SUMMARY
Antibodies mediate natural and vaccine-induced immunity against viral and bacterial pathogens, whereas fungi represent a widespread kingdom of pathogenic species for which neither vaccine nor neutralizing antibody therapies are clinically available. Here, using a multi-kingdom antibody profiling (multiKAP) approach, we explore the human antibody repertoires against gut commensal fungi (mycobiota). We identify species preferentially targeted by systemic antibodies in humans, with Candida albicans being the major inducer of antifungal immunoglobulin G (IgG). Fungal colonization of the gut induces germinal center (GC)-dependent B cell expansion in extraintestinal lymphoid tissues and generates systemic antibodies that confer protection against disseminated C. albicans or C. auris infection. Antifungal IgG production depends on the innate immunity regulator CARD9 and CARD9+CX3CR1+ macrophages. In individuals with invasive candidiasis, loss-of-function mutations in CARD9 are associated with impaired antifungal IgG responses. These results reveal an important role of gut commensal fungi in shaping the human antibody repertoire through CARD9-dependent induction of host-protective antifungal IgG.
Graphical Abstract
In Brief
Doron et al. present evidence that commensal fungi in the human gut shape antibody responses in the blood to protect the host from systemic lethal fungal infections. The innate immunity regulator CARD9 and CARD9+CX3CR1+ macrophages play an important role in antifungal IgG production, which is decreased in CARD9-deficient patients.
INTRODUCTION
The intestinal fungal community (mycobiota) is a highly immuno-reactive component of the microbiota with important implications for host immunity (Bacher et al., 2019; Leonardi et al., 2018; Li et al., 2018; Richard and Sokol, 2019; Shao et al., 2019; Tso et al., 2018). At the same time, the gut mycobiota are a reservoir of evolving fungal species from which serious infections can arise (Fan et al., 2015; Tso et al., 2018; Zhai et al., 2020). Advances in modern medicine, such as immunosuppressive therapy, chemotherapy, and organ transplantation, have contributed to a massive increase in deadly fungal infections affecting millions of people worldwide (Brown et al., 2012). These infections are often caused by systemic translocation of Candida species from the gut (Fan et al., 2015; Zhai et al., 2020). The combination of a limited number of antifungal drugs and global emergence of multidrug-resistant Candida species renders these infections highly lethal, with mortality rates as high as 50% (Brown et al., 2012). Despite years of efforts to produce a much-needed antifungal vaccine through fungal antigen immunization (Casadevall and Pirofski, 2003, 2012; Edwards et al., 2018; Martinez et al., 2004; Montagnoli et al., 2003; Pachl et al., 2006; Wüthrich et al., 2012), fungi still represent a wide-spread kingdom of deadly pathogens for which neither vaccines nor neutralizing antibody therapies are clinically available (Fisher et al., 2012; Garcia-Solache and Casadevall, 2010; Rudkin et al., 2018).
Most antibacterial and antiviral vaccines available today are rooted in boosting antibody-mediated immunity (Plotkin, 2010). Notably, systemic immunoglobulin G (IgG) antibodies with broad specificity for intestinal bacteria are an important component of host protection against systemic bacterial infections of intestinal origin (Ansaldo et al., 2019; Koch et al., 2016). In contrast, IgG antibody responses to several intestinal fungi or their cell wall components are known to occur during gastrointestinal disease (Leonardi et al., 2018, 2020; Riente et al., 2004; Standaert-Vitse et al., 2006; Yang et al., 2017) and are generally perceived as disease markers rather than protective responses. However, antibodies against fungal β-glucan and other fungal antigens have been detected recently in serum of healthy humans (Chiani et al., 2009; Huertas et al., 2017), raising several important questions regarding the function of antifungal antibodies, the species that induce them, and the mechanisms governing their induction and regulation. We hypothesized that fungal species of the gut mycobiota play a critical role in shaping the host antibody repertoire by promoting induction of host-protective antifungal antibodies.
RESULTS
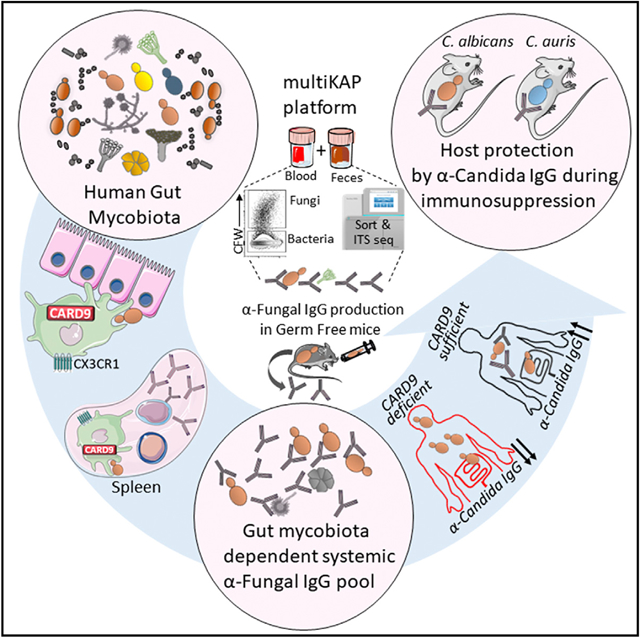
To explore the antibody pool against gut mycobiota, we established a multistep flow cytometry-based technique for multi-kingdom antibody profiling (multiKAP) (Figure 1A) that uses the native reservoir of gut commensal fungi and bacteria in fecal material as potential epitopes for antibody binding. Serum from the same host is applied to the fecal sample to allow systemic antibody binding to intestinal microbes that are also already bound to the luminally secreted antibodies present in the feces (Figure 1A). Live microbes labeled with a DNA-binding dye (SYBR Green; SYBRhi) in mouse feces are distinguished from dead microbes (SYBRlo) in sterilized food and bedding by calibrating the assay against germ-free (GF) mouse feces (Figure 1B). In contrast to previously established microbial flow cytometry-based techniques (Ansaldo et al., 2019; Christmann et al., 2015; Fadlallah et al., 2019; Koch et al., 2016; Palm et al., 2014), multiKAP distinguishes fungi from the more abundant bacteria by their large cell size and the distinct presence of chitin in their cell walls (Figures 1B–1D). Size separation by centrifugation at 900 × g, followed by sorting of antibody-bound fungal and bacterial cells and isolation of their DNA for down-stream deep sequencing/analysis allows assignment of fungal identities, in addition to bacterial identities, in the same multiKAP assay (Figures 1D and 1E).
Figure 1. Systemic IgG antibodies bind gut mycobiota.
(A–E) MultiKAP assay. Serum and feces are co-incubated to allow serum IgG to bind to gut microbial epitopes.
(A and B) Fecal microbes are distinguished as a Sybrhi population that is absent in similarly stained GF feces (B).
(C) Antibody-reactive microbiota (fungi and bacteria) analysis is performed by flow cytometry, and then fungi (SybrhiCFW+) and bacteria (SybrhiCFW+) are enriched in two respective fractions (large and small) through size separation by 900 × g centrifugation.
(D) Antibody-bound fungi and bacteria from each sample are sorted by fluorescence-activated cell sorting (FACS), and DNA is isolated and used in downstream applications such as PCR-based quantification and fungal (ITS1) and bacterial (16S) analysis through amplicon-based deep-sequencing.
(E) Quantitative PCR-based assessment of the enriched fungal or bacterial population; Mann-Whitney test. CFW+, n = 4; CFW−, n = 4.
(F and G) Fungi in SPF mouse feces were assessed for IgA, IgG, and IgM binding by multiKAP; no serum incubation (gray), incubation with matched WT SPF mouse serum (black), or B cell-deficient μMT−/− SPF mouse serum (blue). Pooled from two independent experiments; one-way ANOVA followed by Tukey’s multiple comparisons test. −Serum, n = 10; +WT serum, n = 10; +μMT−/− serum, n = 10.
(H–K) Fungi in WT SPF mouse feces were assessed for IgG (H and I) and IgM binding (J and K) by multiKAP after incubation with matched WT SPF (black) or WT GF (blue) serum. Mann-Whitney test. SPF, n = 5; GF, n = 5.
(L) MultiKAP assay with feces from mice from a dedicated controlled environment room at Weill Cornell Medicine (WCM-CE) and Jackson Laboratory (JAX) or Taconic Bioscience (TAC) facilities with (+serum) or without (−serum) serum co-incubation. Pooled from two independent experiments; one-way ANOVA followed by Tukey’s multiple comparisons test. WCM-CE, n = 7; JAX, n = 9; TAC, n = 7.
(M and N) Fungi in SPF WT mouse feces were assessed for IgG1, IgG2b, and IgG3 binding by multiKAP after incubation with matched WT SPF serum. IgG1, n = 5; IgG2b, n = 5; IgG3, n = 5.
Error bars indicate SEM. ns, p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. See also Figure S1 and Table S1.
Co-staining of fecal microbes with Sybr and calcofluor-white (CFW; a chitin-binding dye) followed by flow cytometry analysis determined that 1%–2% of the microbes in mouse feces were fungal cells (SYBRhiCFW+; Figures 1C and S1A). To determine whether the gut mycobiota are coated by antibodies of local secretory and/or systemic origin, we next analyzed the IgA- and IgG-bound fungal (SYBRhiCFW+) and bacterial (SYB-RhiCFW−) populations before and after fecal incubation with serum from the same mouse. Consistent with previously reported luminal IgA reactivity to intestinal bacteria in mice (Bunker et al., 2015; Koch et al., 2016; Palm et al., 2014), roughly half of the intestinal fungi identified by multiKAP were also bound by IgA independent of serum supplementation (Figures 1F and 1G), suggesting that systemic antibodies make a small contribution to the antifungal IgA repertoire in mice. Notably, a population of intestinal fungi were bound by IgG found exclusively in the blood (Figures 1F, 1G, and S1A; +wild-type [WT] serum) because no IgG-bound fraction of the mycobiota was present in the intestinal lumen under steady-state conditions (Figures 1F, 1G, and S1A; −serum). A similar binding pattern was observed when feces from Ig-deficient Rag2−/− mice were used as a gut fungal source to avoid prior IgA epitope coating (Figure S1B). Antifungal IgG binding was absent upon incubation with serum from B cell-deficient μMT−/− mice, which lack systemic antibodies, confirming the specificity of this assay (Figures 1F, 1G, and S1A; +μMT−/− serum). We also detected systemic antifungal IgM antibodies restricted to the blood (Figures 1F and 1G), supporting previous reports of involvement of the natural antifungal IgM repertoire in early immune responses during Pneumocystis or Cryptococcus infection (McClelland et al., 2010; Rapaka et al., 2010). Although antifungal IgM binding was similar upon incubation of specific pathogen-free (SPF) feces with GF and SPF serum, antifungal IgG binding was absent in GF serum-incubated SPF feces (Figures 1H–1K), indicating that induction of antifungal IgG, but not antifungal IgM, is gut microbiota dependent. Mice from different facilities that harbor compositionally different mycobiota (Doron et al., 2019) exhibit a variety of IgG binding, with our facility’s “Ascomycota-high” mouse colony (Doron et al., 2019) and Taconic mice showing significantly higher antifungal IgG than mice from the Jackson Laboratories facility (Figure 1L). This phenomenon was independent of any detectable fungal pathology in the examined colonies or systemic fungal spread (Figures S1F and S1G). Given these findings and the negligible contribution of serum IgA to the antifungal IgA repertoire (Figures 1F and 1G), we focused on analyzing the gut microbiota-dependent systemic antifungal IgG response. Previous studies have observed that the IgG2b and IgG3 subtypes dominate the IgG response to commensal gut bacteria, with the IgG2b isotype binding the largest fraction of the bacteria (Koch et al., 2016). Although we observed a similar abundance of these two IgG isotypes (Figures S1C and S1D; Koch et al., 2016), IgG3 bound the largest fraction of the intestinal fungi (Figures 1M and 1N).
Fungi and bacteria cohabitate in the human intestine, where we found that fungal constituents of the microbiota are more prevalent (~2%) than initially predicted by metagenomics studies (Arumugam et al., 2011; Li et al., 2019; Figures 2A, 2B, and S1E). An initial multiKAP-based assessment of healthy human subjects determined that, despite substantial inter-individual variation, a large fraction of human intestinal fungi is bound by luminal IgA independent of serum addition (Figure 2C). Similar to mice, mycobiota-reactive IgG antibodies in healthy individuals were restricted to the serum (Figure 2C). MultiKAP assessment of these human subjects revealed that, along with a large proportion of all gut fungi bound to IgG and IgA (IgG+IgA+, ~50%) or neither antibody isotype (IgG−IgA−, ~25%), a significant population of fungi was only bound by IgG (IgG+IgA−, ~20%) (Figure 2D). These data demonstrate that a significant fraction of the gut mycobiota is recognized by systemic antifungal IgG antibodies in humans and mice. Furthermore, systemic mycobiota-reactive IgG antibodies are typically present in the serum of mice and healthy individuals, whereas antifungal IgA antibodies are mostly present in the gut.
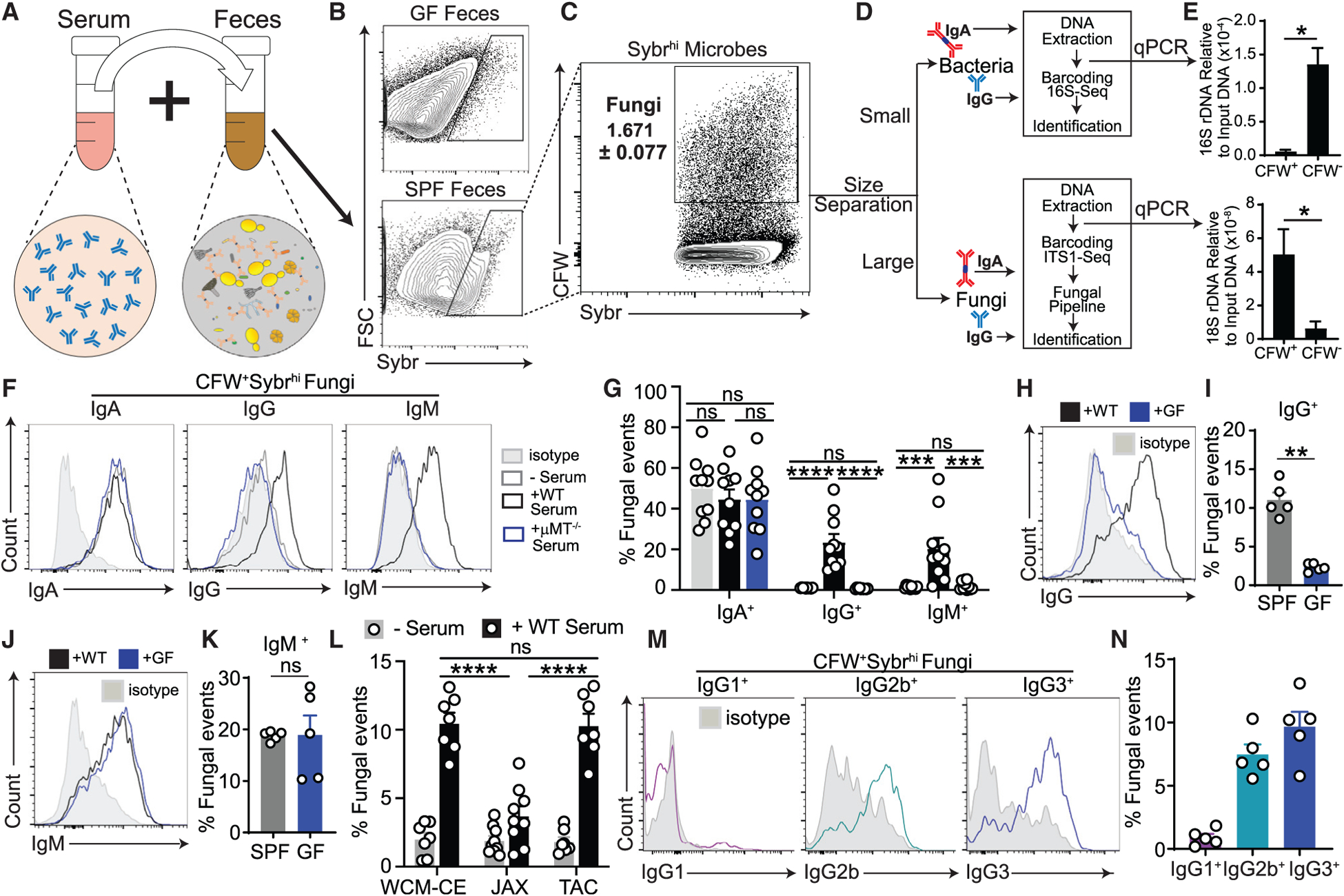
Figure 2. Enrichment of C. albicans among IgG-coated intestinal microbes identifies the predominant driver of systemic antifungal IgG responses in humans.
(A and B) Visualization of commensal fungi and bacteria in human feces stained with Sybr Green (DNA), CFW (fungal cells), and the EUB338 FISH probe (bacteria); the scale bar represents 25 μm (A). The percentages of fungal (CFW+Sybr Green+) and bacterial (EUB338+ Sybr Green+) cells in 6 fecal samples from healthy individuals are shown in (B). Each dot represents an individual feces donor.
(C) Fungi in feces from healthy human donors were assessed for IgA and IgG binding by multiKAP; no serum incubation (gray), incubation with matched serum (black). Mann-Whitney test, n = 13.
(D–G) Combined multiKAP-based sorting and sequencing analysis of antibody-bound species of healthy human mycobiota.
(D) Human feces from healthy donors was incubated with matched serum and assessed for IgA and IgG binding by multiKAP.
(E) Abundance of the top 15 fungal genera in human fecal samples.
(F and G) Relative enrichment of fungal species in antibody-bound fractions, IgG+IgA+ (F) or IgG+IgA− (G), expressed relative to the IgG−IgA− (unbound) population. Confidence interval (CI) values were calculated at the species level. Fungal species represented at 2% or more are depicted. One-sample t test, n = 7. Boxplots represent species’ median and the first to third quartiles, and whiskers represent minimum and maximum values. Starred boxplots indicate fungal species with a mean CI significantly greater or less than 0, indicating preferential enrichment in the antibody-bound or unbound fraction. Non-enriched species are unlabeled.
(H) ELISA-based validation of IgG reactivity against fungal lysates from C. albicans (recognized by antifungal antibodies; F and G) or M. restricta (not recognized by antifungal antibodies; F and G) in healthy human serum. Mann-Whitney test. Human sera, n = 10; each dot represents an individual subject. Each dot represents a serum sample from a healthy individual.
(I) Serum IgG titers in the serum of germ-free (GF) mice monocolonized with the fungal species identified in Figures 2F and 2G, assessed by ELISA. Serum IgG titers of each group are statistically compared relative to uncolonized controls. One-way ANOVA followed by Tukey’s multiple comparisons test. Data are representative of three independent experiments. GF, n = 6; C. albicans, n = 8; S. cerevisiae, n = 4; M. restricta, n = 6; C. cladosporioides, n = 4. Each dot represents an individual mouse. Error bars indicate SEM. ns, p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
(J) Serum IgG reactivity from untreated (GF) or C. albicans-monocolonized GF mice (Ca-exGF) against lysates from C. albicans, S. cerevisiae, A. amstelodami, M. restricta, and M. globosa pure cultures was measured by ELISA. Squares represent individual mice. Heatmap values represent optical density 405 (OD405) measurements. GF, n = 3; Ca-exGF, n = 5. Error bars indicate SEM. ns, p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. See also Figure S2 and Table S1.
To determine specific gut fungi that are preferentially coated or neglected by systemic human IgG antibodies, we coupled the multiKAP technique to ITS sequencing of gut fungal populations that were sorted from fecal material based on antibody binding (Figures 2D, S2A, and S2B). ITS sequencing revealed a variety of commensal, environmental, and food-associated fungi (Figure 2E), consistent with previous studies (Nash et al., 2017; Sokol et al., 2017). As noted above, we distinguished three populations of intestinal fungi (IgG+IgA+, IgG+IgA−, and IgG−IgA−) by multi-KAP, where all luminal IgA-bound fungi were also recognized by systemic IgG. Among these, the IgG-bound mycobiota (IgG+IgA+ and IgG+IgA−) were enriched for C. albicans (as determined by the positive coating index [CI]) (Kau et al., 2015; Figure S2C), whereas gut fungi not bound by either antibody pool (IgG−IgA−) were represented by skin- and food-associated fungal species such as M. restricta and S. cerevisiae, which rarely cause gut-disseminated infections in humans (Brown et al., 2012; Fan et al., 2015; Zhai et al., 2020; Figures 2F–2H and S2D). Other fungi detected by this approach were distributed equally in antibody-bound and unbound fractions and, thus, were not considered to be preferentially coated by anti-fungal antibodies (Figures 2F, 2G, and S2D). These data indicate that the systemic antifungal IgG antibody repertoire in healthy individuals is primarily characterized by preferential specificity to commensal Candida.
Candida is one of the most common genera of fungal commensals in the healthy human gastrointestinal tract (Li et al., 2019; Nash et al., 2017; Sokol et al., 2017) but also harbors the main opportunistic pathogens that cause disseminated fungal infections with high attributable morbidity and mortality in humans (Brown et al., 2012; Fan et al., 2015; Zhai et al., 2020). Among other fungi, C. albicans has been implicated as a potent inducer of Th17 responses in humans and mice; it is bound by host antibodies and, thus, has the potential to induce protective immune responses or trigger inflammation in a context-dependent manner (Bacher et al., 2019; Conti et al., 2009; Huertas et al., 2017; Leonardi et al., 2018; Li et al., 2018; Moreno-Sabater et al., 2020; Shao et al., 2019; Standaert-Vitse et al., 2009). Although most studies of intestinal C. albicans have focused exclusively on innate and Th17-mediated immunity, humoral immunity to this commensal in the gut remains poorly understood. Given our finding that systemic antibodies preferentially coat gut C. albicans, we sought to characterize how systemic antibody responses to this intestinal fungus are mediated.
IgG antibody responses to intestinal microbes can originate by antigen-specific responses to a distinct organism or can be derived from pre-existing cross-reactive specificities (Ansaldo et al., 2019; Koch et al., 2016; Slack et al., 2009). To investigate these possibilities and identify gut fungi that induce systemic antifungal IgG responses, we performed fungal monocolonization of GF mice that lacked antibody responses to fungi before colonization (Figures 2I, 1H, and 1I). Among fungal species identified by multiKAP, C. albicans induced the most potent systemic IgG antibody response (Figure 2I). Notably, antibodies generated by intestinal colonization with C. albicans were not cross-reactive with environmental, food-derived, or skin-resident fungi such as Saccharomyces, Aspergillus, and Malassezia spp. (Figure 2J), suggesting a potential specificity restricted to resident fungal antigens. These data further indicate that intestinal fungal colonization can induce systemic antifungal IgG antibodies that are not derived from pre-existing cross-reactive specificities and implicate C. albicans as the predominant intestinal fungal species triggering this response.
A better understanding of fungus-specific antibody induction is obscured by coexistence of fungi with a dynamic bacterial community whose composition profoundly influences colonization of, and immune responses induced by, newly introduced bacterial or fungal commensals (Fan et al., 2015; Hand et al., 2012; Kim et al., 2019; Leonardi et al., 2020; Zhai et al., 2020). Furthermore, mycobiota are variable, essential components of the gut microbial communities of SPF mice (Doron et al., 2019; Leonardi et al., 2018; Li et al., 2018), and systemic antifungal IgG antibodies are present in their serum during homeostasis (Figures 1F–1L). To avoid the influence of these confounding factors in our investigation into the mechanisms of antifungal antibody induction, we utilized altered Schaedler flora (ASF)-colonized mice that carry a defined set of bacteria but are mycobiota-free (Li et al., 2018; Schaedler et al., 1965). In this setting, intestinal colonization with C. albicans (preferentially coated by systemic IgG antibodies) induced strong production of systemic antifungal IgG antibodies without priming unspecific antibacterial IgG response, whereas S. cerevisiae (preferentially not coated by systemic IgG antibodies) induced a limited IgG response (Figures 3A and S3A). These findings further support the notion that specific gut fungi might play a dominant role in induction of systemic antifungal IgG antibodies.
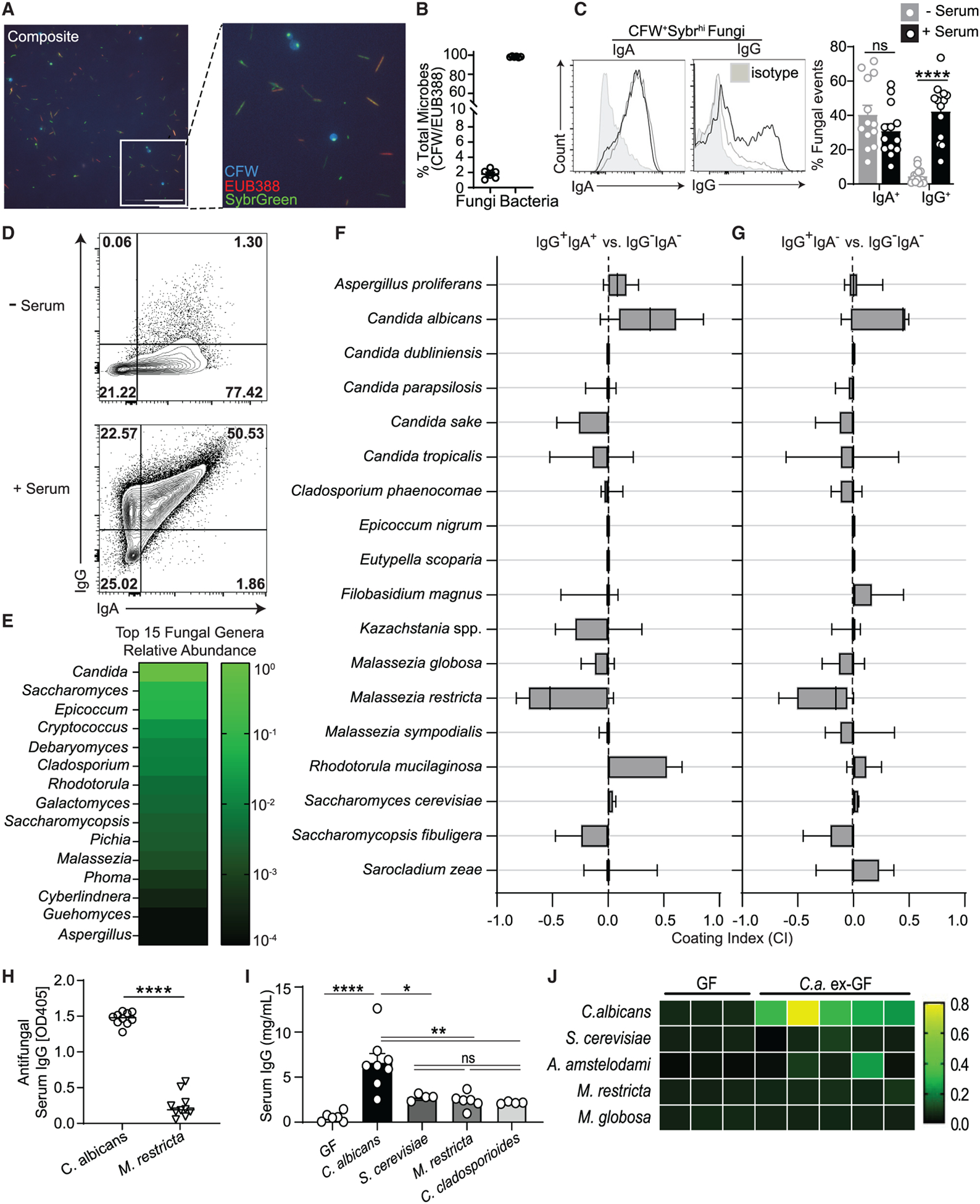
Figure 3. Intestinal C. albicans elicits extra-intestinal GC-dependent IgG+ B cell responses.
Mycobiota-free altered Schaedler flora (ASF) mice were orally gavaged with PBS (ASF), C. albicans (+Ca), or S. cerevisiae (+Sc).
(A) Serum anti-C. albicans IgG antibody production in Ca-colonized ASF mice (A, left) and serum ASCA production in Sc-colonized ASF mice (A, right) compared with ASF controls, as measured by ELISA.
(B) qPCR-based 18S rDNA assessment of spleens from ASF mice colonized for 2 weeks with C. albicans or S. cerevisiae or left untreated.
(C) Frequency of IgG+IgM− among CD19+CD4− B cells in PPs (top), mLNs (center), and spleen (bottom), analyzed by flow cytometry (pooled from two independent experiments).
(D) Frequency of GC-B cells among CD19+CD4− B cells in the spleen, analyzed by flow cytometry. One-way ANOVA followed by Tukey’s multiple comparisons test. ASF, n = 8; +Ca, n = 10; +Sc, n = 10. Data are pooled from three independent experiments.
Each dot represents an individual mouse. Error bars indicate SEM. ns, p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. See also Figure S3 and Table S1.
IgG responses to intestinal bacteria have been attributed to gut-associated lymphoid tissue (GALT) (Ansaldo et al., 2019; Koch et al., 2016; Teng et al., 2016) and extra-intestinal systemic locations (Barbet et al., 2018; Zeng et al., 2016). To determine sites of B cell expansion and IgG class switch recombination (CSR) upon intestinal fungal colonization, gut-distal tissue (spleen) and GALT (mesenteric lymph nodes [mLNs] and Peyer’s patches [PPs]) were obtained from fungus-colonized (C. albicans-colonized [+Ca] and S. cerevisiae-colonized [+Sc]) and fungus-free ASF (ASF) mice. Notably, C. albicans gut colonization induced a robust IgG CSR in the spleen but not in PPs or mLNs (Figure 3C). On the other hand, S. cerevisiae induced a limited CSR response at these sites (Figure 3C), consistent with its low capacity to induce systemic IgG antibodies (Figure 3A). These results were not attributable to any noticeable systemic fungal spread or acute reaction (Macpherson et al., 2000) to C. albicans colonization because no live fungi were detectable in multiple organs, whereas anti-C. albicans IgG antibodies appeared at 2 weeks and persisted after the initial colonization event (Figures S3D and S3E). Generation of high-specificity IgG is often driven by expansion of germinal center B (GC-B) cells (Barbet et al., 2018; Teng et al., 2016), which undergo many cycles of somatic hypermutation (SHM) that are associated with high-affinity and high titers of antibodies (Victora and Nussenzweig, 2012). Consistent with its ability to drive CSR, gut C. albicans induced potent splenic GC-B cell expansion and differentiation accompanied by transfer of C. albicans antigens (as assessed by sensitive DNA-based assay) to the spleen (Figures 3B and 3D). No significant differences were detected in the frequency of total splenic B cells between experimental groups (Figures S4B and S4C). Notably, adoptive transfer of antibody-producing splenic B cells from +Ca mice, but not antibody-deficient B cells or B cells from GF mice, protected highly susceptible μMT−/− mice against systemic candidiasis, suggesting a protective role of B cell-mediated immunity against fungal infection (Figures 4A, 4B, S4A, and S4B). These data suggest that the gut commensal C. albicans induces systemic IgG production through expansion of GC-B cells in the spleen.
Figure 4. Antibodies induced through intestinal Candida colonization confer protection against gut-derived and blood-borne systemic candidiasis caused by C. albicans or C. auris.
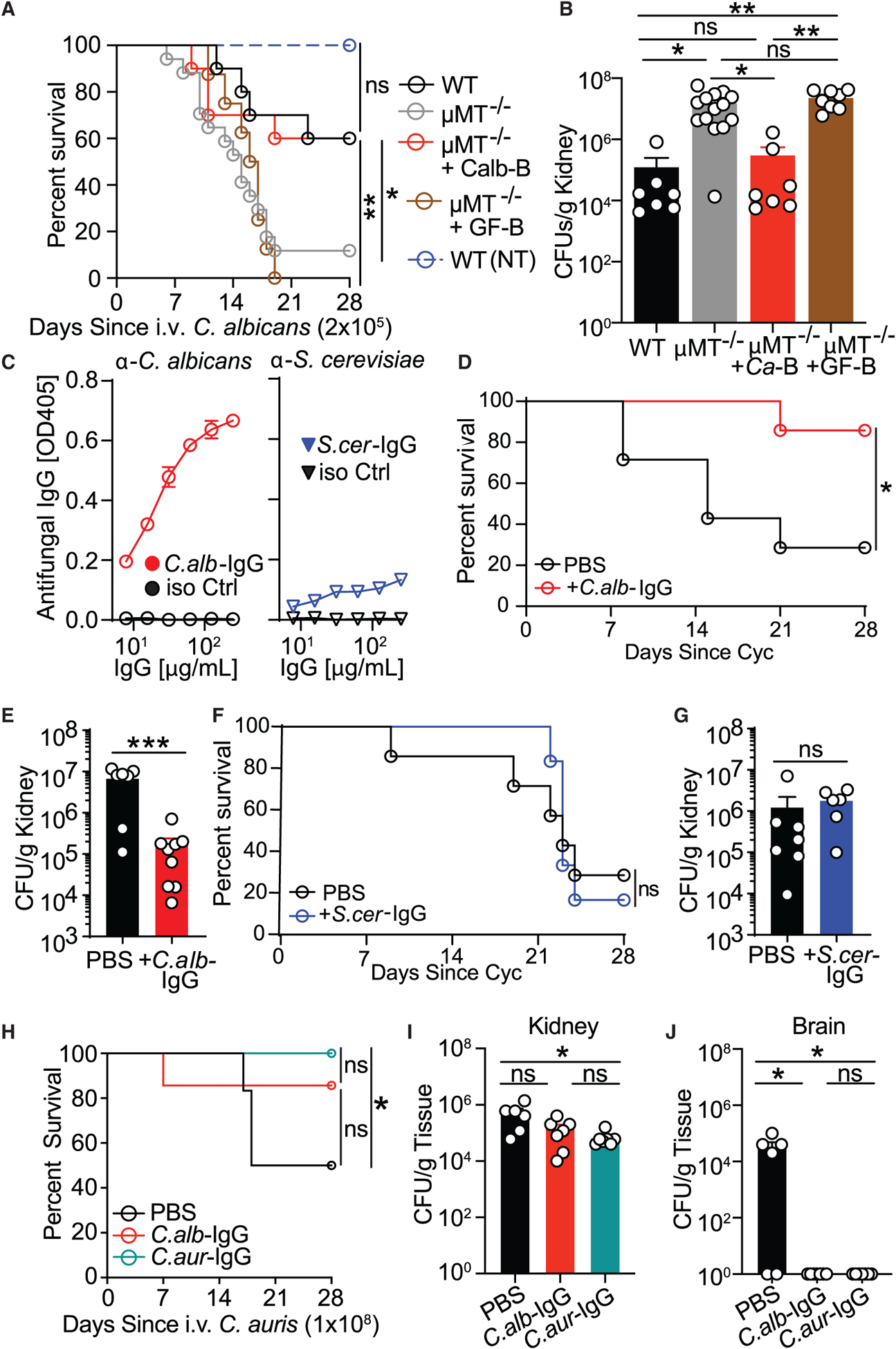
(A and B) WT mice (WT), μMT−/− mice (μMT−/−), and μMT−/− mice adoptively transferred splenic B cells from C. albicans-monocolonized GF mice (mMT−/− +Ca-B) or untreated GF mice (μMT−/− +GF-B) were infected intravenously (i.v.) with C. albicans. A control group of WT mice was left uninfected (WT, NI). Disease morbidity (A) and C. albicans systemic spread to kidney tissue (B) were assessed; log rank Mantel-Cox test (A) and one-way ANOVA followed by Tukey’s multiple comparisons test (B). WT, n = 7; μMT−/−, n = 14; μMT−/− +Ca-B, n = 7; μMT−/− +GF-B, n = 8; WT (NI), n = 5. Data are representative of three independent experiments.
(C) Antibody reactivity assessment against C. albicans and S. cerevisiae lysates of purified serum IgG antibodies from C. albicans- (C.alb-IgG, left panel) and S. cerevisiae- (S.cer-IgG, right panel) monocolonized GF mice, respectively measured by ELISA.
(D and E) C.alb-IgG protection against cyclophosphamide (Cyc)-induced systemic fungal dissemination in SPF mice intestinally colonized with C. albicans (treatment scheme depicted in Figure S4B). Shown are disease morbidity (D) and systemic C. albicans spread into kidney tissue (H). Statistical analysis was performed using log rank Mantel-Cox test (D) or Mann-Whitney test (E). PBS, n = 7; +C.alb-IgG, n = 9.
(F and G) Lack of protection by S.cer-IgG against Cyc-induced systemic fungal dissemination in SPF mice intestinally colonized with C. albicans (treatment scheme depicted in Figure S4B). Shown are survival (F) and systemic C. albicans spread into kidney tissue (G). ns, p > 0.05; log rank Mantel-Cox test (F) and Mann-Whitney test (G). PBS, n = 7; +C.alb-IgG, n = 6.
(H–J) Intestinal GF colonization-induced C.aur-IgG protects mice against systemic infection with C. auris. The effect of C.alb-IgG and C.aur-IgG was assessed after i.v. infection with C. auris. Shown are disease morbidity (H) and systemic C. auris spread into kidney (I) and brain tissue (J). Statistical analysis was performed using log rank Mantel-Cox test (H) or one-way ANOVA followed by Tukey’s multiple comparisons test (I and J). PBS, n = 6; +C.alb-IgG, n = 7; +C.aur-IgG, n = 7. Data are representative of two independent experiments.
Each dot represents an individual mouse. Error bars indicate SEM. ns, p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. See also Figure S4.
To determine the functional role of systemic antifungal IgG, we purified IgG antibodies from serum of GF mice, which normally lack systemic anti-commensal IgG antibodies in the absence of fungal or bacterial antigen exposure (Figures 1H and 1I), intestinally monocolonized with C. albicans (C.alb-IgG) or S. cerevisiae (S.cer-IgG) (Figure 4C). Antifungal antibodies obtained from C. albicans ex-GF mice have strong reactivity with C. albicans, whereas IgG antibodies from serum of S. cerevisiae-monocolonized mice demonstrate weaker reactivity with S. cerevisiae (Figures 4C and 2J).
We next explored the role of these antibodies during immunosuppression, where, among different fungi, Candida species are the main cause of morbidity and mortality in people, with the gut often being the fungus source (Brown et al., 2012; Zhai et al., 2020). Thus, C.alb-IgG or S.cer-IgG antibodies were transferred into SPF mice intestinally colonized with C. albicans and treated with the chemotherapy drug cyclophosphamide (Cyc) to induce immunosuppression and facilitate gut-to-system fungal dissemination and death (Figure S4B; Koh et al., 2008). In addition to severe leukopenia (Koh et al., 2008), Cyc treatment led to a dramatic decrease in antifungal IgG antibodies in the serum of mice (Figure S4C). Notably, C.alb-IgG transfer into these mice led to nearly complete survival 4 weeks after systemic fungal dissemination and significantly decreased kidney fungal burdens (Figures 4D and 4E), whereas S.cer-IgG transfer did not reduce infection mortality (Figures 4F and 4G). Candida auris is an emerging multidrug-resistant fungal pathogen that is a serious global health threat and is the cause of multiple nosocomial outbreaks worldwide that are associated with unusually high mortality rates (Eyre et al., 2018). Thus, we used our GF colonization protocol to generate antibodies against this multidrug-resistant fungal pathogen and test their efficacy. Notably, such generated antibodies (C.aur-IgG) protected mice against intravenous C. auris infection and death. C.alb-IgG antibody transfer was also protective but to a lower extent (diminishing only brain-disseminated candidiasis), suggesting some level of antibody selectivity even among fungi belonging to the same species (Figures 4H–4J). These results demonstrate that systemic antibodies to mycobiota generated through the gastrointestinal route protect the host against deadly systemic fungal infections of intestinal or non-intestinal origin. Moreover, this protection demonstrates the powerful effect of antifungal antibody immunity in the face of broad cellular immunity depression by chemotherapeutics.
Among genetic factors associated with fungal infections, CARD9 deficiency is the only known human genetic disorder that specifically leads to recurrent severe mucosal and systemic fungal infections without conferring bacterial, parasitic, or viral susceptibility (Cao et al., 2015; Glocker et al., 2009; Lionakis et al., 2014). CARD9 is required to activate nuclear factor κB (NF-κB)-based signaling downstream of the receptors Dectin-1, Dectin-2, and Mincle in innate immune cells, which, in turn, induces cytokine production and triggers cellular immunity against fungi (Cao et al., 2015; Drummond et al., 2015; LeibundGut-Landmann et al., 2007; Pérez de Diego et al., 2015; Roth and Ruland, 2013). A common polymorphism in CARD9 is among the strongest genetic risk factors associated with Crohn’s disease, where increased anti-S. cerevisiae IgG antibodies (ASCA IgG) are a clinical marker of disease severity (Evans et al., 2011; Jostins et al., 2012; Rivas et al., 2011). Furthermore, CARD9 deficiency has been associated with gut fungal dysbiosis (Lamas et al., 2016), revealing CARD9 signaling as a critical link between gut mucosal immunity and intestinal fungi. Interestingly, Card9-deficient (Card9−/−) mice that were highly susceptible to systemic candidiasis originating from the gut upon Cyc treatment were unable to induce systemic antifungal IgG antibodies despite a high systemic Candida burden (Figures 5A–5C) and normal development of T and B cell subsets (Hsu et al., 2007; Figures S4H and S4I). In contrast, Card9-sufficient (Card9+/+) mice that were more resistant to fungal infection with intestinal origin were able to generate systemic IgG antibodies against C. albicans (Figures 5A–5C). MultiKAP analysis of antifungal IgG antibodies demonstrated that the presence of a systemic antifungal IgG repertoire was strikingly dependent on the presence of CARD9 signaling (Figures 5D and 5E). These results suggest an unexpected role of CARD9 in induction of systemic anti-fungal IgG responses to intestinal fungi.
Figure 5. CARD9 is critical for the GC-dependent generation of antifungal IgG antibodies.
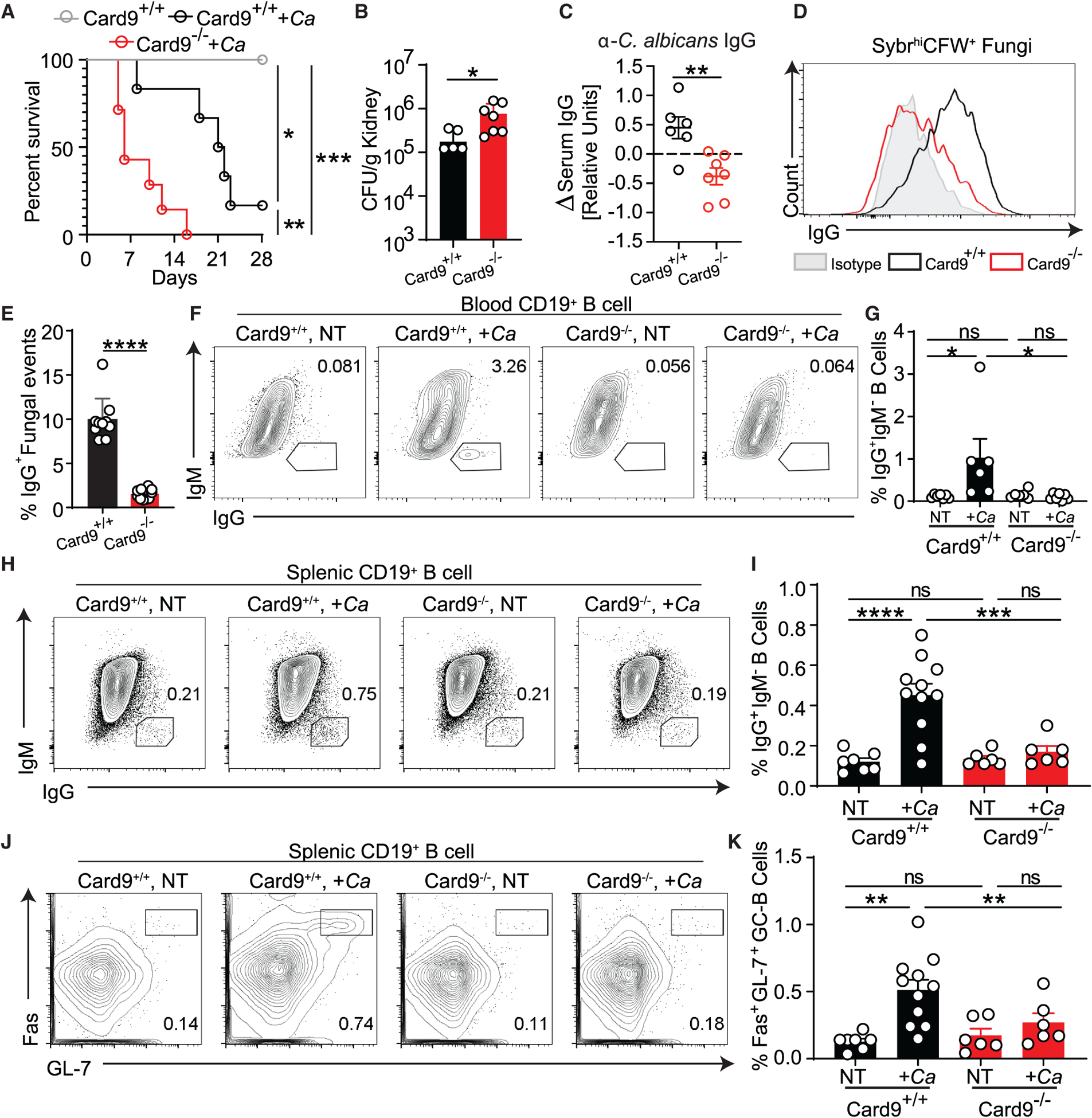
(A–C) Cyc-induced gut-to-system fungal dissemination was induced in Card9+/+ and Card9−/− mice.
(A and B) Disease morbidity assessed by survival (A) and C. albicans systemic spread to the kidneys (B). C. albicans detection failed in one mouse, which was excluded from the analysis.
(C) Change in anti-C. albicans serum IgG after mice became moribund (14 days after infection) relative to pre-infection titers.
(D and E) MultiKAP analysis of anti-fungal antibodies in healthy Card9+/+ and Card9−/− mice.
(A–E) Statistical analysis was performed using log rank Mantel-Cox test (A) or Mann-Whitney test (B, C, and E). Card9+/+, n = 6; Card9+/+ +Ca, n = 6; Card9−/− +Ca, n = 7. Data are representative of two independent experiments.
(F and G) Systemic IgG and B cell responses of Card9+/+ and Card9−/− mice 2 weeks after intraperitoneal injection with C. albicans. The B cell compartment in blood samples from individual mice was analyzed by flow cytometry. One-way ANOVA followed by Tukey’s multiple comparisons test. Card9+/+ NT, n = 7; Card9+/+ +Ca, n = 6; Card9−/− NT, n = 6; Card9−/− +Ca, n = 6. Data are representative of two independent experiments.
(H–K) Systemic IgG and B cell responses of Card9+/+ and Card9−/− mice after intraperitoneal injection with C. albicans. The B cell compartment in spleen samples from individual mice was analyzed by flow cytometry. One-way ANOVA followed by Tukey’s multiple comparisons test. Card9+/+ NT, n = 7; Card9+/+ +Ca, n = 11; Card9−/− NT, n = 6; Card9−/− +Ca, n = 6. Data are representative of three independent experiments.
Each dot represents an individual mouse. Error bars indicate SEM. ns, p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. See also Figure S4.
To investigate the mechanisms behind the observed CARD9-mediated antifungal IgG response, we intraperitoneally delivered heat-killed C. albicans to circumvent the extreme susceptibility of Card9−/− mice to systemic candidiasis (Drummond et al., 2015; Lionakis et al., 2014; Figures 5F–5K, S4D, and S4E). We found that IgG+ B cell frequency among circulating B cells in the blood only increased significantly in response to C. albicans in Card9+/+ mice (Figures 5F and 5G). In contrast to Card9+/+ mice, Card9−/− mice were unable to induce an efficient anti-C. albicans IgG antibody response, and this corresponded with defective IgG+ CSR and GC-B cell expansion in the spleen (Figures 5H–5K) but was not due to B cell-intrinsic or developmental defects in the absence of CARD9 (Figures S4F and S4G). These data indicate that the GC-dependent systemic antifungal IgG antibody response is dependent on CARD9.
Single-cell RNA sequencing (RNA-seq) analysis of a previously published dataset (Smillie et al., 2019) determined that CARD9 expression in humans is restricted to several populations of phagocytes, such as macrophages, monocytes, and dendritic cells (DCs; Figure 6A). In mice, these populations include CD103+ conventional DCs (CD11b− cDC1s and CD11b+ cDC2s) and CX3CR1+ mononuclear phagocytes (MNPs), cells described previously as providing an interface between the gut microbiota and anti-commensal immunoglobulin induction (Flores-Langarica et al., 2018; Kim et al., 2018; Koscsó et al., 2020; Takaki et al., 2018). Because CX3CR1+ MNPs and the cDC2 subset represent the vast majority of C-type lectin receptor (CLR)/CARD9-expressing antigen-presenting cells, and a loss-of-function mutation in CX3CR1 is associated with reduced antifungal IgG antibodies in humans (Li et al., 2018), we next set out to identify the potential involvement of CX3CR1+ MNPs and cDC2 in induction of IgG with specificity for intestinal fungi. We crossed transgenic Cd11c-Cre mice with inducible floxed CX3CR1 (Cx3cr1DTR) or cDC2 master regulator IRF4 (IRF4fl/fl) mice to generate models of inducible CX3CR1+ MNPs (ΔCX3CR1) and cDC2 (ΔIRF4) depletion, respectively (Figures S5A–S5D). MultiKAP analysis of serum IgG binding revealed that, relative to their respective Cre− littermates, CX3CR1+ MNPs, but not cDC2 depletion, abrogated the systemic anti-fungal IgG (Figures 6B and 6D). Intestinal colonization with C. albicans led to a reduction in IgG+ frequency in the splenic B cell compartment of ΔCX3CR1 but not ΔIRF4 mice or littermate controls (Figures 6C and 6E). To assess whether Syk-Card9-mediated antifungal signaling on CX3CR1+ MNPs is necessary for induction of antifungal humoral responses, we crossed transgenic Cx3cr1-cre/ERT2+/− mice with a strain carrying an inducible floxed spleen tyrosine kinase (Syk; Sykfl/fl) to conditionally block Syk-dependent, Card9-mediated antifungal signaling in CX3CR1+ MNPs (CX3CR1ΔSyk mice). We found that CX3CR1+ MNP-specific targeting of this pathway significantly reduced systemic antifungal IgG in CX3CR1ΔSyk mice (Figure 6F). Furthermore, the increased frequency of splenic IgG+ B cells observed in C. albicans-colonized littermates was not observed in CX3CR1ΔSyk mice (Figure 6G). Notably, a comparison of antifungal IgG binding in αβT cell-deficient TCRβ−/− demonstrated significant loss of antifungal IgG binding relative to αβT cell-sufficient counterparts (TCRβ+/+), suggesting that CX3CR1+ MNPs mediate antifungal IgG induction through T cell-dependent mechanisms (Figure 6H). These data pinpoint CX3CR1+ MNPs as mediators of commensal fungus-induced systemic antifungal IgG.
Figure 6. Syk-CARD9-dependent signaling in CX3CR1+ MNPs is critical for induction of antifungal IgG antibody responses.
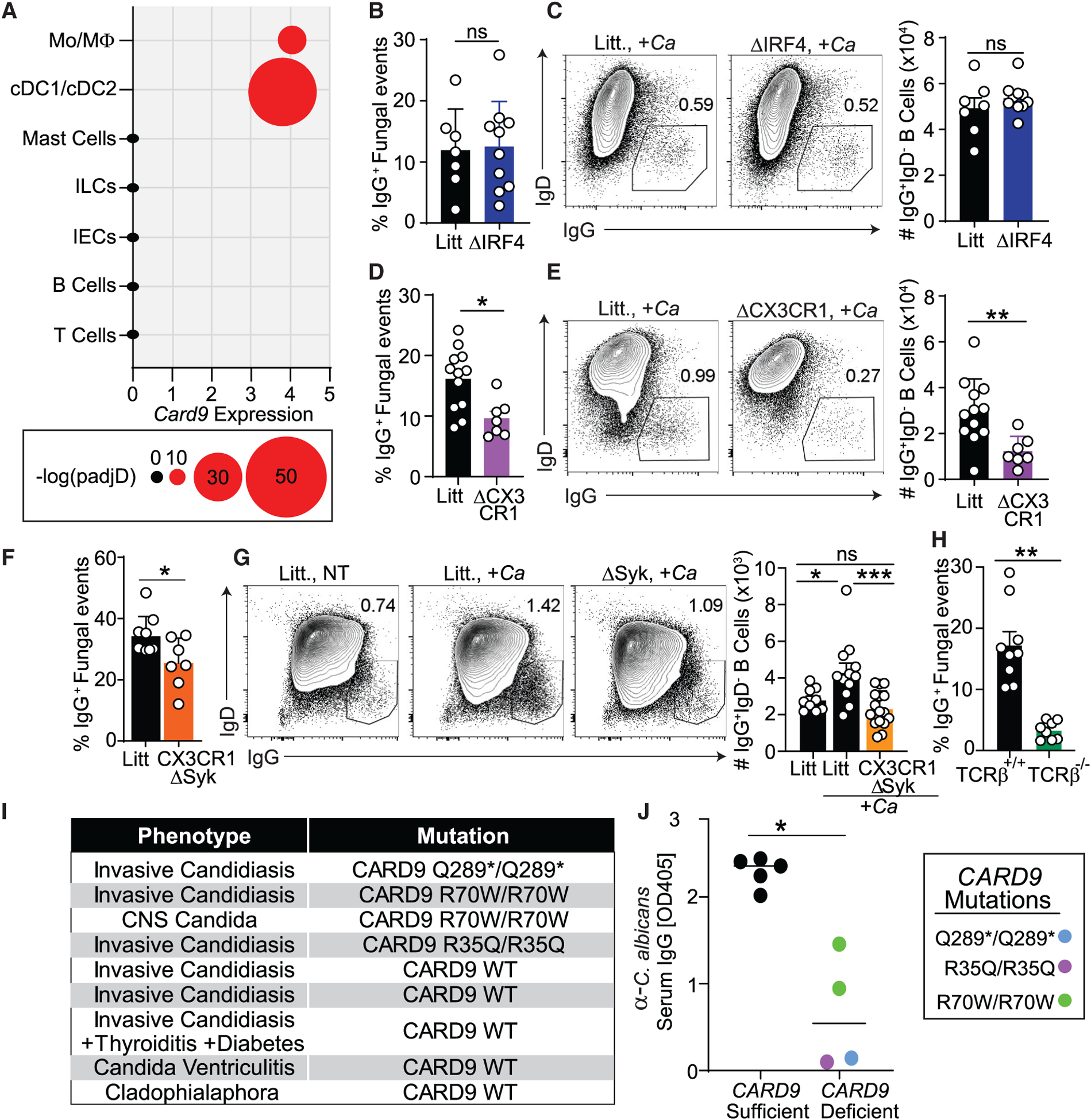
(A) Single-cell RNA-seq analysis of CARD9 expression in human biopsies from a single-cell RNA-seq dataset from Smillie et al. (2019).
(B and C) Systemic IgG and B cell responses in ΔIRF4 or littermates (Litt) upon intestinal colonization with C. albicans. Data were collected from two independent experiments. Statistical analyses were performed using Mann-Whitney test. Litt, n = 7; ΔIRF4, n = 10.
(B) MultiKAP analysis of anti-fungal antibodies in ΔIRF4 and littermate mice.
(C) Frequency and absolute numbers of IgG+IgD− among splenic CD19+CD4− B cells analyzed by flow cytometry.
(D and E) Systemic IgG and B cell responses in ΔCX3CR1 mice and Litt upon intestinal colonization with C. albicans. Data were collected from two independent experiments. Statistical analyses were performed using Mann-Whitney test. Litt, n = 12; ΔCX3CR1, n = 7.
(D) MultiKAP analysis of anti-fungal antibodies in ΔCX3CR1 and Litt mice.
(E) Frequency and absolute numbers of IgG+IgD− among splenic CD19+CD4− B cells analyzed by flow cytometry.
(F) Feces of healthy CX3CR1ΔSyk and Litt were assessed by multiKAP for systemic antifungal IgG binding after incubation with matched sera. Data were collected from two independent experiments; Mann-Whitney test. Litt, n = 8; CX3CR1ΔSyk, n = 7.
(G) Frequency and absolute numbers of IgG+IgD− among splenic CD19+CD4− B cells analyzed by flow cytometry in CX3CR1ΔSyk and Litt mice after oral gavage with C. albicans. Data were collected from three independent experiments; Mann-Whitney test. Litt NT, n = 9; Litt +Ca, n = 12; CX3CR1ΔSyk +Ca, n = 16.
(H) MultiKAP analysis of anti-fungal antibodies in healthy TCRβ+/+ and TCRβ−/− mice. Mann-Whitney test TCRβ+/+, n = 9; TCRβ−/−, n = 8. Data are representative of two independent experiments.
(I and J) Anti-Candida IgG antibody titers (J) assessed by ELISA in sera from individuals with candidiasis who were non-carriers or carriers of CARD9 loss-of-function mutations (Q289*, R35Q, R70W) (I). Non-carriers: CARD9 sufficient, n = 5; carriers: CARD9 deficient, n = 4. Each dot represents an individual human subject; Mann-Whitney test.
Error bars indicate SEM. ns, p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. See also Figures S5.
Having established the necessity of Syk-Card9 signaling for induction of antifungal antibodies in mice, we next explored whether a similar role of this pathway is conserved in humans. So far, over 22 different human homolog CARD9 mutations have been identified that abrogate the Th17 response without affecting total representation of T or B cells (Corvilain et al., 2018), but whether these mutations affect systemic antifungal antibody induction remains unclear. We collected serum from individuals with systemic candidiasis representing non-carriers or carriers of CARD9 mutations (Q289*, R35Q, R70W; Figure 6I) that have been associated previously with Candida-induced meningoencephalitis or colitis (Figure S5E; Lanternier et al., 2015). Individuals with systemic candidiasis mounted a robust antibody response to C. albicans (Figures 6I and 6J). In contrast, carriers of mutant CARD9 alleles produced significantly lower titers of anti-C. albicans IgG antibodies (Figures 6I and 6J). These data implicate CARD9 as a critical mediator of systemic anti-fungal IgG antibody responses to the gut commensal C. albicans during health and during invasive fungal disease.
DISCUSSION
Although several Candida species, including C. albicans, are largely regarded as commensals that can evolve and benefit from the intestinal niche (Liang et al., 2019; Tso et al., 2018; Witchley et al., 2019), any benefit to the host from these fungi is still unclear. Our findings outline a model where these gut fungal species play a key role in induction and therapeutic efficacy of host-protective IgG antibodies. The gut mycobiota elicit GC-dependent IgG+ B cell expansion through CARD9 signaling in CX3CR1+ macrophages to produce antifungal IgG that is protective against systemic fungal infection. Although we did not detect systemic fungal spread, intestinal colonization with C. albicans, but not with S. cerevisiae, induced a strong IgG CSR in the spleen. CSR was accompanied by transfer of C. albicans cell constituents (as assessed by a sensitive DNA-based assay) to this gut-distal organ and was abrogated upon depletion of CX3CR1+ MNPs. Furthermore, a previous study has reported that systemic antibodies to specific fungal antigens can arise in mice upon intestinal C. albicans colonization (Huertas et al., 2017). These findings suggest that low-grade physiological fungal dissemination, reported previously for other commensals (Macpherson et al., 2000; Zeng et al., 2016), or gut-distal transport of fungal antigens and PAMPs by migratory phagocytes might play a role in distal induction of systemic anti-fungal antibodies to specific fungal commensals. In humans, we found that loss-of-function mutations in CARD9 that predispose individuals to severe fungal infections are associated with loss of antifungal antibody responses. Immunosuppression, the most common cause of systemic fungal infection in humans (Brown et al., 2012; Koh et al., 2008; Zhai et al., 2020), led to a dramatic decrease in antifungal IgG antibodies in serum of mice associated with rapid gut-to-system C. albicans translocation and death, whereas supplementation of systemic antifungal IgG against intestinal C. albicans was protective. Furthermore, anti-fungal IgG induced through gastrointestinal colonization was protective against systemic infection with multidrug-resistant C. auris, an emerging pathogen for which limited therapies are currently available. Thus, systemic IgG antibodies to the gut mycobiota appear to confer protective antifungal immunity and some degree of cross-specificity (to similar ligands expressed by different fungal species or to cross-reactive ligands) that may be protective against more than one fungal pathogen. Because fungi comprise a widespread kingdom of deadly pathogens for which only a handful of therapies (several antifungal drugs) are clinically available (Fisher et al., 2012; Garcia-Solache and Casadevall, 2010), the pathways activated in the host by the gut mycobiota outlined in our study may warrant further exploration for development of new approaches to antifungal vaccine or neutralizing antibody generation. Furthermore, the human native antifungal antibody pool can be explored for therapeutic purposes in susceptible immunosuppressed individuals, individuals with cancer, and organ transplant recipients or as a potential source of defense against emerging fungal threats.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Iliyan D. Iliev (iliev@med.cornell.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All sequencing data are accessible under NCBI archives under the BioProject ID PRJNA630985. All other data needed to evaluate the conclusions in the manuscript are available within the main text or supplementary materials.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
7–8-week-old wild-type (WT) C57BL/6J, Ighmtm1Cgn/J (μMT−/−), Cd11c-cre, Irf4fl/fl, Cx3cr1tm (DTR)Litt/J (Cx3cr1DTR), Syk fl/fl, Cx3cr1tm2.1(cre/ERT2)Jung/J (Cx3cr1-cre/ERT2+/+), Tcrbtm1Mom/J, Rag2tm1.1Cgn/J mice were purchased from the Jackson Laboratories (Bar Harbor, ME). Card9−/− mice were kindly provided by Dr. Xin Lin (Department of Molecular and Cellular Oncology, University of Texas, Houston, TX, USA). The Cx3cr1DTR mice were subsequently bred with hemizygous Cd11c-cre+/− mice to allow for selective depletion of CD11c+ CD11b+ CX3CR1+ MNPs upon administration of diphtheria toxin (DT). For depletion, such generated mice (Cd11c-cre+/− Cx3cr1DTR mice) and their respective littermate controls (Cd11c-cre−/− Cx3cr1DTR) were injected for three consecutive days with 100ng of DT i.p followed by maintenance injection every other day for the length of the experiments. Depleted mice were designated as ΔCX3CR1 mice. Irf4fl/fl mice were bred to hemizygous Cd11c-cre+/− mice to allow for genetic ablation of CD11c+CD11b+CD103+ dendritic cells (designated as ΔIrf4 mice) and the generation of littermate controls. Sykfl/fl mice were bred to Cx3cr1-cre/ERT2+/+ to allow for the selective depletion of Syk in CX3CR1 expressing cells upon administration of tamoxifen. Cre-ERT recombinase in Cx3cr1-cre/ERT2+/− mice was induced through injection with tamoxifen (TAM, Sigma) that was dissolved in sunflower oil (Sigma) to a final concentration of 10 mg/ml, and 100μl/mouse were injected i.p. every second day. Control littermates received 100 μl sunflower oil i.p every second day. Littermates were randomly assigned to experimental groups. Animals were used between 8 and 16 weeks of age. Males and females were used in approximately equal ratios. Mice were housed under specific pathogen-free (SPF) conditions unless otherwise described at Weill Cornell Medicine and experiments were performed after prior approval by the Institutional Animal Care and Use Committee of Weill Cornell Medicine.
Germ-free (GF) C57BL/6 mouse colonies were maintained within sterile vinyl isolators at Weill Cornell Medicine’s gnotobiotic mouse facility. Altered Schaedler Flora (ASF) mice were generated by colonizing GF mice with the defined ASF community (Schaedler et al., 1965) and allowing 5 generations of breeding before use.
Human samples and CARD9 genotyping
Fecal and corresponding serum samples were obtained from healthy de-identified individuals following Institutional-Review-Board-approved protocols from the JRI IBD Live Cell Bank Consortium at Weill Cornell Medicine. Plasma and peripheral blood samples from patients with history of proven invasive candidiasis were collected following informed consent and IRB approval, in accordance with the Helsinki Declaration (Corvilain et al., 2018). DNA samples from human peripheral blood were used for CARD9 genotyping as previously described (Corvilain et al., 2018). Plasma samples from homozygous carriers of either wild-type (n = 5) or loss of function (n = 4) CARD9 alleles were processed for serological evaluation performed by ELISA as described below.
Fungal strains
Fungal strains were obtained from the American Type Culture Collection (Manassas, VA). Candida albicans SC5314 (ATCC® MYA-2876) and Saccharomyces cerevisiae (ATCC® MYA-796) were cultured in aerobic conditions on Sabouraud Dextrose Broth (SDB; EMD Chemicals) at 37°C overnight, while Cladosporium cladosporioides (ATCC® 38810) and C. auris (CDC0389) were cultured in SD agar at 30°C for 5–7 days and 24 h, respectively. Malassezia restricta (ATCC® MYA-4611) was plated on Dixon agar (HiMedia Laboratories) coated with 2–3 mL of olive oil and grown at 30°C for 3–5 days.
METHOD DETAILS
Multi-kingdom antibody profiling (multiKAP)
For multiKAP, fecal pellets of 7–8 weeks old mice or human feces were collected. For multiKAP analysis across multiple facilities, samples were collected from age-matched mouse colonies born in our facility (WCM-CE) or from mice raised in Taconic Biosciences (TAC) or the Jackson Laboratory (JAX) 48–72 h after arriving in our facility. Samples were diluted to 25mg feces/mL in PBS, thoroughly homogenized, and filtered through a 35 μm strainer-capped tube (Falcon, 50mL). Resulting fecal slurries were incubated for 90 min at 30°C with serum from the same mouse or the same human subject diluted at 1:20 (mouse serum) or 1:100 (human serum) in blocking buffer (1% bovine serum albumin (BSA, MP Biomedicals)/0.05% sodium azide (Sigma)). After 30-minutes of blocking with staining buffer (2% BSA/0.05% sodium azide), mouse samples were stained with 1:500 mIgA-PE (Thermofisher) and/or 1:200 mIgG-AF647 (Jackson Immunoresearch) antibodies for 45 min at 4°C. Human samples were stained with 1:200 hIgA-PE (Miltenyi) and/or 1:400 hIgG-AF647 (Southern) antibodies. Stained pellets were further fixed in 4% formalin, followed by staining with 1:1000 SYBR-Green (Invitrogen)/1:500 calcofluor white (Sigma), and total fungal and bacterial content was analyzed by flow cytometry. Size separation was achieved by centrifugation at 900 × g for 10 min. The resulting large (fungal) and small (bacterial) IgA- and/or IgG-stained fractions were used for downstream applications such as flow cytometry analysis and/or sorting and/or DNA isolation. In a variation of the method, size separation was performed as a first step followed by serum incubation and antibody staining as above. For IgG isotype multiKAP assays, samples were stained with mIgG1-FITC (Thermofisher), mIgG2b-FITC (Thermofisher), or mIgG3-FITC (BD Phar-Mingen). To avoid spectral overlap between FITC and Sybr-Green dye when FITC-labeled antibodies were used, DNA staining was performed with propidium iodide (PI; Thermofisher) or DAPI (Thermofisher) as an alternative DNA-binding dyes.
Fungal and bacterial cells sorting, and DNA isolation for multiKAP
Antibody bound (IgG+IgA+, IgG+IgA−, IgG−IgA+) and unbound (IgG−IgA−) fungal and bacterial fractions were sorted on FACSAria III (BD Bioscience) with a 100 μm flow cell and processed for DNA isolation and microbiota qPCR and sequencing (see Table S1 for primers).
DNA isolation, fungal, and bacterial quantitative PCR analysis
Sorted antibody-bound and unbound fecal fungal and bacterial fractions, or input feces were treated with lyticase (bacterial fractions were left untreated), followed by DNA isolation using QIAGEN’s QIAmp DNA mini kit as in Doron et al. (2019). Fungal and bacterial DNA was quantified in each sample by RT-PCR using fungal 18S and bacterial 16S rDNA-specific primers (Table S1) as in Leonardi et al. (2018) and Li et al. (2018).
Mycobiota library generation, sequencing, and analysis
Fungal ITS1–2 regions in rDNA isolated from sorted antibody-bound or unbound fecal fractions were amplified by PCR using the following primers: ITS1F CTTGGTCATTTAGAGGAAGTAA; ITS2R GCTGCGTTCTTCATCGATGC. Primers were modified to include forward (5′ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-[locus-specific sequence]) and reverse (5′ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-[locus-specific sequence]) sequencing adaptors (Table S1). ITS1–2 amplicons were generated with 35 cycles using Invitrogen AccuPrime PCR reagents (Carlsbad, CA). In a second PCR reaction, amplicons from each sample were uniquely indexed using Illumina Nextera XT v2 barcoded primers (San Diego, CA), (Tang et al., 2015). Quality control of all libraries were conducted by qPCR, DNA 1000 Bioanalyzer (Agilent), and Qubit (Life Technologies) to validate and quantify library construction prior to preparing a paired end flow cell. Clonal bridge amplification (Illumina) was performed using a cBot (Illumina). 2 × 250 bp sequencing-by-synthesis was performed on Illumina MiSeq platform (Illumina).
Raw FASTQ ITS1 sequencing data were enriched for high quality reads by removing both the adaptor sequence by cutadapt v1.4.1 or any reads that lacked the proximal primer sequence. Sequence reads were then quality-trimmed by truncating reads below an average quality score of 20 over a 3 base pair (bp) sliding window and removing reads shorter than 100bp. These enriched high-quality reads were aligned to the Targeted Host Fungi (THF) ITS1 database (Tang et al., 2015) using BLAST v2.2.22 and the pick_otus.py pipeline in the QIIME v1.9.1 wrapper with an identity percentage ≥ 97% for OTU picking. The alignment results were tabulated across all reads using the accession identifier of the ITS reference sequences as surrogate OTUs and using a Perl script. ITS sequencing data are available at NCBI Sequence Read Archive (SRA) under the Bioproject ID PRJNA630985.
Fungal Lysate Preparation
Fungal lysates were prepared by fixing pure fungal cultures in 4% paraformaldehyde (PFA, diluted in PBS) for 60 min. Fungal suspensions were then washed 3 times by pelleting at 900 × g for 2 min, aspirating the supernatant, and resuspending the pellet in molecular-grade water. Fungi were treated with a cycle of 3 × (10 min on dry ice, 10 minutes 75°C). Finally, fungal suspensions were sonicated for 8 cycles of 15sec on/30sec off. Fungal debris was centrifuged at 4000 rpm for 2 minutes, after which lysate-containing supernatant was used for ELISA plate coating.
Antifungal ELISA assay
Antifungal antibody titers in serum were measured by enzyme-linked immunosorbent assay (ELISA). Fungal lysates were coated on a 96-well high-binding polystyrene plates (Corning) after diluting 1:50 in 50mM carbonate-bicarbonate buffer (pH 9.5). Saccharomyces cerevisiae mannan (Sigma) or bacterial flagellin from S. typhimurium (Invivogen) were coated at 5 μg/well and 0.1 μg/well in carbonate-bicarbonate buffer, respectively. Plates for total IgG measurement were coated with anti-mouse-IgG-IgA-IgM (Sigma) or anti-human Ig (Southern) diluted 1:2000 in coating buffer. Mouse serum was then applied to plates diluted at 1:20 and 1:250 in BSA for antigen-specific and total IgG assays, respectively. Human serum was applied at dilutions of 1:4000 and 1:10000 for antigen-specific and total IgG assays, respectively. Total IgG titers were measured against serial dilution standards of mouse (Bethyl Laboratories) and human (Southern) unconjugated IgG antibodies. Plates were developed using alkaline phosphatase-conjugated anti-mouse or anti-human IgG as previously described (Leonardi et al., 2018). Plates were read at OD 405 nm on a microtiter plate reader (Menlo Park, CA).
Assessment of the adaptive responses to gut fungal colonization
GF mice were orally gavaged with PBS or with 1×108 C. albicans, S. cerevisiae, M. restricta or with C. cladosporioides. ASF mice were orally gavaged with either PBS (ASF control) or with 1×108 C. albicans or S. cerevisiae yeast cells. Alternatively, cefoperazone (0.4 mg/ml; Sigma) was provided to SPF mice ad libitum in drinking water for the length of the colonization experiments followed by 1×108 C. albicans or S. cerevisiae yeast cells colonization via oral gavage. Mice were sacrificed at the described time points for blood, feces, and organ collection. B cell and T cell compartments were assessed by flow cytometry. Total, fungi-specific or flagellin-specific IgG antibody responses in serum collected at the beginning, during, and at the end of the experiment were assessed by ELISA or used in mutiKAP assays.
Isolation of spleen, mesenteric lymph nodes (mLN), Peyer’s patches (PP) and blood lymphocytes
Four to six PP were excised from the ileum of each mouse and placed into Hank’s Balanced Salt Solution (HBSS) medium (Thermofisher), supplemented with 2 mM EDTA, and were shaken for 10 min at 37°C, followed washing and by mincing with scissors in a medium consisting of RPMI 1640 (Thermofisher), 5% FBS, 100 IU/ml penicillin and 100 μg/ml streptomycin (Thermofisher) and filtered through a nylon mesh (70μm). Spleens and mLNs were processed similarly and mesh-filtered, after which spleens were each treated with 1 mL of 1x RBC lysis buffer (Biolegend) for 2 min. Processed tissues were once again filtered through mesh, washed twice in PBS, and resuspended in cold PBS supplemented with 2% BSA for antibody staining.
Blood samples were prepared for lymphocyte isolation by transferring 50μL of blood into 1.7mL Eppendorf tubes containing 5μL citrate dextrose solution (ACD; Sigma) and mixed by pipetting to prevent coagulation. Blood samples are then treated with 200μL of 1 × RBC lysis buffer for 5 min, washed, and resuspended in cold PBS supplemented with 2% BSA for antibody staining.
Antibodies, FISH probes and flow cytometry analysis of the host response
Cell suspensions prepared as described above were blocked with CD16/CD32 (Mouse BD Fc Block, 2.4G2, BD Biosciences). For B cell compartment analysis, suspensions were stained with antibodies against CD45 (30-F11, Tonbo), CD4 (GK1.5, eBioscience), CD19 (6D5, Biolegend), IgD (11–26c.2a, Biolegend), IgG (polyclonal, Jackson ImmunoResearch), IgM (II/41, Thermofisher), CD95 (DX2, Biolegend), and GL7 antigen (GL7, Biolegend). Dead cells were excluded with eBioscience Fixable Viability Dye eFluor 455UV (Thermofisher). In phagocyte depletion experiments, suspensions were stained with antibodies against CD45 (30-F11, Tonbo), I-A/I-E a (M5/114.15.2, Biolegend), CD11c (N418, Biolegend), CD11b (M1/70, Thermofisher), CX3CR1 (SA011F11, Biolegend), and CD103 (2E7, Thermofisher). Dead cells were excluded with eBioscience Fixable Viability Dye eFluor 506 (Thermofisher). Bacterial FISH staining was performed with Cy5-labeled EUB338 5’-GCTGCCTCCCGTAGGAGT-3’ probe (Integrated DNA Technologies; Table S1). Images were acquired under an inverted Nikon Eclipse Ti microscope (Nikon). Flow cytometry was performed using a LSRFortessa (BD Biosciences) and data were analyzed with FlowJo software (TreeStar).
Systemic candidiasis models and B cell adoptive transfer
Pure cultures of C. albicans grown in SDB at 37°C were washed in PBS, resuspended at 5×105 yeast cells/mL PBS, and were injected at days 0 and 14 in the tail vein of 5–7 weeks old WT and μMT−/− mice (1×105 yeast cells/mouse). In rescue experiments splenic B cells from 8-week-old GF mice, C. albicans-monocolonized ex-GF or Aicda−/− mice were purified by MACS negative sorting (Miltenyi), and adoptively transferred (2×107 B cells/mouse) into recipient μMT−/− mice two days (day 2) before systemic infection with C. albicans. Pure cultures of C. auris grown in SDB at 30°C were washed in PBS, resuspended at 5×108 yeast cells/mL PBS, and were injected at days 0 and 14 in the tail vein of 5–7 weeks old WT (1×108 yeast cells/mouse). At day 1 and 10 mice received i.p. injection of 400μg C.aur-IgG, C.alb-IgG, or sterile PBS. Disease progression was tracked for 28 days in all experimental groups. Moribund mice were sacrificed for serum, kidney, and brain tissue collection.
μMT−/− mice were treated for five days with 0.5 mg/ml fluconazole (Sigma), 1 mg/ml Penicillin G (Sigma), and 2 mg/ml streptomycin sulfate (Research Products International) ad libitum in the drinking water. Fluconazole was withdrawn and mice were orally gavaged with 1×108 C. albicans cells. Splenic B cells from 8-week-old CARD9+/+ or CARD9−/− mice were purified by MACS negative sorting (Miltenyi), and adoptively transferred (2×107 B cells/mouse) into recipient μMT−/− mice one day after the intestinal colonization with C. albicans. Serum was collected weekly; mice were sacrificed at 4 weeks for intestinal tissue and spleen collection.
Serum IgG purification from monocolonized ex-GF mice
GF were weaned after 4 weeks and orally gavaged with 1×108 C. albicans, C. auris or S. cerevisiae cells cultured from pure isolates overnight at 37°C. Between ages of 8–12 weeks, blood was collected every 3 days from each respective group and pooled. After 1 h of room temperature incubation to allow for coagulation the blood was centrifuged for 10 min at 6000 × g. The top layer of serum was transferred to a separate Eppendorf tube and stored at −20°C until purification. IgG was purified from pooled serum using the Protein G UltraLink™ Columns (Pierce). Purified IgG samples were desalted through buffer exchange to PBS using Amicon®Ultra-15 Centrifugal Filter tubes (Millipore). After measuring the IgG concentration and assessing specificity, pure IgG suspensions (C.alb-IgG, C.aur-IgG or S.cer-IgG) were adjusted with cold sterile PBS to 4 mg/mL and stored at −20°C until use. Pure IgG suspensions were delivered intraperitoneally for a single dose of 400μg IgG.
Gut-disseminating candidiasis model
Mice were treated for five days with 0.5 mg/ml fluconazole (Sigma), 1 mg/ml Penicillin G (Sigma), and 2 mg/ml streptomycin sulfate (Research Products International) ad libitum in the drinking water. Fluconazole was withdrawn and mice were orally gavaged with 1×108 C. albicans cells three times spaced across five days, followed by i.p. injection of 400μg C.alb-IgG, S.cer-IgG, or sterile PBS (Figure S4B). Immunosuppression and systemic fungal dissemination were induced by i.p. injections of 150mg/kg mouse weight of cyclophosphamide (Cyc; R&D Systems) on days 0, 2, and 4, followed by a second series of injections on days 14, 16, and 18. Weight was measured, and moribund mice were sacrificed for serum, liver, and kidney tissue collection.
Single-cell RNA seq analysis
To assess the expression of CARD9 among immune cells in humans we mined a published single-cell RNA sequencing (scRNA-seq) dataset obtained from colon biopsies from healthy individuals in (Smillie et al., 2019). Plots were created using the published differentially expressed genes for cell subsets, lineages, or sub-clusters in healthy tissue, relative to all other cells. These sequencing data are accessible under the Single Cell Portal (https://singlecell.broadinstitute.org/single_cell) under the Accession Number SCP259.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics were computed using GraphPad Prism version 8 (GraphPad Software) using the Mann-Whitney test (which does not assume normal distribution) for two groups. One-way ANOVA with Tukey’s or Kruskal-Wallis and Dunn’s for multiple comparisons was used for three or more groups. Statistical details specific to experiments are reported in the figure legends. Graphs display mean values with error bars that correspond to the standard error of the mean (mean ± SEM). The p value(s) reported in the figure legends are the likelihood(s) of observing the effect size(s), if the null hypothesis of zero difference is true. They are denoted as: not significant (ns) p ≥ 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
ITS1 Sequencing Analysis of FACS-sorted mycobiota
Relative abundances at the species level were analyzed by R (v3.4.4) with the Phyloseq (1.22.3) and Vegan (2.5–3) packages. Species representing less than 2% of total reads among all samples were grouped into the “Other” category. Sorted populations from one individual were excluded due to contamination. For each species, the coating index (CI) was calculated using previously determined (Kau et al., 2015), shown in Figure S2C. Enrichment in antibody-bound or antibody-unbound populations was tested for each species by one-sample t test in GraphPad Prism (GraphPad Software) to determine whether mean CI values deviated significantly from 0.
Supplementary Material
Figure S1. MultiKAP analysis of IgG binding in mice and microbial visualization of human feces or gut-distal organs of SPF mice, related to Figures 1 and 2
(A) Fungi in SPF mouse feces were assessed for IgA-, IgG-, and IgM-MFI by multiKAP. No serum incubation (gray), incubation with matched WT SPF mouse serum (black), or B cell-deficient μMT−/− SPF serum (blue). − Serum, n = 5; +WT Serum, n = 5; +μMT−/− Serum, n = 5.
(B) Sera from RAG2−/−, n = 5; μMT−/−, n = 5; or WT (SPF), n = 7 mice were assessed for IgG binding to fecal samples from RAG2−/− mice by multiKAP; ANOVA.
(C-D) Bacteria in SPF WT mouse feces were assessed for IgG1-, IgG2b-, and IgG3-binding by multiKAP after incubation with matched WT SPF serum. In each plot, the isotype control is represented by the light gray curve with filled-in area. IgG1, n = 5; IgG2b, n = 5; IgG3, n = 5.
(E) Individual fluorescent microscopy channels of composite image in Figure 2A.
(F-G) Live fungi are not present in gut-distal organs of SPF mice from different facilities and fungal infection-related kidney pathologies do not develop in SPF mice. Organs were harvested from 8-week-old mice from our WCM-CE facility and the Jackson Laboratory Facility (JAX). Mice undergoing the systemic candidiasis were used as positive controls of infection. Indicated tissues were assessed for fungal CFU counts (F) and by H&E staining (G). Cellular infiltrates in the infected area (dark blue) are visible on the kidneys of mice with systemic candidiasis. Scale bars in G represent 100 μm.
Figure S2. Microbiota gating strategy in human feces, isotype control staining, and relative abundance analysis of Ig-coated or -uncoated intestinal fungi, related to Figure 2
(A) DAPI staining of GF mice, WT mice, and healthy humans.
(B) Representative plots of isotype control staining of human feces incubated with serum distinguishing antibody bound and unbound gates within the microbial gate.
(C) Equation for the calculation of CI values represented in Figures 2F and 2G. For each species, Ig+ represents the relative abundance in the antibody-bound population (IgG+IgA+ for Figure 2F, IgG+IgA− for Figure 2G), while Ig− represents the relative abundance in the antibody-unbound population. Output values range between 1 (only represented in the antibody-bound population), and −1 (only represented in the antibody-unbound population), with a value of 0 if a fungal species is not enriched in either sorted population.
(D) Relative abundance of representative species in Figures 2F and 2G for each of the three sorted populations used for CI calculation. Boxplots represent species’ median and the first to third quartile, while whiskers represent minimum and maximum values. Fecal samples from healthy individuals, n = 7.
Figure S3. Systemic anti-C. albicans IgG antibodies appear 2 weeks after intestinal colonization but do not arise because of systemic fungal spread or acute B cell reaction to gut C. albicans, related to Figure 3
(A) Serum was collected from ASF mice or ASF mice colonized with either C. albicans or S. cerevisiae and assessed by ELISA for anti-bacterial flagellin IgG antibodies. Pooled from two independent experiments. ASF, n = 8; +Ca, n = 10; +Sc, n = 10. ns-p ≥ 0.05, one-way ANOVA followed by Tukey’s multiple comparisons test.
(B-C) Comparative analysis of the CD19+CD4− B cell compartment in the spleens of ASF mice colonized with C. albicans or S. cerevisiae as described in Figure 3. Each dot represents individual mice.
(D-E) Mycobiota-free altered Schaedler flora-colonized (ASF) mice were orally gavaged with 1×108 C. albicans and fecal samples and sera were collected before colonization 0, 1, 3, 7, and 14, and 28 days after colonization, groups of mice were sacrificed at each time point to assess extra-intestinal and gut-associated tissues for C. albicans CFUs (D). Sera were assessed for anti-C. albicans anti-IgG titers (E). 0, n = 5; 1, n = 4; 3, n = 3; 7, n = 4; 14, n = 6; 28, n = 6. Data representative of two independent experiments. Boxplots represent median and the first to third quartile, while whiskers represent minimum and maximum values. *p < 0.05, **p < 0.01, Mann-Whitney, comparison to day 0.
Figure S4. Antifungal antibodies are protective in systemic candidiasis, but the extreme susceptibility of CARD9−/− mice cannot be overcome by otherwise effective eradication strategies such as fluconazole treatment, C.alb-IgG treatment, or decreased fungal inoculum, related to Figures 4 and 5
(A) WT mice (WT) or μMT−/− mice adoptively transferred with splenic B cells from Aicda−/− mice (μMT−/− +Aicda−/− B Cells) were intravenously (i. v.) infected with 2×105 C. albicans. Percent survival was assessed, log-ranked Mantel-Cox test. WT, n = 6; μMT−/− +Aicda−/− B Cells, n = 6.
(B) Schematic of the gut-disseminated candidiasis model.
(C) Serum anti-C. albicans IgG antibody decreases in titers upon cyclophosphamide (Cyc)-induced systemic fungal dissemination as measured by ELISA (related to Figures 4D–4G). Paired two-tailed t test, n = 6. Each pair of dots represents individual mice. Data represents two individual experiments.
(D-E) Card9+/+ and Card9−/− mice were intravenously (i. v.) infected with 2×104 C. albicans. Four days later Card9−/− mice were treated with i. p. PBS, 400 μg C.alb-IgG, or provided 0.5 g/L fluconazole in drinking water. A control group of Card9+/+ mice was left uninfected. Percent Survival (D) and C. albicans systemic spread to kidney tissue (E) were assessed. Log-ranked Mantel-Cox test (D) and one-way ANOVA followed by Tukey’s multiple comparisons test (E). Card9+/+ i. p. PBS, n = 13; Card9−/− i. p. PBS, n = 7; Card9−/− i. p. 400 μg C.alb-IgG, n = 11; Card9−/− +Fluconazole, n = 5. Data representative of two independent experiments.
(F-G) Card9−/− mice inability to produce efficient anti-C. albicans IgG antibody response is not due to B cell intrinsic or developmental defect in the absence of CARD9. μMT−/− mice adoptively transferred splenic B cells from Card9+/+ or Card9−/− mice before oral gavaged with C. albicans for two weeks, after which serum anti-C. albicans IgG antibody titers were measured by ELISA (F) and the frequency of IgG+ IgM− among CD19+CD4− B cells in spleens (G) were analyzed by flow cytometry. Mann-Whitney test. Card9+/+, n = 4; Card9−/−, n = 5. Each dot represents individual mice.
(H-I) CD4+ T cell and CD19+ B cell Compartment Analyses in Card9+/+ and Card9−/− mice in response to intraperitoneal challenge with C. albicans. Splenic CD4+ T cell and CD19+ B cell frequencies of Card9+/+ and Card9−/− mice with or without intraperitoneal injection with C. albicans analyzed by flow cytometry. One-way ANOVA followed by Tukey’s multiple comparisons test. Card9+/+ NT, n = 7; Card9+/+ +Ca, n = 11; Card9−/− NT, n = 6; Card9−/− +Ca, n = 6. Data representative of three independent experiments.
Error bars indicate SEM ns-p ≥ 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Figure S5. In vivo depletion of CARD9-expressing phagocyte subsets and human CARD9 SNP map, related to Figure 6
(A) Representative flow cytometry plots and quantification of CD45+MHC-II+CD11c+ phagocyte subsets in ΔCX3CR1 (Cd11c-cre+/− Cx3cr1DTR + DT) and littermate (Litt., Cd11c-cre−/− Cx3cr1DTR + DT) mice. CD11b+CX3CR1+ MNPs (top) and CD11b+CD103+ cDC2s (bottom). Mann-Whitney test. Litt, n = 12; ΔCX3CR1, n = 7. Data representative of two independent experiments.
(B) Representative flow cytometry plots and quantification of CD45+MHC-II+CD11c+ phagocyte subsets in ΔIrf4 (Cd11c-cre+/− Irf4fl/fl) or littermate (Litt., Cd11c-cre−/− Irf4fl/fl) mice. CD11b+CX3CR1+ MNPs (top) and CD11b+CD103+ cDC2s (bottom). Mann-Whitney test. Litt, n = 7; ΔIRF4, n = 10. Data representative of two independent experiments.
(C-D) Frequency of GC-B cells among CD19+CD4− B cells in the spleen analyzed by flow cytometry of mice represented in Figure S8A (C) and S8B (D). One-way ANOVA followed by Tukey’s multiple comparisons test. Mann-Whitney test.
(E) Map of human CARD9 gene and the location of three represented SNPs: Q289* (nonsense mutation), R70W (missense), and R35Q (missense). Each dot represents individual mice. Error bars indicate SEM ns-p ≥ 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat IgG1 kappa Isotype Control (eBRGI), PE | Thermo Fisher Scientific (eBioscience™) | Cat # 12-4301-82; RRID: AB_470047 |
| IgA Monoclonal Antibody (mA-6E1), PE | Thermo Fisher Scientific (eBioscience™) | Cat # 12-4204-82; RRID: AB_465917 |
| Rat IgG2a, k Isotype Control (R35-95), FITC | BD PharMingen™ | Cat # 554688 |
| anti-Mouse IgG1 Secondary Antibody (M1-14D12), FITC | Thermo Fisher Scientific (eBioscience™) | Cat # 11-4015-80; RRID: AB_11043272 |
| anti-Mouse IgG2b Secondary Antibody (m2b-25G4), FITC | Thermo Fisher Scientific (eBioscience™) | Cat # 11-4220-80; RRID: AB_2572494 |
| Rat anti-Mouse IgG3 (R40-82), FITC | BD PharMingen™ | Cat # 553403 |
| Alexa Fluor® 647 ChromPure Goat IgG, whole molecule | Jackson Immunoresearch | Cat # 005-600-003 RRID: AB_2337016 |
| Alexa Fluor® 647 AffiniPure Goat Anti-Mouse IgG, Fcγ fragment specific | Jackson Immunoresearch | Cat # 115-605-071 RRID: AB_2338909 |
| Rat IgG2a kappa Isotype Control (eBR2a), PE-Cyanine7 | Thermo Fisher Scientific (eBioscience™) | Cat # 25-4321-82 RRID: AB_470200 |
| IgM Monoclonal Antibody (Il/41), PE-Cyanine7 | Thermo Fisher Scientific (eBioscience™) | Cat # 25-5790-82 RRID: AB_469655 |
| Isotype Control Antibody, mouse IgG1 (IS5-21F5), PE | Miltenyi Biotec | Cat #130-113-200 |
| IgA Antibody, anti-human (IS11-8E10), PE | Miltenyi Biotec | Cat # 130-113-476 |
| Goat IgG-Alexa Fluor® 647 | Southern Biotech | Cat #0109-31 |
| Goat Anti-Human IgG-Alexa Fluor® 647 | Southern Biotech | Cat # 2040-31 |
| CD16/CD32 monoclonal antibody (93), eBioscience | Thermo Fisher Scientific (eBioscience™) | Cat #14-0161-81; RRID: AB_467132 |
| eBioscience™ Fixable Viability Dye eFluor™ 455UV | Thermo Fisher Scientific (Invitrogen™) | Cat #65-0868-18 |
| Brilliant Violet 650™ anti-mouse CD45 Antibody (30-F11) | Biolegend | Cat # 103151; RRID: AB_2565884 |
| Brilliant Violet 605™ anti-mouse CD4 Antibody (RM4-5) | Biolegend | Cat # 100548; RRID: AB_2563054 |
| Alexa Fluor® 700 anti-mouse CD19 Antibody (6D5) | Biolegend | Cat # 115527; RRID: AB_493734 |
| BV510 Rat Anti-Mouse IgD (11-26c.2a) | BD Horizon™ | Cat #563110 |
| IgM Monoclonal Antibody (Il/41), FITC | Thermo Fisher Scientific (eBioscience™) | Cat # 11-5790-81; RRID: AB_465244 |
| PE-Cy™7 Hamster Anti-Mouse CD95 (Jo2) | BD PharMingen™ | Cat # 557653 |
| Pacific Blue™ anti-mouse/human GL7 Antigen (T and B cell Activation Marker) Antibody | Biolegend | Cat # 144613; RRID: AB_2563291 |
| eBioscience Fixable Viability Dye eFluor 506 | Thermo Fisher Scientific (eBioscience™) | Cat #65-0866-14 |
| Brilliant Violet 605™ Anti-mouse I-A/I-E a (M5/114.15.2) | BioLegend | Cat # 107639; RRID: AB_2565894 |
| Anti-mouse CD11b monoclonal antibody (M1/70), PE-Cyanine7 | Thermo Fisher Scientific (eBioscience™) | Cat # 25-0112-82; RRID: AB_469588 |
| Anti-mouse CX3CR1 antibody (SA011F11), BV421 | BioLegend | Cat #149023; RRID: AB_2565706 |
| Anti-mouse CD11c antibody (N418), Alexa Fluor 700 | BioLegend | Cat # 117319; RRID: AB_528735 |
| CD103 (Integrin alpha E) Monoclonal Antibody (2E7) PerCP-eFluor 710 | Thermo Fisher Scientific (eBioscience™) | Cat # 46-1031-82; RRID: AB_2573704 |
| Anti-Mouse IgG IgA IgM (H+L antibody) produced in goat | Sigma-Aldrich (Millipore) | Cat # SAB3701043-2MG |
| Goat Anti-Human Ig-UNLB | Southern Biotech | Cat # 2010-01; RRID: AB_2687525 |
| Mouse Reference Serum | Bethyl Laboratories | Cat # RS10-101 |
| Human IgG-UNLB | Southern Biotech | Cat # 0150-01; RRID: AB_2794075 |
| Alkaline Phosphotase AffiniPure F(ab’)2 Fragment Goat Anti-Mouse IgG, Fcγ fracgment specific | Jackson Immunoresearch | Cat # 115-056-071; RRID: AB_2338553 |
| Alkaline Phosphatase AffiniPure F(ab’)2 Fragment Goat Anti-Human IgG, Fcγ fragment specific | Jackson Immunoresearch | Cat # 109-056-098; RRID: AB_2337618 |
| Bacterial and fungal strains | ||
| Altered Schaedler Flora (ASF) | N/A | PMID: 14325473 |
| Candida albicans SC5314 | ATCC | ATCC MYA-2876) |
| Saccharomyces cerevisiae Meyen ex E.C. Hansen | ATCC | ATCC MYA-796 |
| Cladosporium cladosporioides | ATCC | ATCC 38810 |
| Malassezia restricta | ATCC | ATCC MYA-4611 |
| Candida auris | CDC | CDC0389 |
| Chemicals, peptides, and recombinant proteins | ||
| Sybr Green Nucleic Acid Gell Stain - 10,000X concentrate in DMSO | Thermo Fisher Scientific (Invitrogen™) | Cat # S7563 |
| Calcofluor White Stain | Sigma-Aldrich (Millipore) | Cat # 18909 |
| DAPI (4’, 6-Diamidino-2-Phenyldole, Dihydrochloride) | Thermo Fisher Scientific (Invitrogen™) | Cat # D1306 |
| Propidium Iodide - 1.0 mg/mL Solution in Water | Thermo Fisher Scientific (Invitrogen™) | Cat # P3566 |
| Bovine Albumin Fraction V | MP Biomedicals | Cat# IC820451 |
| Hanks’ balanced salt solution (HBSS) | Sigma | Cat # H4385 |
| Fetal Bovine Serum | Corning | Cat# MT35016CV |
| RPMI 1640 with L-glutamine | Corning | Cat # 45000-396 |
| Sabouraud dextrose broth | VWR | Cat# 89406-400 |
| Sabouraud 4% dextrose agar | VWR | Cat # EM1.05438.0500 |
| Penicillin-Streptomycin (10,000 U/mL) | Thermo Fisher Scientific | Cat # 15140122 |
| Tween® 80 (Polysorbate) | VWR | Cat # 97061-674 |
| Cefoperazone sodium salt | Sigma | Cat # C4292-5G |
| Cyclophosphamide | Thermofisher Scientific (Tocris Bioscience™) | Cat #40-915-0 |
| RBC Lysis Buffer (10X) | Biolegend | Cat # 420301 |
| Citrate-dextrose solution (ACD) | Sigma-Aldrich (Millipore) | Cat # C3821-50ML |
| Fluconazole | Sigma-Aldrich (Millipore) | Cat # PHR1160-1G |
| Penicillin G sodium salt | Sigma-Aldrich (Millipore) | Cat # PENNA-10MU |
| Streptomycin Sulfate, 25 Grams | Research Products International | Cat #3810-74-0 |
| Gentamycin Sulfate, 1 Gram | Research Products International | Cat # 1405-41-0 |
| Standard flagellin from S. typhimurium | Invivogen | Cat # tlrl-stfla |
| Mannan from Saccharomyces cerevisiae | Sigma-Aldrich (Millipore) | Cat # M7504-100MG |
| pNPP Powder | Thermo Fisher Scientific (Invitrogen™) | Cat # 002201 |
| Critical commercial assays | ||
| B Cell Isolation Kit, Mouse | Miltenyi | Cat # 130-090-862 |
| Protein G UltraLink™ Columns, 2mL | Thermofisher Scientific (Pierce™) | Cat #53127 |
| Amicon Ultra-15 Centrifugal Filter Unit | Sigma-Aldrich (Millipore) | Cat # UFC900308 |
| Deposited data | ||
| ITS Sequencing | NCBI |
https://www.ncbi.nlm.nih.gov/sra/docs/ Bioproject ID: PRJNA630985 |
| Experimental models: organisms/strains | ||
| Mouse: C57BL/6 | The Jackson Laboratory | JAX# 000664 |
| Mouse: B6.129S2-Ighmtm1Cgn/J | The Jackson Laboratory | JAX# 002288 |
| Mouse: B6.Cg-Tg(Itgax-cre)1-1Reiz/J | The Jackson Laboratory | JAX# 008068 |
| Mouse: B6.129S1-Irf4tm1Rdf/J | The Jackson Laboratory | JAX# 009380 |
| Mouse: B6N.129P2-Cx3cr1tm3(DTR)Litt/J | The Jackson Laboratory | JAX# 025629 |
| Mouse: B6.129P2-Syktm1.2Tara/J | The Jackson Laboratory | JAX# 017309 |
| Mouse: B6.129P2(Cg)-Cx3cr1 tm2.1(cre/ERT2)Litt/WganJ | The Jackson Laboratory | JAX# 021160 |
| Mouse: B6.129-Card9tm1Xlin/J | Xin Lin Laboratory | N/A |
| Mouse: B6(Cg)-Rag2tm1.1Cgn/J | The Jackson Laboratory | JAX# 008449 |
| Mouse: B6.129P2-Tcrbtm1Mom/J | The Jackson Laboratory | JAX# 002118 |
| Oligonucleotides and FISH probe | ||
| 18S forward primer for qPCR: 5′-GGRAAACTCACCAGGTCCAG −3′ | Liu et al., 2012 | N/A |
| 18S reverse primer for qPCR: 5′- GSWCTATCCCCAKCACGA −3′ | Liu et al., 2012 | N/A |
| Probe 5′-TGGTGCATGGCCGTT-3′ | Liu et al., 2012 | N/A |
| 16S rRNA Eubacteria Probe EUB338: 5′- /5Cy5/GCT GCC TCC CGT AGG AGT −3′ | Daims et al., 1999 | N/A |
| Software and algorithms | ||
| GraphPad Prim 8.3.0 (538) | GraphPad Software | N/A |
| RStudio Desktop 1.1.463 | https://rstudio.com/ | N/A |
| R version 3.4.4 (2018-03-15) | https://www.r-project.org | N/A |
| FlowJo LLC 10.1. | Becton Dickinson | N/A |
Highlights.
Multi-kingdom antibody profiling reveled systemic IgG repertoires against gut fungi
Candida albicans is the major inducer of antifungal IgG (αF-IgG) in mice and humans
Gut-induced anti-Candida IgG protects against systemic C. albicans and C. auris
CARD9+CX3CR1+ phagocytes mediate αF-IgG that is decreased in CARD9-deficient patients
ACKNOWLEDGMENTS
We thank members of the Iliev laboratory for critical review of the manuscript. We thank Jean-Laurent Casanova of the Laboratory of Human Genetics of Infectious Diseases at Rockefeller University and Howard Hughes Medical Institute for help with the human genetics study of CARD9. We thank Jayanta Chaudhuri and Tobias Hohl for discussions and for providing a key reagent. We thank all contributing members of the JRI IBD Live Cell Bank Consortium and the Microbiome Core Laboratory of Weill Cornell Medicine. Support for sample acquisition through the JRI IBD Live Cell Bank is provided by the JRI, the Jill Roberts Center for IBD, Cure for IBD, the Rosanne H. Silbermann Foundation, and the Weill Cornell Medicine Division of Pediatric Gastroenterology and Nutrition. Research in the Iliev laboratory is supported by the National Institutes of Health (DK113136, DK121977, and AI137157), the Leona M. and Harry B. Helmsley Charitable Trust, the Crohn’s and Colitis Foundation, the Irma T. Hirschl Career Scientist Award, pilot project funding from the Center for Advanced Digestive Care (CADC), the Jill Roberts Institute (JRI) for Research in IBD, and the Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases Award.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.01.016.
DECLARATION OF INTERESTS
Cornell University has filed a provisional patent application covering inventions described in this manuscript.
REFERENCES
- Ansaldo E, Slayden LC, Ching KL, Koch MA, Wolf NK, Plichta DR, Brown EM, Graham DB, Xavier RJ, Moon JJ, and Barton GM (2019). Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 364, 1179–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, et al. ; MetaHIT Consortium (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher P, Hohnstein T, Beerbaum E, Röcker M, Blango MG, Kaufmann S, Röhmel J, Eschenhagen P, Grehn C, Seidel K, et al. (2019). Human Anti-fungal Th17 Immunity and Pathology Rely on Cross-Reactivity against Candida albicans. Cell 176, 1340–1355.e15. [DOI] [PubMed] [Google Scholar]
- Barbet G, Sander LE, Geswell M, Leonardi I, Cerutti A, Iliev I, and Blander JM (2018). Sensing Microbial Viability through Bacterial RNA Augments T Follicular Helper Cell and Antibody Responses. Immunity 48, 584–598.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, and White TC (2012). Hidden killers: human fungal infections. Sci. Transl. Med 4, 165rv13. [DOI] [PubMed] [Google Scholar]
- Bunker JJ, Flynn TM, Koval JC, Shaw DG, Meisel M, McDonald BD, Ishizuka IE, Dent AL, Wilson PC, Jabri B, et al. (2015). Innate and Adaptive Humoral Responses Coat Distinct Commensal Bacteria with Immunoglobulin A. Immunity 43, 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Conway KL, Heath RJ, Rush JS, Leshchiner ES, Ramirez-Ortiz ZG, Nedelsky NB, Huang H, Ng A, Gardet A, et al. (2015). Ubiquitin Ligase TRIM62 Regulates CARD9-Mediated Anti-fungal Immunity and Intestinal Inflammation. Immunity 43, 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, and Pirofski L-A (2003). Exploiting the redundancy in the immune system: vaccines can mediate protection by eliciting ‘unnatural’ immunity. J. Exp. Med 197, 1401–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, and Pirofski LA (2012). Immunoglobulins in defense, pathogenesis, and therapy of fungal diseases. Cell Host Microbe 11, 447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiani P, Bromuro C, Cassone A, and Torosantucci A (2009). Anti-β-glucan antibodies in healthy human subjects. Vaccine 27, 513–519. [DOI] [PubMed] [Google Scholar]
- Christmann BS, Abrahamsson TR, Bernstein CN, Duck LW, Mannon PJ, Berg G, Björkstén B, Jenmalm MC, and Elson CO (2015). Human seroreactivity to gut microbiota antigens. J. Allergy Clin. Immunol 136, 1378–86.e1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, Ho AW, Hai JH, Yu JJ, Jung JW, et al. (2009). Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J. Exp. Med 206, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvilain E, Casanova J-L, and Puel A (2018). Inherited CARD9 Deficiency: Invasive Disease Caused by Ascomycete Fungi in Previously Healthy Children and Adults. J. Clin. Immunol 38, 656–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H, Brühl A, Amann R, Schleifer KH, and Wagner M (1999). The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol 22, 434–444. [DOI] [PubMed] [Google Scholar]
- Doron I, Leonardi I, and Iliev ID (2019). Profound mycobiome differences between segregated mouse colonies do not influence Th17 responses to a newly introduced gut fungal commensal. Fungal Genet. Biol 127, 45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RA, Collar AL, Swamydas M, Rodriguez CA, Lim JK, Mendez LM, Fink DL, Hsu AP, Zhai B, Karauzum H, et al. (2015). CARD9-Dependent Neutrophil Recruitment Protects against Fungal Invasion of the Central Nervous System. PLoS Pathog. 11, e1005293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JE Jr., Schwartz MM, Schmidt CS, Sobel JD, Nyirjesy P, Schodel F, Marchus E, Lizakowski M, DeMontigny EA, Hoeg J, et al. (2018). A Fungal Immunotherapeutic Vaccine (NDV-3A) for Treatment of Recurrent Vulvovaginal Candidiasis-A Phase 2 Randomized, Double-Blind, Placebo-Controlled Trial. Clin. Infect. Dis 66, 1928–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DM, Spencer CCA, Pointon JJ, Su Z, Harvey D, Kochan G, Oppermann U, Dilthey A, Pirinen M, Stone MA, et al. ; Spondyloarthritis Research Consortium of Canada (SPARCC); Australo-Anglo-American Spondyloarthritis Consortium (TASC); Wellcome Trust Case Control Consortium 2 (WTCCC2) (2011). Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet 43, 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre DW, Sheppard AE, Madder H, Moir I, Moroney R, Quan TP, Griffiths D, George S, Butcher L, Morgan M, et al. (2018). A Candida auris Outbreak and Its Control in an Intensive Care Setting. N. Engl. J. Med 379, 1322–1331. [DOI] [PubMed] [Google Scholar]
- Fadlallah J, Sterlin D, Fieschi C, Parizot C, Dorgham K, El Kafsi H, Autaa G, Ghillani-Dalbin P, Juste C, Lepage P, et al. (2019). Synergistic convergence of microbiota-specific systemic IgG and secretory IgA. J. Allergy Clin. Immunol 143, 1575–1585.e4. [DOI] [PubMed] [Google Scholar]
- Fan D, Coughlin LA, Neubauer MM, Kim J, Kim MS, Zhan X, Simms-Waldrip TR, Xie Y, Hooper LV, and Koh AY (2015). Activation of HIF-1α and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat. Med 21, 808–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, and Gurr SJ (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Langarica A, Cook C, Müller Luda K, Persson EK, Marshall JL, Beristain-Covarrubias N, Yam-Puc JC, Dahlgren M, Persson JJ, Uematsu S, et al. (2018). Intestinal CD103+CD11b+ cDC2 Conventional Dendritic Cells Are Required for Primary CD4+ T and B Cell Responses to Soluble Flagellin. Front. Immunol 9, 2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Solache MA, and Casadevall A (2010). Global warming will bring new fungal diseases for mammals. MBio 1, e00061–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glocker E-O, Hennigs A, Nabavi M, Schäffer AA, Woellner C, Salzer U, Pfeifer D, Veelken H, Warnatz K, Tahami F, et al. (2009). A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N. Engl. J. Med 361, 1727–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagán AJ, Pepper M, Maynard CL, Elson CO 3rd, and Belkaid Y (2012). Acute gastro-intestinal infection induces long-lived microbiota-specific T cell responses. Science 337, 1553–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-MS, Zhang Y, You Y, Wang D, Li H, Duramad O, Qin X-F, Dong C, and Lin X (2007). The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat. Immunol 8, 198–205. [DOI] [PubMed] [Google Scholar]
- Huertas B, Prieto D, Pitarch A, Gil C, Pla J, and Díez-Orejas R (2017). Serum Antibody Profile during Colonization of the Mouse Gut by Candida albicans: Relevance for Protection during Systemic Infection. J. Proteome Res 16, 335–345. [DOI] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. ; International IBD Genetics Consortium (IIBDGC) (2012). Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau AL, Planer JD, Liu J, Rao S, Yatsunenko T, Trehan I, Manary MJ, Liu T-C, Stappenbeck TS, Maleta KM, et al. (2015). Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci. Transl. Med 7, 276ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-I, Song J-H, Ko H-J, Kweon M-N, Kang C-Y, Reinecker H-C, and Chang S-Y (2018). CX3CR1+ Macrophages and CD8+ T Cells Control Intestinal IgA Production. J. Immunol 201, 1287–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SG, Becattini S, Moody TU, Shliaha PV, Littmann ER, Seok R, Gjonbalaj M, Eaton V, Fontana E, Amoretti L, et al. (2019). Microbiota-derived lantibiotic restores resistance against vancomycin-resistant Enterococcus. Nature 572, 665–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, Reiner GL, Lugo KA, Kreuk LS, Stanbery AG, Ansaldo E, Seher TD, Ludington WB, and Barton GM (2016). Maternal IgG and IgA Antibodies Dampen Mucosal T Helper Cell Responses in Early Life. Cell 165, 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh AY, Köhler JR, Coggshall KT, Van Rooijen N, and Pier GB (2008). Mucosal damage and neutropenia are required for Candida albicans dissemination. PLoS Pathog. 4, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koscsó B, Kurapati S, Rodrigues RR, Nedjic J, Gowda K, Shin C, Soni C, Ashraf AZ, Purushothaman I, Palisoc M, et al. (2020). Gut-resident CX3CR1hi macrophages induce tertiary lymphoid structures and IgA response in situ. Sci. Immunol 5, eaax0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas B, Richard ML, Leducq V, Pham H-P, Michel M-L, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, et al. (2016). CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med 22, 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanternier F, Mahdaviani SA, Barbati E, Chaussade H, Koumar Y, Levy R, Denis B, Brunel AS, Martin S, Loop M, et al. (2015). Inherited CARD9 deficiency in otherwise healthy children and adults with Candida species-induced meningoencephalitis, colitis, or both. J. Allergy Clin. Immunol 135, 1558–1568.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeibundGut-Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, Tsoni SV, Schweighoffer E, Tybulewicz V, Brown GD, Ruland J, and Reis e Sousa C (2007). Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat. Immunol 8, 630–638. [DOI] [PubMed] [Google Scholar]
- Leonardi I, Li X, Semon A, Li D, Doron I, Putzel G, Bar A, Prieto D, Rescigno M, McGovern DPB, et al. (2018). CX3CR1+ mononuclear phagocytes control immunity to intestinal fungi. Science 359, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi I, Paramsothy S, Doron I, Semon A, Kaakoush NO, Clemente JC, Faith JJ, Borody TJ, Mitchell HM, Colombel JF, et al. (2020). Fungal Trans-kingdom Dynamics Linked to Responsiveness to Fecal Microbiota Transplantation (FMT) Therapy in Ulcerative Colitis. Cell Host Microbe 27, 823–829.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Leonardi I, Semon A, Doron I, Gao IH, Putzel GG, Kim Y, Kabata H, Artis D, Fiers WD, et al. (2018). Response to Fungal Dysbiosis by Gut-Resident CX3CR1+ Mononuclear Phagocytes Aggravates Allergic Airway Disease. Cell Host Microbe 24, 847–856.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XV, Leonardi I, and Iliev ID (2019). Gut Mycobiota in Immunity and Inflammatory Disease. Immunity 50, 1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S-H, Anderson MZ, Hirakawa MP, Wang JM, Frazer C, Alaalm LM, Thomson GJ, Ene IV, and Bennett RJ (2019). Hemizygosity Enables a Mutational Transition Governing Fungal Virulence and Commensalism. Cell Host Microbe 25, 418–431.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionakis MS, Netea MG, and Holland SM (2014). Mendelian genetics of human susceptibility to fungal infection. Cold Spring Harb. Perspect. Med 4, a019638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, Kachur S, Dwan MG, Abraham AG, Aziz M, Hsueh PR, Huang YT, Busch JD, Lamit LJ, Gehring CA, et al. (2012). FungiQuant: a broad-coverage fungal quantitative real-time PCR assay. BMC Microbiol. 12, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, and Zinkernagel RM (2000). A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 288, 2222–2226. [DOI] [PubMed] [Google Scholar]
- Martinez LR, Moussai D, and Casadevall A (2004). Antibody to Cryptococcus neoformans glucuronoxylomannan inhibits the release of capsular antigen. Infect. Immun 72, 3674–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland EE, Nicola AM, Prados-Rosales R, and Casadevall A (2010). Ab binding alters gene expression in Cryptococcus neoformans and directly modulates fungal metabolism. J. Clin. Invest 120, 1355–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli C, Bozza S, Bacci A, Gaziano R, Mosci P, Morschhäuser J, Pitzurra L, Kopf M, Cutler J, and Romani L (2003). A role for antibodies in the generation of memory antifungal immunity. Eur. J. Immunol 33, 1193–1204. [DOI] [PubMed] [Google Scholar]
- Moreno-Sabater A, Autaa G, Sterlin D, Jerbi A, Villette R, Holm JB, Parizot C, Selim S, Senghor Y, Ghillani-Dalbin P, et al. (2020). Systemic anti-commensal response to fungi analyzed by flow cytometry is related to gut mycobiome ecology. Microbiome 8, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, Stewart CJ, Metcalf GA, Muzny DM, Gibbs RA, et al. (2017). The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 5, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachl J, Svoboda P, Jacobs F, Vandewoude K, van der Hoven B, Spronk P, Masterson G, Malbrain M, Aoun M, Garbino J, et al. ; Mycograb Invasive Candidiasis Study Group (2006). A randomized, blinded, multi-center trial of lipid-associated amphotericin B alone versus in combination with an antibody-based inhibitor of heat shock protein 90 in patients with invasive candidiasis. Clin. Infect. Dis 42, 1404–1413. [DOI] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al. (2014). Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez de Diego R, Sánchez-Ramón S, López-Collazo E, Martínez-Barricarte R, Cubillos-Zapata C, Ferreira Cerdán A, Casanova J-L, and Puel A (2015). Genetic errors of the human caspase recruitment domain-B-cell lymphoma 10-mucosa-associated lymphoid tissue lymphoma-translocation gene 1 (CBM) complex: Molecular, immunologic, and clinical heterogeneity. J. Allergy Clin. Immunol 136, 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin SA (2010). Correlates of protection induced by vaccination. Clin. Vaccine Immunol 17, 1055–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaka RR, Ricks DM, Alcorn JF, Chen K, Khader SA, Zheng M, Plevy S, Bengtén E, and Kolls JK (2010). Conserved natural IgM antibodies mediate innate and adaptive immunity against the opportunistic fungus Pneumocystis murina. J. Exp. Med 207, 2907–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard ML, and Sokol H (2019). The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol 16, 331–345. [DOI] [PubMed] [Google Scholar]
- Riente L, Chimenti D, Pratesi F, Delle Sedie A, Tommasi S, Tommasi C, Bombardieri S, and Migliorini P (2004). Antibodies to tissue transglutaminase and Saccharomyces cerevisiae in ankylosing spondylitis and psoriatic arthritis. J. Rheumatol 31, 920–924. [PubMed] [Google Scholar]
- Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, Boucher G, Ripke S, Ellinghaus D, Burtt N, et al. ; National Institute of Diabetes and Digestive Kidney Diseases Inflammatory Bowel Disease Genetics Consortium (NIDDK IBDGC); United Kingdom Inflammatory Bowel Disease Genetics Consortium; International Inflammatory Bowel Disease Genetics Consortium (2011). Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet 43, 1066–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S, and Ruland J (2013). Caspase recruitment domain-containing protein 9 signaling in innate immunity and inflammation. Trends Immunol. 34, 243–250. [DOI] [PubMed] [Google Scholar]
- Rudkin FM, Raziunaite I, Workman H, Essono S, Belmonte R, MacCallum DM, Johnson EM, Silva LM, Palma AS, Feizi T, et al. (2018). Single human B cell-derived monoclonal anti-Candida antibodies enhance phagocytosis and protect against disseminated candidiasis. Nat. Commun 9, 5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaedler RW, Dubos R, and Costello R (1965). The development of the bacterial flora in the gastrointestinal tract of mice. J. Exp. Med 122, 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao T-Y, Ang WXG, Jiang TT, Huang FS, Andersen H, Kinder JM, Pham G, Burg AR, Ruff B, Gonzalez T, et al. (2019). Commensal Candida albicans Positively Calibrates Systemic Th17 Immunological Responses. Cell Host Microbe 25, 404–417.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack E, Hapfelmeier S, Stecher B, Velykoredko Y, Stoel M, Lawson MAE, Geuking MB, Beutler B, Tedder TF, Hardt W-D, et al. (2009). Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science 325, 617–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, Herbst RH, Rogel N, Slyper M, Waldman J, et al. (2019). Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 178, 714–730.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Leducq V, Aschard H, Pham H-P, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, et al. (2017). Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standaert-Vitse A, Jouault T, Vandewalle P, Mille C, Seddik M, Sendid B, Mallet JM, Colombel JF, and Poulain D (2006). Candida albicans is an immunogen for anti-Saccharomyces cerevisiae antibody markers of Crohn’s disease. Gastroenterology 130, 1764–1775. [DOI] [PubMed] [Google Scholar]
- Standaert-Vitse A, Sendid B, Joossens M, François N, Vandewalle-El Khoury P, Branche J, Van Kruiningen H, Jouault T, Rutgeerts P, Gower-Rousseau C, et al. (2009). Candida albicans colonization and ASCA in familial Crohn’s disease. Am. J. Gastroenterol 104, 1745–1753. [DOI] [PubMed] [Google Scholar]
- Takaki H, Kure S, Oshiumi H, Sakoda Y, Suzuki T, Ainai A, Hasegawa H, Matsumoto M, and Seya T (2018). Toll-like receptor 3 in nasal CD103+ dendritic cells is involved in immunoglobulin A production. Mucosal Immunol. 11, 82–96. [DOI] [PubMed] [Google Scholar]
- Tang J, Iliev ID, Brown J, Underhill DM, and Funari VA (2015). Mycobiome: Approaches to analysis of intestinal fungi. J. Immunol. Methods 421, 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Klinger CN, Felix KM, Bradley CP, Wu E, Tran NL, Umesaki Y, and Wu HJ (2016). Gut Microbiota Drive Autoimmune Arthritis by Promoting Differentiation and Migration of Peyer’s Patch T Follicular Helper Cells. Immunity 44, 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso GHW, Reales-Calderon JA, Tan ASM, Sem X, Le GTT, Tan TG, Lai GC, Srinivasan KG, Yurieva M, Liao W, et al. (2018). Experimental evolution of a fungal pathogen into a gut symbiont. Science 362, 589–595. [DOI] [PubMed] [Google Scholar]
- Victora GD, and Nussenzweig MC (2012). Germinal centers. Annu. Rev. Immunol 30, 429–457. [DOI] [PubMed] [Google Scholar]
- Witchley JN, Penumetcha P, Abon NV, Woolford CA, Mitchell AP, and Noble SM (2019). Candida albicans Morphogenesis Programs Control the Balance between Gut Commensalism and Invasive Infection. Cell Host Microbe 25, 432–443.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich M, Deepe GS Jr., and Klein B (2012). Adaptive immunity to fungi. Annu. Rev. Immunol 30, 115–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A-M, Inamine T, Hochrath K, Chen P, Wang L, Llorente C, Bluemel S, Hartmann P, Xu J, Koyama Y, et al. (2017). Intestinal fungi contribute to development of alcoholic liver disease. J. Clin. Invest 127, 2829–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, Inohara N, and Núñez G (2016). Gut Microbiota-Induced Immuno-globulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity 44, 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai B, Ola M, Rolling T, Tosini NL, Joshowitz S, Littmann ER, Amoretti LA, Fontana E, Wright RJ, Miranda E, et al. (2020). High-resolution mycobiota analysis reveals dynamic intestinal translocation preceding invasive candidiasis. Nat. Med 26, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. MultiKAP analysis of IgG binding in mice and microbial visualization of human feces or gut-distal organs of SPF mice, related to Figures 1 and 2
(A) Fungi in SPF mouse feces were assessed for IgA-, IgG-, and IgM-MFI by multiKAP. No serum incubation (gray), incubation with matched WT SPF mouse serum (black), or B cell-deficient μMT−/− SPF serum (blue). − Serum, n = 5; +WT Serum, n = 5; +μMT−/− Serum, n = 5.
(B) Sera from RAG2−/−, n = 5; μMT−/−, n = 5; or WT (SPF), n = 7 mice were assessed for IgG binding to fecal samples from RAG2−/− mice by multiKAP; ANOVA.
(C-D) Bacteria in SPF WT mouse feces were assessed for IgG1-, IgG2b-, and IgG3-binding by multiKAP after incubation with matched WT SPF serum. In each plot, the isotype control is represented by the light gray curve with filled-in area. IgG1, n = 5; IgG2b, n = 5; IgG3, n = 5.
(E) Individual fluorescent microscopy channels of composite image in Figure 2A.
(F-G) Live fungi are not present in gut-distal organs of SPF mice from different facilities and fungal infection-related kidney pathologies do not develop in SPF mice. Organs were harvested from 8-week-old mice from our WCM-CE facility and the Jackson Laboratory Facility (JAX). Mice undergoing the systemic candidiasis were used as positive controls of infection. Indicated tissues were assessed for fungal CFU counts (F) and by H&E staining (G). Cellular infiltrates in the infected area (dark blue) are visible on the kidneys of mice with systemic candidiasis. Scale bars in G represent 100 μm.
Figure S2. Microbiota gating strategy in human feces, isotype control staining, and relative abundance analysis of Ig-coated or -uncoated intestinal fungi, related to Figure 2
(A) DAPI staining of GF mice, WT mice, and healthy humans.
(B) Representative plots of isotype control staining of human feces incubated with serum distinguishing antibody bound and unbound gates within the microbial gate.
(C) Equation for the calculation of CI values represented in Figures 2F and 2G. For each species, Ig+ represents the relative abundance in the antibody-bound population (IgG+IgA+ for Figure 2F, IgG+IgA− for Figure 2G), while Ig− represents the relative abundance in the antibody-unbound population. Output values range between 1 (only represented in the antibody-bound population), and −1 (only represented in the antibody-unbound population), with a value of 0 if a fungal species is not enriched in either sorted population.
(D) Relative abundance of representative species in Figures 2F and 2G for each of the three sorted populations used for CI calculation. Boxplots represent species’ median and the first to third quartile, while whiskers represent minimum and maximum values. Fecal samples from healthy individuals, n = 7.
Figure S3. Systemic anti-C. albicans IgG antibodies appear 2 weeks after intestinal colonization but do not arise because of systemic fungal spread or acute B cell reaction to gut C. albicans, related to Figure 3
(A) Serum was collected from ASF mice or ASF mice colonized with either C. albicans or S. cerevisiae and assessed by ELISA for anti-bacterial flagellin IgG antibodies. Pooled from two independent experiments. ASF, n = 8; +Ca, n = 10; +Sc, n = 10. ns-p ≥ 0.05, one-way ANOVA followed by Tukey’s multiple comparisons test.
(B-C) Comparative analysis of the CD19+CD4− B cell compartment in the spleens of ASF mice colonized with C. albicans or S. cerevisiae as described in Figure 3. Each dot represents individual mice.
(D-E) Mycobiota-free altered Schaedler flora-colonized (ASF) mice were orally gavaged with 1×108 C. albicans and fecal samples and sera were collected before colonization 0, 1, 3, 7, and 14, and 28 days after colonization, groups of mice were sacrificed at each time point to assess extra-intestinal and gut-associated tissues for C. albicans CFUs (D). Sera were assessed for anti-C. albicans anti-IgG titers (E). 0, n = 5; 1, n = 4; 3, n = 3; 7, n = 4; 14, n = 6; 28, n = 6. Data representative of two independent experiments. Boxplots represent median and the first to third quartile, while whiskers represent minimum and maximum values. *p < 0.05, **p < 0.01, Mann-Whitney, comparison to day 0.
Figure S4. Antifungal antibodies are protective in systemic candidiasis, but the extreme susceptibility of CARD9−/− mice cannot be overcome by otherwise effective eradication strategies such as fluconazole treatment, C.alb-IgG treatment, or decreased fungal inoculum, related to Figures 4 and 5
(A) WT mice (WT) or μMT−/− mice adoptively transferred with splenic B cells from Aicda−/− mice (μMT−/− +Aicda−/− B Cells) were intravenously (i. v.) infected with 2×105 C. albicans. Percent survival was assessed, log-ranked Mantel-Cox test. WT, n = 6; μMT−/− +Aicda−/− B Cells, n = 6.
(B) Schematic of the gut-disseminated candidiasis model.
(C) Serum anti-C. albicans IgG antibody decreases in titers upon cyclophosphamide (Cyc)-induced systemic fungal dissemination as measured by ELISA (related to Figures 4D–4G). Paired two-tailed t test, n = 6. Each pair of dots represents individual mice. Data represents two individual experiments.
(D-E) Card9+/+ and Card9−/− mice were intravenously (i. v.) infected with 2×104 C. albicans. Four days later Card9−/− mice were treated with i. p. PBS, 400 μg C.alb-IgG, or provided 0.5 g/L fluconazole in drinking water. A control group of Card9+/+ mice was left uninfected. Percent Survival (D) and C. albicans systemic spread to kidney tissue (E) were assessed. Log-ranked Mantel-Cox test (D) and one-way ANOVA followed by Tukey’s multiple comparisons test (E). Card9+/+ i. p. PBS, n = 13; Card9−/− i. p. PBS, n = 7; Card9−/− i. p. 400 μg C.alb-IgG, n = 11; Card9−/− +Fluconazole, n = 5. Data representative of two independent experiments.
(F-G) Card9−/− mice inability to produce efficient anti-C. albicans IgG antibody response is not due to B cell intrinsic or developmental defect in the absence of CARD9. μMT−/− mice adoptively transferred splenic B cells from Card9+/+ or Card9−/− mice before oral gavaged with C. albicans for two weeks, after which serum anti-C. albicans IgG antibody titers were measured by ELISA (F) and the frequency of IgG+ IgM− among CD19+CD4− B cells in spleens (G) were analyzed by flow cytometry. Mann-Whitney test. Card9+/+, n = 4; Card9−/−, n = 5. Each dot represents individual mice.
(H-I) CD4+ T cell and CD19+ B cell Compartment Analyses in Card9+/+ and Card9−/− mice in response to intraperitoneal challenge with C. albicans. Splenic CD4+ T cell and CD19+ B cell frequencies of Card9+/+ and Card9−/− mice with or without intraperitoneal injection with C. albicans analyzed by flow cytometry. One-way ANOVA followed by Tukey’s multiple comparisons test. Card9+/+ NT, n = 7; Card9+/+ +Ca, n = 11; Card9−/− NT, n = 6; Card9−/− +Ca, n = 6. Data representative of three independent experiments.
Error bars indicate SEM ns-p ≥ 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Figure S5. In vivo depletion of CARD9-expressing phagocyte subsets and human CARD9 SNP map, related to Figure 6
(A) Representative flow cytometry plots and quantification of CD45+MHC-II+CD11c+ phagocyte subsets in ΔCX3CR1 (Cd11c-cre+/− Cx3cr1DTR + DT) and littermate (Litt., Cd11c-cre−/− Cx3cr1DTR + DT) mice. CD11b+CX3CR1+ MNPs (top) and CD11b+CD103+ cDC2s (bottom). Mann-Whitney test. Litt, n = 12; ΔCX3CR1, n = 7. Data representative of two independent experiments.
(B) Representative flow cytometry plots and quantification of CD45+MHC-II+CD11c+ phagocyte subsets in ΔIrf4 (Cd11c-cre+/− Irf4fl/fl) or littermate (Litt., Cd11c-cre−/− Irf4fl/fl) mice. CD11b+CX3CR1+ MNPs (top) and CD11b+CD103+ cDC2s (bottom). Mann-Whitney test. Litt, n = 7; ΔIRF4, n = 10. Data representative of two independent experiments.
(C-D) Frequency of GC-B cells among CD19+CD4− B cells in the spleen analyzed by flow cytometry of mice represented in Figure S8A (C) and S8B (D). One-way ANOVA followed by Tukey’s multiple comparisons test. Mann-Whitney test.
(E) Map of human CARD9 gene and the location of three represented SNPs: Q289* (nonsense mutation), R70W (missense), and R35Q (missense). Each dot represents individual mice. Error bars indicate SEM ns-p ≥ 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Data Availability Statement
All sequencing data are accessible under NCBI archives under the BioProject ID PRJNA630985. All other data needed to evaluate the conclusions in the manuscript are available within the main text or supplementary materials.


