Abstract
Ribosomal protein synthesis is a central process of the modern biological world. Because the ribosome contains proteins itself, it is very important to understand its precursor and evolution. Small ribozymes have demonstrated the principle of “RNA world” hypothesis, but protein free peptide ligase remains elusive. In this report, we have identified two fragments in the peptidyl transfer center that can synthesize a 9-mer poly-lysine in a solution contains Mg2+. This result is deduced from isotope-shifting in high resolution MS. To our best knowledge, this is the longest peptide oligo that can be synthesized by a pure ribozyme. Via single molecule FRET experiments, we have demonstrated the ligase mechanism was probably by substrate proximity via dimerization. We prospect that these RNA fragments can be useful to synthesize template free natural and non-natural peptides, to be model system for peptidyl transfer reaction mechanism and can shed light to the evolution of ribosome.
Keywords: Peptidyl transferase activity, Peptide ligase, LUCA (Last universal common ancestor), Single molecule FRET, High resolution MS
1. Introduction
The ribosome is the universal molecular device that synthesizes all proteins in cells. These proteins are responsible for executing every major cellular function in the modern biological world. Ribosomes have two subunits (30S and 50S), each containing 20–50 proteins and 1–3 RNA polymers. Since modern ribosomes are made of the proteins that they synthesize, there must have existed protein-free ancestors that exhibited peptide ligase activity. The origin of peptide ligase is one of the most fundamental evolution questions to understand the transition from the “ancient RNA-” to the “modern protein-” worlds [1,2]. Before the divergence of the three life domains, the common ancestor of all organisms, LUCA (last universal common ancestor), contained nearly complete components for protein translation, implying that the development of the ribosome is an early event in the history of evolution. The lack of primitive intermediates also makes the origin of the ribosome elusive [3].
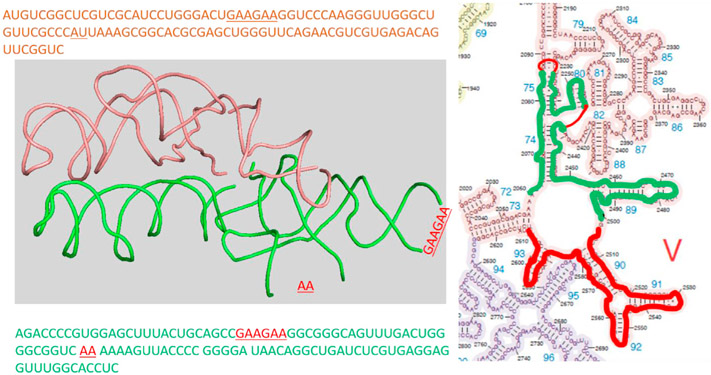
Structural and biochemical assays have indicated that the peptidyl transfer center of the ribosome is the most ancient component and contains no protein [4,5]. However, the search for an RNA-only peptide ligase is still unsuccessful [6,7]. Nevertheless, very small non-rRNA ribozymes prepared through in vitro selection can catalyze peptide bond between short aminoacylated RNA oligos [8,9]. In addition, aminoacyl minihelix, which is more similar to current tRNA substrates, can form peptide bond with puromycin-containing oligos that complemented to the CCA sequence of the minihelix [10]. These results demonstrated the feasibility of RNA-only peptide ligase. However, truncated and in vitro selected constructs of longer RNA chains from the domain V of the large subunit rRNA 23S, in which the peptidyl transfer center resides [11], were inactive of peptide bond formation. In the center of domain V, approximately 180 nucleotides formed a two-fold rotation symmetry where each half forms H-bond with the A- and P-site tRNAs, respectively [12]. These two fragments formed similar secondary and tertiary structures with little sequence homologs (Fig. 1). Furthermore, residues G2553, G2252/G2253 (bacterial numbering) in the A- and P-loops formed H-bonds with the tRNAs to position them for optimized reaction orientation [13,14].
Fig. 1.
Short rRNA fragments at the peptidyl transfer with a 2-fold rotated symmetry that are outlined red (ptc1b) and green (ptc1a). The left panel shows the 1st and tertiary structures from 4wpo; the right panel shows the 2nd structure. In ptc1a piece (green), two stem loops are truncated and replaced with short oligos from ptc1b (red lines).
Although proximity is not sufficient for peptide bond formation, it is a precondition [10]. Thus, we searched for the protein-free ligase using single molecule FRET detection. By attaching rRNA chains to a glass surface under a microscope, proximal binding of tRNAs is easy to detect without ambiguity. Surprisingly, we have found that a 108 nt rRNA scaffold (residues 2503–2610, named ptc1b and outlined red in Fig. 1 [15]) alone can bring two labeled tRNA 3′-fragments into very close distance, which is indicated by a FRET efficiency of 0.6. Further FRET experiments on labeled ptc1b suggested that closeness of the tRNA fragments was due to rRNA dimerization, which depended on Mg2+. By superimposing this rRNA piece into its symmetric counterpart in domain V (pdb ID 4wpo), we identified another rRNA scaffold (green outline in Fig. 1, named ptc1a). It contains 2060 to 2501 fragments, with two stem loops truncated and filled with short RNA oligos guided by the superposition. Both sequences are shown in Fig. 1. We then tested the ligase activities of the homo/hetero rRNA dimers with charged Lysine-tRNAlysine after RNaseT digestion. Surprisingly, we identified formation of a 9-mer poly-lysine. This result is corroborated by mass-shifting of product incorporating N15,N15-labled lysine in high resolution Mass Spec. We found that rRNA alone can synthesize not just single peptide bonds but multiple ones, suggesting co-appearance and co-evolution of peptides and RNAs in the primordial world. The template-free protein synthesis seems to suggest a module-based evolution hypothesis, in which 3D structure interactions without sequence constrain drive the evolution [16]. The simple RNA scaffolds also provided an easy-to-manipulate model to study the chemical mechanism of the peptidyl transfer reactions, which remains a fundamental challenge [17]. Finally, we project that this simple template-free, proofreading-free protein polymerase will be useful to synthesize non-natural peptides for bioengineering applications.
2. Materials and methods
All chemicals were purchased from Millipore Sigma unless stated otherwise.
2.1. RNA scaffolds
The 5′-biotinylated ptc1b RNA and ptc1a RNA molecules are purchased from IDTDNA. The 3′-ends are labeled with the “3′ EndTag™ DNA End Labeling System” from the Vector@Laboratories. The Cy3/Cy5-maleimide dyes are purchased from GE Life Sciences. The labeling efficiency is approximately 50% based on spectrophotometer absorptions.
2.2. tRNA charging and Rnase T1 digestion
The tRNAlysine powder is purchased from chemical-block.com and is charged with normal- (Millipore-Sigma), C14- (PerkinElmer), or N15,N15- (Millipore-Sigma) labeled lysine as described previously [18]. The charging efficiency is approximately 1000 pmol/A260 based on absorption and C14 radioactivity. Afterward, 4 A260 of charged tRNAlysine (theoretical amount 1800 pmol/A260) is digested with 1000 units of RNase T1 (thermofisher) under denature condition following the manufacture’s manual. The ACCCACCA-lysine piece is first phenol extracted and precipitated, and re-dissolved with water. Then it is enriched with the Monarch® RNA Cleanup kit. First, the RNA oligos >25 nt is captured by the Monarch® binding column by adding 1x volume of ethanol. Then another 1x volume of ethanol is added to the run-through liquid, and loaded onto a new filter to bind the smaller pieces <25 nt. After washing, the RNA-lysine piece is eluted with 50 μl of RNase-free water. The expected RNA-lysine pieces (ACCCACCA-lysN14,N14 and -lys N15,N15) are supported by high resolution MALDI MS (Mass Spectrometry facility, University of Alabama), and named “RNA-lys” (Fig. 3A1).
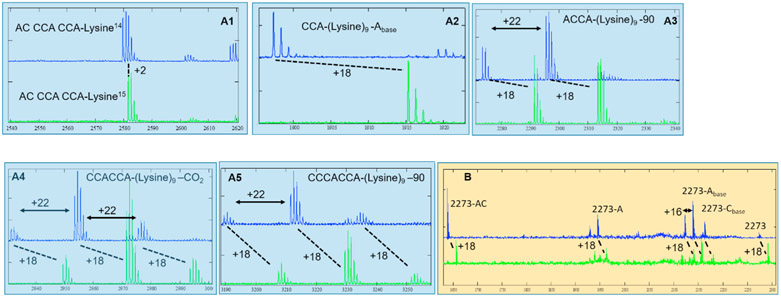
Fig. 3.
High resolution MS of poly-lysine of N14,N14- (blue trace) and N15,N15- (green trace) labeled lysine monomers. Shifting of 18 suggests 9 lysine residues. The exact masses and assignments are shown in Tables S1 and S2. (A1-5) Mass spectrum of lysine monomer and oligos. (B) Tandem MS/MS on 2273 and 2291 peaks.
2.3. RNA-lysine labeling
The RNA-lysine piece was labeled in two positions: at the lysine moiety and the 5′-end of the RNA. Labeling at the lysine moiety. Cy3 or Cy5 NHS-dyes (GE Life Sciences) were dissolved in DMSO and added directly into the RNA-lysine solution mentioned above (final dye concentration ~ 500 μM from 10 mM stocks). The labeling reaction was incubated at 37 °C for 2 h. And the excess dye was removed via a G25 size-exclusive column. Then the Cy3-or Cy5-labeled RNA-lysine was precipitated and resuspended in 10–20 μl of RNase-free water. Labeling at the 5′-end was prepared with the “5′ EndTag™ DNA/RNA End Labeling System” from the Vector@Laboratories. The labeling efficiency of both position is approximately (0.6 μM of dye)/(A260 of RNA).
2.4. Single molecule FRET experiments
The total internal reflection fluorescence microscope (TIRF) was based on a Nikon Eclipse Ti2-E inverted microscope with two autoturrets and two CMOS cameras as described before [18]. The top turret reflects the laser for TIRF illumination, and the bottom turret split the FRET signals into two CMOS cameras based on wavelengths. The sample holder was described previously [19]. For all measurements, sample concentrations are in the range of 10–100 nm. An oxygen scavenger cocktail (3 mg/mL glucose, 100 mg/mL glucose oxidase, 48 mg/mL catalase, and 2 mM trolox) was added to the channel before imaging to prevent photo bleaching.
3. Results and discussion
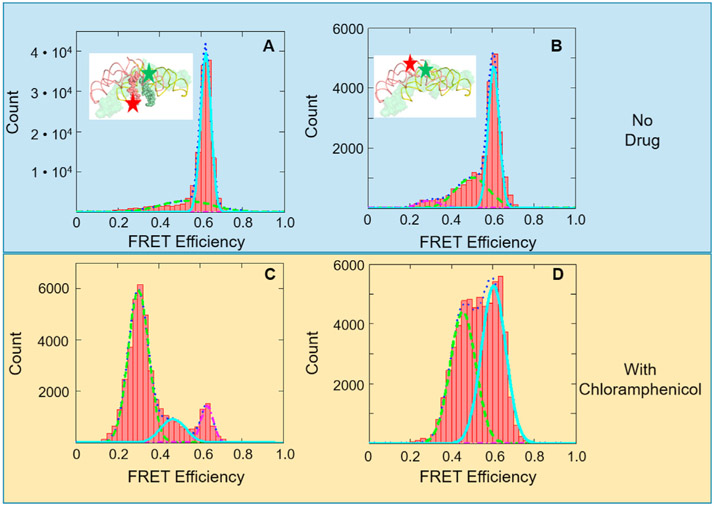
First, we have observed high FRET efficiency between Cy3/Cy5-labled RNA-lys in the presence of ptc1b. The reason ptc1b was chosen, instead of the total 180 nt from the literature [12], was due to the complicated folding for longer RNAs. The sequence of ptc1b reproduced the same secondary structure as in the ribosome, while the 180 nt chain did not (Mfold Web Server). As shown in Fig. 2A, the Cy3 and Cy5 dyes are located at the aminoacyl and the 5′-terminal of RAN-lys, respectively. 1 μM of ptc1b was incubated with 0.5 μM of Cy3/Cy5-labeld RNA-lys in the presence of 0 or 15 mM MgCl2. The mixture was incubated at 37 °C for 10 min and diluted with either water or 15 mM MgCl2 solution at room temperature. This solution was loaded into the sample cell and attached to the surface via biotin-streptavidin interaction. Typical FRET images and representative time-lapse traces of donor and acceptor fluorescent intensities are shown in Fig. S1. The FRET efficiencies are calculated as Iacceptor/(Idonor + Iacceptor), in which Ixx is the fluorescence intensity in the donor or acceptor channel. In the presence of Mg2+, a FRET species centered at 0.62 was observed, which corresponds to 50 Å (R0 = 55 Å) [20]. From the X-ray structure (4wpo), the amino acid of A-site tRNA to the 5′- of ACCCACCA at P-site (tRNAlys sequence) is approximately 43 Å. This result suggested that the ptc1b can dimerize to mimic the two-fold symmetric peptidyl transfer center; and the tRNA fragments bind to the dimer in a similar way as tRNAs binding to the ribosome.
Fig. 2.
FRET efficiency histograms of tRNA (oligo)-tRNA (oligo) or rRNA-rRNA interactions. (A) FRET efficiency histogram of 5′- and aminoacyl labeled RNA-lys pieces. The relative Cy3/Cy5 positions are shown in the insert. (B) FRET efficiency histogram of 3′- labeled ptc1b. The relative Cy3/Cy5 positions are shown in the insert. (C) and (D) FRET efficiency histograms of (A) and (B) in the presence of 1 mM chloramphenicol, respectively.
Secondly, the ptc1b/a dimerization hypothesis was tested. The ptc1b is labeled on the 3′-ends (C2610) with Cy3 and Cy5, respectively. Their dimerization is directly monitored with FRET. As predicted, FRET between Cy3-ptc1b and Cy5-ptc1b (Fig. 2), exhibited a high FRET species centered at 0.60. Without Mg2+, this species disappeared (Fig. S1), indicating that the dimerization also needs Mg2+. This FRET value corresponds to 51.4 Å, where the theoretical distance from the structure is about 23 Å (by aligning ptc1b to its hypothetical symmetric counterpart, insert of Fig. 2A). This discrepancy indicates a looser structure in the RNA-only dimers than in the well-packed single-chain peptidyl transfer center. Variation of linker length, quantum yields and orientation may contribute to the difference as well. We then synthesized ptc1a molecule, in which most of the sequence were from rRNA, but two long stem loop structures were replaced with short oligos, based on its alignment with ptc1b (Fig. S2). The ptc1a piece was also labeled at the 3′-end, and hetero-dimer formation of ptc1a/b was detected. The FRET efficiencies are almost the same in the hetero dimer experiment (Fig. S3). These high FRET species (ptc1b homo-dimer and the ptc1a/b hetero-dimer) are probably the reasons to bring the RNA-lys species together that generated the high FRET signals in Fig. 2A.
Thirdly, the 0.62 FRET species between RNA-lys in Fig. 2A was diminished by chloramphenicol, but the 0.60 FRET species between ptc1b homodimer in Fig. 2B was not (Fig. 2C and D). Chloramphenicol is an antibiotic that binds to the same location as the aminoacyl moiety of A-site tRNA [21]. If the prospected structures are correct in Fig. 2A and B, then adding chloramphenicol should interfere the high FRET species in 2A, but not in 2B. This is because chloramphenicol directly competes the binding pocket with RNA-lys, but does not compete with the ptc1a/b dimerization without the short tRNA fragments.
We observed this effect in Fig. 2C and D. The 0.62 FRET species diminished while a 0.3 FRET species emerged in the RNA-lys experiments, indicating inhibition of RNA-lys binding. Meanwhile, the 0.6 FRET species remained unchanged in the ptc1a/b dimerization experiments. Furthermore, the small peak at 0.5 FRET species in Fig. 2B is significantly larger when drug is present in Fig. 2D, which means chloramphenicol may have induced more dimers in a more loose structure. Fig. 2C and D suggest that the structural assignments of the FRET species in Fig. 2A and B are reasonable.
Fourthly, we have found that a 9-mer poly-lysine oligo is formed by the ptc dimers. In these experiments, 1 μM of ptc1b homo-dimer or ptc1a/b hetero-dimer was incubated with l5 mM MgCl2 and 1 μM of RNA-lys, in which normal or N15,N15-labeled lysine was used. The mixtures were incubated at 37 °C for 1hr or 3 h (no difference was detected in mass spectrometry analysis), run through a G25 desalting column, and analyzed with high resolution MALDI (all the exact masses are shown in Table S1). Multiple mass peaks exhibited +l8 shift in comparison of the N14 (blue trace) to the N15 (green trace) labeled experiments, showing incorporation of 9 lysine residues (2 Da mass shift per residue). Tandem MS/MS spectra were obtained on the 2273 peak and its +18 shifting peak in the normal and N15, N15-labeled experiments, respective (Fig. 3B). Again, these spectra showed consistent +18 shifting at multiple positions, indicating there were 9-mer lysine in the 2273/2291 peaks. Mass calculation indicated that these peaks correspond to CCA-K9 (Fig. 3A2), ACCA-K9 (Fig. 3A3), CCACCA-K9 (Fig. 3A4), CCCACCA-K9 (Fig. 3A5). For the MS/MS of 2273 peak, species losing A or C base on the riboses and losing complete A- and AC- nucleotides are observed. Although the starting material was confirmed to be ACCCACCA-K (Fig. 3A1, material and methods), we did not observe the product of 9-mer poly-lysine on the 8-nt fragment. It is probably due to the loss of the terminal A residue, either during the reaction incubation or MALDI measurements [22].
In summary, short rRNA fragments, containing the same sequences as those at the peptidyl transfer center, dimerized and brought tRNA fragments to proximity. High resolution mass spectrometry analysis indicated that these dimers have synthesized not one but eight peptide bonds to form a 9-mer RNA-K9 molecule out of the Lysine-charged tRNA fragment (ACCCACCA-K). Poly-lysine has shown to adopt β-sheets structure, which intercalates well into the double helix grooves [16,23]. The ancient molecules tRNA synthetases and ribosome reserved these interactions. The tRNA synthetases depend on their β-sheets structures to charging tRNAs [24], while some ribosomal proteins bind to rRNA’s grooves. For example L15 and S11 proteins demonstrated such interaction (Figure S4). This relatively long oligo synthesis we have observed suggests that primitive peptide ligase may have higher capacity than we thought. Therefore, a peptide/nucleic acid co-evolution route is possible, as suggested in the literature [25]. Theoretic study has suggested that optimizing the interaction between RNA’s double helix and peptide’s β-sheet could drive the evolution of both species without sequencing fidelity [16]. Indeed, diverse RNA sequences can form similar structural modules [26,27]. The ptc1a and 1b molecules in this report are examples. Alternatively, the discovery of RNA-only self-replicators strongly supports the “RNA world” theory, which is another possibility [28,29].
One potential application of our finding is to mutate these simple rRNA fragments to test hypothetical catalytic residues at the peptidyl center. Although a major catalytic mechanism of the ribosome is to position the substrates at proximity, it remains unclear whether any chemical catalytic mechanism is involved. Several highly conserved residues have been tested, but none seem indispensable [30-32]. Using this short rRNA fragment that can be readily mutated and transcribed in vitro, we expect that some chemical mechanism of the peptidyl transfer reaction will be revealed, although this system is overly simplified and primitive.
We also propose that this simple peptide ligase can be a starting point in bioengineering to synthesize non-natural peptides without mRNA templates, which is difficult to incorporate by the native ribosome and is seriously pursued for drug development [33]. Synthesis of a 9-mer peptide oligo is a promising starting point to engineer a more powerful peptide ligase to make longer peptide chains without overcoming the rigorous selection and proofreading steps of the native ribosome. Therefore, unnatural/mixed peptide chains can be synthesized in the future.
Supplementary Material
Acknowledgements
This research is supported by the the Welch foundation (grant number E-1721) and NIGMS (grant number 1 R01 GM111452) to Y.W.
We thank Dr. Qiaoli Liang of University of Alabama, Department of Chemistry for her excellent collection of the mass spectra.
Footnotes
Declaration of competing interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2021.01.036.
References
- [1].Crick FH, The origin of the genetic code, J. Mol. Biol 38 (1968) 367–379, 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- [2].Gilbert W, Origin of life: the RNA world, Nature 319 (1986), 10.1038/319618a0, 618–618 [DOI] [Google Scholar]
- [3].Fox GE, Origin and evolution of the ribosome, Cold Spring Harb. Perspect. Biol 2 (2010) a003483, 10.1101/cshperspect.a003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nissen P, Hansen J, Ban N, Moore PB, Steitz TA, The structural basis of ribosome activity in peptide bond synthesis, Science 289 (2000) 920–930, 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- [5].Hsiao C, Lenz TK, Peters JK, Fang PY, Schneider DM, Anderson EJ, Preeprem T, Bowman JC, O’Neill EB, Lie L, Athavale SS, Gossett JJ, Trippe C, Murray J, Petrov AS, Wartell RM, Harvey SC, Hud NV, Williams LD, Molecular paleontology: a biochemical model of the ancestral ribosome, Nucleic Acids Res. 41 (2013) 3373–3385, 10.1093/nar/gkt023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Noller HF, Hoffarth V, Zimniak L, Unusual resistance of peptidyl transferase to protein extraction procedures, Science 256 (1992) 1416–1419, 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- [7].Khaitovich P, Tenson T, Mankin AS, Green R, Peptidyl transferase activity catalyzed by protein-free 23S ribosomal RNA remains elusive, RNA 5 (1999) 605–608, 10.1017/s1355838299990295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang B, Cech TR, Peptidyl-transferase ribozymes: trans reactions, structural characterization and ribosomal RNA-like features, Chem. Biol 5 (1998) 539–553, 10.1016/s1074-5521(98)90113-2. [DOI] [PubMed] [Google Scholar]
- [9].Zhang B, Cech TR, Peptide bond formation by in vitro selected ribozymes, Nature 390 (1997) 96–100, 10.1038/36375. [DOI] [PubMed] [Google Scholar]
- [10].Tamura K, Schimmel P, Oligonucleotide-directed peptide synthesis in a ribosome- and ribozyme-free system, Proc. Natl. Acad. Sci. U. S. A 98 (2001) 1393–1397, 10.1073/pnas.98.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Anderson RM, Kwon M, Strobel SA, Toward ribosomal RNA catalytic activity in the absence of protein, J. Mol. Evol 64 (2007) 472–483, 10.1007/s00239-006-0211-y. [DOI] [PubMed] [Google Scholar]
- [12].Agmon I, Bashan A, Zarivach R, Yonath A, Symmetry at the active site of the ribosome: structural and functional implications, Biol. Chem 386 (2005) 833–844, 10.1515/BC.2005.098. [DOI] [PubMed] [Google Scholar]
- [13].Kim DF, Green R, Base-pairing between 23S rRNA and tRNA in the ribosomal A site, Mol. Cell 4 (1999) 859–864, 10.1016/s1097-2765(00)80395-0. [DOI] [PubMed] [Google Scholar]
- [14].Samaha RR, Green R, Noller HF, A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome, Nature 377 (1995) 309–314, 10.1038/377309a0. [DOI] [PubMed] [Google Scholar]
- [15].Noller HF, Kop J, Wheaton V, Brosius J, Gutell RR, Kopylov AM, Dohme F, Herr W, Stahl DA, Gupta R, Waese CR, Secondary structure model for 23S ribosomal RNA, Nucleic Acids Res. 9 (1981) 6167–6189, 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Carter CW Jr., Kraut J, A proposed model for interaction of polypeptides with RNA, Proc. Natl. Acad. Sci. U. S. A 71 (1974) 283–287, 10.1073/pnas.71.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Green R, Lorsch JR, The path to perdition is paved with protons, Cell 110 (2002) 665–668, 10.1016/s0092-8674(02)00965-0. [DOI] [PubMed] [Google Scholar]
- [18].Altuntop ME, Ly CT, Wang Y, Single-molecule study of ribosome hierarchic dynamics at the peptidyl transferase center, Biophys. J 99 (2010) 3002–3009, 10.1016/j.bpj.2010.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lin R, Wang Y, Automated smFRET microscope for the quantification of label-free DNA oligos, Biomed. Optic Express 10 (2019) 682–693, 10.1364/BOE.10.000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S, Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase, Nature 419 (2002) 638–641, 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- [21].Bulkley D, Innis CA, Blaha G, Steitz TA, Revisiting the structures of several antibiotics bound to the bacterial ribosome, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 17158–17163, 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Joyner JC, Keuper KD, Cowan JA, Analysis of RNA cleavage by MALDI-TOF mass spectrometry, Nucleic Acids Res. 41 (2013) e2, 10.1093/nar/gks811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dzwolak W, Ravindra R, Nicolini C, Jansen R, Winter R, The diastereomeric assembly of polylysine is the low-volume pathway for preferential formation of beta-sheet aggregates, J. Am. Chem. Soc 126 (2004) 3762–3768, 10.1021/ja039138i. [DOI] [PubMed] [Google Scholar]
- [24].Ribas de Pouplana L, Schimmel P, Two classes of tRNA synthetases suggested by sterically compatible dockings on tRNA acceptor stem, Cell 104 (2001) 191–193, 10.1016/s0092-8674(01)00204-5. [DOI] [PubMed] [Google Scholar]
- [25].Piette B, Heddle JG, A peptide-nucleic acid replicator origin for life, Trends Ecol. Evol 35 (2020) 397–406, 10.1016/j.tree.2020.01.001. [DOI] [PubMed] [Google Scholar]
- [26].Miao Z, Westhof E, RNA structure: advances and assessment of 3D structure prediction, Annu. Rev. Biophys 46 (2017) 483–503, 10.1146/annurev-biophys-070816-034125. [DOI] [PubMed] [Google Scholar]
- [27].Hermann T, Westhof E, Non-Watson-Crick base pairs in RNA-protein recognition, Chem. Biol 6 (1999) R335–R343, 10.1016/s1074-5521(00)80003-4. [DOI] [PubMed] [Google Scholar]
- [28].Wilson DS, Szostak JW, In vitro selection of functional nucleic acids, Annu. Rev. Biochem 68 (1999) 611–647, 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- [29].Tjhung KF, Shokhirev MN, Horning DP, Joyce GF, An RNA polymerase ribozyme that synthesizes its own ancestor, Proc. Natl. Acad. Sci. U. S. A 117 (2020) 2906–2913, 10.1073/pnas.1914282117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Muth GW, Chen L, Kosek AB, Strobel SA, pH-dependent conformational flexibility within the ribosomal peptidyl transferase center, RNA 7 (2001) 1403–1415. PMID: 11680845 PMCID: PMC1370184. [PMC free article] [PubMed] [Google Scholar]
- [31].Youngman EM, Brunelle JL, Kochaniak AB, Green R, The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release, Cell 117 (2004) 589–599, 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- [32].Polacek N, Gaynor M, Yassin A, Mankin AS, Ribosomal peptidyl transferase can withstand mutations at the putative catalytic nucleotide, Nature 411 (2001) 498–501, 10.1038/35078113. [DOI] [PubMed] [Google Scholar]
- [33].Young DD, Schultz PG, Playing with the molecules of life, ACS Chem. Biol 13 (2018) 854–870, 10.1021/acschembio.7b00974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.