Abstract
The Pelagophyceae are marine stramenopile algae that include Aureoumbra lagunensis and Aureococcus anophagefferens, two microbial species notorious for causing harmful algal blooms. Despite their ecological significance, relatively few genomic studies of pelagophytes have been carried out. To improve understanding of the biology and evolution of pelagophyte algae, we sequenced complete mitochondrial genomes for A. lagunensis (CCMP1510), Pelagomonas calceolata (CCMP1756), and five strains of Aureoc. anophagefferens (CCMP1707, CCMP1708, CCMP1850, CCMP1984, and CCMP3368) using Nanopore long-read sequencing. All pelagophyte mitochondrial genomes assembled into single, circular mapping contigs between 39,376 bp (P. calceolata) and 55,968 bp (A. lagunensis) in size. Mitochondrial genomes for the five Aureoc. anophagefferens strains varied slightly in length (42,401–42,621 bp) and were 99.4–100.0% identical. Gene content and order were highly conserved between the Aureoc. anophagefferens and P. calceolata genomes, with the only major difference being a unique region in Aureoc. anophagefferens containingDNA adenine and cytosine methyltransferase (dam/dcm) genes that appear to be the product of lateral gene transfer from a prokaryotic or viral donor. Although the A. lagunensis mitochondrial genome shares seven distinct syntenic blocks with the other pelagophyte genomes, it has a tandem repeat expansion comprising ∼40% of its length, and lacks identifiable rps19 and glycine tRNA genes. Laterally acquired self-splicing introns were also found in the 23S rRNA (rnl) gene of P. calceolata and the coxI gene of the five Aureoc. anophagefferens genomes. Overall, these data provide baseline knowledge about the genetic diversity of bloom-forming pelagophytes relative to nonbloom-forming species.
Keywords: mitochondrial genome, Stramenopila, Pelagophyceae, lateral gene transfer, evolution
Significance
Pelagophytes are marine microalgae that can cause harmful algal blooms (HABs). Although nuclear and mitochondrial genomes have been sequenced for the pelagophyte Aureococcus anaphagefferens, very little sequence data exist for other species and the molecular basis of HAB formation is poorly understood. We have sequenced the mitochondrial DNA (mtDNA) of five Aureococcus anaphagefferens strains, as well as that of Aureoumbra lagunensis and Pelagomonas calceolata. The A. anaphagefferens genomes are noteworthy in possessing DNA methyltransferase genes of apparent viral origin not found in the other two species—this is potentially significant given that viruses are known to mediate HAB formation and collapse. Our dataprovide insight into fine-scale variation of A. anaphagefferens mtDNAs relative to those of their closest non-HAB-forming species.
Introduction
The pelagophytes are marine, mostly picoplanktonic algae that branch within the stramenopiles (heterokonts). Two species of pelagophytes, Aureoumbra lagunensis and Aureococcus anophagefferens, are well known for their ability to form brown tides, a type of harmful algal bloom (HAB) that can cause significant damage to ecosystems and negatively impact fisheries (Sieburth et al. 1988; DeYoe et al. 1997; Gobler and Sunda 2012). Aureococcus anophagefferens HABs have occurred annually in estuaries along the northeast and mid-Atlantic coasts of the United States since 1985, and more recently have been observed off the coasts of South Africa and China (Gobler and Sunda 2012; Zhang et al. 2012). Aureoumbra lagunensis blooms have occurred intermittently in lagoons near Texas, Florida, and Cuba since 1990 (Gobler and Sunda 2012; Gobler et al. 2013; Hall et al. 2018). Given that both of the pelagophyte HABs occur in anthropogenically modified environments—likely due to the organism’s abilities to utilize particular nutrients (Gobler and Sunda 2012)—it seems probable that their respective bloom regions will continue to expand further in the future.
Aureoumbra lagunensis and Aureoc. anophagefferens are small (2–4 μm), spherical, nonmotile unicellular algae (Sieburth et al. 1988; DeYoe et al. 1997; Gobler and Sunda 2012). Despite their morphological similarities, they are genetically distinct (DeYoe et al. 1997). One notable difference between these two HAB-forming algae is that Aureoc. anophagefferens is known to associate with Aureoc. anophagefferens Virus (AaV), a nucleocytoplasmic large DNA virus (Gastrich et al. 1998; Moniruzzaman et al. 2014, 2016). AaV has been suggested to influence Aureoc. anophagefferens bloom dynamics, including their formation and collapse (Moniruzzaman et al. 2018), and the AaV genome contains several genes acquired from its host, suggesting that AaV and Aureoc. anophagefferens have coevolved (Moniruzzaman et al. 2014).
Despite their obvious ecological importance, relatively little molecular data are available across the diversity of pelagophytes, limiting our knowledge of the molecular biology and evolution of this class as a whole. Based on 18S ribosomal DNA (rDNA) sequence data, the interspecies phylogenetic relationships of Pelagophyceae have been estimated and show that the HAB-forming species Aureoc. anophagefferens and A. lagunensis are not closely related and in fact belong to different orders within the class Pelagophyceae (Wetherbee et al. 2015). Other pelagophyte species not known to form HABs, such as P. calceolata (Andersen et al. 1993), appear more closely related to the HAB-forming taxa. On the basis of 18S rDNA and plastid RuBiSCO genes, Bailey and Andersen (1999) suggested that Aureoc. anophagefferens blooms are genetically uniform and not comprised of crypticspecies. A recent study of Aureoc. anophagefferens diversity in China by Tang et al. (2019) lends further support to this notion.
Gobler et al. (2011) published the first nuclear genome of a pelagophyte alga, that of Aureoc. anophagefferens CCMP1984. Transcriptomes have since been sequenced for both Aureoc. anophagefferens (Frischkorn et al. 2014) and A. lagunensis,as well as P. calceolata and Pelagococcus subviridis as a part of the Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP) (Keeling et al. 2014; Caron et al. 2017). However, although the plastid (chloroplast) genomes of both A. lagunensis and Aureoc. anophagefferens have been sequenced (Ong et al. 2010), the mitochondrial genome of Aureoc. anophagefferens (CCMP1984) was only recently published (Liu et al. 2020).
Mitochondria are organelles derived from an α-proteobacterial endosymbiont. A reduced version of the endosymbiont’s genome is still maintained in almost all mitochondria and mitochondria-related organelles (see Roger et al.2017 and references therein). Across eukaryote lineages, the size, structure, and gene content of mitochondrial genomes varies quite widely (e.g., Smith2016). Targeted sequencing of mitochondrial genomes from undersample lineages (e.g., Ševčíková et al. 2016; Kim et al. 2018) has allowed for a greater understanding of the evolutionary history of mitochondrial genomes across specific groups of taxa. However, with only a single mitochondrial genome available for all pelagophyte algae, we are missing a large chunk of its diversity.
We have sequenced complete mitochondrial genomes for the pelagophytes A. lagunensis (CCMP1510), P. calceolata (CCMP1756), and five strains of Aureoc. anophagefferens (CCMP1707, CCMP1708, CCMP1850, CCMP1984, and CCMP3368). These data allowed us to assess gene content and synteny, identify strain- and species-specific genomic features and, more generally, better understand how the mitochondrial genome has evolved over the course of pelagophyte and stramenopile evolution. Our Aureoc. anophagefferens data help define strain diversity in this important HAB-forming species.
Materials and Methods
Cell Culturing and DNA Extraction
Aureoumbra lagunensis CCMP1510, P. calceolata CCMP1756, and Aureoc. anophagefferens (CCMP1707, CCMP1708, CCMP1850, CCMP1984, and CCMP3368) cultures were obtained from the National Center for Marine Algae and Microbiota (NCMA, East Boothbay, ME). Aureoumbra lagunensis was cultured axenically in h/2 media prepared with artificial seawater (Guillard 1975), whereas Aureoc. anophagefferens strains were maintained in axenic or uni-eukaryotic cultures in L1-Si media made with artificial seawater. All A. lagunensis and Aureoc. anophagefferens cultures were grown at 20°C under 100 µmol quanta m−2s−1 light on a 12 h : 12 h light : dark cycle. Similarly, P. calceolata was grown axenically in L1-Si media prepared with artificial seawater at 22°C and under an identical lighting regime.
Total genomic DNA was extracted from both A. lagunensis and P. calceolata using the DNeasy PowerSoil Pro Kit (Qiagen, Hilden, Germany) with the following modifications: cells were isolated from 100 ml of liquid culture in mid-exponential growth phase by centrifugation at 4°C for 5 min at 5,000×g and cells were lysed by alternating between 5-min inversion at room temp and 5-min incubation at 67°C for three iterations after the addition of lysis buffer. Total genomic DNA was extracted from Aureoc. anophagefferens strains using the DNeasy PowerSoil Kit (Qiagen, Hilden, Germany) with the following modifications: cells were isolated from 50 ml of liquid culture in mid-exponential growth phase by centrifugation at 4°C for 5 min at 5,000×g and cells were lysed by inversion for 15 min after the addition of solution C1. DNAs extracted from Aureoc. anophagefferens and A. lagunensis samples were additionally purified using a modified CTAB protocol (Clark 1992). Prior to long read sequencing, DNA fragments below 25 kb were reduced in the DNA extracted from P. calceolata using a Short Read Eliminator (SRE) Kit (Circulomics, Baltimore, MD).
Genome Sequencing and Assembly
All long-read based sequencing libraries were prepared following the SQK-LSK109 1 D genomic DNA protocol and sequenced on FLO-MIN106 flow cells on the MinION device (Oxford Nanopore Technologies, Oxford, UK) for 48 h. Aureococcus anophagefferens strain CCMP1984 was sequenced on its own on a single flowcell. Other samples were barcoded and sequenced simultaneously on a single flow cell as follows: A. lagunensis and P. calceolata (EXP-NBD104); Aureoc. anophagefferens strains CCMP1707 and CCMP1708 (EXP-NBD104); Aureoc. anophagefferens strains CCMP1850 and CCMP3368 (EXP-NBD103). Illumina short-read sequencing was performed for all pelagophyte strains at Génome Québec (Montreal, Canada) on the NovaSeq (CCMP1707, CCMP1708, CCMP1756, CCMP1510) or HiSeq (CCMP1984, CCMP1850, CCMP3368) platforms using PCR-free, 150-bp paired-end libraries.
Long-read sequence data from barcoded samples were demultiplexed using Deepbinner (v0.2.0) and all raw data were basecalled using Guppy (v3.3.0; Oxford Nanopore Technologies) prior to trimming adaptors using Porechop (v0.2.1; https://github.com/rrwick/Porechop; last accessed February 16, 2021). Datasets were filtered using filtlong (v0.2.0; https://github.com/rrwick/Filtlong; last accessed February 16, 2021) to remove reads below 10 kb in length or with a phred score less than Q7. Nanopore long reads were assembled using Flye (v2.6) (Kolmogorov et al. 2019) with overlap parameters set to 10,000 bp for A. lagunensis and P. calceolata, and 6,500 bp for all Aureoc. anophagefferens strains. Initial genome assemblies were corrected using long-read sequence data with Nanopolish (v0.9.0; https://github.com/jts/nanopolish; last accessed February 16, 2021) and Illumina short-read sequence data with Pilon (v1.2.3) (Walker et al. 2014) as well as by manual curation. Prior to correction, Illumina sequence reads were trimmed using Trimmomatic (v0.39) (Bolger et al. 2014).
Mitochondrial Genome Annotation
For each organism, contigs corresponding to the mitochondrial genome were readily identified by assessing contig size and GC content, and were confirmed via BlastN searches against the nt database (Altschul et al. 1990). To confirm that mitochondrial contigs were circular mapping, long reads were mapped to candidate contigs using ngmlr (v0.2.7) (Sedlazeck et al. 2018) and assessed manually. Preliminary mitochondrial gene annotations were performed using GeSeq (Tillich et al. 2017) with the published mitochondrial genome from Aureoc. anophagefferens CCMP1984 (Liu et al. 2020) as reference and using ARAGORN (v1.2.38) (Laslett and Canback 2004) for tRNA gene predictions. 5S rRNA (rn5) genes were annotated using Rfam (v13.0) (Kalvari et al. 2017) and based on similarity to other ochrophyte 5S rRNA genes (Valach et al. 2014; Liu et al. 2020). All gene boundaries were checked manually for accuracy and corrected as necessary using TBlastX and BlastP (Altschul et al. 1990) alignments as a guide. Intergenic regions were carefully scanned for additional open reading frames (ORFs) that may have been missed during initial gene identification by GeSeq using TBlastX and BlastP. All manual curations were made using Geneious Prime (v2020.1.2; http://www.geneious.com/; last accessed February 16, 2021). Repeats were identified using Tandem Repeats Finder (v4.09) (Benson 1999) and the EMBOSS palindrome application (Rice et al. 2000) (http://emboss.bioinformatics.nl/cgi-bin/emboss/palindrome; last accessed February 16, 2021). Annotations were oriented arbitrarily to begin with coxI on the forward strand. Whole mitochondrial-genome alignments for the pelagophytes were generated using progressiveMauve with default settings (v1.1.1) (Darling et al. 2010). In the case of ORFs in A. lagunensis that were not annotated by GeSeq or BLAST, synteny and sequence similarity to pelagophyte homologs were assessed in order to assign gene annotations.
Phylogenetic Analysis
To investigate the evolutionary history of mitochondrial genes encoded only in Aureoc. anophagefferens or P. calceolata, phylogenies were generated by first retrieving the top 1,000 homologs below an e-value cut-off of 1e-10 from the nr database, the Tara Global Oceans Viromes dataset (Roux et al. 2016), and the transcriptome-based MMETSP dataset (Keeling et al. 2014; Caron et al. 2017) using BlastP (Altschul et al. 1990). Sequences were then aligned using MAFFT-linsi (v7.3.1) (Katoh and Standley 2013) and ambiguously aligned regions were removed using BMGE with the BLOSUM30 scoring matrix (v1.1) (Criscuolo and Gribaldo 2010). Maximum likelihood (ML) phylogenetic analyses were performed using IQ-tree (v1.5.5) (Nguyen et al. 2015) with the substitution model determined to best fit the data by standard model selection using the Bayesian information criterion (BIC) (Kalyaanamoorthy et al. 2017) and 100 standard bootstrap replicates.
Results
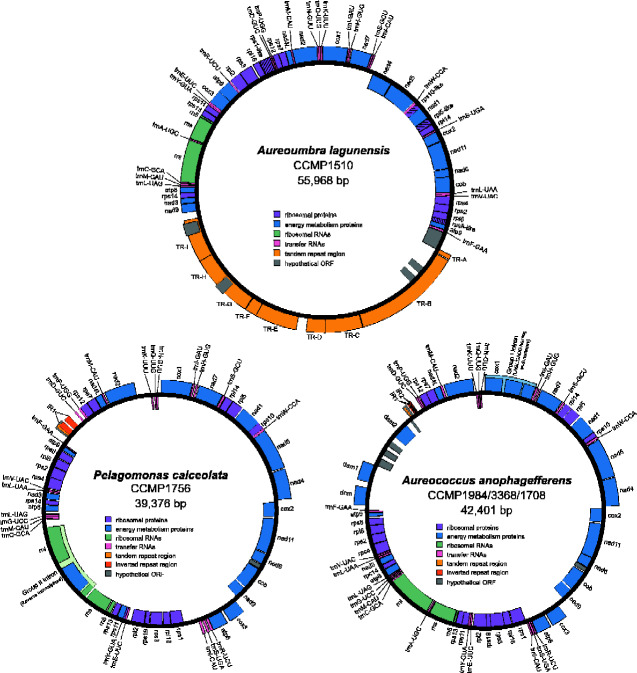
Mitochondrial genomes for all three pelagophyte species assembled into single, circular mapping contigs ranging in length from 39,376 bp (P. calceolata) (fig. 1) to 55,968 bp (A. lagunensis) (fig. 1) (table 1). The Aureoc. anophagefferens mitochondrial genome for strains CCMP1707, CCMP1984, and CCMP3368 assembled at 42,401 bp and were identical to the recently published sequence for the reference strain CCMP1984 (Liu et al. 2020) (fig. 1). The mitochondrial genomes of the two other Aureoc. anophagefferens strains (CCMP1708 and CCMP1850) were found to vary slightly in length and sequence, ranging from 99.4–99.9% sequence identity to CCMP1984 (table 1 and supplementary fig. S1, Supplementary Material online). Sequence differences between the strains are the result of a handful of SNPs in both noncoding and coding regions as well as variations in copy number of a tandem repeat (table 2). The size increase of the A. lagunensis mitochondrial genome relative to the other pelagophytes is largely attributed to an increase in repetitive elements. Over 40% of its genome (∼22 kb) consists of nine distinct tandem repeat blocks localized to a single region (fig. 1). These blocks are comprised of repeat units ranging from 60 to 284 bp in length and are present in 2–117 copies (supplementary table S1, Supplementary Material online). None of the tandem repeat unit sequences were found to show similarity to the single tandem repeat block identified in Aureoc. anophagefferens or P. calceolata. The increase in tandem repeats in A. lagunensis results in a mitochondrial genome that is only 61.2% coding, in contrast to P. calceolata and Aureoc. anophagefferens whose genomes are 84.9% and 86.0% coding, respectively.
Fig. 1.
Circular maps of the mitochondrial genome of Aureoumbra lagunensis (CCMP1510), Pelagomonas calceolata (CCMP1756), and Aureococcus anophagefferens (CCMP1984/3368/1708). Genes transcribed in the clockwise direction are shown as blocks on the outside of the circle, whereas genes transcribed in the counter-clockwise direction are shown as blocks on the inside of the circle. Genes are color-coded according to functional categories as indicated in the legend. Hash marks in an individual gene block indicates that a gene is particularly divergent and has been annotated based on the presence of an ORF, nucleotide alignments, and synteny with other pelagophyte mitochondrial genomes.
Table 1.
Summary of Pelagophyte Mitochondrial Genomes
| Species | Length (bp) | GC (%) | Number of genes |
Overlap (bp) | Percent coding (%)a | |||
|---|---|---|---|---|---|---|---|---|
| tRNA | rRNA | mRNA | Hypo-thetical | |||||
| Aureoumbra lagunensis CCMP1510 | 55,968 | 29.3 | 22 | 3 | 33 | 5 | 94 | 61.2 |
| Pelagomonas calceolata CCMP1756 | 39,376 | 35.6 | 23 | 3 | 35 | 2 | 47 | 84.9 |
|
Aureococcus anophagefferens CCMP1984 CCMP3368 CCMP1708 |
42,401 | 34.7 | 23 | 3 | 38 | 6 | 73 | 86.0 |
| CCMP1850 | 42,621 | 34.6 | 23 | 3 | 33 | 4 | 73 | 86.0 |
| CCMP1707 | 42,586 | 34.7 | 23 | 3 | 32 | 4 | 73 | 86.0 |
Percent coding was determined using protein-coding genes as well as rRNA and tRNA genes.
Table 2.
Intraspecies Variation in the Mitochondrial Genomes of Aureococcus anophagefferens Strains CCMP1707, CCMP1708, CCMP1850, CCMP1984, and CCMP3368
| CCMP1850 | CCMP1707 | ||
|---|---|---|---|
| No. differences/% identity to |
CCMP1984 CCMP1708 CCMP3368 |
235/99.4% | 198/99.5% |
| CCMP1707 | 45/99.9% | — | |
| CCMP1850 | — | 45/99.9% | |
|
Silent mutations (relative to CCMP1984) |
Non-coding regions | 4 | 4 |
| Coding regions | 5 | 5 | |
| Missense mutations | 3 | 1 | |
| Nonsense mutations | 1 | 0 | |
| –dcm gene truncated, split into two partial dcm genes | |||
| Indels | 2 | 2 | |
|
–1-bp deletion resulting in complete viral methyltransferase domain –tandem repeat expansion (17-bp repeat×19 copies, CCMP1984 has 6 copies) resulting in longer hypothetical protein |
–1-bp deletion resulting in complete viral methyltransferase domain –tandem repeat expansion (17-bp repeat×17 copies) resulting in longer hypothetical protein |
Note.—All variation is presented relative to strain CCMP1984. Where SNPs/indels have consequences on the overall structure of a protein-coding gene, the resulting changes are noted.
A core mitochondrial gene set consisting of 33 protein-coding genes was identified in all three pelagophyte species (supplementary table S2, Supplementary Material online). Aureoumbra lagunensis clearly lacks one ribosomal protein gene present in both Aureoc. anophagefferens and P. calceolata (rps19)and four ribosomal protein genes in A. lagunensis were found in particularly divergent forms (rpl5, rps1, rps10,and rps8) that were annotated on the basis of synteny and ORF length due to low sequence similarity. Homologs with putative amino-terminal mitochondrial targeting signals to these missing/divergent mitochondrial genes could not be identified in contigs assigned to the A. lagunensis nuclear genome (see below).
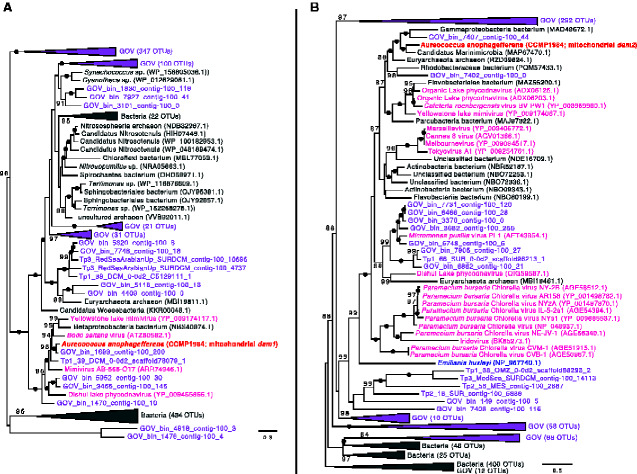
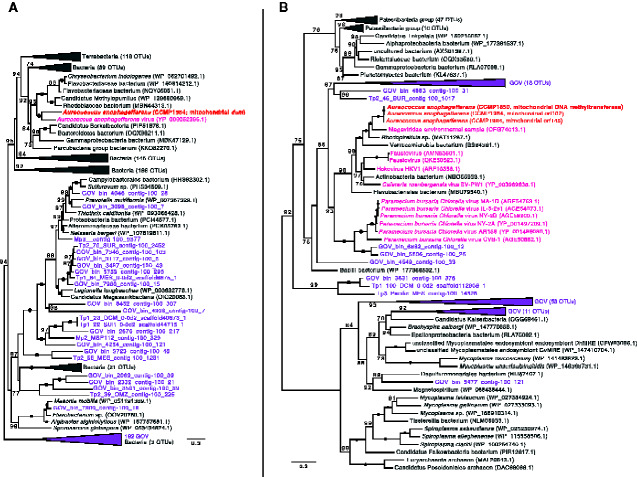
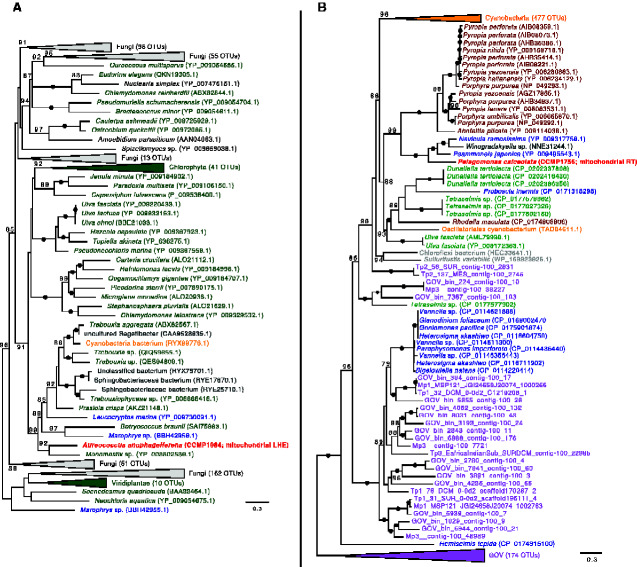
Additional protein-coding genes in the pelagophyte mitochondrial genomes include a set of hypothetical ORFs, one of which is conserved between Aureoc. anophagefferens and P. calceolata (orf79), as well as a suite of putative DNA adenine and cytosine methyltransferase genes in Aureoc. anophagefferens (dam1, dam2, dcm). The dam1, dam2, dcm, and hypothetical ORFs unique to Aureoc. anophagefferens are located in a single region of the mitochondrial genome flanked by a tRNA gene (trnF(GAA)) on one side and the inverted repeat-containing region on the other (fig. 1 and supplementary fig. S1, Supplementary Material online). Our Aureoc. anophagefferens assemblies include long reads that span this unique region and upstream/downstream native mitochondrial genes (long reads spanning the entire mitochondrial genome are also present). Phylogenetic analyses revealed that the genes in this area are most closely related to various bacterial and viral genes (figs. 2 and 3), including a homolog in the algal virus AaV in the case of the dcm gene (fig. 3A), and have no or very few obvious homologs in the organellar or nuclear genomes of other eukaryotes. Notably, most of the interstrain variation in the mitochondrial genomes of Aureoc. anophagefferens is localized to this region. The coxI gene of all Aureoc. anophagefferens strains contains a group I intron that encodes a LAGLIDADG Homing Endonuclease (LHE) (as identified previously by Liu et al. 2020) that is closely related to LHEs found in the mitochondrial genomes of various chlorophyte algae and fungi (fig. 4A).
Fig. 2.
Maximum-likelihood phylogeny of (A) dam1 and (B) dam2 genes. Aureococcus anophagefferens sequences are bolded and colored red. Bacterial/archaeal homologs are indicated in gray, whereas viral homologs are indicated in pink (nr database) or purple (Tara Global Ocean Viromes [GOV] dataset). Other colors represent eukaryotic homologs. Numbers in brackets indicate the number of homologs collapsed at a given node. Numbers on branches indicate standard bootstrap support; only values higher than 80 are shown and black circles indicate maximal support for a particular node. The tree is midpoint rooted and was inferred using 238 (dam1) and 268 (dam2) unambiguously aligned sites. The scale bar shows the number of inferred amino acid substitutions per site.
Fig. 3.
Maximum-likelihood phylogeny of (A) dcm and (B) DNA methyltransferase genes. Aureococcus anophagefferens sequences are bolded and colored red. Bacterial/archaeal homologs are indicated in gray, whereas viral homologs are indicated in pink (nr database) or purple (Tara Oceans Global Ocean Viromes [GOV] dataset). Numbers in brackets indicate the number of homologs collapsed at a given node. Numbers on branches indicate standard bootstrap support; only values higher than 70 are shown. The tree is midpoint rooted and was inferred using 304 (dcm) and 368 (DNA methyltransferase) unambiguously aligned sites. The scale bar shows the inferred number of amino acid substitutions per site.
Fig. 4.
Maximum-likelihood phylogeny of (A) the group I intron LAGLIDADG homing endonuclease (LHE) encoded in coxI in Aureococcus anophagefferens and (B) the group II intron encoded reverse transcriptase (RT) in the rnl gene in Pelagomonas calceolata. Pelagophyte sequences are bolded and colored red. Cyanobacterial homologs are indicated in orange, whereas other bacterial/archaeal homologs are indicated in gray. Viral homologs are indicated in pink (nr database) or purple (Tara Global Ocean Viromes [GOV] dataset). Other colors represent eukaryotic homologs. Numbers in brackets indicate the number of homologs collapsed at a given node. Numbers on branches indicate standard bootstrap support; only values higher than 70 are shown and black circles indicate maximal support for a particular node. The tree is midpoint rooted and was inferred using 159 (LHE) and 587 (RT) unambiguously aligned sites. The scale bar shows the number of inferred amino acid substitutions per site.
All the pelagophyte mitochondrial genomes analyzed herein possess three rRNA genes (including a 5S rRNA gene) and the same core set of 22 tRNA genes (supplementary table S3, Supplementary Material online), including three CAU anticodons (one of which was interpreted as trnI(CAU))and no trnT. The sole exception was A. lagunensis, whose genome lacks the gene trnG(UCC), and does not contain any other glycine tRNA. A group II intron encoding a reverse transcriptase/maturase was identified in the rnl gene of P. calceolata that is closely related to similar rnl intron-encoded reverse transcriptase genes in the mitochondrial genomes of some diatoms and red algae, as well as in cyanobacteria (fig. 4B). More distantly related homologs to the reverse transcriptase/maturase were identified in the GOV dataset, which contains sequences derived from the Tara Global Oceans Viromes dataset (Roux et al. 2016) (fig. 4B).
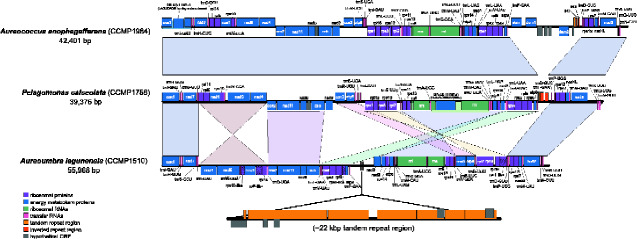
The mitochondrial genomes of Aureoc. anophagefferens and P. calceolata share near perfect synteny (fig. 5); with exception of the unique dam/dcm region in Aureoc. anophagefferens, they have identical gene arrangements along their entire length and can be aligned in a single syntenic block.In contrast, A. lagunensis shares a number of similar syntenic blocks of genes with the other two pelagophytes, but the arrangement of these blocks is rather different (fig. 5). Seven larger syntenic blocks were identified, including three that have been inverted and several which have been reordered. In A. lagunensis,genes were evenly encoded on the positive and negative strands in two distinct regions, whereas in Aureoc. anophagefferens and P. calceolata, genes are slightly more scattered over both the positive and negative strands.
Fig. 5.
Synteny map of pelagophyte mitochondrial genomes. Whole mitochondrial genomes were linearized starting at the coxI gene and aligned using progressiveMauve. Corresponding synteny blocks are shown in the same color. Mitochondrial genomes of Aureococcus anophagefferens and Pelagomonas calceolata align fully along their entire length in a single synteny block. For simplicity, a single Aureoc. anophagefferens mitochondrial genome is shown and the ∼22-kb tandem repeat region unique to Aureoumbra lagunensis is shown below its linear map. Hash marks in an individual gene block indicates that a gene is particularly divergent and has been annotated based on the presence of an ORF, nucleotide alignments, and synteny to other pelagophytes.
Discussion
Gene Presence/Absence of Ribosomal Protein and tRNA Genes
The A. lagunensis mitochondrial genome possesses four putative ribosomal protein genes (rpl5, rps1, rps10, and rps8) that are particularly divergent relative to their homologs in Aureoc. anophagefferens and P. calceolata. Although these A. lagunensis genes share only ∼20–30% amino acid identity to homologs in the other pelagophytes, they are each positioned within syntenic blocks and encode proteins of similar lengths. Furthermore, with the exception of rps1, these genes are found ubiquitously (rps8) or nearly ubiquitously (rpl5 and rps10) across the stramenopiles (Ševčíková et al. 2016). All things considered, it seems likely that the hypothetical ribosomal proteins in A. lagunensis are orthologous but divergent versions of the known ribosomal proteins in related pelagophytes (Huynen et al. 2000; Tamames 2001). The presence of an rps1-like gene in A. lagunensis and a clear rps1 in P. calceolata suggests that within the stramenopiles, rps1 is a unique feature of pelagophytes as a whole and not just Aureoc. anophagefferens. The rps1 gene is not commonly found in the mitochondrial genomes of most eukaryotic lineages (Kannan et al. 2014) and is presumed to have been lost on multiple occasions across eukaryotes and within stramenopiles (Ševčíková et al. 2016).
Homologs to mitochondrial rps19 are present in all stramenopiles surveyed thus far (Ševčíková et al. 2016) and could be clearly identified in the pelagophytes Aureoc. anophagefferens and P. calceolata. The absence of rps19 in the mitochondrial genome of A. lagunensis suggests that this gene has been lost at some point since this species diverged from the last common ancestor shared with Aureoc. anophagefferens and P. calceolata. As there are no additional mitochondrial genomes available from other members of the order to which A. lagunensis belongs (Sarcinochrysidales), the exact point at which rps19 was lost cannot be pinpointed with certainty. Furthermore, we found no evidence that the “missing” rps19 mitochondrial gene resides in the nuclear genome in A. lagunensis,or indeed evidence of mitochondrial targeting signals on eukaryotic (i.e., nonmitochondrial) homologs in the nuclear genome. As such, it appears that the A. lagunensis mitochondrion functions without this particular ribosomal protein or imports a nucleus-encoded copy that we were unable to identify.
With the exception of a missing glycine tRNA gene in A. lagunensis, all three pelagophyte species examined herein have identical mitochondrial tRNA gene sets encoding tRNAs for all standard amino acids except threonine. A gene for tRNA-Thr is not encoded in any other stramenopile mitochondrial genome except for a single eustigmatophyte species (Monodopsis) and is thought to have already been lost from the organelle in the common ancestor of all stramenopiles (Burger and Nedelcu 2012; Ševčíková et al. 2016). A gene for tRNA-Gly, on the other hand, has been found in all stramenopile mitochondrial genomes examined thus far (Ševčíková et al. 2016) and appears to be a unique loss in A. lagunensis. As the amino acid threonine (and glycine in the case of A. lagunensis) is still present in pelagophyte mitochondrial-encoded proteins (∼5% of amino acid residues in both cases), it is likely that nuclear-encoded tRNAs are imported into mitochondria from the cytosol, as occurs across the eukaryotic tree (Salinas-Giegé et al. 2015). Even when there is a corresponding mitochondrial-encoded tRNA, some species have been shown to import a cytosolic version of a given tRNA into the organelle (e.g., Vinogradova et al. 2009). The decreased use of glycine in mitochondrial protein-coding sequences in A. lagunensis (4.8% of sites) compared with that of the other pelagophytes (6.3% of sites) may be related to the lack of a mitochondrial encoded tRNA-Glyand import of cytosolic tRNA-Gly; the mitochondrial genome could have evolved by fine-tuning codon usage to reflect availability of tRNA-Gly rather than adopting cytosolic tRNA import (Salinas-Giegé et al. 2015).
Three CAU anticodons were identified in all the pelagophyte mitochondrial genomes considered here, one of which likely corresponds to trnI(CAU)rather than trnM(CAU), as has been suggested in a variety of stramenopiles, including Aureoc. anophagefferens (Ševčíková et al. 2016;Cai and Scofield 2020). Transcribed trnI(CAU)is thought to be posttranscriptionally modified from cytosine to lysidine at the first position of the anticodon such that it acts as trnI(AUA)(Grosjean and Björk 2004; Lang et al. 2012).
Presence of Dam and DcmGenes in Aureococcus anophagefferens
We have shown that a unique ∼6.8-kb region in the Aureoc. anophagefferens mitochondrial genome contains a set of dam and dcm genes, as well as a hypothetical protein(s) with homology to a DNA methyltransferase and other hypothetical ORFs (note that our sequence data include numerous long reads that span this entire region and indeed the entire mitochondrial genome). Dam and dcm genes are generally unique to bacteria and viruses, although one has been identified in the mitochondrial genome of the bloom-forming haptophyte alga Emiliania huxleyi (Sánchez-Puerta et al. 2004) and some streptophyte green algae (Turmel et al. 2013). Intriguingly, Aureoc. anophagefferens has multiple dam genes and a dcm gene, whereas the aforementioned mitochondrial genomes have either a single dam or dcm gene. It is worth noting that most of the intraspecies sequence variation observed across the five Aureoc. anophagefferens mitochondrial genomes sequenced herein is localized to the dam/dcm-containingregion. This region is also flanked by a tRNA gene and an inverted repeat region on either end (figs. 1 and 5 and supplementary fig. S1, Supplementary Material online), both of which have been linked to the insertion of foreign DNA (e.g., Juhas et al. 2009).
Phylogenetic analysis of each of these dam and dcm genes strongly suggests that they are the result of one or more lateral gene transfer (LGT) events (figs. 2 and 3), either directly to the mitochondrial genome or by an initial transfer to the nuclear genome followed by subsequent relocation to the organellar genome. Although homologs of each of these genes are almost exclusively found in various bacteria and viruses, their precise origin(s) in Aureoc. anophagefferens is unclear. The genome of AaV—the dsDNA virus that infects Aureoc. anophagefferens—has a dcm gene that branches robustly with the Aureoc. anophagefferens mitochondrial homolog (fig. 3A), but this is not in the case for the other genes. The Aureoc. anophagefferens and AaV dcm genes are the only closely related, nonprokaryotic sequences that we could identify, suggesting that the mitochondrial homolog was obtained from a bacterium, but it remains unclear if the gene was first picked up by AaV and then transferred to Aureoc. anophagefferens during viral infection or vice versa. Interestingly, a single base-pair mutation in strain CCMP1850 from G to A at position 497 results in an early stop codon and thus a truncated version of the dcm gene product; the latter third of the putative protein is encoded by a separate, stand-alone ORF (with its own start codon). Although a dcm gene has been observed in the mitochondrial genomes of two streptophyte algae (Klebsormidium flaccidum and Microspora stagnorum) that are thought to be a result of a viral-to-host LGT (Turmel et al. 2013), the dcm gene found in Aureoc. anophagefferens is not at all closely related to these green algal homologs (data not shown) and appears to have a separate origin.
Although dam1 and dam2 genes are not found in the AaV genome, close homologs were identified in the genomes of a number of other large double-stranded DNA (dsDNA) viruses as well as in various bacterial species and in metagenomics-derived viral contigs present in the Tara Oceans Global Ocean Viromes (GOV) dataset (Roux et al. 2016) (fig. 2). Interestingly, as noted above there is a dam2 homolog in the mitochondrial genome of E. huxleyi, which was suggested to be a result of LGT when it was originally identified (Sánchez-Puerta et al. 2004). As is the case for Aureoc. anophagefferens, E. huxleyi has its own dsDNA virus that is thought to play a role in controlling its bloom dynamics (e.g., Bratbak et al. 1993). As such, it was previously speculated that the dam2 gene in E. huxleyi could control the virulence of its virus and possibly other pathogens, or be related to mitochondrial genome replication or gene expression (Sánchez-Puerta et al. 2004), processes known to involve adenine methylation in Escherichia coli (Heithoff et al. 1999). It is thus possible that the dam genes play similar functional roles in Aureoc. anophagefferens, although there is no evidence for this at the present time.
Liu et al. (2020) noted that the Aureoc. anophagefferens CCMP1984 mitochondrial genome encodes a putative DNA methyltransferase in which the N and C terminal portions are the product of two separate ORFs, with a single T insertion at position 294 presumed to have given rise to the split gene. In addition to confirming the results of Liu et al. for CCMP1984, the same “split” genotype was observed here in the mitochondrial genomes of CCMP3368 and CCMP1708. Notably, however, strains CCMP1850 and CCMP1707 possess an intact T294 insertion-lacking version of the gene, as predicted to exist by Liu et al. (2020), thus giving rise to a single ORF that encodes a partial type II restriction m6 adenine DNA methyltransferase domain. As with the dam and dcm genes mentioned above, molecular phylogenies show that this DNA methyltransferase gene is closely related to homologs in dsDNA viruses, in viral-derived contigs in the GOV dataset, and in bacteria, with specific branching patterns suggesting that it is the result of LGT directly from a virus (that may have originally obtained the gene from a bacterial species) or from a bacterium after a series of LGTs between multiple bacteria and viruses (fig. 3B). Although homologs to most of the methyltransferase genes are not present in the genome of the AaV isolate that was sequenced (Moniruzzaman et al. 2014), it is possible that the Aureoc. anophagefferens genes were acquired from AaV isolates that have not yet been sampled. The plethora of dam and dcm homologs we retrieved from the GOV database make this a distinct possibility (figs. 2 and 3).
Mitochondrial Encoded Introns
Group I and II introns are mobile elements that can move readily both within and between genomes. We found a group I intron encoding a LAGLIDADG homing endonuclease (LHE) in the mitochondrial coxI gene of Aureoc. anophagefferens. Phylogenetic analysis of the LHE shows that it is closely related to homologs in green algae and fungi, many of which are also found in the mitochondrial coxI gene (fig. 4A). That said, outside of green algae and fungi, an LHE homolog has thus far only been found in the mitochondrial coxI gene of the centrohelid Marophrys sp., the katablepharid Leucocryptos marina, and a few bacteria. Although group II introns have been identified in the mitochondrial coxI gene (as well as the rnl gene) of various stramenopiles, group I introns are relatively rare (Guillory et al. 2018). It is unclear whether or not this intron is found only in Aureoc. anophagefferens (among stramenopiles) due to recurrent loss or LGT. Its presence is not obviously an invasion from a nuclear version of the LHE, as a homolog in the nuclear genome was not readily identified in our Aureoc. anophagefferens genomic data or that of the other pelagophytes examined here (data not shown). A lateral transfer of a coxI group II intron was previously suggested to have occurred from diatoms to raphidophytes belonging to the genus Chattonella (Kamikawa et al. 2009; Guillory et al. 2018); a similar lateral transfer may have occurred here, but with a group I intron in the coxI gene between green algae and Aureoc. anophagefferens. Interestingly, homing endonucleases have been identified in the genomes of “giant” viruses (e.g., Gallot-Lavallée et al. 2017; Deeg et al. 2018), including two putative HNH homing endonucleases in the Aureoc. anophagefferens virus genome (Moniruzzaman et al. 2014). Although homologs of the LHE encoded in the Aureoc. anophagefferens mitochondrial genomewere not identified in any published viral genome or in the GOV dataset, it is possible that this gene was acquired from a giant virus that has not yet been sampled, or that has lost the corresponding homolog.
A group II intron was identified in the mitochondrial rnl gene of P. calceolata; this intron encodes a reverse transcriptase/maturase. Similar group II introns have been identified in the mitochondrial genomes of various stramenopiles, including in the rnl gene of diatoms (Guillory et al. 2018). Group II introns are also common in the mitochondrial rnl gene of red algae (Yang et al. 2015) and particularly common in coxI, as mentioned above, where they have been identified in red algae (Yang et al. 2015), diatoms (Guillory et al. 2018), and raphidophycean flagellates (Chattonella spp.; Kamikawa et al. 2009). Phylogenetic analysis of the reverse transcriptase encoded in P. calceolata’s mitochondrial rnl intron shows that it is most closely related to a reverse transcriptase encoded in the rnl gene of the diatoms Psammoneis japonica (Guillory et al. 2018) and Navicula ramosissima (An et al. 2016) and the bacterium Winogradskyella sp., as well as in the red algae Pyropia spp. (fig. 4B). Numerous close homologs are also found in cyanobacterial species, suggesting (in combination with its sparse distribution among eukaryotes) that this intron was initially acquired in a eukaryote lineage via LGT from cyanobacteria, either the result of multiple recent lineage-specific acquisitions or a single acquisition followed by eukaryote–eukaryote LGTs (unrelated to plastid establishment). It is also conceivable that the reverse transcriptase-encoding intron was transferred from the plastid to the mitochondrial genome (on one or multiple occasions) and/or recurrently lost in the majority of plastid-bearing lineages. Interestingly, more distantly related reverse transcriptase homologs were identified in viral contigs present in the GOV dataset; none of these viral sequences, however, are closely related to the P. calceolata reverse-transcriptase gene and all branch together at the base of the tree.
Genome Structure
Mitochondrial gene order is perfectly conserved between Aureoc. anophagefferens and P. calceolata (fig. 5), reflecting their close phylogenetic relationship within the same phylogenetic order (Pelagomonadales). In comparison, several genome rearrangements have occurred since these organisms diverged from the more distantly related pelagophyte A. lagunensis (order Sarcinochrysidales). Seven blocks of fully collinear genes between A. lagunensis and P. calceolata/Aureoc. anophagefferens were identified, each with two to sixcore protein-coding genes. The relative arrangement of these blocks suggests that they are the result of three inversions and at least two translocations. Other mitochondrial comparative genomic studies of stramenopile lineages have shown similar patterns in the number of gene rearrangements between classes of algae, noting highly conserved or identical gene order within a given order (Ševčíková et al. 2016; Liu et al. 2019, 2020).
Other than the unique dcm/dam gene-containing region found in Aureoc. anophagefferens (discussed above), the only substantial difference between the pelagophyte mitochondrial genomes is the addition of a large tandem repeat region in A. lagunensis. This region consists of nine distinct repeat units that together comprise ∼40% of the A. lagunensis mitochondrial genome. Although this is an unusual feature compared with the other pelagophytes investigated here, similar observations have been made in other algal lineages. For example, large tandem repeat regions have been identified in the mitochondrial genomes of algal species ranging in length from 3.3 kb in the diatom Synedra acus (10.6% of its mitochondrial genome) (Ravin et al. 2010) to 9.3 kb in the haptophyte Chrysochromulina tobin (28% of its mitochondrial genome) (Hovde et al. 2014) and 35.4 kb in the diatom Phaeodactylum tricornutum (45.5% of its mitochondrial genome) (Secq and Green 2011). In any given case the exact origin(s) of these repeats is usually unclear, but it is generally assumed that they are the product of strand slippage during recombination, as the repeats are direct, with few inversions. Once in place, the repeat tract can expand or contract over short evolutionary timescales, and serve as a hot spot for genome rearrangements, as observed here in the pelagophytes.
Conclusion
The formation of harmful algal blooms by pelagophyte algae is a problem of increasing ecological and economic significance (Gobler and Sunda 2012; Hall et al. 2018; Tang et al. 2019). Answers to the “who,” “how,” and “why” questions of bloom establishment and collapse rely on a detailed understanding of pelagophyte biology and the viruses that prey upon them. Laboratory experiments have shown strain-specific differences in resistance to viral infection in the pelagophyte Aureoc. anophagefferens (e.g., Gobler et al. 2007), and genome sequences provide an important reference point for understanding these differences. We have analyzed the mitochondrial genomes of five different Aureoc. anophagefferens strains, three of which have identical sequences. Interestingly, all five genomes contain DNA adenine and cytosine methyltransferase genes with distinct viral affinities, genes that are absent in the mitochondrial genomes of P. calceolata and A. lagunensis (and indeed any other known stramenopiles). Exactly how these genes came to reside in the Aureoc. anophagefferens genome is unclear, as is their biological relevance—these are important topics for future research. Nevertheless, the presence of viral genes in Aureoc. anophagefferens provides further evidence for host-viral coevolution in pelagophytes, as suggested previously by the presence of host genes in the AaV genome (Moniruzzaman et al. 2014). Moving forward, nuclear genomic data from multiple Aureoc. anophagefferens strains as well as P. calceolata, A. lagunensis,and other pelagophytes will be needed to provide a much stronger foundation for understanding pelagophyte-specific bloom dynamics and, more generally, harmful algal blooms caused by other algae.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council of Canada (RGPIN 05871-2014 to J.M.A., CGS-D to S.J.S., and USRA to M.L.), the Gordon and Betty Moore Foundation (GBMF5782 to J.M.A.) and the Killam Trusts (to S.J.S.). We thank three anonymous reviewers for their helpful comments on an earlier version of this manuscript.
Data Availability
Mitochondrial genomes for Aureoc. anophagefferens (CCMP1984, CCMP3368, CCMP1707, CCMP1708, CCMP1850),A. lagunensis (CCMP1510), and P. calceolata (CCMP1756) are available in the GenBank database under accessions: MW438348, MW438347, MW438345, MW4383446, MW438344, MW438350, and MW438349, respectively. Raw sequence datasets are available in the SRA repository under accession numbers: SRR13386523 (CCMP1984), SRR13386499 (CCMP3368), SRR13386516 (CCMP1707), SRR13386522 (CCMP1708), SRR13378193 (CCMP1850), SRR13386728 (CCMP1510), and SRR13386796 (CCMP1756).
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- An SM, et al. 2016. Complete mitochondrial genome of biraphid benthic diatom, Navicula ramosissima (Naviculales, Bacillariophyceae). Mitochondrial DNA B. 1(1):549–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen RA, Saunders GW, Paskind MP, Sexton JP.. 1993. Ultrastructure and 18s rRNA gene sequence for Pelagomonas calceolata gen. et sp. nov. and the description of a new algal class, the pelagophyceae classis nov. J Phycol. 29(5):701–715. [Google Scholar]
- Bailey JC, Andersen RA.. 1999. Analysis of clonal cultures of the brown tide algae Aureococcus and Aureoumbra (pelagophyceae) using 18s rRNA, rbcl, and rubisco spacer sequences. J Phycol. 35(3):570–574. [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratbak G, Egge J, Heldal M.. 1993. Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar Ecol Prog Ser. 93:39–48. [Google Scholar]
- Burger G, Nedelcu AM.. 2012. Mitochondrial genomes of algae In: Bock R, Knoop V, editors. Genomics of chloroplasts and mitochondria. Advances in photosynthesis and respiration series.Vol. 35. London: Springer. p. 127–157. [Google Scholar]
- Cai G, Scofield SR.. 2020. Mitochondrial genome sequence of Phytophthora sansomeana and comparative analysis of Phytophthora mitochondrial genomes. PLoS One 15(5):e0231296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron DA, et al. 2017. Probing the evolution, ecology and physiology of marine protists using transcriptomics. Nat Rev Microbiol. 15(1):6–20. [DOI] [PubMed] [Google Scholar]
- Clark CG. 1992. DNA purification from polysaccharide-rich cells. In: Lee JJ, Soldo AT,editors.Protocols in protozoology.Vol. 1. Kansas: Allen Press. p. D-3.1-2. [Google Scholar]
- Criscuolo A, Gribaldo S.. 2010. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 10(1):210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT.. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5(6):e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeg CM, Chow C-ET, Suttle CA.. 2018. The kinetoplastid-infecting Bodo saltans virus (BsV), a window into the most abundant giant viruses in the sea. Elife 7:e33014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoe HR, et al. 1997. Description and characterization of the algal species Aureoumbra lagunensis gen. et sp. nov. and referral of Aureoumbra and Aureococcus to the Pelagophyceae. J Phycol. 33(6):1042–1048. [Google Scholar]
- Frischkorn KR, Harke MJ, Gobler CJ, Dyhrman ST.. 2014. De novo assembly of Aureococcus anophagefferens transcriptomes reveals diverse responses to the low nutrient and low light conditions present during blooms. Front Microbiol. 5:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallot-Lavallée L, Blanc G, Claverie J-M.. 2017. Comparative genomics of Chrysochromulina Ericina virus and other microalga-infecting large DNA viruses highlights their intricate evolutionary relationship with the established Mimiviridae family. J Virol. 91(14):e00230–e00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastrich M, Anderson RO, Benmayor SS, Cosper EM.. 1998. Ultrastructural analysis of viral infection in the brown-tide alga, Aureococcus anophagefferens (Pelagophyceae). Phycologia 37(4):300–306. [Google Scholar]
- Gobler CJ, et al. 2013. Expansion of harmful brown tides caused by the pelagophyte, Aureoumbra lagunensis DeYoe et Stockwell, to the US east coast. Harmful Algae. 27:29–41. [Google Scholar]
- Gobler CJ, Sunda WG.. 2012. Ecosystem disruptive algal blooms of the brown tide species, Aureococcus anophagefferens and Aureoumbra lagunensis. Harmful Algae. 14:36–45. [Google Scholar]
- Gobler CJ, et al. 2011. Niche of harmful alga Aureococcus anophagefferens revealed through ecogenomics. Proc Natl Acad Sci U S A. 108(11):4352–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobler CJ, Anderson OR, Gastrich MD, Wilhelm SW.. 2007. Ecological aspects of viral infection and lysis in the harmful brown tide alga Aureococcus anophagefferens. Aquat Microb Ecol. 47:25–36. [Google Scholar]
- Grosjean H, Björk GR.. 2004. Enzymatic conversion of cytidine to lysidine in anticodon of bacterial tRNAIle – an alternative way of RNA editing. Trends Biochem. Sci. 29(4):165–168. [DOI] [PubMed] [Google Scholar]
- Guillard RRL. 1975. Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH,editors. Culture of marine invertebrate animals. New York: Plenum Press. p. 29–60. [Google Scholar]
- Guillory WX, et al. 2018. Recurrent loss, horizontal transfer, and the obscure origins of mitochondrial introns in diatoms (Bacillariophyta). Genome Biol Evol. 10:evy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall NS, et al. 2018. Consortial brown tide − picocyanobacteria blooms in Guantánamo Bay, Cuba. Harmful Algae. 73:30–43. [DOI] [PubMed] [Google Scholar]
- Heithoff DM, Sinsheimer RL, Low DA, Mahan MJ.. 1999. An essential role for DNA adenine methylation in bacterial virulence. Science 284(5416):967–970. [DOI] [PubMed] [Google Scholar]
- Hovde BT, et al. 2014. The mitochondrial and chloroplast genomes of the haptophyte Chrysochromulina tobin contain unique repeat structures and gene profiles. BMC Genomics 15(1):604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen M, Snel B, Lathe W, Bork P.. 2000. Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res. 10(8):1204–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, et al. 2009. Genomic islands: tools of bacterial horizontal gene transfer and evolution. Fems Microbiol Rev. 33(2):376–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I, et al. 2017. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 46:gkx1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh B, Wong TK, Haeseler AV, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikawa R, et al. 2009. Mitochondrial group II introns in the raphidophycean flagellate Chattonella spp. suggest a diatom-to-Chattonella lateral group II intron transfer. Protist 160(3):364–375. [DOI] [PubMed] [Google Scholar]
- Kannan S, Rogozin IB, Koonin EV.. 2014. MitoCOGs: clusters of orthologous genes from mitochondria and implications for the evolution of eukaryotes. BMC Evol Biol. 14(1):237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, et al. 2014. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12(6):e1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yoon H, Yi G, Shin W, Archibald JM.. 2018. Comparative mitochondrial genomics of cryptophyte algae: gene shuffling and dynamic mobile genetic elements. BMC Genomics 19(1):275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA.. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 37(5):540–546. [DOI] [PubMed] [Google Scholar]
- Lang BF, Lavrov D, Beck N, Steinberg SV.. 2012. Mitochondrial tRNA Structure, Identity, and Evolution of the Genetic Code. In: Bullerwell C, editor. Organelle Genetics. Berlin: Springer. p. 431–474.
- Laslett D, Canback B.. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Liu S, Huang T, Chen N.. 2020. Construction and comparative analysis of mitochondrial genome in the brown tide forming alga Aureococcus anophagefferens (Pelagophyceae, Ochrophyta). J Appl Phycol. 32(1):441–450. [Google Scholar]
- Liu F, Zhang Y, Bi Y, Chen W, Moejes FW.. 2019. Understanding the evolution of mitochondrial genomes in phaeophyceae inferred from mitogenomes of Ishige okamurae (Ishigeales) and Dictyopteris divaricata (Dictyotales). J Mol Evol. 87(1):16–26. [DOI] [PubMed] [Google Scholar]
- Moniruzzaman M, et al. 2016. Diversity and dynamics of algal Megaviridae members during a harmful brown tide caused by the pelagophyte, Aureococcus anophagefferens. FEMS Microbiol Ecol. 92(5):fiw058. [DOI] [PubMed] [Google Scholar]
- Moniruzzaman M, et al. 2014. Genome of brown tide virus (AaV), the little giant of the Megaviridae, elucidates NCLDV genome expansion and host–virus coevolution. Virology 466:60–70. [DOI] [PubMed] [Google Scholar]
- Moniruzzaman M, Gann ER, Wilhelm SW.. 2018. Infection by a giant virus (AaV) induces widespread physiological reprogramming in Aureococcus anophagefferens CCMP1984 – a harmful bloom algae. Front Microbiol. 9:752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong HC, et al. 2010. Analyses of the complete chloroplast genome sequences of two members of the pelagophyceae: aureococcus anophagefferens CCMP1984 and Aureoumbra lagunensis CCMP1507. J Phycol. 46(3):602–615. [Google Scholar]
- Ravin NV, et al. 2010. Complete sequence of the mitochondrial genome of a diatom alga Synedraacus and comparative analysis of diatom mitochondrial genomes. Curr Genet. 56(3):215–223. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A.. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16(6):276–277. [DOI] [PubMed] [Google Scholar]
- Roger AJ, Muñoz-Gómez SA, Kamikawa R.. 2017. The origin and diversification of mitochondria. Curr Biol. 27(21):R1177–R1192. [DOI] [PubMed] [Google Scholar]
- Roux S, et al. 2016. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537(7622):689–693. [DOI] [PubMed] [Google Scholar]
- Salinas-Giegé T, Giegé R, Giegé P.. 2015. tRNA biology in mitochondria. Int J Mol Sci. 16(3):4518–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Puerta MV, Bachvaroff TR, Delwiche CF.. 2004. The complete mitochondrial genome sequence of the haptophyte Emiliania huxleyi and its relation to heterokonts. DNA Res. 11(1):1–10. [DOI] [PubMed] [Google Scholar]
- Secq M-P-L, Green BR.. 2011. Complex repeat structures and novel features in the mitochondrial genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana. Gene 476(1-2):20–26. [DOI] [PubMed] [Google Scholar]
- Sedlazeck FJ, et al. 2018. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 15(6):461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ševčíková T, et al. 2016. A comparative analysis of mitochondrial genomes in eustigmatophyte algae. Genome Biol Evol. 8(3):705–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth JMcN, Johnson PW, Hargraves PE.. 1988. Ultrastructure and ecology of Aureococcus anophageferens gen. et sp. nov. (chrysophyceae): the dominant picoplankton during a bloom in Narragansett Bay, Rhode Island, summer 1985. J Phycol. 24(3):416–425. [Google Scholar]
- Smith DR. 2016. The past, present and future of mitochondrial genomics: have we sequenced enough mtDNAs? Brief Funct Genomics. 15(1):47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamames J. 2001. Evolution of gene order conservation in prokaryotes. Genome Biol. 2(6):research0020.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YZ, et al. 2019. 3,000 km and 1,500-year presence of Aureococcus anophagefferens reveals indigenous origin of brown tides in China. Mol Ecol. 28(17):4065–4076. [DOI] [PubMed] [Google Scholar]
- Tillich M, et al. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C.. 2013. Tracing the evolution of streptophyte algae and their mitochondrial genome. Genome Biol Evol. 5(10):1817–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valach M, Burger G, Gray MW, Lang BF.. 2014. Widespread occurrence of organelle genome-encoded 5S rRNAs including permuted molecules. Nucleic Acids Res. 42(22):13764–13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova E, Salinas T, Cognat V, Remacle C, Maréchal-Drouard L.. 2009. Steady-state levels of imported tRNAs in Chlamydomonas mitochondria are correlated with both cytosolic and mitochondrial codon usages. Nucleic Acids Res. 37(5):1521–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetherbee R, Gornik SG, Grant B, Waller RF.. 2015. Andersenia, a genus of filamentous, sand-dwelling Pelagophyceae from southeastern Australia. Phycologia 54(1):35–48. [Google Scholar]
- Yang EC, et al. 2015. Highly conserved mitochondrial genomes among multicellular red algae of the florideophyceae. Genome Biol Evol. 7(8):2394–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q-C, et al. 2012. Emergence of brown tides caused by Aureococcus anophagefferens Hargraves et Sieburth in China. Harmful Algae. 19:117–124. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mitochondrial genomes for Aureoc. anophagefferens (CCMP1984, CCMP3368, CCMP1707, CCMP1708, CCMP1850),A. lagunensis (CCMP1510), and P. calceolata (CCMP1756) are available in the GenBank database under accessions: MW438348, MW438347, MW438345, MW4383446, MW438344, MW438350, and MW438349, respectively. Raw sequence datasets are available in the SRA repository under accession numbers: SRR13386523 (CCMP1984), SRR13386499 (CCMP3368), SRR13386516 (CCMP1707), SRR13386522 (CCMP1708), SRR13378193 (CCMP1850), SRR13386728 (CCMP1510), and SRR13386796 (CCMP1756).