Graphical abstract
Keywords: SARS-CoV-2, COVID-19, Mpro, MTase, Thiazolide Derivatives
Abstract
Humans around the globe have been severely affected by SARS-CoV-2 and no treatment has yet been authorized for the treatment of this severe condition brought by COVID-19. Here, an in silico research was executed to elucidate the inhibitory potential of selected thiazolides derivatives against SARS-CoV-2 Protease (Mpro) and Methyltransferase (MTase). Based on the analysis; 4 compounds were discovered to have efficacious and remarkable results against the proteins of the interest. Primarily, results obtained through this study not only allude these compounds as potential inhibitors but also pave the way for in vivo and in vitro validation of these compounds.
1. Introduction
Coronaviruses are the members of order Nidovirales and the subfamily Coronaviridae. These viruses are the etiological cause of severe human and animal infections, which leads to disturbance not only in the respiratory tract but also in the digestive tract. After evaluating the genomic structures of COVID 19, it was found out that it belongs to genera Beta coronavirus. The identification of strain CoV originally called 2019-nCoV was done during an outbreak of unusual viral pneumonia in Wuhan, China [1].
This outbreak was later recognized as a pandemic, which affected people around the globe. Moreover, due to severe global health threats, changes in climate and ecology, increased interactions of human with animals and current emergence, the new CoV outbreaks are thought to be unavoidable. And thus, effective therapies and vaccines against CoVs are required to be developed urgently and effectively. Coronaviruses (CoVs) are single-stranded RNA viruses consisting of the largest genome of the size 3.17 kb, among all RNA viruses. This largest genome consists of six to ten open reading frames. The genetic material of CoVs is found to be susceptible to the frequent recombination process, leading to the production of new strains with altered virulence [2]. Till date, seven known strains of human CoVs includes; 229E, NL63, OC43, HKU1, Middle East respiratory syndrome (MERS)-CoV, severe acute respiratory syndrome (SARS)-CoV, and 2019-novel coronavirus (nCoV). Out of these seven strains, three strains; SARS-CoV, MERS-CoV, and 2019-nCoV are proved to be highly pathogenic strains and have caused endemic of severe CoV disease [3]. Although the reservoir of (SARS)-CoV is unknown, yet bats and subsequent spread to Himalayan palm civets are hypothesized [4]. The outbreak of (SARS)-CoV, firstly occurred in Guangdong province of China in 2003. The second outbreak of (MERS)-CoV occurred during 2012 in Saudi Arabia, MERS-CoV is transmitted through camels and it’s found to be of zoonotic origin in Middle East [4].
Although these previous two attacks of CoV were known to cause milder diseases yet they were highlighted to have the adaptive potential to changing environmental conditions. This adaptive potential lead to their classification as “emerging viruses” and that’s why it is necessary to have a familiarity about their structure, metabolic pathways and physiology of CoV-associated diseases. Hence, leading to the more possibilities would be there to distinguish possible drug targets [5].
The RNA genome, packed in the nucleocapsid protein and being covered by an envelope, has seven conserved genes: ORF1a, ORF1b, S, OEF3, E, M, and N in 5′ to 3′ direction [1]. Almost two-third part of the RNA genome has been covered by ORF1a/b and produces two viral replicase proteins, known as the polyprotein PP1a and the polyprotein PP1b. These polyproteins are further getting auto proteolytically processed into sixteen mature non-structural (Nsp1-16) proteins [5]. These proteins form a replicase/transcriptase complex. While the other part of the genome encodes the structural proteins being produced by the mRNA, i.e., spike (S), envelope (E), membrane (M), and nucleocapsid (N), and other accessory proteins or putative adjuvant factors. The trimeric spike protein, membrane protein, envelop proteins and nucleocapsid protein are the most strategic structural proteins of CoV. Hemagglutinin esterase glycoprotein (HE) is another protein being expressed by a few strains of CoV [6]. A protease enzyme; Mpro or 3CLpro, plays an essential role in the replication and maturation life cycle of COVID-19. The novel coronavirus is responsible for the severe infection with special reference to the involvement of both upper and lower respiratory tract and digestive system. The main symptoms of infections are common cold, pneumonia, bronchiolitis, rhinitis, pharyngitis and sinusitis, as well as occasional diarrhea [1].
COVID-19 being part of coronaviruses is among quickly spreading viruses, and that’s why instead of following individual strategy for drug designing, a reasonable and attractive strategy is required to develop broad-spectrum inhibitory drugs for this virus. This type of drug developing strategy would help to develop drugs that can provide better first-line defence against current and future emerging CoV-associated threats and ailments. The over-activation of T cells and macrophages is induced by SARS-CoV-2 is another aspect of its pathology, which leads to the cytokine storms [7]. The cytokine storm is considered fatal, especially in the lower lungs of patients with severe SARS-CoV-2 infection. T cells, upon activation, require an increased amount of methionine and cannot survive at low methionine level. Limiting the amount of methionine can prevent the cytokines storm; this can be done through the body methionase, an enzyme responsible for the breakdown of methionine. Methionase is found to be very stable and strong even at a temperature of 50 °C [5].
All three genetic clusters have been represented by homology models and displayed highly frequent mutations in the genome sequence of CoV [5]. In order to develop wide-spectrum drugs, the requirement of conserved target sequences within the whole genus coronavirus is inevitable. And that’s why extensive research has been done to discover the possible potent target. This directed to the identification of Mpro (molecular weight 34 kDa) which is the main CoV protease and is involved in the overall replication and transcription. Moreover, by comparing four crystal structures it was found that Mpro shares highly conserved substrate-recognition pockets [2]. Moreover, none of the parts of COVID-19 genome has been found to be more exposed than its main protease, which highlights Mpro as the best target for the SARS-CoV-2 replication [1]. This important drug target; cysteine protease known as 3C-like proteinase (3CLpro), plays a central and essential role in viral replication and transcription through extensive proteolysis of polyprotein replicases, pp1a and pp1ab. Although this 3C-like proteinase is found in coronavirus replication polyprotein it’s separated by its proteolytic activity and formation of a homodimer with an active centre per unit. At least 11 interdomain sites of polyproteins; PP1a and PP1b are found to be cleaved by it, leading to the production of functional proteins including RNA-directed RNA polymerase, helicase, exoribonuclease, endoribonuclease, and 2′-O-ribose methyltransferases. These properties of 3CLpro, definitely make it an important drug target for SARS-CoV-2 [8].
Being a positive sense, single-stranded RNA virus, the genomic RNA is used by SARS-CoV-2 for translation and replication [7]. In order to bring about proper RNA replication and translation, the methylation of viral RNA cap is required. However, there are two apparent methylation sites present in coronavirus RNA; one is essentially required for the replication and translation while the other site is required to help viral RNA in order to escape from the host immune system that can degrade the RNA in the absence of cap methylation [3]. The non-structural coronavirus (nsp) proteins, especially nsp10, nsp14 and nsp16, appear to be methyltransferase RNA or methyltransferase-related proteins [6]. The universal donor of methyl; uses S-adenosylmethionine (SAM), is being used by Methyltransferase for the transfer of methyl groups to the viral genome. The synthesis of SAM chiefly depends upon the presence of methionine, which suggests that the low level of methionine will stop methyltransferase from completing its response [9]. The transfer of the methyl group to SAM doesn’t only methylate the molecule but also produces S-adenosyl homocysteine (SAH); this reaction is catalyzed through an enzyme methionine adenosyl transferase (MAT). Moreover, the ratio of SAM to SAH is also important as methyltransferase reaction also cannot be preceded in low ratio. As a result, the viral genome becomes prone to degradation [2].
In order to recognize and characterize two cap methyltransferases (MTases), guanine-N7-MTase and ribose-2′-O-MTase of coronaviruses, the extensive study of RNA cap formation and methylation has been done previously [10]. This suggests that the novel coronavirus SARS-CoV-2 contains Protease Mpro. Wu et al, initially determined the pneumonia virus from the seafood market of Wuhan, city of china and studied it through multiple sequence alignment with known SARS protease. As a result, they successfully published a genome sequence (NCBI genome ID MN908947, GenBank MN908947.3) showing highly accurate similarity with SARS crystal structure [11]. Another protein found to be responsible for viral replication and expression in the host cell is a non-structural protein 16 known as 2′-O-ribose methyltransferase or 2′-OMTase. The 5′-terminal cap structure (m7GpppN) of viral mRNA is important for efficient splicing, core export, translation and stability. 2′-OMTase protects the viral mRNA from getting recognized by the host cell, hence prevents the activation of the immune response against it [12]. Therefore, the role of this protein suggests it to be another potential drug target against SARS-CoV-2. Before carrying out scientific laboratory experiments the cost-effective and predictive computational approaches are being used through in silico research. This, in turn, provides the researchers with meaningful outcomes, through certain biomedical regime and databases [13].
In this study, computational approaches are utilized to identify the potent inhibiting candidates of Main protease and Methyltransferase of SARS-CoV-2. Through molecular docking and DFT-based computations, reactivity and binding of compounds are analyzed with the targeted receptors, while ADMET properties are computed to represent the suitability of selected compounds for human administration.
2. Materials and methods
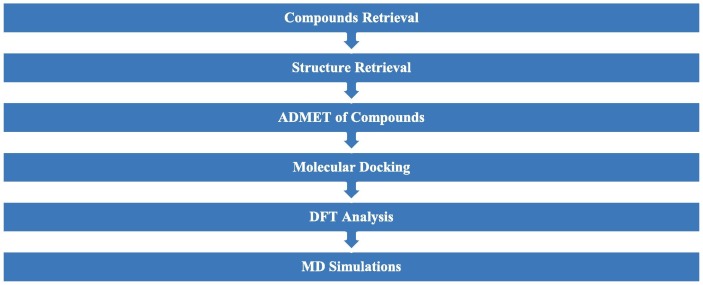
The overall flow of the methodology is shown in Fig. 1 . Previously, these approaches have been validated in various studies [14], [15], [16], [17], [18].
Fig. 1.
Flowchart of methodology.
2.1. Selection of compounds
In this study, we selected a total of 19 thiazolides derivatives, as reported by [19], and are shown in Table S1. Thiazolides i.e. 2-hydroxyaroyl-N-(thiazol-2-yl)-amides are known to be biologically active compounds are related to nitazoxanide, and belong to the family of salicylamides of 2-aminothiazoles [20]. These compounds comprise a new class antiviral drugs, which have a significant potential against a broad range of viruses, both DNA and RNA. Nitazoxanide was the first thiazolide and is known for the treatment of chronic hepatitis C [21]. Besides this, the recent generation i.e. the second generation of thiazolides are reported to have a significant potential for treating chronic hepatitis B and C, enteric viruses, herpes viruses and influenza A [22]. Nitazoxanide can effectively treat influenza A and B in cell culture trials and clinical trials. Nitazoxanide was initially approved as an antiprotozoal in the United States and many countries around the world [20], [23]. More importantly, it is the only product approved for treatment recent apicomplexan protozoa, Cryptosporidium parvum and against rotavirus. In the United States, it is used as a broad-spectrum antiviral drug for the treatment of viral respiratory infections [19]. According to reports, nitazoxanide is also effective in clinical trials for the treatment of rotavirus and norovirus, as well as the combination of pegylated interferon with and without ribavirin in the treatment of chronic hepatitis C [19], [22]. The structures of these compounds were sketched in ACD ChemSketch [24] and 3D energy optimization was performed in Discovery Studio [25], before further analysis.
2.2. ADMET analysis
To evaluate the drug-likeliness of the selected compounds, their ADME (Absorption, Distribution, Metabolism, Excretion) profile and toxicity were analyzed. For this purpose: SwissADME web server and PreADMET online server were used [26], [27]. SwissADME is an online web-based service that provides information regarding the solubility, GI absorption, BBB penetration and Lipinski rule’s violation associated with certain Thiazolide Derivates, while PreADMET server, and is also an online tool which chiefly provides information about the toxicity and carcinogenicity associated with the compounds of interest.
2.3. Retrieval of protein structures
Proteins of interest for this study were Main Protease and Methyltransferase. 3D structures of these proteins were available online and were downloaded from RCSB: Protein Databank (Methyltransferase: 6 W61; Main Protease: 6 W63). The visualization and processing of these structures (Removal of water and ligands) were done through UCSF Chimera [28].
2.4. Molecular docking, binding affinity and Ki values
Molecular docking was necessary, to analyze the binding affinity and calculate the inhibitory constant Ki (μM) of the screened Thiazolide Derivates with the target proteins. For this, AutoDock Tools and AutoDock Vina were utilized, through which the addition of hydrogen bonds to prepare the receptors and the torsion adjustments along with modification of ligands were carried out [29], [30]. For ligand–protein interactions, designing of grid box was also carried out through the specification of x, y and z dimensions. The output files were visualized through Discovery Studio [25]. Threshold values/ cut-off values were also set to get the complexes with best binding affinities. Binding energies were estimated and Ki values were calculated through Eq (1) where docking energy, temperature and gas constant is denoted by ΔG, T and R, respectively.
| (1) |
2.5. DFT analysis
The compounds with highest binding affinity were further subjected to reactivity evaluations which were carried out through Density Functional Theory analysis. Density functional theory (DFT) is an efficient computational tool which provides insight about the reactivity and efficiency of the screened Thiazolide Derivates against the targeted proteins. For DFT analysis, the highest occupied molecular orbital (HOMO) and lowest unoccupied molecular (LUMO) energies were used and the band energy gap (ΔE) was calculated through the expression EHOMO-ELUMO. For this purpose that input files were created through a chemical an analyzer tool named Avogadro, while for the energy calculation ORCA program was used. Basis set which was used for calculations was def2-SV(P), as reported by [31]. The employment of B3LYP exchange–correlation functional was done, this targeted functional is a hybrid exchange–correlation functional, more precisely a combination of Hartree-Fock exact exchange functional and any other density functional and defined as:
| (2) |
Is the generalized gradient approximation while the correlation functional of Le yang-Parr is denoted by. The local density approximation is denoted by . Moreover, a0 = 0:20, ax = 0:72, and ac = 0:81 [32].
2.6. Molecular dynamics simulation
To determine the stability of the resulting complexes and the binding potential of ligands, MD simulations were carried out. Besides molecular docking, this computational tool provides a way to investigate the major conformational changes as well as the stability of ligand–protein complexes. Moreover, it provides a way to re-score the resultant complexes in term of their binding affinities. Molecular Dynamics Simulation was carried out through Groningen Machine for Chemical Simulations (GROMACS) v 5.0 [33] and Root means square deviation (RMSD) values along with the radius of gyrations (Rg) were computed. The CGenFF server was used for ligand topology generation as it provides a stream file (CHARMM topology) for the ligands as output [34]. As the MD simulations were subjected to run in GROMACS, therefore, for conversion to GROMACS format, the cgenff_charmm2gmx.py script was used. To initiate the molecular dynamics, a cubic box was generated while keeping protein in middle while for solvation, spc216 water molecules were added in the system. A force field named OPLS-AA (Optimized Potential for Liquid Simulation-All Atoms) was applied [35]. This solvated system was neutralized by adding counter ions of Na+ and Cl−. At the next step, this system was subjected to energy minimization with the steepest descent method, keeping step limit as 50000. Later on, constant Number Volume and Temperature (NVT), and constant Number Pressure and Temperature (NPT) equilibrations were performed with 1 atm pressure. The simulations were performed at four temperature values i.e. at 300 K, 310 K (approx. normal human body temperature), 320 K, and 330 K. Explicit water molecules were also added and for all simulations, standard pH of 7.0 was considered. This set of constraints was selected due to keeping the simulations similar to the human biological system. The duration for both equilibrations was 1 ns whereas the algorithm for application of force field used in both equilibrations was Particle Mesh Ewald (PME) with a cubic interpolation implementation [36], as reported by [37]. While performing equilibrations, the hydrogen bonds were re-adjusted with the help of Linear Constraint Solver (LINCS) technique [38]. The final Production MD simulation was performed for 50 ns, keeping the method same as equilibrations. The analysis of the results was performed using rms and gyrate utilities of the GROMACS. Based on RMSD, the graph was plotted using Graphing Advanced Computation and Exploration (GRACE) of data [39].
3. Results
3.1. Screening of compounds based upon pharmacological properties
Based on the activities reported in the previous literature 19 compounds were selected to carry out this study. These selected Thiazolide derivatives were further analyzed through ADMET analysis. In order to carry out this screening process, screening criteria was set as: solubility = high, GI absorption = high or moderate, Lipinski’s violation = 0, blood–brain barrier permeability = no, and toxicity = zero/nil. Through the screening criteria for their ADMET properties, 16 out of 19 qualified for performing the further analysis (Table 1 ).
Table 1.
ADMET Properties of selected Thiazolide Derivates.
| Compounds | ESOL Class | GI Absorption | BBB Penetration | Lipinski violations | Toxicity | Carcinogenicity |
|---|---|---|---|---|---|---|
| Comp1 | Soluble | High | No | 0 | Non-Toxic | Non-Carcinogenic |
| Comp2 | Soluble | High | No | 0 | Non-Toxic | Non-Carcinogenic |
| Comp3 | Soluble | High | No | 0 | Non-Toxic | Non-Carcinogenic |
| Comp4 | Soluble | High | No | 3 | Non-Toxic | Non-Carcinogenic |
| Comp5 | Soluble | High | No | 0 | Non-Toxic | Non-Carcinogenic |
| Comp6 | Soluble | High | No | 2 | Non-Toxic | Non-Carcinogenic |
| Comp7 | Soluble | High | No | 3 | Non-Toxic | Non-Carcinogenic |
| Comp8 | Soluble | High | No | 0 | Non-Toxic | Non-Carcinogenic |
| Comp9 | Soluble | High | No | 0 | Non-Toxic | Non-Carcinogenic |
| Comp10 | Soluble | High | No | 0 | Non-Toxic | Non-Carcinogenic |
| Comp11 | Soluble | High | No | 0 | Non-Toxic | Non-Carcinogenic |
| Comp12 | Soluble | High | No | 0 | Non-Toxic | Non-Carcinogenic |
| Comp13 | Soluble | High | No | 0 | Non-Toxic | Non-Carcinogenic |
| Comp14 | Soluble | High | No | 0 | Non-Toxic | Non-Carcinogenic |
| Comp15 | Soluble | High | No | 0 | Non-Toxic | Non-Carcinogenic |
| Comp16 | Soluble | High | No | 0 | Non-Toxic | Non-Carcinogenic |
| Comp17 | Soluble | High | No | 0 | Non-Toxic | Non-Carcinogenic |
| Comp18 | Soluble | High | No | 0 | Non-Toxic | Non-Carcinogenic |
| Comp19 | Soluble | High | No | 0 | Non-Toxic | Non-Carcinogenic |
Thus, due to Lipinski’s rules violations, Comp4, Comp6 and Comp7 were excluded from docking.
3.2. Molecular docking analysis
The screened Thiazolide Derivates were prepared for molecular docking with the target proteins of SARS-CoV-2. Our target proteins, retrieved through RCSB were Main Protease (PDB ID 6w63) and Methyltransferase (PDB ID 6w61). These structures were further prepared for the docking and cut of the value of ≥− 6.5 kcal/mol was applied to get the best complexes. For screened compounds, the binding affinities and inhibitory constant (Ki) values are reported in Table 2 .
Table 2.
Binding affinities and Ki values for docking of screened compounds against MTase and Mpro.
| Compound Name | MTase |
Mpro |
||
|---|---|---|---|---|
| Binding Energies (kcal/mole) | Ki (µM) | Binding Energies (kcal/mole) | Ki (µM) | |
| Comp1 | −6.8 | 10.23 | −6.7 | 12.12 |
| Comp2 | −7.0 | 7.30 | −6.3 | 23.82 |
| Comp3 | −6.4 | 20.12 | −6.0 | 39.54 |
| Comp5 | −6.9 | 8.64 | −6.0 | 39.54 |
| Comp8 | −6.6 | 14.35 | −6.8 | 10.23 |
| Comp9 | −7.1 | 6.16 | −6.6 | 14.35 |
| Comp10 | −6.8 | 10.23 | −6.2 | 28.20 |
| Comp11 | −6.5 | 16.99 | −6.5 | 16.99 |
| Comp12 | −7.0 | 7.30 | −6.4 | 20.12 |
| Comp13 | −7.2 | 5.21 | −6.1 | 33.40 |
| Comp14 | −7.1 | 6.16 | −6.4 | 20.12 |
| Comp15 | −6.9 | 8.64 | −6.6 | 14.35 |
| Comp16 | −7.1 | 6.16 | −7.5 | 3.14 |
| Comp17 | −7.2 | 5.21 | −6.7 | 12.12 |
| Comp18 | −7.0 | 7.30 | −6.4 | 20.12 |
| Comp19 | −6.5 | 16.99 | −6.7 | 12.12 |
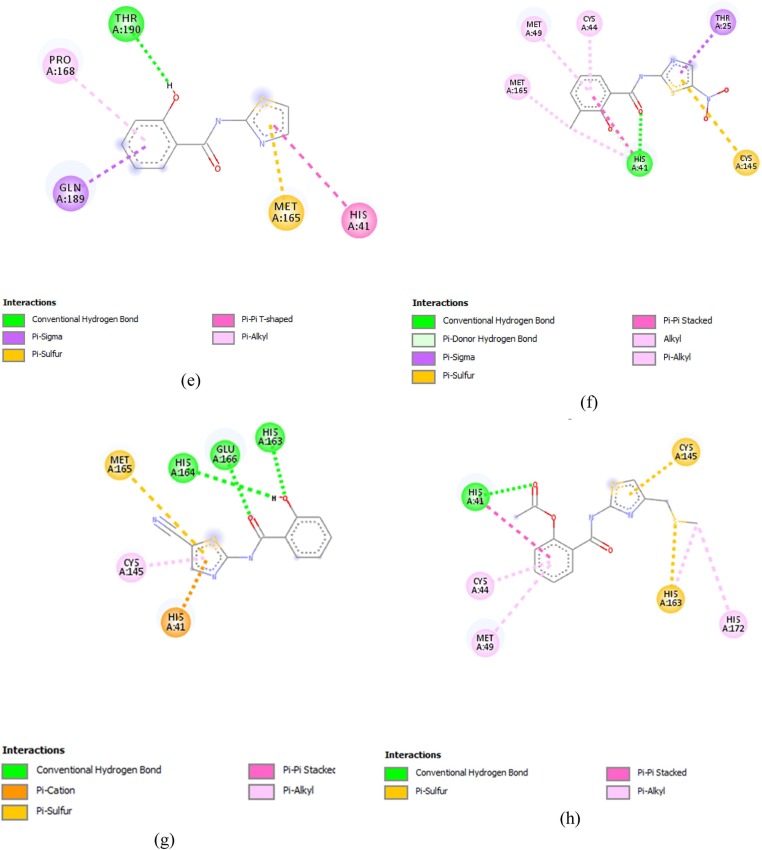
3.2.1. Molecular docking results for methyltransferase
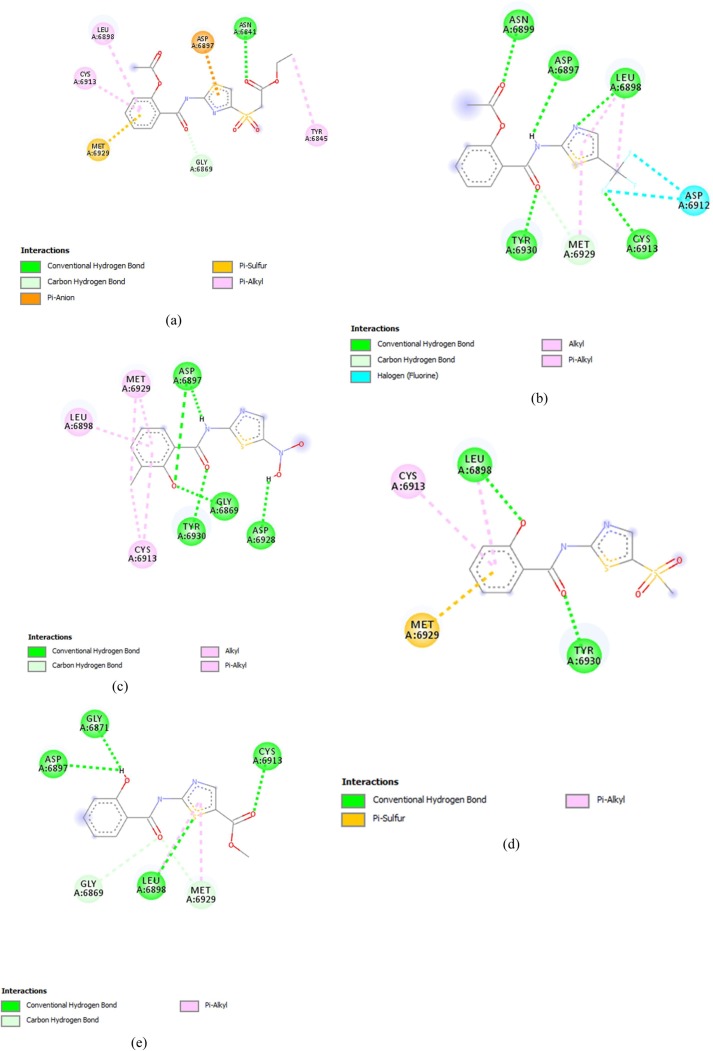
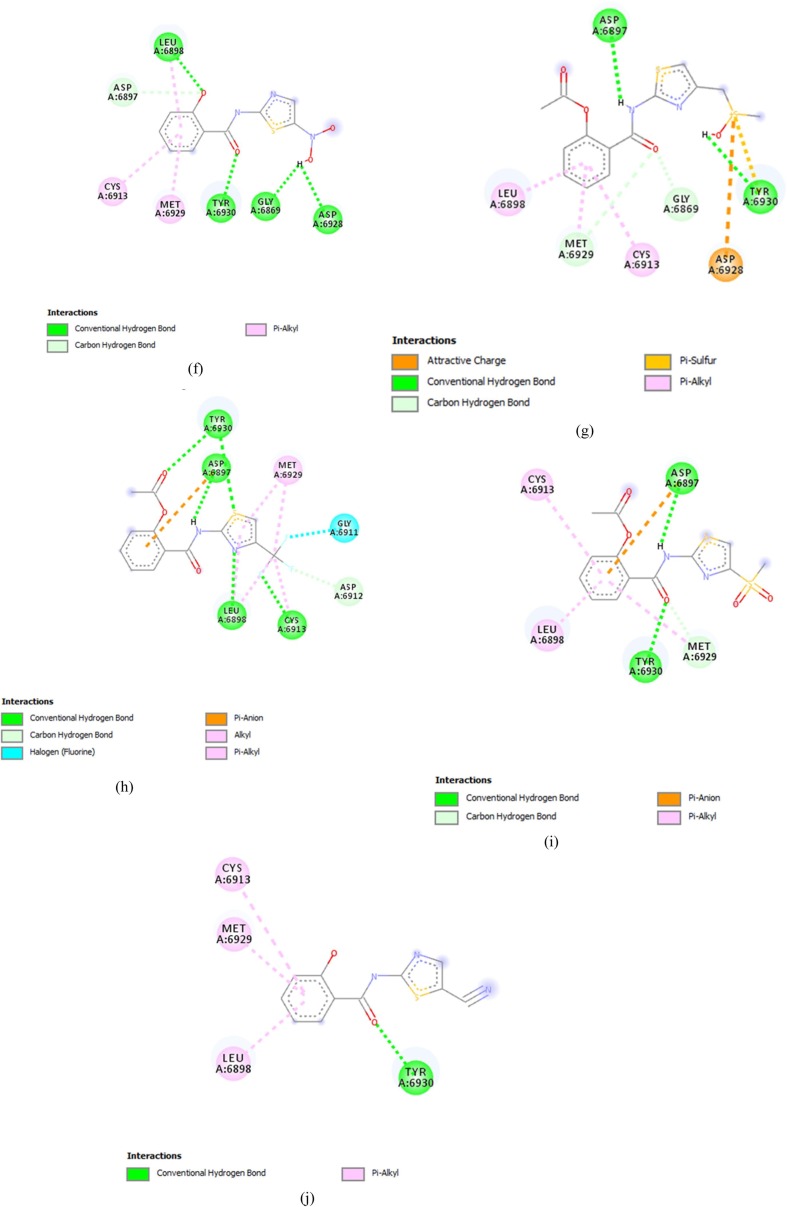
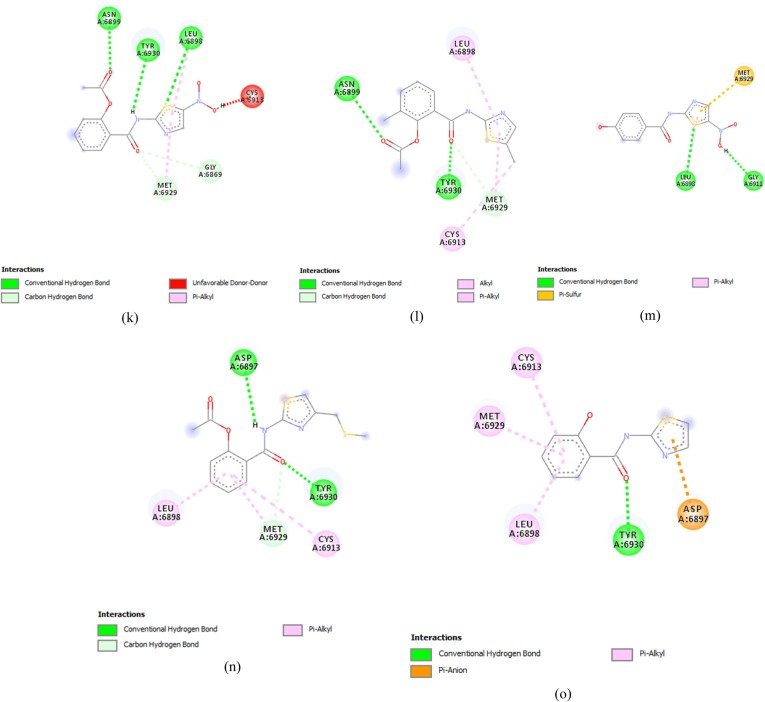
Comp13 formed conventional hydrogen bonds with ASN6897, along with a carbon-hydrogen bond with GLY6867, Pi anion bond with ASP6897, Pi- sulfur bond with MET6929 and Pi alkyl bond with CYS6913, LEU6898 and TYR6845 (Fig. 2 (a)). While comp17 formed conventional hydrogen bonds with ASN6899, ASP6897, LEU6898, CYS6913 and TYR6930 along with halogen bonds with ASP6912 and carbon-hydrogen bond with MET6929, Alkyl bond was also made with LEU6898 and MET6929(Fig. 2(b)).
Fig. 2.
Thiazolide Derivates displaying promising results against targeted receptor: methyltransferase with binding affinities ≥−6.5 kcal/mol. (a) Comp13 (b) Comp17 (c) Comp9 (d) Comp14 (e) Comp16 (f) Comp2 (g) Comp12 (h) Comp18 (i) Comp5 (j) Comp15 (k) Comp1 (l) Comp10 (m) Comp8 (n) Comp11 (o) Comp19.
Comp9 made five conventional bonds with ASP6897, TYR6930, GLY, and ASP along with alkyl and pi-alkyl bonds with MET6929, LEU6898 and CYS6913 (Fig. 2(c)). Comp14 formed a conventional hydrogen bond with LEU6813 and TYR6930 along with Pi Alkyl bond with LEU6898 and CYS6913. Pi sulfur bond was also formed with MET6929 (Fig. 2(d)). Comp 16 made conventional hydrogen bonds with GLY6817, CYS6913, LEU6898 and ASP6897 along with a Pi- alkyl bond with MET6929. Carbon hydrogen bonds were also made with GLY6869 and MET6929 (Fig. 2(e)).
Comp2 displayed conventional hydrogen bonds with LEU6898, TYR6930, GLY6869 and ASP6928 while Pi-alkyl bond was formed with MET6929 and CYS6913 along with a carbon-hydrogen bond with ASP6897(Fig. 2(f)). Comp12 formed a conventional hydrogen bond with ASP6897 and TYR6930, along with a Pi sulfur bond with ASP6928. Carbon hydrogen bond was also formed with MET6929 and GLY6869 along with pi alkyl bonds with LEU6898 and CYS6913. The active charge was also observed at TYR6930 (Fig. 2(g)). Comp18 formed conventional hydrogen bonds with TYR6930, ASP6897, LEU6898 and CYS6913. Carbon hydrogen bind was also formed with ASP6897, while halogen bond was formed with GLY6911. Alkyl and Pi-alkyl bonds were formed with MET6929 and Pi-anion was formed with ASP6897 (Fig. 2(h)).
Comp5 formed the conventional hydrogen bond with TYR 6930 and ASP6897, along with pi alkyl bond with CYS6913. And LEU6898 and carbon-hydrogen bond with MET6929 (Fig. 2(i)). While, Comp15, formed the conventional hydrogen bond with TYR6930 and Pi Alkyl bond with LEU6898, MET6929 and CYS6913 (Fig. 2(j)).
Comp1 formed conventional hydrogen bonds with LEU6898, ASN6898 and TYR6930 along with a carbon-hydrogen bond with MET6929 and GLY6869. An un-favourable donor bond was also formed with CYS6913 (Fig. 2(k)). On the other hand, Comp10 made conventional hydrogen bonds with TYR6930 and ASN6899 along with a carbon-hydrogen bond with MET6929 and Alkyl and Pi Alkyl bonds with CYS6913 and LEU6898 (Fig. 2(l)).
Comp8 interacted through the formation of a conventional hydrogen bond with GLY6911 and LEU6898, along with a Pi sulfur bond with MET6929 (Fig. 2(m)). Comp11 formed the conventional hydrogen bonds ASP6897 and TYR6930 and Pi-alkyl bond with LEU and CYS6913 along with a carbon-hydrogen bond with MET6929 (Fig. 2(n)). On the other hand, Comp19 made the conventional hydrogen bonds with TYR6930 and Pi alkyl bonds with LEU6930, MET6929 and CYS6913 along with a Pi-Anion bond with ASP6897 (Fig. 2(o)).
Comp3, on the other hand, failed to cross the threshold value in the terms of its binding affinity and made conventional hydrogen bond with TYR6930 along with Alkyl and Pi Alkyl bonds with LEU6898 and CYS6913. It also displayed the formation of carbon-hydrogen bond with MET6929.and GLY6869 .
3.2.2. Molecular docking of the main protease
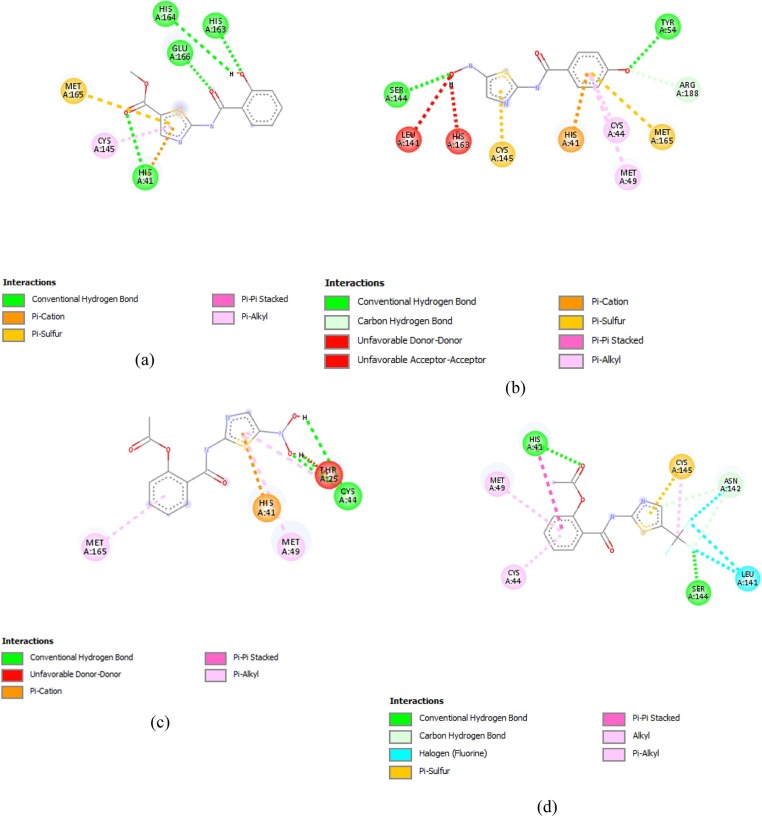
A total of eight compounds i.e. Comp16, Comp8, Comp1, Comp17, Comp19, Comp9, Comp15 and Comp11, displayed binding affinities ≥−6.5 kcal/mol.
Comp16 formed the conventional hydrogen bonds with HIS164, HIS163, HIS41 and GLU166, Along with Pi cation and Pi sulfur bond with MET165. It also formed Pi-alkyl bond with CYS145 (Fig. 3 (a)). Comp8 made the conventional hydrogen bonds with SER144 and TYR54, along with carbon-hydrogen bonds with ARG188. It also made Pi cation bond with HIS41 along with Pi sulfur bond with MET165 and CYS145 and Pi alkyl bonds with CYS44 and MET49. Un-favourable acceptor–acceptor and un-favourable donor–donor bond were also formed with HIS143 and LEU162 (Fig. 3(b)).
Fig. 3.
Thiazolide Derivates displaying promising results against targeted receptor: main protease with binding affinities ≥−6.5 kcal/mol. (a) Comp16 (b) Comp8 (c) Comp1 (d) Comp17 (e) Comp19 (f) Comp9 (g) Comp15 (h) Comp11.
Comp1 formed conventional hydrogen bonds with CYS44. A Pi-Alkyl bond with MET49 and MET165, along with Pi-carbon bond with HIS41 (Fig. 3(c)). While, Comp17 made conventional hydrogen bond with SER144 and HIS41 along with Alkyl and Pi Alkyl bonds with CYS44, MET49 and HIS41 respectively. A halogen bond and Pi sulfur bond was also witnessed with the LEU144 and CYS145 respectively. A carbon-hydrogen bond was also made with ASN142 (Fig. 3(d)). Comp19 made conventional hydrogen bond with THR190 along with a Pi sigma bond with GLN189, a Pi-Alkyl bond with PRO168, a Pi-Pi T-shaped bond with HIS41 and a Pi sulfur bond with MET165 (Fig. 3(e)).
Comp9 formed a conventional hydrogen bond with HIS41, Alkyl and Pi alkyl bond with MET165, CYS44 and MET49. A Pi sigma bond was also formed THR25 and Pi sulfur bond with CYS145 was also formed (Fig. 3(f)). On the other hand, Comp15 formed three conventional hydrogen bonds with GLU166, HIS164 and HIS163. Pi sulfur and Pi cation were also formed with MET16 5 and HIS41, respectively (Fig. 3(g)).
Comp11 made conventional hydrogen bond and Pi-Pi stacked bond with HIS41, along with Pi-alkyl bonds with CYS44, HIS172 and MET49. It also made Pi-sulfur bonds with CYS44 and HIS163 (Fig. 3(h)).
3.3. DFT analysis
To analyze the reactivity of these screened compounds, DFT analysis was executed. The whole process was done in terms of band energy gap and molecular orbital descriptors. The low difference of ELUMO and EHOMO exhibited the low band energy gaps which depict the higher reactivity of a compound with the receptor. The lower band energy gap reflects higher reactivity of compounds as the ELUMO and EHOMO are responsible for the charges transferred in a chemical reaction [40]. Through results, it was observed that band energy gaps i.e. reactivity of the compound were in correlation with binding affinities and the same trend was observed in both. For Methyltransferase, the high reactivity shown by compounds was 0.114–0.159 kcal/mol (Table 3 ). Among these complexes, the complex made by Comp13 showed the highest reactivity. For the Main protease, compounds exhibited reactivity between 0.115 and 0.131 kcal/mol (Table 4 ). The complex made by comp16 showed the highest reactivity.
Table 3.
DFT results for compounds showing binding affinity ≥ threshold value against Methyltransferase.
| Compound | ELUMO (kcal/mol) | EHOMO (kcal/mol) | Band energy gap (ΔE) (kcal/mol) |
|---|---|---|---|
| Comp13 | −0.274 | −0.389 | 0.114 |
| Comp17 | −0.280 | −0.395 | 0.116 |
| Comp9 | −0.262 | −0.380 | 0.118 |
| Comp14 | −0.275 | −0.395 | 0.120 |
| Comp16 | −0.266 | −0.390 | 0.124 |
| Comp2 | −0.293 | −0.418 | 0.125 |
| Comp12 | −0.303 | −0.428 | 0.125 |
| Comp18 | −0.237 | −0.369 | 0.132 |
| Comp5 | −0.242 | −0.375 | 0.133 |
| Comp15 | −0.273 | −0.417 | 0.144 |
| Comp1 | −0.302 | −0.449 | 0.148 |
| Comp10 | −0.235 | −0.386 | 0.151 |
| Comp8 | −0.281 | −0.432 | 0.151 |
| Comp11 | −0.258 | −0.409 | 0.151 |
| Comp19 | −0.256 | −0.415 | 0.159 |
Table 4.
DFT results for compounds showing binding affinity ≥ threshold value against Main Protease.
| Compound | ELUMO (kcal/mol) | EHOMO (kcal/mol) | Band energy gap (ΔE) (kcal/mol) |
|---|---|---|---|
| Comp16 | −0.236 | −0.351 | 0.115 |
| Comp8 | −0.274 | −0.391 | 0.116 |
| Comp1 | −0.304 | −0.421 | 0.118 |
| Comp17 | −0.288 | −0.407 | 0.118 |
| Comp19 | −0.238 | −0.359 | 0.120 |
| Comp9 | −0.273 | −0.401 | 0.127 |
| Comp15 | −0.271 | −0.400 | 0.129 |
| Comp11 | −0.251 | −0.382 | 0.131 |
3.4. MD simulation
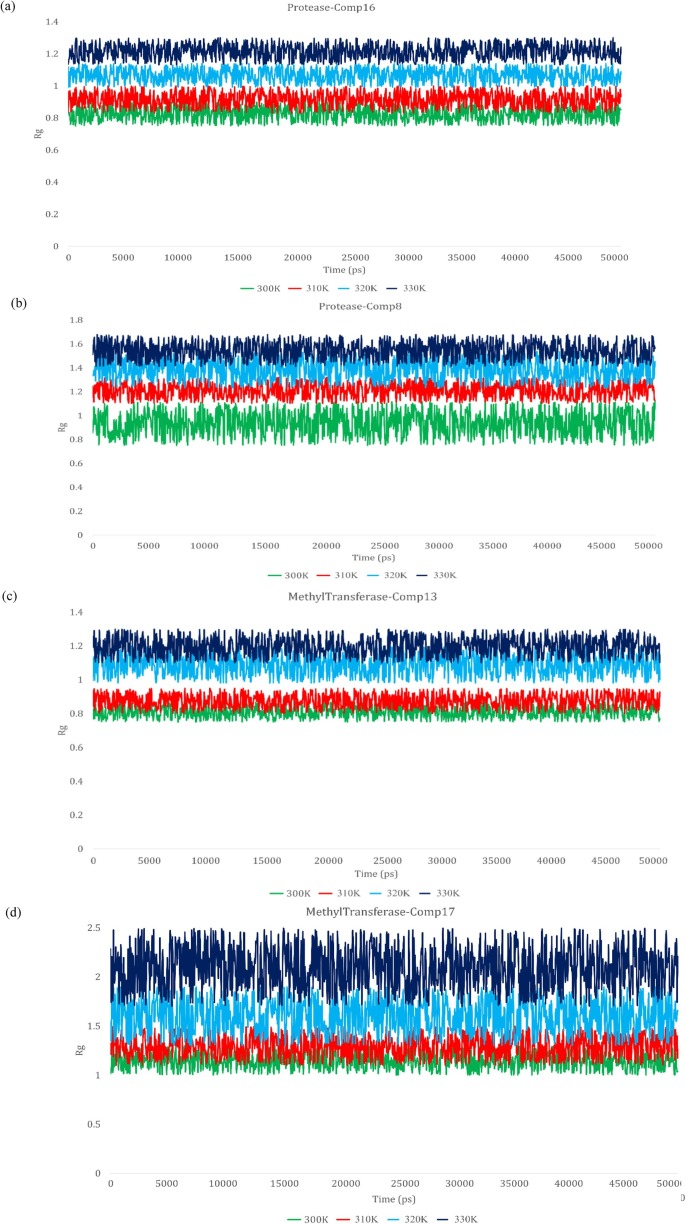
By a thorough computation of the radius of gyration and RMSD scores, compactness and stability of compounds were determined. These computations were executed to monitor the changes in the state of complexes before and after MD Simulations. To execute this analysis, the top two complexes were selected for each receptor. This selection was based upon the results of reactivity obtained through DFT results and band energy gap. It is pertinent to mention that for MD simulation, ligands are chosen based on molecular docking results. However, in this study, it was observed that the results of DFT and molecular docking studies were correlated and represented the same trend. If ligands had been chosen based on molecular docking results, same complexes would have been selected. Thus, to keep continuation in the pipeline of method and streamline the process, the ligands were selected from the previous step i.e., DFT study.
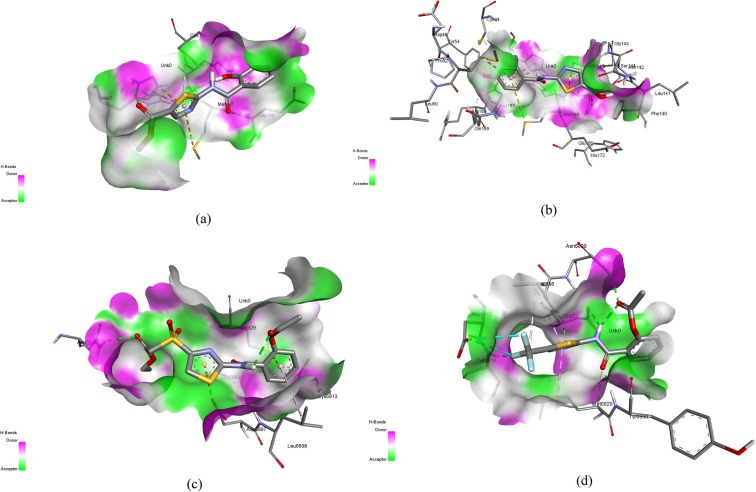
The radius of gyration graphs actually depicts the compactness and folding of the structure. The higher fluctuations in the gyration values depict lower stability and compactness of folding of structure. However, in the present study, fluctuations depicted by the graphs were less and the Rg value for each complex ranged between 0.8 Å and 2.5 Å for both receptors, depicting the compactness in complexes (Fig. 4 ). Similarly, the purpose of computing RMSD was to depict the structural differences in backbones of the structure, preMD and postMD. This depicted the thermostability of the complexes, thus, the RMSD values were represented in terms of numeric representations to provide a better understanding. RMSD values reported in Table 5 , depicted high stability as these values ranged between 0.75 Å and 2.94 Å. The 3D plots for the interactions of Mpro-Comp16, Mpro-Comp8, MTase-Comp13, and MTase-Comp17 are shown in Fig. 5 .
Fig. 4.
Rg graph of experimental ligands with the targeted receptor (a) Mpro-Comp16 (b) Mpro -Comp8 (c) MTase-Comp13 (d) MTase-Comp17.
Table 5.
Average RMSD values for complexes of compounds passing threshold at temperature 300 K, 310 K, 320 K and 330 K.
| Complexes | Average RMSD at various temperatures (Å) |
|||
|---|---|---|---|---|
| 300 K | 310 K | 320 K | 330 K | |
| Mpro-Comp16 | 0.75 | 0.98 | 1.15 | 1.30 |
| Mpro-Comp8 | 1.09 | 1.26 | 1.44 | 1.78 |
| MTase-Comp13 | 0.88 | 1.07 | 1.29 | 1.52 |
| MTase-Comp17 | 1.70 | 1.93 | 2.72 | 2.94 |
Fig. 5.
3D interaction diagrams for (a) Mpro-Comp16 (b) Mpro -Comp8 (c) MTase-Comp13 (d) MTase-Comp17.
4. Discussion
To screen certain antiviral compounds, three approaches are utilized by scientists, generally for the inhibition of infection. These approaches include; (a). The analysis of presently existing antiviral molecules and compounds to sort out their effects on viral replication and packaging, (b). Advancement of novel agents and (c). High throughput screening of compounds effective against transcriptional machinery of certain cell lines. Out of these tactics, the high throughput approach is more promising as it helps to screen and evaluate large libraries of compounds having drug likeliness. Moreover, it provides the facility for screening the already existing drugs to support repurposing efforts, which leads to the discovery of the new functions of already known drugs [41].
Thiazolides is a new class of antiviral drugs that were discovered in the mid-1970s and had nitrothiazole derivates in abundance. Thiazolides i.e. 2-hydroxyaroyl-N-(thiazol-2-yl)-amides are known to be biologically active compounds are related to nitazoxanide, and belong to the family of salicylamides of 2-aminothiazoles [42]. This class of antiviral drugs has been found to have a wide range of activities against DNA and RNA viruses. If we take an extensive look at it, various parasitic diseases were found to be treated by nitroimidazoles. This took to an advanced and better treatment of infections such as trichomoniasis, ameiasis, and giardiasis. Intestinal parasitic infections caused by nematodes were also reported to be treated by benzimidazole carbamate derivatives, which dramatically contributed to the treatment [43].
A compound named praziquantel was found to be effective against intestinal cestode infections, chiefly caused by taenia solium. With the development of nitazoxanide, efforts were made to check its effectiveness against AIDS-related cryptosporidiosis which didn’t only prove beneficent against AIDS but this effort lead to the antiviral activity of nitazoxanide against a wide range of RNA and DNA viruses such as HBV and HCV. This chemical was further subjected to clinical studies in treating chronic hepatitis B and C and rotavirus and stimulated the search for the second generation of thiazoles [44]. Nitazoxanide was initially approved as an antiprotozoal in the United States and many countries around the world [20], [23]. More importantly, it is the only product approved for treatment recent apicomplexan protozoa, Cryptosporidium parvum and against rotavirus. In the United States, it is used as a broad-spectrum antiviral drug for the treatment of viral respiratory infections. According to reports, nitazoxanide is also effective in clinical trials for the treatment of rotavirus and norovirus, as well as the combination of pegylated interferon with and without ribavirin in the treatment of chronic hepatitis C [19].
Thiazolides has also been analyzed to check their activity against other viruses such as vesicular stomatitis virus, herpes simplex and presented auspicious outcomes. Absorption, distribution, metabolism and excretion analysis revealed that thiazolides are well absorbed in human, and administration of thiazolides denuded their good absorption and metabolic stability and rapid de-acylation. In rats, toxicity analysis revealed no toxicity, weak positive Ames test and high probability of QT prolongation [45]. As an absolute, this new class of promising antiviral is favoured by the safety profile, and their good bioavailability, metabolic stability, non-mutagenicity, low toxicity and low cardiotoxicity have shown them to be safe for c consideration. Primarily, some thiazolides derivative has exhibited better activity and could become new drugs in the market.
Advancement and discovery of novel drugs is a time consuming, costly and parlous process. Standard costs for drug discovery ranges from 0.8 to 1.0 billion USD and takes nearly 14 years to ultimately introduce a drug into the market, as it requires various steps to analyze the effectivity, drug-likeliness and most importantly the safety. Even though, since past decade the investment for the development of drugs has been increased yet due to low efficiency and high failure rate, positive resultants are very much less as compared to the investment. As a whole, certain approaches have always been required to not only lessen the investment but also to shorten the time which it takes for the analyses of the compound; chiefly for its drug-likeliness, prediction and examination. The solution has been forwarded by the computer-aided drug designing, as rapid development in the field of combinatorial chemistry and computational technologies that provide an environment to accelerate the drug discovery. It didn’t only help to obtain potent compounds within weeks but also had made it easy to search new chemicals, optimize certain molecular libraries for their diversity and construct high-quality data sets [14], [15], [16], [17], [18].
The most important constituent of traditional drug discovery is the investigation of promising compounds for pharmacokinetics, metabolism and potential toxicity. An in silico technique, the ADMET analysis; being abbreviated as absorption, distribution, metabolism, excretion, and toxicity analyses, has made it easy to evaluate these properties much earlier and has accelerated the process of drug discovery by excluding the compounds before any preclinical research. During the last three decades, due to dramatic growth in the power and availability of computers, ease in the availability of small molecules and databases and needs of molecular biology and structural based drug discovery lead to the field of molecular docking [46]. Being performed between the small molecule and targeted macromolecule, the relieving molecular docking soft wares pinpoint to understand and predict the structural recognition, prediction of binding affinity energetically along with the binding modes. It has also revolutionized the field of drug discovery by providing facilities such as; optimization of lead compounds, the study of activation energies, searching of a potential lead compound through virtual screening, mutagenesis studies, assisting x-ray crystallography, studies of chemical mechanisms and designing of certain libraries [47].
Based on molecular descriptors and physicochemical properties, virtual screening aims at searching the hits and lead compounds. However, molecular docking helps to provide the idea of how a ligand is interacting with a target through three-dimensional representations of these interactions Certain molecular docking tools are available such as FlexX, GOLD, ICM, AutoDock, etc. If we look deeply, then molecular docking assist in the prediction of binding energies, shape and chemical complementarities, which isn’t enough to finalize a compound as a potent drug, hence certain efficient tool for solving a real-world problem, optimization algorithm with multiple objectives and results are always required [48].
This problem has been solved to some extent by the laws governing the behaviour of electrons that at the other hand has this possibility to predict the performance of materials under study. This has become achievable by another computational tool called Density function theory analysis, which is based upon quantum mechanics. This technique analyzes the electronic structures and calculates the orbital energy values. It uses HOMO and LUMO energies to provide information about structural stability. Another very advanced and most important tool is the MD simulation, which doesn’t only provide insight into natural dynamics to bring the biomolecular structure alive but also provides information about the confirmations of a molecule. It helps in the understanding of the thermally accessible molecules and explores the conformational spaces put forward through docking. Furthermore, by combining experimental data and general properties of molecular structure MD simulation put forward the understanding of equilibrium properties such as free energies, fully solvated membrane protein complexes [49]. By keeping the previously reported properties of thiazolides, certain derivatives were used in our study. An in silico research was executed through the above-mentioned techniques, to evaluate the selected thiazolides for their activity against Mpro and MTase of COVID-19.
5. Conclusion
The present study against SARS-CoV-2 Main Protease and Methyltransferase suggests that based upon ADMET parameters to the stability accessed by MD simulation, four different thiazolides derivatives were found as potential inhibitors of targeted proteins. Out of these, two thiazolides derivates; methyl 2-(2-hydroxybenzamido)-1,3-thiazole-5-carboxylate and N-[5-(dihydroxyamino)-1,3-thiazol-2-yl]-4-hydroxybenzamide against the Main protease, and two thiazolides; ethyl {2-[2-(acetyloxy)benzamido]-1,3-thiazole-4-sulfonyl}acetate and 2-{[5-(trifluoromethyl)-1,3-thiazol-2-yl]carbamoyl}phenyl acetate against MTase came forward as the best and dependable inhibitors of target proteins. As a whole, these evaluated compounds can further be accessed through in vitro and in vivo research to adjudicate their efficaciousness and can be used in future as a drug against this devastating virus. Altogether this study provides a cost-effective and economically practical solution for the treatment of SARS-CoV-2.
CRediT authorship contribution statement
Nouman Rasool: Conceptualization, Writing - review & editing, Supervision. Farkhanda Yasmin: Resources, Data curation, Investigation. Shalini Sahai: Validation, Writing - review & editing. Waqar Hussain: Software, Validation, Formal analysis, Methodology, Project administration. Hadiqa Inam: Writing - original draft, Methodology, Visualization. Arooj Arshad: Writing - original draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cplett.2021.138463.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Xu Z., Peng C., Shi Y., Zhu Z., Mu K., Wang X., Zhu W. Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. BioRxiv. 2020 [Google Scholar]
- 2.A. Zhavoronkov, B. Zagribelnyy, A. Zhebrak, V. Aladinskiy, V. Terentiev, Q. Vanhaelen, D.S. Bezrukov, D. Polykovskiy, R. Shayakhmetov, A. Filimonov, Potential non-covalent SARS-CoV-2 3C-like protease inhibitors designed using generative deep learning approaches and reviewed by human medicinal chemist in virtual reality, 2020.
- 3.Liu X., Wang X.-J. Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. J. Gen. Genomics. 2020;47:119. doi: 10.1016/j.jgg.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang K.-O., Kim Y., Lovell S., Rathnayake A.D., Groutas W.C. Antiviral drug discovery: norovirus proteases and development of inhibitors. Viruses. 2019;11:197. doi: 10.3390/v11020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woźniak Ł., Skąpska S., Marszałek K. Ursolic acid—a pentacyclic triterpenoid with a wide spectrum of pharmacological activities. Molecules. 2015;20:20614–20641. doi: 10.3390/molecules201119721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X., Zhang B., Jin Z., Yang H., Rao Z. The crytal structure of 2019-nCoV main protease in complex with an inhibitor N3. RCSB Protein Data Bank. 2020 [Google Scholar]
- 7.Gil C., Ginex T., Maestro I., Nozal V., Barrado-Gil L., Cuesta-Geijo M.A., Urquiza J., Ramírez D., Alonso C., Campillo N.E. COVID-19: Drug targets and potential treatments. J. Med. Chem. 2020 doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- 8.Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., Michalska K., Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020 doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viswanathan T., Arya S., Chan S.-H., Qi S., Dai N., Misra A., Park J.-G., Oladunni F., Kovalskyy D., Hromas R.A. Structural basis of RNA cap modification by SARS-CoV-2. Nat. Commun. 2020;11:1–7. doi: 10.1038/s41467-020-17496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saxena A. Drug targets for COVID-19 therapeutics: ongoing global efforts. J. Biosci. 2020;45:1–24. doi: 10.1007/s12038-020-00067-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.R. Vivek-Ananth, A. Rana, N. Rajan, H.S. Biswal, A. Samal, In silico identification of potential natural product inhibitors of human proteases key to SARS-CoV-2 infection, arXiv preprint arXiv:2006.00652, 2020. [DOI] [PMC free article] [PubMed]
- 13.Shekhar C. In silico pharmacology: computer-aided methods could transform drug development. Chem. Biol. 2008;15:413–414. doi: 10.1016/j.chembiol.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Hussain W., Amir A., Rasool N. Computer-aided study of selective flavonoids against chikungunya virus replication using molecular docking and DFT-based approach. Struct. Chem. 2020:1–12. [Google Scholar]
- 15.Hussain W., Rasool N., Khan Y.D. Insights into machine learning-based approaches for virtual screening in drug discovery: existing strategies and streamlining through FP-CADD. Curr. Drug Discov. Technol. 2020 doi: 10.2174/1570163817666200806165934. [DOI] [PubMed] [Google Scholar]
- 16.Qaddir I., Majeed A., Hussain W., Mahmood S., Rasool N. An in silico investigation of phytochemicals as potential inhibitors against non-structural protein 1 from dengue virus 4. Brazil. J. Pharmac. Sci. 2020;56 [Google Scholar]
- 17.Rasool N., Akhtar A., Hussain W. Insights into the inhibitory potential of selective phytochemicals against Mpro of 2019-nCoV: a computer-aided study. Struct. Chem. 2020 doi: 10.1007/s11224-020-01536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasool N., Majeed A., Riaz F., Hussain W. Identification of novel inhibitory candidates against two major Flavivirus pathogens via CADD protocols: in silico analysis of phytochemical binding, reactivity, and pharmacokinetics against NS5 from ZIKV and DENV. Struct. Chem. 2020;31:2189–2204. [Google Scholar]
- 19.Stachulski A.V., Santoro M.G., Piacentini S., Belardo G., Frazia S.L., Pidathala C., Row E.C., Berry N.G., Iqbal M., Allman S.A. Second-generation nitazoxanide derivatives: thiazolides are effective inhibitors of the influenza A virus. Future Med. Chem. 2018;10:851–862. doi: 10.4155/fmc-2017-0217. [DOI] [PubMed] [Google Scholar]
- 20.Korba B.E., Montero A.B., Farrar K., Gaye K., Mukerjee S., Ayers M.S., Rossignol J.-F. Nitazoxanide, tizoxanide and other thiazolides are potent inhibitors of hepatitis B virus and hepatitis C virus replication. Antiviral Res. 2008;77:56–63. doi: 10.1016/j.antiviral.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Rossignol J.F., La Frazia S., Chiappa L., Ciucci A., Santoro M.G. Thiazolides, a new class of anti-influenza molecules targeting viral hemagglutinin at the post-translational level. J. Biol. Chem. 2009;284:29798–29808. doi: 10.1074/jbc.M109.029470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haffizulla J., Hartman A., Hoppers M., Resnick H., Samudrala S., Ginocchio C., Bardin M., Rossignol J.-F., U.N.I.C.S. Group Effect of nitazoxanide in adults and adolescents with acute uncomplicated influenza: a double-blind, randomised, placebo-controlled, phase 2b/3 trial. Lancet Infect. Dis. 2014;14:609–618. doi: 10.1016/S1473-3099(14)70717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.La Frazia S., Ciucci A., Arnoldi F., Coira M., Gianferretti P., Angelini M., Belardo G., Burrone O.R., Rossignol J.-F., Santoro M.G. Thiazolides, a new class of antiviral agents effective against rotavirus infection, target viral morphogenesis, inhibiting viroplasm formation. J. Virol. 2013;87:11096–11106. doi: 10.1128/JVI.01213-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter A.D. ACS Publications; 1997. ACD/ChemSketch 1.0 (freeware)- ACD/ChemSketch 2.0 and its tautomers, dictionary. [Google Scholar]
- 25.D. Studio, version 2.5, Accelrys Inc.: San Diego, CA, USA, 2009.
- 26.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S., Lee I., Kim H., Chang G., Chung J., No K. The PreADME Approach: Web-based program for rapid prediction of physico-chemical, drug absorption and drug-like properties. EuroQSAR 2002 Design. Drugs Crop Protect.: Process. Probl. Solut. 2003;2003:418–420. [Google Scholar]
- 28.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 29.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasool N., Husssain W., Khan Y.D. Revelation of enzyme activity of mutant pyrazinamidases from Mycobacterium tuberculosis upon binding with various metals using quantum mechanical approach. Comput. Biol. Chem. 2019;83:107108. doi: 10.1016/j.compbiolchem.2019.107108. [DOI] [PubMed] [Google Scholar]
- 32.Rasool N., Hussain W. Three major phosphoacceptor sites in HIV-1 capsid protein enhances its structural stability and resistance against the inhibitor: explication through molecular dynamics simulation. Mole. Dock. DFT Anal. Combinat. Chem. High Throughput Screen. 2020;23:41–54. doi: 10.2174/1386207323666191213142223. [DOI] [PubMed] [Google Scholar]
- 33.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- 34.Zhu S. Validation of the generalized force fields GAFF, CGenFF, OPLS-AA, and PRODRGFF by testing against experimental osmotic coefficient data for small drug-like molecules. J. Chem. Inform. Model. 2019;59:4239–4247. doi: 10.1021/acs.jcim.9b00552. [DOI] [PubMed] [Google Scholar]
- 35.Jorgensen W.L., Maxwell D.S., Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996;118:11225–11236. [Google Scholar]
- 36.Cheatham T.I., Miller J., Fox T., Darden T., Kollman P. Molecular dynamics simulations on solvated biomolecular systems: the particle mesh Ewald method leads to stable trajectories of DNA, RNA, and proteins. J. Am. Chem. Soc. 1995;117:4193–4194. [Google Scholar]
- 37.Kousar K., Majeed A., Yasmin F., Hussain W., Rasool N. Phytochemicals from selective plants have promising potential against SARS-CoV-2: investigation and corroboration through molecular docking, MD simulations, and quantum computations. BioMed Res. Int. 2020;2020 doi: 10.1155/2020/6237160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hess B., Bekker H., Berendsen H.J., Fraaije J.G. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 39.P. Turner, XMGRACE, Version 5.1. 19, Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology, Beaverton, OR, 2005.
- 40.Rasool N., Jalal A., Amjad A., Hussain W. Probing the pharmacological parameters, molecular docking and quantum computations of plant derived compounds exhibiting strong inhibitory potential against NS5 from Zika virus. Brazil. Arch. Biol. Technol. 2018;61 [Google Scholar]
- 41.Lobanov V. Using artificial neural networks to drive virtual screening of combinatorial libraries. Drug Disc. Today: BIOSILICO. 2004;2:149–156. [Google Scholar]
- 42.Fox L.M., Saravolatz L.D. Nitazoxanide: a new thiazolide antiparasitic agent. Clin. Infect. Dis. 2005;40:1173–1180. doi: 10.1086/428839. [DOI] [PubMed] [Google Scholar]
- 43.Rossignol J., Maisonneuve H., Cho Y. Nitroimidazoles in the treatment of trichomoniasis, giardiasis, and amebiasis. Int. J. Clin. Pharmacol. Therapy Toxicol. 1984;22:63. [PubMed] [Google Scholar]
- 44.J.-F. Rossignol, E.B. Keeffe, Thiazolides: a new class of drugs for the treatment of chronic hepatitis B and C, 2008. [DOI] [PubMed]
- 45.Hemphill A., Mueller J., Esposito M. Nitazoxanide, a broad-spectrum thiazolide anti-infective agent for the treatment of gastrointestinal infections. Expert Opin. Pharmacother. 2006;7:953–964. doi: 10.1517/14656566.7.7.953. [DOI] [PubMed] [Google Scholar]
- 46.Hu L., Benson M.L., Smith R.D., Lerner M.G., Carlson H.A. Binding MOAD (mother of all databases) Proteins Struct. Funct. Bioinf. 2005;60:333–340. doi: 10.1002/prot.20512. [DOI] [PubMed] [Google Scholar]
- 47.Pozzan A. Molecular descriptors and methods for ligand based virtual high throughput screening in drug discovery. Curr. Pharm. Des. 2006;12:2099–2110. doi: 10.2174/138161206777585247. [DOI] [PubMed] [Google Scholar]
- 48.Jain A., Shin Y., Persson K.A. Computational predictions of energy materials using density functional theory. Nat. Rev. Mater. 2016;1:1–13. [Google Scholar]
- 49.Hansson T., Oostenbrink C., van Gunsteren W. Molecular dynamics simulations. Curr. Opin. Struct. Biol. 2002;12:190–196. doi: 10.1016/s0959-440x(02)00308-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.