Abstract
Objective
Large clinical databases are increasingly used for research and quality improvement. We describe an approach to data quality assessment from the General Medicine Inpatient Initiative (GEMINI), which collects and standardizes administrative and clinical data from hospitals.
Methods
The GEMINI database contained 245 559 patient admissions at 7 hospitals in Ontario, Canada from 2010 to 2017. We performed 7 computational data quality checks and iteratively re-extracted data from hospitals to correct problems. Thereafter, GEMINI data were compared to data that were manually abstracted from the hospital’s electronic medical record for 23 419 selected data points on a sample of 7488 patients.
Results
Computational checks flagged 103 potential data quality issues, which were either corrected or documented to inform future analysis. For example, we identified the inclusion of canceled radiology tests, a time shift of transfusion data, and mistakenly processing the chemical symbol for sodium (“Na”) as a missing value. Manual validation identified 1 important data quality issue that was not detected by computational checks: transfusion dates and times at 1 site were unreliable. Apart from that single issue, across all data tables, GEMINI data had high overall accuracy (ranging from 98%–100%), sensitivity (95%–100%), specificity (99%–100%), positive predictive value (93%–100%), and negative predictive value (99%–100%) compared to the gold standard.
Discussion and Conclusion
Computational data quality checks with iterative re-extraction facilitated reliable data collection from hospitals but missed 1 critical quality issue. Combining computational and manual approaches may be optimal for assessing the quality of large multisite clinical databases.
Keywords: data quality; data validation; clinical databases; data extraction, electronic medical records
INTRODUCTION
Routinely collected clinical and administrative health data are increasingly being used in large databases for research and quality improvement.1–4 In many health systems, the electronic data in hospitals are stored in a complex array of repositories with limited central oversight or standardization. The data extraction process may result in data errors. Ensuring data quality post-extraction is challenging but important,5,6 particularly given the impact that large clinical databases might have on quality improvement, research, and policy.
There are several widely cited frameworks used to assess data quality.7–9 A systematic review by Weiskopf and colleagues identified 5 key data quality dimensions: completeness, correctness, concordance, plausibility, and currency.10 The methodological approach to examining each dimension remains challenging and poorly described.11 Manual data validation may be important to ensure data quality.12 This typically involves manually abstracting a subset of data directly from the source and comparing this to electronically extracted data.13 However, manual chart reviews are resource-intensive13–15 and difficult to scale as databases grow in size.6 Computational data quality assessment has been proposed as an alternative method of ensuring data quality.16 Computational data quality assessment techniques have been explored by groups such as the National Patient-Centred Clinical Research Network, and may include application of thresholds to ensure plausibility (eg, birth date cannot be a future date), visual inspection of linear plots (eg, to assess temporal trends), and outlier detection formulas.7,17,18
OBJECTIVE
Our objective was to describe the iterative combination of computational data quality assessment followed by manual data validation in a large, real-world, clinical database, the General Medicine Inpatient Initiative (GEMINI). The purpose of this data quality assessment was to identify data quality issues arising from data extract-transform-load and transfer processes required to create the GEMINI database. This case study offers insights for approaches to assess data quality in large datasets based on routinely collected clinical data.
METHODS
Overall approach
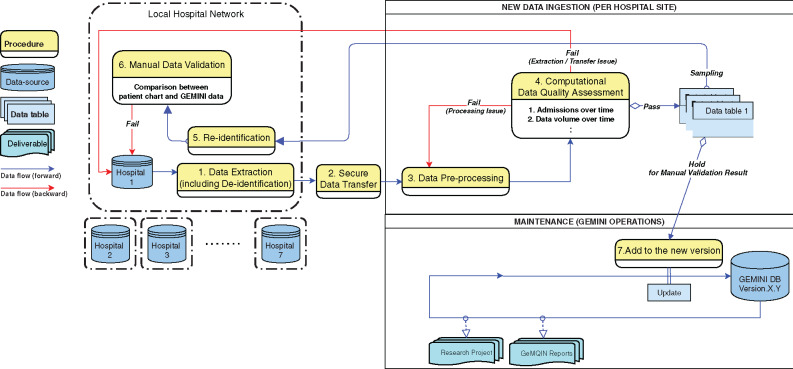
The creation of the GEMINI database involved (Figure 1): 1) extracting and deidentifying data from participating hospitals, 2) transferring it to a central site, 3) processing data centrally (eg, standardizing variables across sites), 4) performing computational data quality assessment for structured (non-narrative) variables, 5) resolving identified data quality issues, which sometimes required data to be reidentified to investigate problems; and, finally, 6) manually validating a sample of selected structured (nonnarrative) variables before 7) updating the GEMINI database.
Figure 1.
Data flows and processes for the GEMINI database. The following steps are involved in preparing the GEMINI database: 1. Data are extracted from hospitals and deidentified. 2. Data are securely transferred to the central site. 3. Data are processed, including standardization and reformatting of variables. 4. Data undergo computational data quality checks, which are manually reviewed by a data analyst to flag potential issues. Data processing issues are resolved within the central site by addressing the problematic processing step. Data extraction/transfer issues are addressed at the hospital sites and may require data re-extraction. 5. Data are sampled from the central repository and sent back to the hospital site for reidentification. 6. Manual data validation is performed using the reidentified sample. Data quality issues are investigated and resolved as possible. 7. Finally, data are added to the GEMINI database with all data quality issues either resolved or clearly documented to inform future use. GeMQIN: General Medicine Quality Improvement Network.
We define computational data quality assessment as a series of data quality checks based on visualizations and tabulations of data that are reviewed manually to assess whether data are complete and plausible. We define manual data validation as a comparison between data in the GEMINI database and data that are manually collected from individual patient records. Both computational data quality assessment and manual data validation sought to identify data quality issues arising from extract-transform-load and transfer processes, rather than issues integral to the data itself (eg, errors in data entry into medical records). Our efforts focused on structured data variables rather than narrative (free text) data.
Setting
The General Medicine Inpatient Initiative (GEMINI) database collects administrative and clinical data for all patients admitted to or discharged from the general medicine inpatient service of 7 hospital sites affiliated with the University of Toronto in Toronto and Mississauga, Ontario, Canada.19 The 7 hospitals include 5 academic health centers and 2 community-based teaching sites. At the time of this manuscript, data had been collected for 245 559 hospitalizations with discharge dates between April 1, 2010 and October 31, 2017.
GEMINI supports both research and quality improvement applications. The Ontario General Medicine Quality Improvement Network (GeMQIN)20 uses data from the GEMINI database to create confidential individualized audit-and-feedback reports for eligible physicians at participating hospital sites.21 The first version of these reports included 6 quality indicators (hospital length-of-stay, 30-day readmission, inpatient mortality, use of radiology tests, use of red blood cell transfusions, and use of routine blood work), and directly informed frontline quality improvement efforts. Our data quality assessment efforts prioritized data variables that were included in these physician audit-and-feedback reports.
Ethics
Research Ethics Board approval was obtained from each participating institution.
Data extraction, transfer, and processing
Data extraction occurred over 2 cycles: 1) patients discharged between April 1, 2010 and March 31, 2015 and 2) patients discharged between April 1, 2015 and October 31, 2017.4 A wide range of administrative and clinical data were extracted from a variety of data sources (Table 1): patient demographics, diagnostic codes, intervention procedure codes, admission and discharge times, cost of admission, test results from biochemistry, hematology, and microbiology laboratories, radiology reports, in-hospital medication orders, vital signs, and blood transfusions. A template document describing requested data formats and standards was provided to each hospital site to facilitate data extraction. Data were extracted by local hospital staff and stored locally in shared network folders.
Table 1.
GEMINI database structure
| Source | Data table | Description |
|---|---|---|
| Discharge Abstract Databasea | Admission | Inpatient administrative information |
| Discharge Abstract Databasea | Diagnosis | Inpatient discharge diagnoses, classified using ICD-10 codes. |
| Discharge Abstract Databasea | Intervention | Inpatient interventions, classified based on CCI code |
| Discharge Abstract Databasea | Case Mix Group | Patient risk groups based on case mix grouping methodology from the Canadian Institute for Health Information |
| Discharge Abstract Databasea | Special Care Unit | Use of critical care and other special care units |
| Discharge Abstract Databasea | HIG Weight | Patient risk groups based on Ontario-specific grouping methodology |
| National Ambulatory Care Reporting Systema | ED Admission | Emergency department administrative information |
| National Ambulatory Care Reporting Systema | ED Consults | Consultations performed in the emergency department |
| National Ambulatory Care Reporting Systema | ED Diagnosis | Diagnoses based on emergency department visit, classified using ICD-10 codes. |
| National Ambulatory Care Reporting Systema | ED Intervention | Interventions in the emergency department, classified based on CCI code |
| National Ambulatory Care Reporting Systema | ED Case Mix Group | Emergency department case mix grouping methodology from the Canadian Institute for Health Information |
| Hospital ADT system | Admission-Discharge | Admission date, time, and service information |
| Hospital ADT system | Transfers | In-hospital room transfer information |
| Hospital ADT system | Physicians | Physician information for each admission |
| Hospital Electronic Information System | Echocardiography | Echocardiography data |
| Hospital Electronic Information System | Laboratory | Hematology and biochemistry laboratory data |
| Hospital Electronic Information System | Microbiology | Microbiology laboratory data |
| Hospital Electronic Information System | Pharmacy | In-hospital medication orders |
| Hospital Electronic Information System | Radiology | Radiology data |
| Hospital Electronic Information System | Transfusion | Blood transfusion data |
| Hospital Electronic Information System | Vitals | Vital signs data |
Abbreviations: ADT, admission, discharge, transfer; CCI, Canadian Classification of Health Interventions; HIG, Health Based Allocation Model (HBAM) Inpatient Group; ED, Emergency Department; ICD-10, 10th revision of the International Statistical Classification of Diseases and Related Health Problems.
Data are extracted from hospital sites based on what each hospital site reports to the Canadian Institute for Health Information for the Discharge Abstract Database and National Ambulatory Care Reporting System.
After data were extracted, GEMINI staff deidentified the data at each hospital site and then securely transferred the data to a central repository. Deidentification was performed by removing personally identifying variables from the dataset (medical record number, encounter number, date of birth, first/middle/last name, postal code, health card number, and emergency room registration number).22 The remaining dataset included no direct patient identifiers. Each patient admission was assigned a unique identifier that allowed for integration of various data tables at the central data repository and reidentification at the local hospital site for the purpose of manual validation.23 A secure hash algorithm was applied to each patient’s provincial health insurance number, which allowed us to link encounters for the same patient across multiple institutions.24
In the central repository, GEMINI data were processed and organized into 21 linkable data tables from each hospital site in order to group data into related categories (Table 1). Data were processed to ensure consistent formatting across hospital sites. Data quality was then iteratively improved by computational data quality assessment with re-extraction as needed.
Computational data quality assessment
The computational data quality assessments consisted of 7 checks. Checks 1–4 were designed to identify errors in data completeness associated with data extraction and transfer procedures and were applied to each data table. Checks 5–7 consisted of detailed inspection of select variables in each table (eg, sodium test result values in laboratory data table). Each check assessed different dimensions of data quality (Table 2). The checks are described in detail below:
Table 2.
Summary of computational data quality checks
| Check | Name | Level | Description | Quality Dimensions Assessed |
|---|---|---|---|---|
| 1 | Patient admissions over time | Data table | Proportion of all GEMINI patient admissions that are present in the data table, by date | Completeness |
| 2 | Data volume over time | Data table | Data volume per data table, by date | Completeness |
| 3 | Admission-specific data volume over time | Data table | Data volume per patient admission, by date |
Completeness Plausibility |
| 4 | Distribution of data in relation to admission and discharge times | Data table | Data volume in relation to admission time and discharge time |
Completeness Plausibility |
| 5 | Variable presence over time | Variable | Heat map of missing data for each variable, by date |
Completeness Plausibility |
| 6 | Specific variable presence over time | Variable | Proportion of data within a data table that pertains to a certain variable after data processing, by date |
Completeness Plausibility |
| 7 | Accuracy check | Variable |
Categorical variables: report frequencies Numeric variables: report measures of central tendency and distribution |
Correctness Concordance Plausibility |
Note: Seven computational data quality assessment checks were conducted on entire data tables of specific variables prior to manual validation to assess different dimensions of data quality (reported in a systematic review by Weiskopf and colleagues).10 We describe each check in detail under the Methods section. A data analyst visually reviewed each check to identify issues (unexpected results, outliers, disruptions in trends), which then underwent a stepwise approach including any data re-extraction, communication with site-specific IT personnel, and manual chart review to correct the issue.
Admissions over time: We examined the number of patient admissions meeting the inclusion criteria at each hospital site, which were subsequently used to extract data from source systems. We produced a histogram of the proportion of patient admissions that are contained in each data table over time, based on discharge date. This explored whether data for patient admissions were systematically missing from any data table. (Supplementary Appendix S2Figure 1)
Data volume over time: We examined the amount of data (as measured by number of rows/observations for each data table) by producing a histogram of total data volume by date and time (if available). In contrast to Check 1, this check assessed missing data, rather than missing patient admissions. Temporal patterns of missingness as well as variations in trends were carefully inspected. (Supplementary Appendix S2Figure 2)
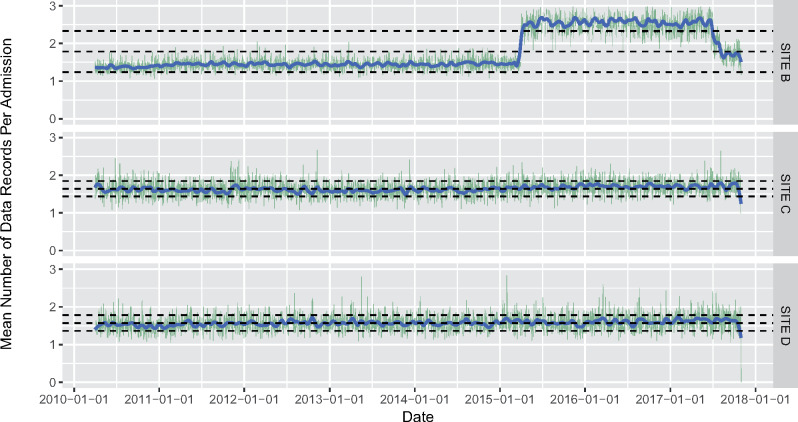
Admission-specific data volume over time: We examined the total volume of data (as measured by number of rows/observations for each data table) per patient admission to eliminate variation in data volume that may be driven by variations in the number of patient admissions over time. We produced a line graph (with a moving average) of data volume per patient admission by date and time (if available). (Figure 2)
Distribution of data in relation to admission and discharge times: We examined the distribution of date and time labels on variables in each data table compared to the patient’s admission and discharge date and time (eg, the difference in time between a radiology test and the patient’s admission date and time). This examined whether date and time information collected were plausible and ensured that we were not missing data from a specific portion of a patient’s hospitalization (eg, emergency department stay). (Figure 3)
Overall variable presence over time: We inspected the missingness of every variable from each data table using a heat map. The y-axis was populated with the list of variables in each data table and the x-axis was populated with each row of data in each table, sequenced by admission date. (Supplementary Appendix S2Figure 3). This allowed us to identify temporal patterns and clustering in variable missingness.
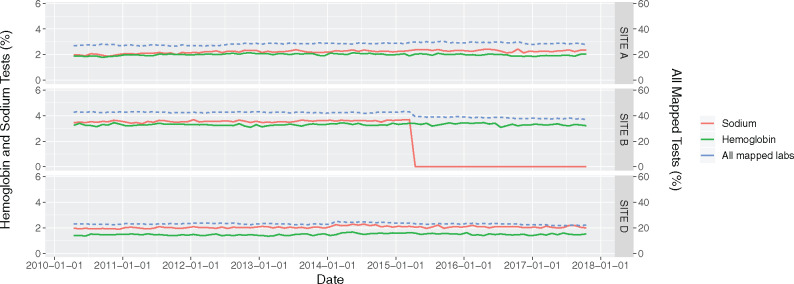
Specific variable presence over time: We assessed the quality of data categorization and standardization for a specific variable. Because naming conventions differ at each hospital site (eg, for radiology or laboratory tests) and can change within hospital sites over time, data are manually mapped to categories or standard naming conventions to permit multisite analysis. For each variable, we produced a line graph of the proportion of mapped data volume at each hospital site by date. Visual inspection of data volume over time allowed us to identify problems with mapping and standardization. (Figure 4)
Plausibility check: Each variable was inspected to ensure it contained plausible values. For categorical variables, we examined frequency tables. For numerical interval or continuous variables, we computed distributions and measures of central tendency (minimum, mean, median, maximum, and interquartile range). Specific data tables such as laboratory tests required further ad-hoc analysis. For example, for each laboratory test type, we inspected the proportion of non-numeric test results, measurement units, and the numerical distribution of test result values.
Figure 2.
Data volume of radiology tests per patient admission by date at 3 sites. This is an example of Computational Data Quality Check 3: The x-axis represents the date and the y-axis represents the mean number of radiology tests per patient admission. Sites C and D represent the expected situation, where the number of radiology procedures per patient admission (green lines) and the moving average (blue line) are relatively consistent over time, and within the prespecified thresholds (bottom and top grey dotted lines represent 1 standard deviation away from the overall mean). The sharp increase in the average number of tests at Site B between 2015 and 2017 identifies a potential issue, as this level of increase is implausible. Re-extraction did not correct the issue initially. Subsequent manual investigation identified that the extraction had inadvertently included canceled radiology tests. After successful data re-extraction, without the inclusion of canceled tests, the aberrancy in the plot was no longer present.
Figure 3.
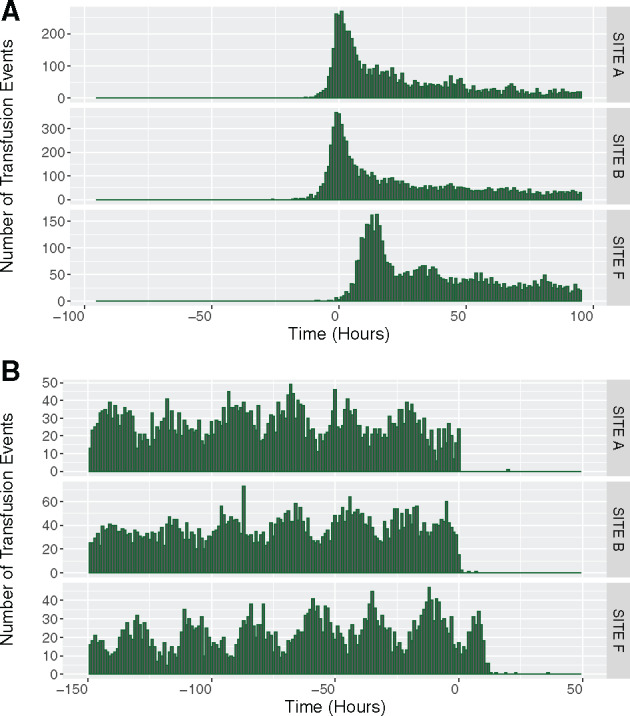
(A) Distribution of the time between transfusion events and the time of admission across 3 hospital sites. (B). Distribution of the time between transfusion events and the time of discharge across 3 hospital sites. This is an example of Computational Data Quality Check 4: The x-axis represents the time of the transfusions event in relation to the admission (Figure 3A—negative values indicating a time before admission date and time) and discharge (Figure 3B—positive values indicate a time after discharge date and time). The y-axis represents the volume of transfusion events that occurred at the time on the x-axis. The plots demonstrate that at Site F, there are no transfusion events prior to admission (at Time < 0 Hours). Upon first glance, this suggested missing emergency department transfusion data (ie, patient in the emergency room and receives a transfusion prior to the admission order) but the issue could not be corrected after re-extraction efforts and discussions with the site data extraction team. The plots also indicate that Site F conducted many transfusions post-discharge (at Time > 0 Hours), which would be unlikely. The combination of time from admission, time to discharge, and subsequent manual investigation uncovered a time shift for transfusion data at Site F. This time shift was then corrected in the GEMINI database.
Figure 4.
Volume of hemoglobin and sodium laboratory test data over time. This is an example of Computational Data Quality Check 6: The x-axis represents the date. The left y-axis represents the percentage of hemoglobin and sodium tests out of all laboratory tests. The right y-axis represents the percentage of all laboratory data that has been standardized across sites (blue line, many less common laboratory tests had not yet been standardized). There is a sudden decrease in the volume of sodium tests after April 1, 2015, at Site B corresponding to the second round of data extraction. Further investigation revealed that statistical software using default settings mistakenly processed the symbol for sodium “Na” as a null/missing value.
Data analysts manually reviewed each computational quality check to identify any potential issues of concern. Data analysts had graduate (Master’s or PhD) training in biostatistics, epidemiology, or bioinformatics. Clinician investigators were asked to review potential data quality issues and provide input as needed.
Potential data quality issues could be identified as unexpected results, outliers, and disruptions in trends. These potential data quality issues were subsequently investigated through an iterative process. We first examined the original deidentified extracted data table to check if there were any errors with data processing (ie, formatting, mislabeling, etc). We next examined the extracted files at each hospital to determine whether there were any errors with deidentification or transfer (ie, not all files transferred correctly). If the issue remained, we then worked with the hospital site to re-extract the problematic data and we then performed the computational data quality check again. If the issue persisted, a data abstractor manually reviewed data of individual problem cases directly from the patient records in the hospital electronic information system to identify differences between extracted data and the electronic information system. They then reported these differences to the local hospital site IT personnel to help fix the data quality issue. These processes typically led to correction of the data quality issue, after which time the computational data quality check was repeated. If these steps did not correct the data quality issue, the data remained in the GEMINI database but that specific data table was flagged such that it would not be used to inform any quality improvement efforts or academic research. Computational data quality checks occurred in the central data repository and did not require direct access to the source data at individual hospital sites.
Evaluation of computational data quality assessment
To evaluate the utility of this computational data quality assessment process, we report the number of potential data quality issues flagged and how many of these ultimately needed correction or were important to be aware of. We also report whether data quality issues were related to extraction/transfer or central processing. We categorize the potential impact of each data quality issue as small (affecting less than 0.5% of admissions or a single infrequently used variable), large (affecting more than 50% of admissions or a commonly used variable), or moderate (between small and large). Finally, we report the time that data quality analysts spent per data table for each hospital, which includes the time to run the computational check (using quadcore Intel Core i5-8350U with 16 GB of RAM, running on Windows 10 version 1809 using R version 4.0.1) and review the visualization to identify potential issues but not the time required to investigate and resolve data quality issues. Ultimately, manual data validation was used to determine the overall quality of the database after improvement based on computational quality assessment.
Manual data validation
Manual data validation was performed on the GEMINI database after improvement based on computational data quality assessment and iterative re-extraction from hospitals. Manual validation was performed by a medical graduate (MBBS) with 2 years of clinical residency training and who was trained on the use of the electronic medical record at participating hospitals. GEMINI data were compared to data that could be accessed through each hospital’s electronic medical record (ie, the information that clinicians see when providing care), which was taken to be the gold standard. We focused primarily on data that were included in the physician audit-and-feedback reports. Thus, we manually validated data from 6 data tables (laboratory, radiology, physicians, death, transfers, and transfusion). Two cycles of manual validation occurred, corresponding to each cycle of data extraction. Because 2 hospital sites share a single electronic information system, we grouped them together and therefore refer to 6 hospital sites (A-F) for manual data validation.
A data abstractor manually reviewed data for a sample of patient admissions at each of the 6 sites. The data abstractor was not given access to the patient admission data stored in the GEMINI database meaning that they were “blinded” to any expected results. For 2010–2015 data, admissions were sampled randomly and with a more targeted approach to ensure sufficient sampling of rare events (Supplementary Appendix S1):
100 random admissions per site for laboratory data table (specific variables were haemoglobin, sodium, creatinine, calcium, aspartate transaminase, international normalized ratio, and troponin)
100 random admissions per site for radiology data table (specific variables were computed tomography, plain radiography, ultrasound, magnetic resonance imaging, and echocardiography)
200 random admissions per site for physician data table (specific variables were admitting physician name and discharging physician name)
800 selected admissions per site for death, transfers, and transfusions data tables (specific variables being mortality, critical care unit transfer, and red blood cell transfusions, respectively)
For each of the aforementioned data tables, we validated structured non-narrative data. For laboratory, radiology, death, transfer, and transfusion tables, we validated the occurrence of the test or clinical event (death, transfer, or transfusion). For laboratory tests, we also validated the actual value of the test result. For radiology, we validated that an abdominal ultrasound was performed on a certain date, but not the findings of the radiology report. For the physician table, because attending physicians must sign off on admission and discharge notes, we used the physician signature on these notes to validate the extracted admitting/discharging physician data.
Because we performed extensive manual data validation in the first cycle, we adopted a more targeted approach in the second cycle (2015–2017 data). For the second cycle, data were manually abstracted from a random sample of 5 admissions for each physician who would be receiving an audit-and-feedback report (approximately 20–30 physicians at each site corresponding to 100–150 admissions at each site).
The data abstractor recorded the result and date and time for the first (or only) occurrence of each specific variable to be validated in the hospital’s electronic medical record for each patient admission. This was considered the gold standard and was compared against the data in the GEMINI database. To avoid potential human recording errors, the data abstractor double-checked the electronic medical record in the event of a discrepancy between the GEMINI database and the manually recorded data. We report both cycles of data validation together. We calculated the sensitivity, specificity, positive predictive value, and negative predictive value of the GEMINI database for each data table and for individual variables, overall and stratified by hospital site. We also calculated the overall accuracy (true values ÷ total values) for each data table. Analyses were performed using R version 4.0.1.
RESULTS
Computational data quality assessment
Computational data quality assessment flagged 103 potential data quality issues. All of these potential data quality issues were ultimately determined to be real issues that required fixing or were important to recognize for future use of the dataset. Each of the 7 computational data quality assessment checks successfully identified at least 1 data quality issue (Table 3). For example, check 3 (admission-specific data volume over time) identified a sharp implausible increase in radiology tests at Site B that resulted from the inappropriate inclusion of canceled radiology tests in extracted data (Figure 2). Check 4 (distribution of data with admission and discharge times) identified an implausible time shift of transfusion data at Site F, which made it appear as if patients received transfusions after discharge (Figure 3). Check 5 (variable presence over time) identified a data processing and mapping issue whereby the raw hospital sodium test code was interpreted as “null” or “missing” during the second cycle of data extraction. This occurred because the abbreviation, “NA” (Figure 4), which refers to sodium in chemical notation, was interpreted as missing by our statistical software.
Table 3.
Examples of errors identified using computational data quality checks
| Quality Check | Example of Data Quality Issue Identified | Cause of Issue | Manual chart review required to investigate and fix (Yes or No) |
|---|---|---|---|
| 1. Patient admissions over time | Missing ED data extraction for certain patient admissions in 2015 at 1 site (Supplementary Appendix S2Figure 1) | Data extraction/transfer | No |
| 2. Data volume over time | Missing transfusion data during 2 time periods at 1 institution (Supplementary Appendix S2Figure 2) | Data extraction/transfer | No |
| 3. Admission-specific data volume over time | Incorrectly included canceled radiology tests resulting in sharp increase in radiology tests at 1 institution (Figure 1) | Data extraction/transfer | Yes |
| 4. Distribution of data in relation to admission and discharge times | Transfusion data at 1 site was time-shifted, leading to the timestamp on certain transfusions being after patient discharge (Figure 2) | Data extraction/transfer | Yes |
| 5. Overall variable presence over time | Glasgow Coma Scale data at 1 institution was missing (Supplementary Appendix S2Figure 3) | Unclear cause at this time | Yes |
| 6. Specific variable presence over time | At site B, “NA” was used to describe sodium tests. At the data processing stage, statistical software recognized this as a null/missing value. (Figure 3) | Data processing at central repository | No |
| 7. Plausibility check | No quality issues identified | Data extraction/transfer | No |
Note: Each computational data quality assessment check identified a data quality issue, the causes of which were determined to be data extraction/transfer, data processing, or hypothetically local deidentification (though we did not find an example of this). Some issues that were identified by the computational checks required a manual chart review to fix, while others did not.
Of the 103 total issues identified, 90 could be attributed to data extraction/transfer, and 13 to data processing. We provide examples of these issues in Table 3. Most (N = 53, 51%) data quality issues were classified as having a small impact on overall database quality, whereas 10 issues (10%) and 40 issues (39%) would have moderate or large impact, respectively.
The runtime required to perform computational data quality assessment per data table at each hospital site (including loading the data table and generating visualizations or tabulations) ranged from 30 seconds (eg, Special Care Unit data table with 12 224 data points for 2706 admissions) to 5 minutes (eg, lab data table with about 90 million data points for 28 946 admissions). It required approximately 5 minutes of data analyst time per data table per hospital to flag potential issues by reviewing the computational visualizations and tabulations.
Manual data validation
Across all sites, data tables, and both cycles of manual validation, 23 419 data points were manually abstracted from 7488 patient admissions. The specific number of data points for each data table and each specific variable at each site is listed in Supplementary Appendix S2.
Compared to the gold standard of data manually abstracted from an electronic medical record, the GEMINI database was found to be highly sensitive (Table 4), ranging from 95%–100% across data tables, and highly specific, ranging from 99%–100% across data tables. The database was also found to have high positive and negative predictive values, with overall results ranging from 93–100% and 99–100%, respectively, across data tables. The overall accuracy of the database was found to be 98%–100% across data tables.
Table 4.
Manual data validation results for each data table, overall and stratified by site
| Variable | Laboratory | Radiology | Physicians | Death | Transfer | Transfusion | |
|---|---|---|---|---|---|---|---|
| Overall | N | 5648 | 5092 | 2449 | 3814 | 3300 | 3116 |
| Accuracy | 100% | 100% | 98% | 100% | 100% | 98% | |
| Sens. | 100% | 100% | 98% | 100% | 100% | 95% | |
| Spec. | 100% | 100% | a | 100% | 99% | 99% | |
| PPV | 100% | 100% | 100% | 100% | 96% | 93% | |
| NPV | 100% | 100% | a | 100% | 100% | 99% | |
| Hospital Site A | N | 1300 | 1000 | 400 | 800 | 800 | 800 |
| Sens. | 100% | 99% | 94% | 100% | 100% | 100% | |
| Spec. | 100% | 100% | a | 100% | 100% | 100% | |
| PPV | 100% | 100% | 99% | 100% | 100% | 100% | |
| NPV | 100% | 100% | a | 100% | 100% | 100% | |
| Hospital Site B | N | 950 | 820 | 400 | 90 | b | 90 |
| Sens. | 100% | 100% | 98% | 100% | b | 95% | |
| Spec. | 100% | 100% | a | 100% | b | 97% | |
| PPV | 100% | 100% | 100% | 100% | b | 90% | |
| NPV | 100% | 100% | a | 100% | b | 99% | |
| Hospital Site C | N | 960 | 1040 | 560 | 980 | 800 | 982 |
| Sens. | 100% | 100% | 100% | 100% | 100% | 100% | |
| Spec. | 100% | 100% | a | 100% | 99% | 100% | |
| PPV | 100% | 100% | 100% | 100% | 91% | 97% | |
| NPV | 100% | 100% | a | 100% | 100% | 100% | |
| Hospital Site D | N | 860 | 740 | 360 | 280 | 200 | 280 |
| Sens. | 100% | 100% | 98% | 100% | 100% | 66% | |
| Spec. | 100% | 100% | a | 100% | 100% | 95% | |
| PPV | 100% | 100% | 100% | 100% | 100% | 75% | |
| NPV | 100% | 100% | a | 100% | 100% | 93% | |
| Hospital Site E | N | 588 | 707 | 339 | 769 | 700 | 69 |
| Sens. | 100% | 100% | 99% | 100% | 100% | 100% | |
| Spec. | 100% | 100% | a | 100% | 99% | 95% | |
| PPV | 100% | 100% | 99% | 100% | 95% | 80% | |
| NPV | 100% | 100% | a | 100% | 100% | 100% | |
| Hospital Site F | N | 890 | 785 | 390 | 895 | 800 | 895 |
| Sens. | 100% | 99% | 99% | 100% | 100% | 96% | |
| Spec. | 100% | 100% | a | 100% | 100% | 99% | |
| PPV | 100% | 99% | 99% | 100% | 98% | 92% | |
| NPV | 100% | 100% | a | 100% | 100% | 99% |
Note: N=Number of data points, which is distinct from number of patient admissions. For example, at site A, 1000 radiology data points were manually checked, but these came from 200 patient admissions for each type of radiology test.
For physician variables (accuracy of admitting and discharging physician names), specificity and negative predictive value cannot be calculated given the lack of true negatives (ie, every individual should theoretically have an admitting and discharging physician, making the number of true negatives equal to 0).
Site B used paper charts for critical care transfer thus manual data validation was not feasible.
The following formulas/definitions were used:
Sensitivity = true positives ÷ (true positives + false negatives)
Specificity = true negative ÷ (true negatives + false positives)
PPV (positive predictive value) = true positives ÷ (true positives + false positives)
NPV (negative predictive value) = true negatives ÷ (true negatives + false negatives)
True positives = number of admissions for which a test or clinical event occurred on a specific day according to both the hospital electronic medical records and GEMINI database
True negatives = number of admissions for which a test or clinical event did not occur according to both hospital electronic medical records and the GEMINI database
False positives = number of admissions for which a test or clinical event occurred on a specific day according to the GEMINI database, but not according to the electronic medical record
False negatives = number of admissions for which a test or clinical event occurred on a specific day according to the electronic medical record, but not according to the GEMINI database
Manual data validation identified 1 important data quality issue that was not flagged by computational approaches. Specifically, blood transfusion data at Site D had poor sensitivity (66%) and positive predictive value (75%) because of problems with the date and time at which the transfusions reportedly occurred.
DISCUSSION
This article reports the experience of an extensive data quality assessment effort involving a broad range of administrative and clinical data extracted from 7 hospital sites (with 6 different IT systems) over 8 years. We highlight a feasible and pragmatic approach to computational data quality assessment and illustrate how various data quality issues were identified. Thereafter, manual validation of over 23 000 data points found that the GEMINI database had an overall accuracy of 98%–100% compared with hospital electronic medical records. Our experience suggests that although computational data quality checks are effective, they may not identify all important data quality issues. Specifically, we identified crucial data quality issues in blood transfusion data at 1 hospital site that were not detected through computational data quality checks. The GEMINI experience suggests that computational and manual approaches should be used together to iteratively improve and validate databases that are extracted from clinical information systems.
Data quality assessment is crucial before routinely collected data can be used for secondary purposes, such as research or quality improvement.25 One flexible approach is to ensure that data are “fit-for-purpose” by data consumers, which has informed numerous frameworks and models for data quality assessment.7,9,10,26,27 However, less has been published about how to operationalize these approaches, particularly in multisite clinical datasets.7,28,29 Kahn and colleagues describe a conceptual model and a number of computational rules to explore data quality in electronic health record-based research.7 Similarly, van Hoeven and colleagues articulate an approach to assessing the validity of linked data using computational methods and report its application in a specific case using transfusion data.28 Terry and colleagues developed 11 measures of quality for primary health care data extracted from electronic medical records.29 Each of these studies admirably documents the process of operationalizing conceptual data quality frameworks into real-world applications. These studies all focus on computational quality checks. Conversely, Baca and colleagues report a data validation effort of the Axon Registry, a clinical quality data registry, and focus entirely on the validation effort but do not describe computational efforts to assess and improve data quality within the registry.30 We have been unable to find any studies that report the experience of using both computational and manual approaches to data quality assessment. Our study extends the literature by operationalizing broad data quality domains, specifically using computational data checks to inform iterative data quality improvement, and reporting the effectiveness of this approach based on rigorous manual validation across a range of data types and healthcare organizations.
The main implication of our study is that computational quality checks can identify most data quality issues but not all. Blood transfusion data in our study highlight the strengths and weaknesses of computational checks. Computational checks identified that transfusion data were time-shifted at 1 hospital because some blood transfusions were apparently administered after patient discharge, which was implausible. However, computational checks missed major inaccuracies in the dates and times of blood transfusions at another hospital because the errors were not systematic and did not create any discernible patterns. The GEMINI experience suggests that although manual data validation is labor-intensive, it may be necessary to ensure high-quality data. Starting with computational checks and targeting manual data validation with a “fit-for-purpose” strategy (as we did in our second data extraction cycle when we validated a smaller sample based on physician quality reports) may minimize manual workload without sacrificing data quality.
Limitations
Our study is limited in several ways. First, we only performed manual validation on certain data tables, stemming from our “fit-for-purpose”26 approach focused on ensuring high-quality data for physician audit-and-feedback reports. Given that this included a breadth of data tables, we feel that our findings are likely generalizable to other data tables within GEMINI. Second, our approach involved manual assessment (by a data analyst) of computational data quality checks to flag potential data quality issues. Other researchers have generated more automated statistical detection models that aim to computationally detect potential data quality issues, and incorporating these methods is an area of future work for our program.31 Third, our approach to manual data validation cannot address data quality issues at the source system (eg, missing vital signs values related to incomplete clinical documentation) as these are already embedded into the data, which form our “gold standard.” Databases like GEMINI, which rely on routinely generated clinical data, are thus subject to the inherent data quality and accuracy problems that arise in routine clinical care and its documentation. An interesting avenue of future research could be to compare routinely generated clinical data with prospectively gathered data to strengthen understanding of how routine data may be flawed or biased. Fourth, we examined structured data variables but not variables that included narrative text, which may raise a number of additional data quality concerns.32 Finally, we focused on traditional domains of data quality, but future research could further assess nontraditional aspects such as context, representation, and accessibility.27
Lessons learned
Our experience has highlighted several lessons that might inform future work related to data quality in real-world clinical datasets:
Computational data quality assessment can successfully identify numerous data quality issues, particularly for structured data variables, and should be included in the development of health-related datasets.
The aspects of data quality that were most easily assessed computationally were completeness and plausibility; to a lesser extent it was possible to assess concordance and correctness.
Computational approaches were most effective at detecting systematic errors (eg, a large chunk of missing data or all dates/times shifted by a fixed amount). Non-systematic (eg, dates/times inaccurate by varying or random amounts) data quality issues were difficult to identify without some form of manual validation.
Nearly 90% of data quality issues were related to data extraction and transfer from hospitals rather than processing at the central site. Correcting these issues required strong support from staff and leadership at participating hospitals.
Because manual data validation is labor intensive, we were able to reduce the number of data points to be validated by focusing on key data variables through a “fit-for-purpose” approach. We also resolved data quality issues identified through computational checks prior to manual validation, to reduce the need for duplicating manual validation after data issues were corrected.
CONCLUSION
The GEMINI experience highlights the importance of an iterative data quality assessment methodology that sequentially combines computational and manual techniques. Through this approach, GEMINI has achieved highly reliable data extraction and transfer procedures from hospital sites. Our findings demonstrate that computational data quality assessment and manual data validation are complementary, and combining these should be the ideal method to assess the quality of large clinical databases. Future research should focus on methods to reduce the amount of manual validation that is needed, and to assess nontraditional aspects of data quality.
FUNDING
This work was supported by Green Shield Canada Foundation and University of Toronto Division of General Internal Medicine. FR is supported by an award from the Mak Pak Chiu and Mak-Soo Lai Hing Chair in General Internal Medicine, University of Toronto.
AUTHOR CONTRIBUTIONS
AV and FR designed the study and led the study team. AV, SP, and HYJ drafted the manuscript. SP and HYJ analyzed relevant data. VK and DM contributed to the manuscript and data analysis. RK performed manual data validation. YG contributed to the computational data quality assessment checks. JK, LL-S, SR, TT, and AW led data collection from individual study sites. All of the authors contributed to data interpretation, critically revised the manuscript for important intellectual content, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
AAV, HYJ, VK, DM, and FR are employees of Ontario Health. The funders/employers had no role in the study.
Supplementary Material
REFERENCES
- 1. Weiner MG, Embi PJ.. Toward reuse of clinical data for research and quality improvement: the end of the beginning? Ann Intern Med 2009; 151 (5): 359–60. [DOI] [PubMed] [Google Scholar]
- 2. Herrett E, Gallagher AM, Bhaskaran K, et al. Data resource profile: clinical practice research datalink (CPRD). Int J Epidemiol 2015; 44 (3): 827–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Birtwhistle R, Queenan JA.. Update from CPCSSN. Can Fam Physician 2016; 62 (10): 851. [PMC free article] [PubMed] [Google Scholar]
- 4. Ko CY, Hall BL, Hart AJ, Cohen ME, Hoyt DB.. The American College of Surgeons National Surgical Quality Improvement Program: achieving better and safer surgery. Jt Comm J Qual Patient Saf 2015; 41 (5): 199–204. [DOI] [PubMed] [Google Scholar]
- 5. Nicholls SG, Langan SM, Benchimol EI.. Routinely collected data: the importance of high-quality diagnostic coding to research. CMAJ 2017; 189 (33): E1054–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cook JA, Collins GS.. The rise of big clinical databases. Br J Surg 2015; 102 (2): e93–101. [DOI] [PubMed] [Google Scholar]
- 7. Kahn MG, Raebel MA, Glanz JM, Riedlinger K, Steiner JF.. A pragmatic framework for single-site and multisite data quality assessment in electronic health record-based clinical research. Med Care 2012; 50: S21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kahn MG, Callahan TJ, Barnard J, et al. A harmonized data quality assessment terminology and framework for the secondary use of electronic health record data. EGEMS 2016; 4 (1): 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith M, Lix LM, Azimaee M, et al. Assessing the quality of administrative data for research: a framework from the Manitoba Centre for Health Policy. J Am Med Inform Assoc 2018; 25 (3): 224–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weiskopf NG, Weng C.. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inform Assoc 2013; 20 (1): 144–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Batini C, Cappiello C, Francalanci C, Maurino A.. Methodologies for data quality assessment and improvement. ACM Comput Surv 2009; 41 (3): 1–52. [Google Scholar]
- 12. Williamson T, Green ME, Birtwhistle R, et al. Validating the 8 CPCSSN case definitions for chronic disease surveillance in a primary care database of electronic health records. Ann Fam Med 2014; 12 (4): 367–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nissen F, Quint JK, Morales DR, Douglas IJ.. How to validate a diagnosis recorded in electronic health records. Breathe 2019; 15 (1): 64–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee K, Weiskopf N, Pathak J.. A framework for data quality assessment in clinical research datasets. AMIA Annu Symp Proc 2017; 2017: 1080–9. [PMC free article] [PubMed] [Google Scholar]
- 15. Cai L, Zhu Y.. The challenges of data quality and data quality assessment in the big data era. Data Sci J 2015; 14 (0): 2. [Google Scholar]
- 16. Van den Broeck J, Cunningham SA, Eeckels R, Herbst K.. Data cleaning: detecting, diagnosing, and editing data abnormalities. PLoS Med 2005; 2 (10): e267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qualls LG, Phillips TA, Hammill BG, et al. Evaluating foundational data quality in the national patient-centered clinical research network (PCORnet(R)).EGEMS 2018; 6 (1): 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sunderland KM, Beaton D, Fraser J, ONDRI Investigators, et al. The utility of multivariate outlier detection techniques for data quality evaluation in large studies: an application within the ONDRI project. BMC Med Res Methodol 2019; 19 (1): 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Verma AA, Guo Y, Kwan JL, et al. Patient characteristics, resource use and outcomes associated with general internal medicine hospital care: the General Medicine Inpatient Initiative (GEMINI) retrospective cohort study. CMAJ Open 2017; 5 (4): E842–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.HQO. General Medical Quality Improvement Network. 2019. https://www.hqontario.ca/Quality-Improvement/Quality-Improvement-in-Action/The-General-Medicine-Quality-Improvement-Network Accessed October 14, 2019
- 21.HQO. MyPractice: General Medicine. 2019. https://www.hqontario.ca/Quality-Improvement/Practice-Reports/MyPractice-General-Medicine Accessed October 14, 2019
- 22. El Emam K. Guide to the De-identification of Personal Health Information. Boca Raton, FL: CRC Press; 2013. [Google Scholar]
- 23. Neubauer T, Heurix J.. A methodology for the pseudonymization of medical data. Int J Med Inform 2011; 80 (3): 190–204. [DOI] [PubMed] [Google Scholar]
- 24. Dang Q. Secure Hash Standard. Federal Information Processing Standards Publication. Gaithersburg, MD: National Institute of Standards and Technology; 2015. [Google Scholar]
- 25. Dentler K, Cornet R, ten Teije A, et al. Influence of data quality on computed Dutch hospital quality indicators: a case study in colorectal cancer surgery. BMC Med Inform Decis Mak 2014; 14 (1): 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canadian Institute for Health Information, TCDQF. Ottawa, ON: CIHI, 2009. https://www.cihi.ca/sites/default/files/data_quality_framework_2009_en_0.pdf Accessed July 31, 2020
- 27. Wang RY, Strong DM.. Beyond accuracy: what data quality means to data consumers. J Manag Inform Syst 1996; 12 (4): 5–33. [Google Scholar]
- 28. Hoeven LRV, Bruijne MC, Kemper PF, et al. Validation of multisource electronic health record data: an application to blood transfusion data. BMC Med Inform Decis Mak 2017; 17 (1): 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Terry AL, Stewart M, Cejic S, et al. A basic model for assessing primary health care electronic medical record data quality. BMC Med Inform Decis Mak 2019; 19 (1): 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baca CM, Benish S, Videnovic A, et al. Axon Registry(R) data validation: accuracy assessment of data extraction and measure specification. Neurology 2019; 92 (18): 847–58. [DOI] [PubMed] [Google Scholar]
- 31. Ray S, McEvoy DS, Aaron S, Hickman TT, Wright A.. Using statistical anomaly detection models to find clinical decision support malfunctions. J Am Med Inform Assoc 2018; 25 (7): 862–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yadav S, Kazanji N, K CN, et al. Comparison of accuracy of physical examination findings in initial progress notes between paper charts and a newly implemented electronic health record. J Am Med Inform Assoc 2017; 24 (1): 140–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.