Here, we report the identification and coding-complete genome sequence of a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strain (NYI.B1-7.01-21) obtained from a patient with symptoms of COVID-19 who had a recent travel history to the United Kingdom. The sample was tested by the Cayuga Health Systems laboratory as part of New York State’s travel testing guidance and was sequenced at Cornell University after testing positive.
ABSTRACT
Here, we report the identification and coding-complete genome sequence of a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strain (NYI.B1-7.01-21) obtained from a patient with symptoms of COVID-19 who had a recent travel history to the United Kingdom. The sample was tested by the Cayuga Health Systems laboratory as part of New York State’s travel testing guidance and was sequenced at Cornell University after testing positive.
ANNOUNCEMENT
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a member of the genus Betacoronavirus, family Coronaviridae (1), was first identified in Wuhan, China (2). Since its emergence in December 2019, several mutations have been detected, leading to the emergence of multiple genetic lineages. The lineage B.1.1.7 was first detected in September 2020. It is characterized by a large number of mutations and has since been detected in numerous countries around the world (3). As of 25 January 2021, 22 cases of this strain have been identified in the state of New York (4).
The sample characterized here was obtained for routine diagnostics for SARS-CoV-2 on 7 January 2021 from a person returning from a trip to England and presenting symptoms of shortness of breath, fatigue, and fever (institutional review board [IRB] approval number CMC0420EP). Viral RNA was extracted from a saliva sample treated with inactivating medium (containing guanidine hydrochloride) using the QIAamp viral RNA minikit (Qiagen). The sequencing library was prepared for sequencing on an Oxford Nanopore Technologies (ONT) MinION instrument using a modification of the approach developed by the ARTIC Network for sequencing SARS-CoV-2 (https://www.protocols.io/view/ncov-2019-sequencing-protocol-v2-bdp7i5rn). A total of 58 primers were designed using Primer3 (5) in the Geneious Prime 2019 software targeting approximately 1,500-bp and 500-bp products with 100 bp of overlap between different amplicons. Primer sequences are available at dx.doi.org/10.17504/protocols.io.brkxm4xn, and the first-strand synthesis and PCR conditions are available at dx.doi.org/10.17504/protocols.io.br54m88w. Libraries were multiplexed and sequenced on an R9.4 flow cell for 6 h.
Raw reads were base called and demultiplexed with the MinIT device (ONT) and then processed through the artic-ncov2019-medaka conda environment (https://github.com/artic-network/artic-ncov2019). We obtained a 29,713-bp genome with a mean depth of coverage of 2,236.3× and a GC content of 38%. The sequence does not cover 51 bp from the 5′ end and 121 bp from the 3′ end of the reference genome (GenBank accession number NC_045512.2). Mutations were also verified in Geneious, and a table of variations in relation to the reference genome was generated (Table 1).
TABLE 1.
Mutations of SARS-CoV-2 strain NYI.B1-7.01-21 in comparison to the reference strain (GenBank accession number NC_045512.2)
| Gene or region | Nucleotide position | Amino acid change | CDS codon positiona | Nucleotide change |
|---|---|---|---|---|
| 5′ untranslated region | 241 | C → T | ||
| ORF1ab | 913 | 216 | C → T | |
| 2110 | 615 | C → T | ||
| 2485 | 740 | C → T | ||
| 3037 | 924 | C → T | ||
| 3267 | T → I | 1001 | C → T | |
| 5388 | A → D | 1708 | C → A | |
| 5986 | 1907 | C → T | ||
| 6954 | I → T | 2230 | T → C | |
| 7984 | 2573 | T → C | ||
| 11288–11296 | SGF deletion | 3675–3677 | ||
| 14120 | P → L | 4619 | C → T | |
| 14408 | P → L | 4715 | C → T | |
| 14676 | 4804 | C → T | ||
| 15279 | 5005 | C → T | ||
| 16176 | 5304 | T → C | ||
| 19390 | P → S | 6376 | C → T | |
| S | 21765–21770 | HV deletion | 69−70 | |
| 21991–21993 | Y deletion | 144 | ||
| 23063 | N → Y | 501 | A → T | |
| 23271 | A → D | 570 | C → A | |
| 23403 | D → G | 614 | A → G | |
| 23604 | P → H | 681 | C → A | |
| 23709 | T → I | 716 | C → T | |
| 24506 | S → A | 982 | T → G | |
| 24914 | D → H | 1118 | G → C | |
| ORF3a | 25638 | 82 | C → T | |
| ORF8 | 27972 | Q → stop | 27 | C → T |
| 28048 | R → I | 52 | G → T | |
| 28095 | K → stop | 68 | A → T | |
| 28111 | Y → C | 73 | A → G | |
| N | 28280–28282 | D → L | 3 | GAT → CTA |
| 28881–28883 | RG → KR | 203–204 | GGG → AAC | |
| 28977 | S → F | 235 | C → T |
CDS, coding DNA sequence.
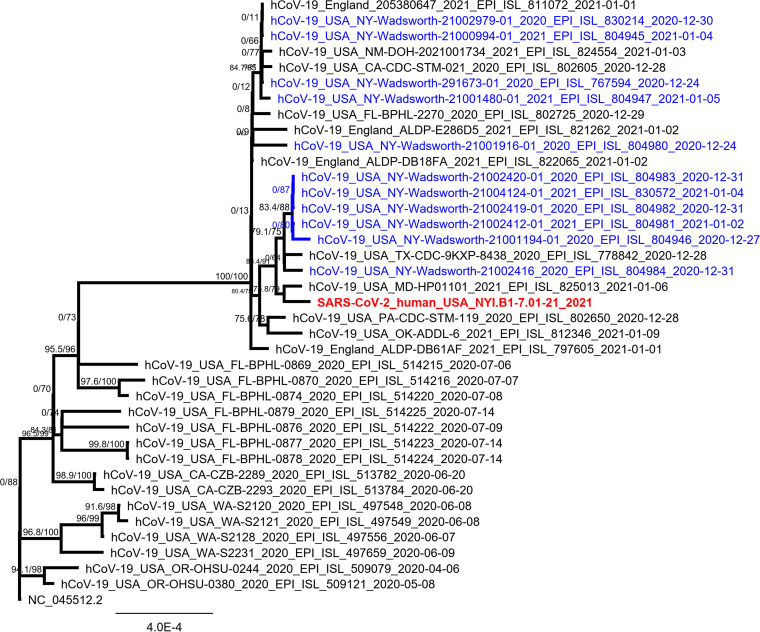
Phylogenetic analysis classified strain NYI.B1-7.01-21 as part of the B.1.1.7 lineage (Fig. 1). Compared to the reference sequence (GenBank accession number NC_045512.2), a total of 34 mutations and deletions were detected, including all the nonsynonymous mutations inferred to occur on the branch leading to the B.1.1.7 lineage. An additional mutation at position 28881 to 28883, GGG > AAC (R203K and G204R), in the N gene may have impacts on the structure and function of the N protein (6, 7). Open reading frame 8 (ORF8) has an additional stop codon, downstream of the Q27 stop. Our analyses show that SARS-CoV-2 NYI.B1-7.01-21 does not cluster with the other B.1.1.7 viruses detected in the state of New York to date, indicating an independent introduction. These observations are consistent with the patient’s travel history to England and highlight the importance of arrival testing and genetic characterization of SARS-CoV-2-positive samples, especially following travel to locations where newly emerging SARS-CoV-2 variants are known to be circulating.
FIG 1.
Phylogenetic tree of strain NYI.B1-7.01-21, highlighted in red in a clade with other B.1.1.7 sequences. Other samples from New York belonging to this lineage are shown in blue. A whole-genome nucleotide sequence alignment was performed with sequences retrieved from GISAID (https://www.gisaid.org) using MAFFT v7.450 (8) with the parameters Auto for algorithm, scoring matrix of 200PAM/k=2, gap open penalty of 1.53, and offset value of 0.123. The phylogenetic tree was constructed using 1,000 bootstrap replicates and the model TIM2+F, selected as the best-fit model by the IQ-TREE Web server (9).
Data availability.
This sequence has been deposited in GenBank under the accession number MW487270. The accession numbers for the raw sequencing reads in the NCBI Sequence Read Archive (SRA) are PRJNA692972 (BioProject), SAMN17373206 (BioSample), and SRR13453793 (SRA).
ACKNOWLEDGMENT
This work and the sequencing capacity were supported in part by NAHLN grant AP20 VS DB000C020.
REFERENCES
- 1.Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D, Perlman S, Poon LLM, Samborskiy DV, Sidorov IA, Sola I, Ziebuhr J. 2020. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2021. New COVID-19 variants. https://www.cdc.gov/coronavirus/2019-ncov/transmission/variant.html. [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2021. US COVID-19 cases caused by variants. https://www.cdc.gov/coronavirus/2019-ncov/transmission/variant-cases.html. [Google Scholar]
- 5.Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 6.Liu CI, Hsu KY, Ruaan RC. 2006. Hydrophobic contribution of amino acids in peptides measured by hydrophobic interaction chromatography. J Phys Chem B 110:9148–9154. doi: 10.1021/jp055382f. [DOI] [PubMed] [Google Scholar]
- 7.Peng TY, Lee KR, Tarn WY. 2008. Phosphorylation of the arginine/serine dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J 275:4152–4163. doi: 10.1111/j.1742-4658.2008.06564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This sequence has been deposited in GenBank under the accession number MW487270. The accession numbers for the raw sequencing reads in the NCBI Sequence Read Archive (SRA) are PRJNA692972 (BioProject), SAMN17373206 (BioSample), and SRR13453793 (SRA).