. 3e). OPN expression corresponding to osteogenic differentiation was higher in the treated group (p < 0.05) (Fig. 3f). For adipogenic differentiation, treated BMSCs after 21 days of induction exhibited a larger number of lipid droplet clusters compared to the untreated control group (Fig. 3g, h). Similarly, PPARγ2 expression was higher in the treated group (p < 0.05) (Fig. 3i). Thus, FN + FGF2 + BMP4 promoted the multi-lineage differentiation of BMSCs.
Fig. 3.
The differentiation potential of control and treated BMSCs. Treated (FN + FGF2 + BMP4) and control BMSCs at passage 5 were induced in chondrogenic, osteogenic, and adipogenic medium for 21 days, followed by staining and qRT-PCR analysis (a–h). Strong staining for all three lineages was observed in treated BMSCs. The qRT-PCR analysis for c ACAN, f OPN, and i PPARγ2 revealed higher expression of lineage-specific markers in treated BMSCs. Statistical significance was accepted at *p < 0.05
Manipulation of microenvironment facilitated metabolic alterations
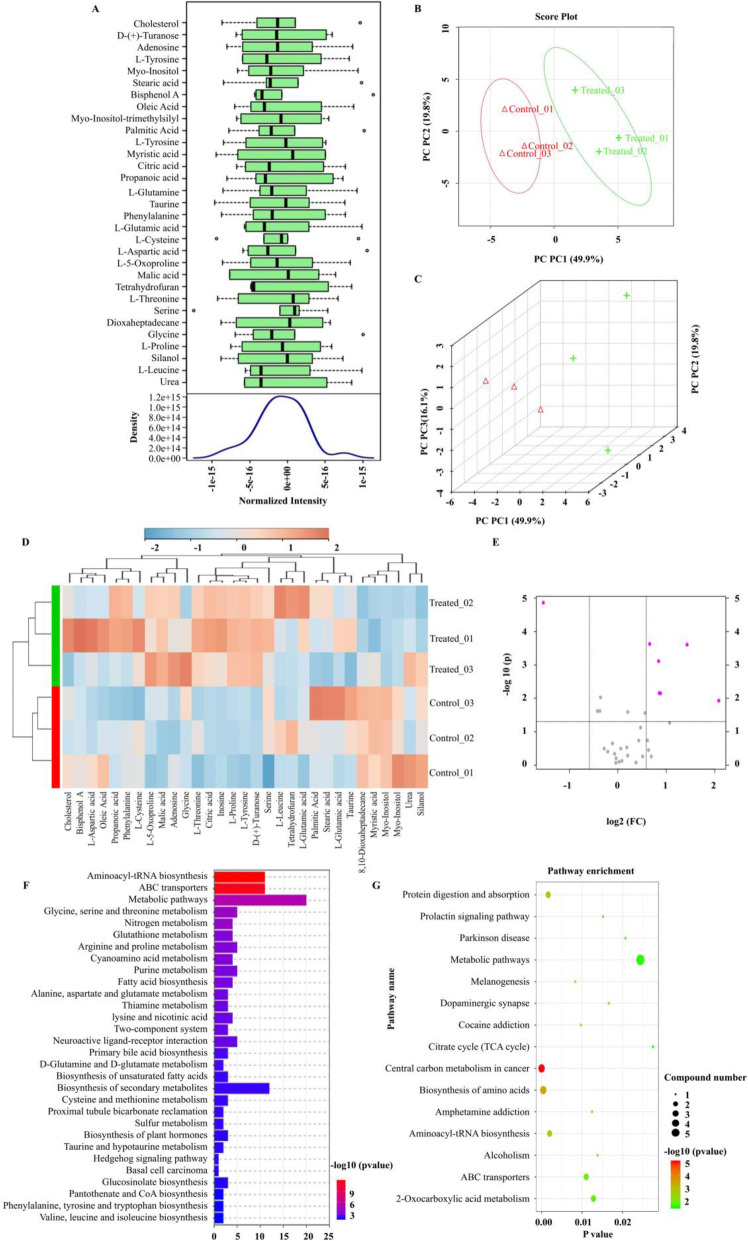
GC-MS was utilized to determine the metabolic cues involved in the maintenance of BMSCs. Row-wise normalization allowed general-purpose adjustment for differences among samples thereby allowing users to manually adjust concentrations based on biological inputs. The density plot analysis based on all samples, indicated that the metabolite data distribution conformed to normal distributions (Fig. 4a). The 2D and 3D scores plot indicated distinct clustering between samples of the control or treatment groups, which met the experimental requirements and could be used for subsequent analysis (Fig. 4b, c). The 31 identified analytes are presented in Table S1. The clustering results were shown in the form of a heatmap (Fig. 4d). Using volcano plot analysis, we also identified 7 differentially abundant metabolites that had a greater than 1.5-fold change in the treated BMSCs compared with the control (Myo-Inositol, L-Proline, D-(+)-Turanose, L-Tyrosine, Malic acid, Citric acid, Inosine) (Fig. 4e). The functional annotations of the abundant metabolites were further examined using KEGG Ontology assignments to classify their relative influence on various biochemical pathways (Fig. 4f, g). Primarily, this included increases in pathways that play a role in metabolic activity, energy production, and potentially protein production.
Fig. 4.
GC-MS analysis of treated BMSCs compared to untreated BMSCs. a Box plots and kernel density plots after normalization. b, c 2 and 3D score plot between all samples. The explained variances were shown in brackets. d Heatmap showed 31 metabolites identified by GC-MS. e Volcano plots demonstrated differential abundance between two groups. The vertical lines corresponded to a 1.5-fold increase and decrease in abundance and the horizontal line represents a p value of 0.05. The colored dots in the plot represent the metabolites that exhibited statistically significant differential abundance compared to controls. f KEGG pathway annotation of the analytes where the horizontal axis represents the number of metabolite pathway associated genes. g KEGG pathway annotation of the differentially abundant analytes. The horizontal axis indicated p value and vertical ordinates were the terms of the pathways. The size of the node indicates the number of metabolite pathway-associated genes matched in the pathways. The degree of color represents −log10 (p value): Logarithmic conversion of Fisher’s exact test p value, indicating the significance of pathway correlations
Proteomic alterations for stemness maintenance of BMSCs
LC-MS/MS was performed to explore the altered proteomes involved in the maintenance of BMSCs. The box plot in Fig. 5a indicated that the distribution of the intensities among all samples was nearly the same. The 3D scores plot indicated a close distance between samples in the same groups, which met the experimental requirements and can be used for subsequent analysis (Fig. 5b). Figure 5c showed the correlation heatmap. The Volcano Plots in Fig. 5d showed differential abundance between samples. We identified 21 differentially abundant proteins that had a greater than 1.5-fold change in the treated BMSCs compared with the control (Table S2 & Fig. 5e). Among these, 7 proteins were upregulated, whereas 14 proteins were downregulated (Table S2). We further determined the functional annotations of the differentially abundant proteins through gene ontology analysis (Fig. 5f). These data indicated that the differentially abundant proteins were associated with extracellular matrix/structure organization, the production of cytoplasmic vesicles, and the proteoglycan or sulfur compound binding. Additionally, KEGG significant enrichment analysis showed differentially abundant proteins involved in multiple signal transduction, including metabolic pathways, autophagy-related pathways, and signaling pathways regulating pluripotency of stem cell (Fig. 5g).
Fig. 5.
Proteomic characterization of treated BMSCs compared to untreated BMSCs. a Box plots to visualize the distributions of a dataset. The box plots in the same figure indicated that the distribution of the intensities among all samples was nearly the same. b 3D score plot for all samples. The explained variances were shown in brackets. c Hierarchical clustering was performed with the h.clust function in a statistical package, and the clustering result was shown in the form of a heatmap. d Correlation analysis could be used to visualize the overall correlations between different samples. e Volcano plots were used to demonstrate differential abundance between two groups. The vertical lines corresponded to 1.5-fold up and down and the horizontal line represented a p value of 0.05. The colored dots in the plot represented the differentially abundant proteins with statistical significance. f Heatmap showed 21 differentially abundant proteins among samples (high relative abundance in red, and low relative abundance in green or black). g Gene ontology (GO) analysis of differentially abundant proteins. The vertical axis represented the gene ratio (the ratio of the gene count in GO terms to the total differentially expressed genes count) and the horizontal axis described the enrichment components. BP biological process, CC cellular component, MF molecular function. h KEGG significant enrichment analysis for the differentially abundant proteins. The horizontal axis indicated p value and vertical ordinates were the terms of the pathways. The size of the node indicated the number of genes matched in the pathways. The degree of color represented −log10 (p value): Logarithmic conversion of Fisher’s exact test p value, indicating the significance of pathway correlations
Biological analysis revealed a potential relationship between metabolic and proteomic cues involved in stemness maintenance
Target genes regulated by 3 transcription factors were identified (Table S3) and 31,850 co-regulated genes (including protein genes and non-coding RNAs) were found by Venn analysis (Table S3 & Fig. S1). Out of the 31,850 co-regulated genes, 31,526 were present in the Ensembl Database, among which 16,129 were coding-protein genes (16,126, excluding repeated ones) (Table S3). These genes were associated with a variety of signaling pathways, among which 17 were known metabolic pathways based on the metabolic results via Venn analysis (Table S4). Additionally, Venn analysis was further performed to correlate these 16,126 co-regulated genes and the 21 genes with noted differences from the proteomic analysis. The results showed that a total of 20 genes were co-expressed. The gene outside of the intersection was HBB, which is a potential regulatory gene of SOX2 and c-MYC (Fig. S2). All these data implied a reciprocal communication between metabolic/proteomic cues of MSCs and their stemness. Furthermore, we identified five common KEGG pathways between the metabolic and proteomic analysis (Table S5), among which the metabolites and differential proteins were correlated in the same pathways (Fig. S3 - Fig. S7).
Discussion
BMSCs are a resource for autologous and allogeneic cell therapies in regenerative medicine [1, 3, 37]. However, BMSCs senesce with prolonged culture in vitro and undergo a decline in stemness and progenitor cell functions such as proliferation and differentiation, thereby limiting their therapeutic applications [6–8]. Some researchers have reported various in vitro treatments that may improve the performance of aged BMSCs [7, 8, 26–28]. Thus, improving the conditions of cell culture and discovering potential signaling cues that regulate BMSC aging may help to maintain the competency of BMSCs.
As noted in recent research, cell reprogramming leading to pluripotency is a gradual process involving the sequential reactivation of pluripotency markers such as OCT4, SOX2, and c-MYC [12, 31, 38–41]. In the present study, following the isolation and characterization of BMSCs, we assessed the stemness markers (OCT4, SOX2, and c-MYC) of these cells and found that they decreased with successive passaging. Notably, the molecules FN, FGF2, and BMP4 could rescue the stemness loss with an individual or synergistic effect, among which the simultaneous introduction of FN, FGF-2, and BMP4 significantly increased the expression of OCT4, SOX2, and c-MYC. Thus, we anticipate that the manipulation of the microenvironment with the ECM component and growth factors will be useful concerning the reverse of the aging BMSCs.
Based on our immunofluorescence staining results, OCT4 along with SOX2 and c-MYC were mainly localized in the nucleus and gradually lost their nuclear localization while being expressed in the cytoplasm of BMSCs after passaging in vitro, indicating the loss of their functional role during this process. Of note, when treated with FN, FGF2, and BMP4, the location of stemness markers translocated from the cytoplasm to nuclear. Interaction of these key transcription factors leads to the establishment of a regulatory loop which controls the self-renewal and differentiation potential of stem cells, implying that the “cocktail treatment” of combined stimulating factors may keep cells in a state of active reprogramming and is critical for maintaining stemness. Consistently, those treated BMSCs formed significant Alcian blue-positive pellets, Alizarin Red-positive mineralized nodules, and Oil Red O-positive lipid droplets. Therefore, the treated BMSCs hold much promise in regenerative medicine, and identification of the underlying molecular mechanisms is an avenue for further research.
Recent advancement of various high-throughput experimental techniques makes it feasible to integrate large amounts of data into a coherent quantitative picture of cell fate control. Once thought to be a mere consequence of the state of a cell, metabolism is now known to play a pivotal role in dictating whether a cell proliferates, differentiates or remains quiescent [42–46]. GC-MS is a predominant tool in the detection of metabolites due to its ability to explore the unique fingerprints that specific cellular processes leave behind [47, 48]. It can analyze metabolite abundance to reveal active metabolic pathways which allow for unique biomarker identification [47, 49–51].
Recent studies have demonstrated that nuclear reprogramming of stem cells through the induction of transcription factors associated with dramatic metabolic remodeling [42–46]. Among 7 differentially abundant metabolites, Proline, Citric acid, Tyrosine and Malic acid are intermediate metabolites of the energy-generating pathways, such as the tricarboxylic acid cycle (TCA cycle), the glutamine metabolism and the proline regulatory axis. All these pathways play a role in metabolic remodeling, which can fuel maintenance of pluripotent state [43, 44]. Besides, Myo-Inositol [52], and Inosine [53] can regulate the antioxidant defenses. As the cellular redox status plays an important role in stem cell biology [54], these two altered metabolites may help preserve stem cell functionality. To understand the biological functions of the abundant metabolites further, KEGG Ontology assignments were conducted to classify their relative influence on various biochemical pathways. The functional identity of the target genes confirmed that these metabolites may have important implications in the maintenance of BMSCs through their effects on metabolic activity, energy production, and potentially protein production. Interestingly, based on the bioinformatics analysis, we identified 17 common pathways in the relation network among these metabolites-associated pathways and the biological pathways associated with 3 transcription factors. All these results suggested that the introduction of FN, FGF-2, and BMP4 can regulate the metabolic pathways, thereby driving cell fate conversions by regulating the transcriptional identity of the stem cells.
Concurrently, the LC-MS proteomic technique was also utilized in our study to characterize the protein complements involved in stemness maintenance. This type of study generates a snapshot of the environment’s proteins and provides an overall view of the potential biochemical pathways being utilized by the cells [55, 56]. Our study identified 21 differentially abundant proteins, among which 20 were predicted to be the targets of 3 transcription factors (OCT4, SOX2, and c-MYC), indicating that the proteins from proteomic analysis participated in the pluripotent fate control of BMSCs. To further understand the orchestrating roles of these 21 abundant proteins in the pluripotency of BMSCs, their potential target gene functions were illustrated using gene ontology and KEGG pathway analysis. The ontology analysis data indicated that 21 differentially abundant proteins were associated with extracellular matrix/structure organization, the production of cytoplasmic vesicles, and the proteoglycan or sulfur compound binding. Additionally, KEGG pathway analysis revealed that target genes of the 21 differentially expressed proteins were involved in multiple signal transductions, including metabolic pathways, autophagy-related pathways, and signaling pathways regulating pluripotency of stem cells. Of note, some of these were consistent with our metabolic results which were mentioned above; thus, we performed bioinformatics analysis to further explore the correlation between metabolic and proteomic profiling. According to our results, we identified five common KEGG pathways between these two analysis techniques. These findings suggest that the importance of metabolism and proteome networks and their reciprocal communication in the preservation of stemness and the modulation of cell-fate decision making.
Proteomic analysis of MSCs could provide detailed information into what processes are occurring in the cells and can be used to monitor their growth and stemness. Concordantly, metabolomic studies can provide a more acute perspective on the dynamic changes of an environment, as the flux of metabolites can provide information on the activity of the cells [42, 47]. Determination of both the metabolomic and proteomic markers could help us fully understand the biological status of the MSCs and provide significant insights into how to maintain their stem phenotype. Further research needs to be conducted to confirm how these metabolic intermediates or proteins contribute to the stemness maintenance during nuclear reprogramming. While GC-MS and LC-MS are highly reliable analytical techniques, using them in tandem with other high-throughput analytical techniques would obtain a more accurate analysis of the results. Despite the limitations of this research due to using only one method of metabolic or proteomic analysis, this study demonstrates that the manipulation of the microenvironment can lead the cells with high passage numbers to a more primitive cell state, which may be related to a specific metabolomic and proteomic profile.
Conclusion
The present study has demonstrated that endogenous expression of stemness markers (OCT4, SOX2, and c-MYC) can be induced by modulation of culture conditions (FN, FGF-2, and BMP4) and deepen our understanding of the contribution made by metabolic and proteomic cues to the regulation of BMSC stemness. Further investigations into the metabolite-protein dynamics and the underlying molecular mechanisms hold great therapeutic promise to direct stem cell fate for tissue regeneration and the development of novel strategies to combat degenerative disorders.
Supplementary Information
Additional file 1: Supplementary Materials and Methods.
Additional file 5: Table S4 and Table S5.
Additional file 6: Supplementary Figures.
Acknowledgements
The authors would like to acknowledge the scientific and technical assistance of Dr. Pawel Sadowski and Mr. Joel Herring, and the facilities at the Central Analytical Research Facility (CARF) operated by the Institute for Future Environments (IFE) at QUT.
Abbreviations
- MSCs
Mesenchymal stem cells
- FN
Fibronectin
- FGF2
Fibroblast growth factor 2
- BMP4
Bone morphogenetic protein 4
- BMSCs
Bone marrow derived MSCs
- GC-MS
Gas chromatography–mass spectrometry
- LC-MS/MS
Liquid chromatography–tandem mass spectrometry
- TCA cycle
Tricarboxylic acid cycle
- DMEM
Dulbecco’s modified Eagle’s medium
- FBS
Fetal bovine serum
- P/S
Penicillin/streptomycin
- qRT-PCR
Quantitative real-time reverse-transcription polymerase chain reaction
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- PFA
Paraformaldehyde
- ACAN
Aggrecan
- OPN
Osteopontin
- PPAγ2
Peroxisome proliferator activated receptor γ2
- GTRD
Gene Transcription Regulation Database
- GO
Gene ontology
Authors’ contributions
LLC and YHZ conceived the idea, designed the experiments, conducted the experiments, analyzed the data, and wrote the manuscript. MC and NK performed the experiments and collected and analyzed the data. XDC did the bioinformatical analysis and wrote parts of the manuscript. HR conducted the experiments and wrote parts of the manuscript. TP analyzed data, reviewed data, and edited the manuscript. ZML and YX designed the experiments, monitored the project progression, and edited the manuscript. The authors read and approved the final manuscript.
Funding
This work was supported by the Queensland University of Technology (QUT) and Guangdong Science and Technology Commission (GDSTC) Collaborative Research Grant Scheme, the Institute of Health and Biomedical Innovation-Injury Prevention and Trauma Management Theme Collaborative Development Grant Scheme, the National Health and Medical Research Council (NHMRC) Early Career Fellowship (Grant No. 1105035), the National Natural Science Foundation of China (NSFC) General Project (Grant No. 31771025), the NSFC Young Scientists Fund (Grant No. 81700969), the Guangdong Basic and Applied Basic Research Foundation (Grant No. 2020A1515010148), and the QUT Boost Scheme.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The bone marrow was obtained from patients (50–60 years old) undergoing elective knee replacement surgery at the Prince Charles Hospital after informed consent was given. The procedure was approved by the Office of Research Ethics and Integrity, Queensland University of Technology (ethics approval number 1400001024).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lingling Chen, Email: chenlling5@mail.sysu.edu.cn.
Morgan Carlton, Email: morgan.carlton@hdr.qut.edu.au.
Xiaodan Chen, Email: chenxd76@mail.sysu.edu.cn.
Navdeep Kaur, Email: n.kaur@uq.edu.au.
Hollie Ryan, Email: h5.ryan@qut.edu.au.
Tony J Parker, Email: a.parker@qut.edu.au.
Zhengmei Lin, Email: linzhm@mail.sysu.edu.cn.
Yin Xiao, Email: yin.xiao@qut.edu.au.
Yinghong Zhou, Email: yinghongzhou@qut.edu.au.
References
- 1.Zhou Y, Fan W, Prasadam I, Crawford R, Xiao Y. Implantation of osteogenic differentiated donor mesenchymal stem cells causes recruitment of host cells. J Tissue Eng Regen Med. 2015;9:118–126. doi: 10.1002/term.1619. [DOI] [PubMed] [Google Scholar]
- 2.Zakrzewski W, Dobrzyński M, Szymonowicz M, Rybak Z. Stem cells: past, present, and future. Stem Cell Res Ther. 2019;10:68. doi: 10.1186/s13287-019-1165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiang Y, Wu C, Wu J, Quan W, Cheng C, Zhou J, et al. In vitro expansion affects the response of human bone marrow stromal cells to irradiation. Stem Cell Res Ther. 2019;10:82. doi: 10.1186/s13287-019-1191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z, Li T, Zhu F, Deng S, Li X, He Y. Regulatory roles of Mir-22/Redd1-mediated mitochondrial Ros and cellular autophagy in ionizing radiation-induced Bmsc injury. Cell Death Dis. 2019;10:227. doi: 10.1038/s41419-019-1373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liesveld JL, Sharma N, Aljitawi OS. Stem cell homing: from physiology to therapeutics. Stem Cells. 2020;38:1241-53. [DOI] [PubMed]
- 6.Baker N, Boyette LB, Tuan RS. Characterization of bone marrow-derived mesenchymal stem cells in aging. Bone. 2015;70:37–47. doi: 10.1016/j.bone.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 7.Abdallah BM, Alzahrani AM, Abdel-Moneim AM, Ditzel N, Kassem M. A simple and reliable protocol for long-term culture of murine bone marrow stromal (mesenchymal) stem cells that retained their in vitro and in vivo stemness in long-term culture. Biol Proced Online. 2019;21:3. doi: 10.1186/s12575-019-0091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondrikov D, Elmansi A, Bragg RT, Mobley T, Barrett T, Eisa N, et al. Kynurenine inhibits autophagy and promotes senescence in aged bone marrow mesenchymal stem cells through the aryl hydrocarbon receptor pathway. Exp Gerontol. 2020;130:110805. doi: 10.1016/j.exger.2019.110805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shou K, Huang Y, Qi B, Hu X, Ma Z, Lu A, et al. Induction of mesenchymal stem cell differentiation in the absence of soluble inducer for cutaneous wound regeneration by a chitin Nanofiber-based hydrogel. J Tissue Eng Regen Med. 2018;12:e867–e880. doi: 10.1002/term.2400. [DOI] [PubMed] [Google Scholar]
- 10.Sui B, Zheng C, Li M, Jin Y, Hu C. Epigenetic regulation of mesenchymal stem cell homeostasis. Trends Cell Biol. 2020;30:97–116. doi: 10.1016/j.tcb.2019.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Darabi S, Tiraihi T, Delshad A, Sadeghizadeh M, Taheri T, Hassoun HK. Creatine enhances transdifferentiation of bone marrow stromal cell-derived neural stem cell into gabaergic neuron-like cells characterized with differential gene expression. Mol Neurobiol. 2017;54:1978–1991. doi: 10.1007/s12035-016-9782-9. [DOI] [PubMed] [Google Scholar]
- 12.Deng Y, Huang G, Chen F, Testroet ED, Li H, Li H, et al. Hypoxia enhances buffalo adipose-derived mesenchymal stem cells proliferation, stemness, and reprogramming into induced pluripotent stem cells. J Cell Physiol. 2019;234:17254-68. [DOI] [PubMed]
- 13.Rust K, Tiwari MD, Mishra VK, Grawe F, Wodarz A. Myc and the Tip60 chromatin remodeling complex control neuroblast maintenance and polarity in drosophila. EMBO J. 2018;37:e98659. [DOI] [PMC free article] [PubMed]
- 14.He N, Chen X, Wang D, Xu K, Wu L, Liu Y, et al. Ve-cadherin regulates the self-renewal of mouse embryonic stem cells via Lif/Stat3 signaling pathway. Biomaterials. 2018;158:34–43. doi: 10.1016/j.biomaterials.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 15.Mas-Bargues C, Vina-Almunia J, Ingles M, Sanz-Ros J, Gambini J, Ibanez-Cabellos JS, et al. Role of P16(Ink4a) and Bmi-1 in oxidative stress-induced premature senescence in human dental pulp stem cells. Redox Biol. 2017;12:690–698. doi: 10.1016/j.redox.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang L, Jurczak KM, Ge L, van Rijn P. High-throughput screening and hierarchical topography-mediated neural differentiation of mesenchymal stem cells. Adv Healthc Mater. 2020;9:e2000117. doi: 10.1002/adhm.202000117. [DOI] [PubMed] [Google Scholar]
- 17.Li CJ, Xiao Y, Yang M, Su T, Sun X, Guo Q, et al. Long noncoding Rna Bmncr regulates mesenchymal stem cell fate during skeletal aging. J Clin Invest. 2018;128:5251–5266. doi: 10.1172/JCI99044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai S, Tsui YP, Tam KW, Shea GK, Chang RS, Ao Q, et al. Directed differentiation of human bone marrow stromal cells to fate-committed Schwann cells. Stem Cell Rep. 2017;9:1097–1108. doi: 10.1016/j.stemcr.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morganti C, Bonora M, Marchi S, Ferroni L, Gardin C, Wieckowski MR, et al. Citrate mediates crosstalk between mitochondria and the nucleus to promote human mesenchymal stem cell in vitro osteogenesis. Cells-Basel. 2020;9:1034. [DOI] [PMC free article] [PubMed]
- 20.Nowwarote N, Sukarawan W, Pavasant P, Foster BL, Osathanon T. Basic fibroblast growth factor regulates phosphate/pyrophosphate regulatory genes in stem cells isolated from human exfoliated deciduous teeth. Stem Cell Res Ther. 2018;9:345. doi: 10.1186/s13287-018-1093-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park J, Lee JH, Yoon BS, Jun EK, Lee G, Kim IY, et al. Additive effect of Bfgf and selenium on expansion and paracrine action of human amniotic fluid-derived mesenchymal stem cells. Stem Cell Res Ther. 2018;9:293. doi: 10.1186/s13287-018-1058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan T, Volckaert T, Chanda D, Thannickal VJ, De Langhe SP. Fgf10 signaling in lung development, homeostasis, disease, and repair after injury. Front Genet. 2018;9:418. doi: 10.3389/fgene.2018.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang W, Liang Q, Du L, Shang L, Wang T, Ge S. Sequential application of Bfgf and Bmp-2 facilitates osteogenic differentiation of human periodontal ligament stem cells. J Periodontal Res. 2019;54:424-34. [DOI] [PubMed]
- 24.Carluccio M, Ziberi S, Zuccarini M, Giuliani P, Caciagli F, Di Iorio P, et al. Adult Mesenchymal stem cells: is there a role for purine receptors in their osteogenic differentiation? Purinergic Signal. 2020;16:263-87. [DOI] [PMC free article] [PubMed]
- 25.Heo JS, Kim HO, Song SY, Lew DH, Choi Y, Kim S. Poly-L-lysine prevents senescence and augments growth in culturing mesenchymal stem cells ex vivo. Biomed Res Int. 2016;2016:8196078. doi: 10.1155/2016/8196078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore L, Skop NB, Rothbard DE, Corrubia LR, Levison SW. Tethered growth factors on biocompatible scaffolds improve stemness of cultured rat and human neural stem cells and growth of oligodendrocyte progenitors. Methods. 2018;133:54–64. doi: 10.1016/j.ymeth.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 27.Bae SH, Ryu H, Rhee KJ, Oh JE, Baik SK, Shim KY, et al. L-ascorbic acid 2-phosphate and fibroblast growth factor-2 treatment maintains differentiation potential in bone marrow-derived mesenchymal stem cells through expression of hepatocyte growth factor. Growth Factors. 2015;33:71–78. doi: 10.3109/08977194.2015.1013628. [DOI] [PubMed] [Google Scholar]
- 28.Lynch K, Pei M. Age associated communication between cells and matrix: a potential impact on stem cell-based tissue regeneration strategies. Organogenesis. 2014;10:289–298. doi: 10.4161/15476278.2014.970089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin F, Ren XD, Pan Z, Macri L, Zong WX, Tonnesen MG, et al. Fibronectin growth factor-binding domains are required for fibroblast survival. J Invest Dermatol. 2011;131:84–98. doi: 10.1038/jid.2010.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, Adachi K, Sheridan MA, Alexenko AP, Schust DJ, Schulz LC, et al. Heightened potency of human pluripotent stem cell lines created by transient Bmp4 exposure. Proc Natl Acad Sci U S A. 2015;112:E2337–E2346. doi: 10.1073/pnas.1504778112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawkins K, Joy S, McKay T. Cell Signalling pathways underlying induced pluripotent stem cell reprogramming. World J Stem Cells. 2014;6:620–628. doi: 10.4252/wjsc.v6.i5.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luz-Madrigal A, Grajales-Esquivel E, McCorkle A, DiLorenzo AM, Barbosa-Sabanero K, Tsonis PA, et al. Reprogramming of the chick retinal pigmented epithelium after retinal injury. BMC Biol. 2014;12:28. doi: 10.1186/1741-7007-12-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li D, Qiu X, Yang J, Liu T, Luo Y, Lu Y. Generation of human lens epithelial-like cells from patient-specific induced pluripotent stem cells. J Cell Physiol. 2016;231:2555–2562. doi: 10.1002/jcp.25374. [DOI] [PubMed] [Google Scholar]
- 34.Sun M, Chi G, Li P, Lv S, Xu J, Xu Z, et al. Effects of matrix stiffness on the morphology, adhesion, proliferation and osteogenic differentiation of mesenchymal stem cells. Int J Med Sci. 2018;15:257–268. doi: 10.7150/ijms.21620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou L, Zou X, Chen L, Li H, Mygind T, Kassem M, et al. Multilineage differentiation of porcine bone marrow stromal cells associated with specific gene expression pattern. J Orthop Res. 2008;26:56–64. doi: 10.1002/jor.20467. [DOI] [PubMed] [Google Scholar]
- 36.Elsafadi M, Manikandan M, Atteya M, Hashmi JA, Iqbal Z, Aldahmash A, et al. Characterization of cellular and molecular heterogeneity of bone marrow stromal cells. Stem Cells Int. 2016;2016:9378081. doi: 10.1155/2016/9378081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hazehara-Kunitomo Y, Hara ES, Ono M, Aung KT, Komi K, Pham HT, et al. Acidic pre-conditioning enhances the stem cell phenotype of human bone marrow stem/progenitor cells. Int J Mol Sci. 2019;20:1097. [DOI] [PMC free article] [PubMed]
- 38.Nefzger CM, Polo JM. Dead-box Rna binding protein Ddx5: not a black-box during reprogramming. Cell Stem Cell. 2017;20:419–420. doi: 10.1016/j.stem.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 39.Loewer S, Cabili MN, Guttman M, Loh Y, Thomas K, Park IH, et al. Large intergenic non-coding Rna-Ror modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bourgeois CF, Auboeuf D. The Rna helicase Ddx5 is a reprogramming roadblock. Stem Cell Investig. 2017;4:79. doi: 10.21037/sci.2017.09.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Pelham-Webb B, Di Giammartino DC, Li J, Kim D, Kita K, et al. Widespread mitotic bookmarking by histone marks and transcription factors in pluripotent stem cells. Cell Rep. 2017;19:1283–1293. doi: 10.1016/j.celrep.2017.04.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Folmes CDL, Terzic A. Energy metabolism in the acquisition and maintenance of stemness. Semin Cell Dev Biol. 2016;52:68–75. doi: 10.1016/j.semcdb.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Bio. 2014;15:243–256. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lisowski P, Kannan P, Mlody B, Prigione A. Mitochondria and the dynamic control of stem cell homeostasis. EMBO Rep. 2018;19:e45432. doi: 10.15252/embr.201745432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salazar-Noratto GE, Luo G, Denoeud C, Padrona M, Moya A, Bensidhoum M, et al. Understanding and leveraging cell metabolism to enhance mesenchymal stem cell transplantation survival in tissue engineering and regenerative medicine applications. Stem Cells. 2020;38:22–33. doi: 10.1002/stem.3079. [DOI] [PubMed] [Google Scholar]
- 46.Wei P, Dove KK, Bensard C, Schell JC, Rutter J. The force is strong with this one: metabolism (over) powers stem cell fate. Trends Cell Biol. 2018;28:551–559. doi: 10.1016/j.tcb.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monteiro MS, Carvalho M, Bastos ML, Guedes DPP. Metabolomics analysis for biomarker discovery: advances and challenges. Curr Med Chem. 2013;20:257–271. doi: 10.2174/092986713804806621. [DOI] [PubMed] [Google Scholar]
- 48.Beale DJ, Pinu FR, Kouremenos KA, Poojary MM, Narayana VK, Boughton BA, et al. Review of recent developments in Gc-Ms approaches to metabolomics-based research. Metabolomics. 2018;14:152. doi: 10.1007/s11306-018-1449-2. [DOI] [PubMed] [Google Scholar]
- 49.Halket JM, Waterman D, Przyborowska AM, Patel RK, Fraser PD, Bramley PM. Chemical derivatization and mass spectral libraries in metabolic profiling by Gc/Ms and Lc/Ms/Ms. J Exp Bot. 2005;56:219–243. doi: 10.1093/jxb/eri069. [DOI] [PubMed] [Google Scholar]
- 50.Semreen MH, Alniss H, Cacciatore S, El-Awady R, Mousa M, Almehdi AM, et al. Gc-Ms based comparative metabolomic analysis of Mcf-7 and Mda-Mb-231 cancer cells treated with tamoxifen and/or paclitaxel. J Proteome. 2020;225:103875. doi: 10.1016/j.jprot.2020.103875. [DOI] [PubMed] [Google Scholar]
- 51.Wan X, Li X, Wang Q, Zheng B, Zhou C, Kang X, et al. Metabolitic profiling of amino acids in paraquat-induced acute kidney injury. Clin Exp Nephrol. 2019;23:474-83. [DOI] [PubMed]
- 52.Pallio G, Micali A, Benvenga S, Antonelli A, Marini HR, Puzzolo D, et al. Myo-inositol in the protection from cadmium-induced toxicity in mice kidney: an emerging nutraceutical challenge. Food Chem Toxicol. 2019;132:110675. doi: 10.1016/j.fct.2019.110675. [DOI] [PubMed] [Google Scholar]
- 53.Teixeira FC, Gutierres JM, Soares MSP, Da Siveira De Mattos B, Spohr L, Do Couto CAT, et al. Inosine protects against impairment of memory induced by experimental model of Alzheimer disease: a nucleoside with multitarget brain actions. Psychopharmacol. 2020;237:811-23. [DOI] [PubMed]
- 54.Kobayashi CI, Suda T. Regulation of reactive oxygen species in stem cells and cancer stem cells. J Cell Physiol. 2012;227:421–430. doi: 10.1002/jcp.22764. [DOI] [PubMed] [Google Scholar]
- 55.Simburger J, Dettmer K, Oefner PJ, Reinders J. Optimizing the swath-Ms-workflow for label-free proteomics. J Proteome. 2016;145:137–140. doi: 10.1016/j.jprot.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 56.Mareddy S, Broadbent J, Crawford R, Xiao Y. Proteomic profiling of distinct clonal populations of bone marrow mesenchymal stem cells. J Cell Biochem. 2009;106:776–786. doi: 10.1002/jcb.22088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Materials and Methods.
Additional file 5: Table S4 and Table S5.
Additional file 6: Supplementary Figures.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.