Abstract
In animal breeding and genetics, the ability to cope with disease, here defined as immune competence (IC), with minimal detriment to growth and fertility is a desired objective which addresses both animal production and welfare considerations. However, defining and objectively measuring IC phenotypes using testing methods which are practical to apply on-farm has been challenging. Based on previously described protocols, we measured both cell-mediated immune response (Cell-IR) and antibody-mediated immune response (Ab-IR) and combined these measures to determine an animal’s IC. Using a population of 2,853 Australian Angus steers and heifers, we compared 2 alternative methods to combine both metrics into a single phenotype to be used as a tool for the genetic improvement of IC. The first method, named ZMEAN, is obtained by taking the average of the individual metrics after subjecting each to a Z-score standardization. The second, ImmuneDEX (IDEX), is a weighted average that considers the correlation between Cell-IR and Ab-IR, as well as the difference in ranking of individuals by each metric, and uses these as weights in the averaging. Both simulation and real data were used to understand the behavior of ZMEAN and IDEX. To further ascertain the relationship between IDEX and other traits of economic importance, we evaluated a range of traits related to growth, feedlot performance, and carcass characteristics. We report estimates of heritability of 0.31 ± 0.06 for Cell-IR, 0.42 ± 0.06 for Ab-IR, 0.42 ± 0.06 for ZMEAN and 0.370 ± 0.06 for IDEX, as well as a unity genetic correlation (rg) between ZMEAN and IDEX. While a moderately positive rg was estimated between Cell-IR and Ab-IR (rg = 0.33 ± 0.12), strongly positive estimates were obtained between IDEX and Cell-IR (rg = 0.80 ± 0.05) and between IDEX and Ab-IR (rg = 0.85 ± 0.04). We obtained a moderately negative rg between IC traits and growth including an rg = −0.38 ± 0.14 between IDEX and weaning weight, and negligible with carcass fat measurements, including an rg = −0.03 ± 0.12 between IDEX and marbling. Given that breeding with a sole focus on production might inadvertently increase susceptibility to disease and associated antibiotic use, our analyses suggest that ImmuneDEX will provide a basis to breed animals that are both highly productive and with an enhanced ability to resist disease.
Keywords: beef cattle, heritability, immune competence, stress, weaning
Introduction
Future livestock industries need novel traits to address societal concerns and to maintain sustainable livestock production. Key among these pressing needs is the ability to simultaneously consider improved productivity and animal welfare outcomes. One such desirable attribute of the animal is improved immune competence (IC) (Wilkie et al., 1999), which describes an animal’s ability to mount an immune response. Enhanced IC has demonstrated health benefits in dairy cattle (Thompson-Crispi et al., 2012; Aleri et al., 2019) and is associated with reduced health-related costs in Angus cattle during feedlot finishing (Hine et al., 2016).
Recently, Hine et al. (2019) developed a method for on-farm assessment of IC in Australian Angus beef cattle which combines measures of an animal’s ability to mount both cell-mediated immune response (Cell-IR) and antibody-mediated immune response (Ab-IR), and reported on phenotypic and genetic variation for immune competence traits and their association with indirect measures of stress-coping ability such as body weight change, flight speed, and temperament. Those preliminary results were based on ~ 1,100 cattle and suggested that IC traits are moderately heritable (h2 ~ 0.30) and favorably genetically correlated with flight speed (rg ~ 0.60).
Using the same animal resource, Dominik et al. (2019) developed a simple function to combine both immune response metrics into a single IC phenotype based on a weighted average as follows: IC = α Cell-IR + (1 − α) Ab-IR; where the weight α varied from 0 to 1. In terms of h2 estimates for IC and its rg with 600-d weight, the authors reported an optimal weight in the range of 0.7 to– 0.8 suggesting a stronger emphasis on Cell-IR. However, the ideal metric would consider both immune competence traits, Cell-IR and Ab-IR, so that selection on the metric achieves improvement in general immune response and does not favor one over the other.
The objective of this study was to expand on that earlier work and (1) report genetic parameter estimates for immune response metrics with a 2.6-fold larger population sample size; (2) compare 2 alternative methods to combine both metrics into a single phenotype, ZMEAN and ImmuneDEX (IDEX), to be used as a tool for the genetic improvement of immune competence; and (3) estimate the relationships between the immune competence phenotypes and a range of 9 commonly measured traits related to body weight, feedlot performance, and carcass characteristics of beef cattle.
Materials and Methods
All experimental procedures were performed in accordance with methodology outlined in an application approved by the Chiswick, Animal Ethics Committee, Armidale, NSW, Australia (application number 12/31).
Animals and phenotypes
A total of 1,913 Angus steers and 940 Angus heifers were included in this study. Animals were born in 2012 (N = 472), 2013 (N = 405), 2016 (N = 1,071), and 2017 (N = 905), representing 11 herds and 195 sires from cohorts 2, 3, 6, and 7 of the Australian Angus Sire Benchmarking Program (ASBP). The ASBP is a major initiative of Angus Australia with support from Meat and Livestock Australia (MLA) and industry partners which aims to generate progeny test data on modern Angus bulls, particularly for hard to measure traits such as feed efficiency, abattoir carcass measurements, meat quality attributes, and female reproduction (https://www.angusaustralia.com.au/sire-benchmarking/about/generalinformation/).
Based on previously described protocols (Hine et al., 2019), animals were assessed to determine their ability to mount both Cell-IR and Ab-IR. In brief, calves were vaccinated with Ultravac 7in1 clostridial and leptospira vaccine (Zoetis) on the day they were weaned. Calves were then given time to respond to the vaccination and Cell-IR was assessed by measuring the magnitude of delayed-type hypersensitivity reactions induced by intradermal injection of the clostridial vaccine in the caudal fold of the tail. The trait is expressed in relation to the skin fold at a control site (saline). To assess Ab-IR the production of tetanus toxoid specific IgG1 antibody in blood between days 8 and 21 postvaccination was measured in optical density units. Immune responses for all animals within a herd testing cohort were assessed on the same day postvaccination.
In addition, for these same animals, phenotypic data were available on a range of traits related to body weight, feedlot performance, and carcass characteristics including weaning weight (WWT), yearling weight (YWT), feedlot average daily gain (ADG), feedlot average daily dry matter intake (DMI), carcass weight (CWT), carcass eye muscle area (EMA), carcass marbling score (Meat Standards Australia, MSA), carcass ossification score (OSS), and carcass subcutaneous rib fat depth (RIB). Feedlot and carcass measurements were available for steers only.
Phenotypic measurements of live body weights, feedlot, and carcass characteristics were taken as described by Torres-Vázquez et al. (2018) using a subset of the cattle employed here and corresponding to the animals for which immune competence phenotypes were available. In brief, WWT and YWT correspond to 200- and 400-d weight, respectively. Steers entered the feedlot at an average age (±SD) of 530.10 ± 78.69 d and measurements of ADG and DMI were taken during the first 10 wk following a 3-wk conditioning period which was not used in the computation of ADG and DMI. Animals were slaughtered at an average age of 732.54 ± 116.66 d. Carcass measurements of MSA and OSS, not included in the study of Torres-Vázquez et al. (2018), were measured as follows: Marling based on MSA was measured in scores ranging from 100 to 1,100 in increments of 10, with higher scores indicating greater marbling (McGilchrist et al., 2019). Ossification scores ranged from 100 to 590 in increments of 10, with lower scores indicating less physiological maturity, being a ameasure of the calcification in the spinous processes in the sacral, lumbar, and thoracic vertebrae (AUS-MEAT, 2005; Watson et al., 2008; Gudex et al., 2019).
Table 1 provides summary statistics for all phenotypes and covariates included in this study. Of the IC phenotyped animals, only steers had phenotypic data for feedlot and carcass traits available. Hence, the lower number of records for these traits. The seemingly large range of values for carcass traits (e.g., 214 to 607 kg for CWT) can be attributed to the fact that some animals were short-fed and some long-fed at feedlot depending on final market destination.
Table 1.
Summary statistics including number of records (N), mean, standard deviation (SD), minimum and maximum for traits and covariates included in this study
| Category/trait1 | N | Mean | SD | Min. | Max. |
|---|---|---|---|---|---|
| Body weight | |||||
| WWT, kg | 2,853 | 241.10 | 49.82 | 102.00 | 402.00 |
| YWT, kg | 1,945 | 389.49 | 78.45 | 183.00 | 692.00 |
| Feedlot | |||||
| ADG, kg/d | 1,788 | 1.63 | 0.33 | 0.530 | 2.900 |
| DMI, kg/d | 1,788 | 14.47 | 2.21 | 3.360 | 22.780 |
| Carcass | |||||
| CWT, kg | 1,745 | 425.33 | 76.28 | 214.00 | 607.00 |
| EMA, cm2 | 1,744 | 89.85 | 11.26 | 60.00 | 128.00 |
| MSA | 1,744 | 486.12 | 123.13 | 160.00 | 1,030.00 |
| OSS | 1,744 | 147.57 | 21.45 | 100.00 | 280.00 |
| RIB, mm | 1,736 | 16.25 | 6.24 | 3.00 | 41.00 |
| Immune competence | |||||
| Cell-IR, log(mm) | 2,853 | 25.18 | 11.11 | -2.303 | 69.236 |
| Ab-IR | 2,853 | 83.73 | 26.77 | 0.000 | 145.954 |
| ZMEAN | 2,853 | -0.00 | 0.77 | -2.696 | 2.448 |
| IDEX | 2,853 | 0.01 | 1.15 | -4.596 | 4.428 |
| Regression covariates | |||||
| WAGE, d | 2,853 | 208.14 | 33.82 | 136.00 | 290.00 |
| YAGE, d | 1,945 | 428.04 | 28.06 | 338.00 | 500.00 |
| FAGE, d | 1,788 | 530.10 | 78.70 | 372.00 | 767.00 |
| CAGE, d | 1,745 | 732.54 | 116.66 | 504.00 | 990.00 |
| IRAGE, d | 2,853 | 207.33 | 34.09 | 118.00 | 310.00 |
| CST, log(mm) | 2,853 | -0.00 | 0.04 | -0.19 | 0.25 |
1WWT, weaning weight; YWT, yearling weight; ADG, average daily gain at feedlot; DMI, average daily dry matter intake at feedlot; CWT, carcass weight; EMA, carcass eye muscle area; MSA, carcass MSA marbling score; OSS, carcass ossification score; RIB, carcass subcutaneous fat depth at the ribs level; Cell-IR, cell-mediated immune response (log-transformed); Ab-IR, antibody-mediated immune response (square root transformed of optical density); ZMEAN, average of standardized Cell-IR and Ab-IR; IDEX, ImmuneDEX; WAGE, weaning age; YAGE, yearling age; FAGE, age at feedlot entry; CAGE, carcass age; IRAGE, age at immune competence testing; CST, change in skin thickness during immune competence testing.
To approximate normality, the IC phenotypes for Cell-IR and Ab-IR were log- and square root transformed, respectively, and well within the values reported by Dominik et al. (2019) and Hine et al. (2019) on a subset of 1,149 animals. These values were used to guide the development of a phenotype that combines both metrics of immune competence into a single value.
Development of ImmuneDEX
Two alternative ways of combining Cell-IR and Ab-IR into a single phenotype were considered. A simulation study was undertaken to examine the impact of the correlation between Cell-IR and Ab-IR on each phenotype and the relationship between them.
The first such phenotype (ZMEAN) was obtained from the average of the 2 individual metrics after subjecting each to a Z-score standardization by subtracting the mean and dividing by the standard deviation:
where ZCELL and ZAB are the Z-score standardization of Cell-IR and Ab-IR, respectively. Standardizing prior to averaging was done to ensure both metrics were in standard units and to stabilize the variance, otherwise the resulting average would be dominated by the most variable metric. Assuming ZCELL and ZAB are independent, ZMEAN was expected to follow a normal distribution with zero mean and a variance equal to 0.5. Also, ZMEAN was expected to be equally correlated with both ZCELL and ZAB. Finally, ZMEAN corresponds to the function evaluated by Dominik et al. (2019) at weight α = 0.5.
ImmuneDEX (IDEX) is an alternative to ZMEAN that considers the correlation (r) between Cell-IR and Ab-IR as well as the difference in ranking (dRank) of individuals for each metric, and uses them as weights in the averaging as follows:
In agreement with the finding of Dominik et al. (2019), IDEX places greater emphasis on Cell-IR. Neither r nor dRank are affected by the standardization of Cell-IR and Ab-IR into ZCELL and ZAB, respectively. At r = 1.0, dRank = 0, and IDEX = ZCELL. On the other extreme, at r = –1.0, dRank = n/2 on average (where n is the sample size or 2,853 in our case) and IDEX = ZCELL/2 on average. However, also at r = –1.0, ZMEAN is uncorrelated with either Cell-IR or Ab-IR. With complete independence at r = 0, IDEX and ZMEAN were on average identical in value for a function of dRank which can range from 0 to (n – 1).
Simulation studies
Two simulation studies were undertaken to understand the behavior of ZMEAN and IDEX and their relationship with each other and with each of the individual components, ZCELL and ZAB.
In the first simulation study, 2 standard normal random variables, representing ZCELL and ZAB, both of dimension 3,000 and with zero mean and unit variance were simulated. The correlation (r) between Cell-IR and Ab-IR ranged from −1.0 to 1.0 at 0.05 increment. At each value of r, the difference in ranking (dRank) as well as the correlation between ZMEAN and IDEX were calculated, and between them and each of the individual components, ZCELL and ZAB. The entire simulation process was repeated 10 times.
In the second simulation study, the focus was on the 3-way relationship between ZMEAN, ZCELL, and ZAB, and between IDEX, ZCELL, and ZAB and for all possible observable values of ZCELL and ZAB. To this end, the sample size n was increased to 1,000,000 but only 5 values of r were explored: −0.75, −0.25, 0, 0.25, and 0.75.
Genetic parameter estimates from real data
For animals with IC phenotypes, we traced back 3 generations of ancestor to generate a pedigree file of 8,216 animals. Phenotyped animals were by 195 sires with an average of 14.63 progeny per sire and ranging from 2 to 59 progeny per sires. There were 2,342 dams and 168 maternal grandsires.
Variance components, heritabilities, genetic, and residual correlations were estimated from bivariate analyses using VCE6 software (Neumaier and Groeneveld, 1998; Groeneveld et al. 2010). For the 13 phenotypes available, all possible 78 bivariate analyses were performed to estimate covariance components.
The bivariate linear mixed-model used to analyze all pair-wise traits contained the fixed effects of contemporary group (CG), age of dam (AOD) at birth of calf in years and the linear covariate of age at measurement. For each phenotype, CG contained animals of the same sex, year and month of birth, property of origin, feeding regime (short or long), and date of measurement. For the IC phenotypes (Cell-IR, Ab-IR, ZMEAN, and IDEX), the logarithm of the fold change in skin thickness (CST) between the test and the control sites on day 0 of testing was included as an additional linear covariate (Hine et al., 2019).
Additionally, the random additive polygenic and residual effects were fitted with assumed distributions N(0, AG) and N(0, IR), respectively, and where A represents the pedigree-based numerator relationship matrix, G is the genetic co-variance matrix, I is an identity matrix, R is the residual variance–covariance matrix, and represents the Kronecker product.
Results and Discussions
Across all 2,853 animals and using raw unadjusted measurements, the correlation between Cell-IR and Ab-IR was moderately positive at 0.19 ± 0.02 and slightly higher for steers (0.22 ± 0.02) than for heifers (0.10 ± 0.03).
ImmuneDEX was developed to provide a comprehensive phenotype that incorporates both metrics of immune competence, Cell-IR and Ab-IR, into a single value that is highly correlated with both metrics even when they themselves might not be correlated, or indeed are negatively correlated with each other. In computing IDEX, is a dissimilarity measure that approaches its maximum at unity as r approaches zero. It acts as a weight on Ab-IR and takes the same value for all samples. In contrast, the term varies with each individual sample except when r = 1 in which case equals zero for all samples.
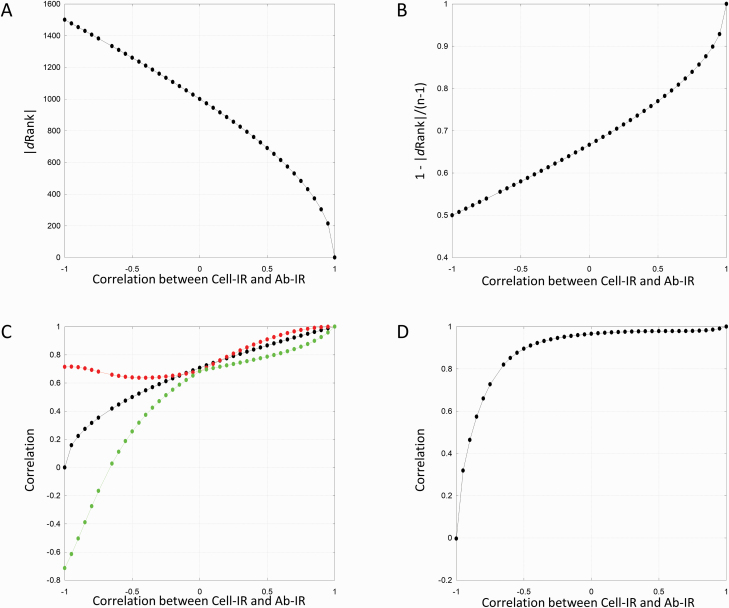
Figure 1 illustrates the inter-relationships among all the individual components of ZMEAN and IDEX as r varies from −1 to 1 at 0.05 increments. Each dot on the plots shown in Figure 1 represents the average of 10 random replicates of the simulation process. At r = 0 (indicating independence between Cell-IR and Ab-IR), averaged 1,000.02 ± 3.57 (Figure 1A) yet values ranging between 0 (no change in ranking) to 2,984 (or near maximum change in ranking) were observed in any single replicate. Also, at r = 0, ZMEAN and IDEX coincided at ~0.7 (Figure 1C) in the way they correlate with Cell-IR and Ab-IR after standardization and, across the 10 replicates, the correlation between ZMEAN and IDEX averaged 0.97 ± 0.00 (Figure 1D). The only point at which ZMEAN and IDEX were more similar was at r = 1 where, as expected, equaled zero (Figure 1A) and ZMEAN and IDEX were perfectly correlated (Figure 1D).
Figure 1.
Behavior of components of ZMEAN and IDEX as a function of the correlation between Cell-IR and Ab-IR: (A) average absolute value of the differential ranking (|dRank|) between Cell-IR and Ab-IR; (B) one minus |dRank| divided by (n − 1) where n is the sample size, i.e. n = 3,000 in these simulations; (C) correlation between ZMEAN and each of Cell-IR and Ab-IR (black profile), between IDEX and Cell-IR (red profile) and between IDEX and Ab-IR (green profile); (D) Correlation between ZMEAN and IDEX.
Regardless of the correlation between Cell-IR and Ab-IR, the correlation between IDEX and Cell-IR was higher than or equal to the correlation between ZMEAN and Cell-IR, and this difference was particularly pronounced when Cell-IR and Ab-IR were negatively correlated (Figure 1C). However, as long as the correlation between Cell-IR and Ab-IR was > –0.5, then the correlation between ZMEAN and IDEX remained > 0.90 (Figure 1D). Therefore, as an IC phenotype, both ZMEAN and IDEX tended to perform similarly whenever Cell-IR and Ab-IR were not strongly negatively correlated with each other. In the extreme scenario where their correlation was –1, ZMEAN became uncorrelated with either immune competence metric, and IDEX selected individuals with high Cell-IR in favor of those with high Ab-IR.
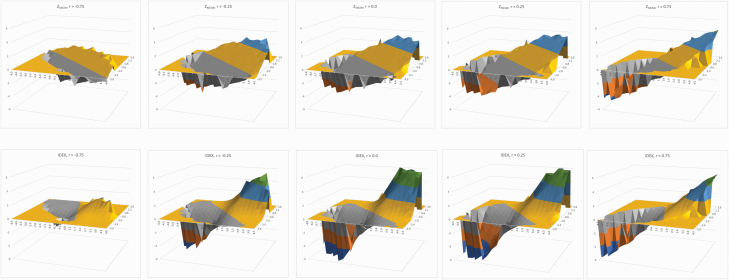
The higher dynamic range of values expected for IDEX as compared with ZMEAN is illustrated in Figure 2. Across the 5 correlation points simulated (–0.75, –0.25, 0, 0.25, and 0.75) the range of values for IDEX (ZMEAN) were 4.22 (3.59), 10.61 (6.08), 13.53 (6.77), 13.44 (7.62), and 11.27 (8.90). These simulated values are in accordance with those observed with the real data where the correlation between Cell-IR and Ab-IR was 0.191 ± 0.018 and the range of values for IDEX and ZMEAN was 9.02 and 5.14, respectively (Table 1). If this variation translates to higher h2 for IDEX and genetic correlation with the two components of immune response, IDEX would become the preferred phenotype.
Figure 2.
Surface plots of values of ZMEAN (upper plots) and IDEX (lower plots) that can be expected as a function of the standardized values of Cell-IR and Ab-IR (width and depth axes, irrespective) and the correlation with each other being from left to right −0.75, −0.25, 0, 0.25, and 0.75.
Table 2 provides estimates of genetic and residual variances for the four IC phenotypes: Cell-IR, Ab-IR, ZMEAN, and IDEX. In agreement with the observed higher range and variation for the raw measurements of IDEX (Table 1), the estimated genetic and residual components of variance for IDEX (0.31 ± 0.05 and 0.54 ± 0.04) were higher than those estimated for ZMEAN (0.15 ± 0.02 and 0.21 ± 0.02).
Table 2.
Estimates ± SE for the genetic (Vg) and residual (Ve) components of variance for the 4 components of immune competence: Cell-IR, Ab-IR, ZMEAN, and IDEX
| Trait1 | V g | V e |
|---|---|---|
| Cell-IR | 19.59 ± 3.94 | 44.05 ± 3.38 |
| Ab-IR | 226.81 ± 36.99 | 314.64 ± 32.16 |
| Z MEAN | 0.15 ± 0.02 | 0.21 ± 0.02 |
| IDEX | 0.31 ± 0.05 | 0.54 ± 0.04 |
1Cell-IR, cell-mediated immune response (log transformed); Ab-IR, antibody-mediated immune response (square root transformed of optical density); ZMEAN, average of standardized Cell-IR and Ab-IR; IDEX, ImmuneDEX.
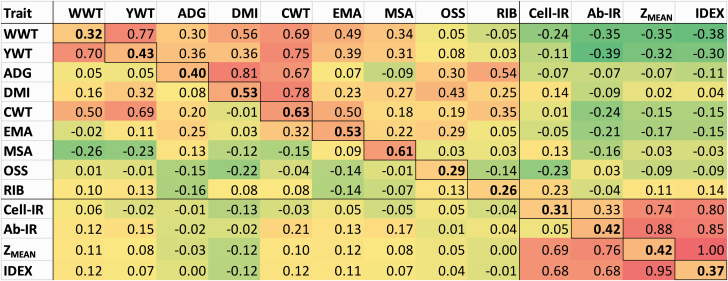
Figure 3 is a heatmap of the estimates of h2, genetic and residual correlations across the 13 traits investigated. Full details of estimates and associated SE are given in Supplementary Table 1 where estimates larger than 2 SE in absolute value are highlighted to facilitate the identification of significant estimates.
Figure 3.
Heatmap of the estimates of heritability (bold, diagonals), genetic (above diagonal), and residual (below diagonal) correlations for growth, feedlot performance, carcass characteristics, and the four components of immune response: Cell-IR, Ab-IR, ZMEAN, and IDEX. The color spectrum goes from bright green for large negative values, to yellow for close to zero values, to bright red for large positive values.
The estimates of h2 for Cell-IR and Ab-IR were 0.31 ± 0.06 and 0.42 ± 0.06, respectively. These values were more accurate and comparatively higher than those recently reported by Hine et al. (2019) of 0.27 ± 0.08 and 0.32 ± 0.09, or those reported by Dominik et al. (2019) of 0.33 ± 0.11 and 0.30 ± 0.10 also for the same traits. The estimated rg between Cell-IR and Ab-IR was 0.33 ± 0.12, more precisely estimated because of the larger sample size and within one SE of the 0.48 ± 0.19 from Hine et al. (2019) or the 0.40 ± 0.22 from Dominik et al. (2019).
Both ZMEAN and IDEX were found to be highly heritable at 0.42 ± 0.06 and 0.37 ± 0.06, respectively. The estimated rg between them was 1.00 ± 0.00, indicating that they are both the same trait at the genetic level. The moderately positive rg estimated between Cell-IR and Ab-IR (0.33 ± 0.12) contrasts with the strong positive estimates obtained between IDEX and Cell-IR (0.80 ± 0.05) and between IDEX and Ab-IR (0.85 ± 0.04). These values anticipate the suitability of ImmuneDEX as a single phenotype to aid in the genetic improvement of immune competence and potentially general disease resistance in the current population.
Our estimates of h2 and correlations among feedlot and carcass traits presented in Figure 3 are in close agreement with those reported by Torres-Vázquez et al. (2018) using a subset of the Australian Angus cattle employed here. For instance, our high estimate of h2 for CWT of 0.63 ± 0.11 matches with the 0.66 ± 0.09 of Torres-Vázquez et al. (2018), and our low estimate of rg between ADG and EMA of 0.07 ± 0.13 matches with the 0.06 ± 0.15 of Torres-Vázquez et al. (2018).
Genetic parameters for feedlot traits have also been reported in a crossbred population of 687 heifers with Angus influence by Freetly et al. (2020) who reported a heritability estimate of 0.53 ± 0.12 for ADG and of 0.84 ± 0.12 for DMI with an rg estimate of 0.86 ± 0.07 between the traits. Our estimates for the same 3 parameters were 0.40 ± 0.08, 0.53 ± 0.09, and 0.81 ± 0.07, respectively (Supplementary Table 1). For carcass traits, the review of Ríos Utrera and Van Vleck (2004) reported average heritability estimates for carcass weight, backfat thickness, longissimus muscle area, and marbling score of 0.40, 0.36, 0.40, and 0.37, respectively. These values are in contrast with the heritability estimates reported here of 0.63 ± 0.11, 0.26 ± 0.07, 0.53 ± 0.09, and 0.61 ± 0.09 for CWT, RIB, EMA, and MSA, respectively (Supplementary Table 1). For both feedlot and carcass traits in an F1 population of 464 steers, compared to estimates reported here (given in brackets), Nkrumah et al. (2007) reported h2 estimates for ADG, DMI, CWT, and EMA of 0.59 ± 0.17 (0.40 ± 0.08), 0.54 ± 0.15 (0.53 ±0.09), 0.33 ± 0.14 (0.63 ± 0.11), and 0.45 ± 0.15 (0.53 ±0.09) respectively. Similarly, the authors reported estimates of rg between ADG and DMI of 0.87 ± 0.09 (0.81 ± 0.07), between DMI and CWT of 0.66 ± 0.17 (0.78 ± 0.06), and between DMI and EMA of 0.21 ± 0.13 (0.23 ± 0.12).
Of note from the heatmap of Figure 3 and Supplementary Table 1, is the negative rg between the IC traits and the growth traits. For instance, an rg = –0.38 ± 0.14 was estimated between IDEX and WWT. This was not an unexpected result as it is well documented that selection for productivity, with no emphasis on health and fitness traits, has increased susceptibility to disease in many species of food-producing animals (Rauw et al., 1998). Although productivity and disease resistance traits are often unfavorably genetically correlated, such correlations may be weak for resistance to certain diseases. For instance, results from Crawford et al. (2016) suggested that selection for lower pulmonary arterial pressure should have minimal influence on the growth performance of yearling Angus bulls. These findings highlight the importance of considering both economically important production traits and health and fitness traits as part of a breeding objective. However, closer to zero values were observed for correlations between IC phenotypes and carcass fat measurements, including an rg = –0.03 ± 0.12 between IDEX and MSA marbling score, and rg = 0.14 ± 0.16 between IDEX and RIB.
The linear function used by Dominik et al. (2019) to capture the optimal weighting for computing a weighted average of Cell-IR and Ab-IR was not evaluated in this study. That was an important piece of research as it demonstrated that both components of the immune competence are unlikely to attract an equal weight in any given scenario. However, the function requires an optimization process with the dual purpose of maximizing the h2 estimate and improving the genetic correlation with a trait in the breeding objective. Therefore, in the current study, re-evaluating that function was considered numerically unpractical and unlikely to result in a linear combination of Cell-IR and Ab-IR that was better than either IDEX or ZMEAN. Nevertheless, as more IC phenotypes are currently being produced, further research is warranted to calibrate the function that combines both IC metrics as well as the analytical model for the estimation of genetic parameters and breeding value prediction. For instance, given that the IC phenotypes were taken around weaning, a model containing maternal components could be explored to test for the relative importance of maternal genetics and maternal permanent environmental effects.
Taken together, our analyses suggest that ImmuneDEX will provide a tool to underpin long-term genetic strategies aimed at improving the immune competence of animals in production systems which in turn is expected to reduce the incidence of disease and our reliance on antibiotics to treat disease. Such a phenotype will allow the producers to put selection emphasis on the immune competence of animals and to breed animals that are both highly productive and have an enhanced ability to resist disease challenges in their production environments.
Supplementary Material
Acknowledgments
This work was co-funded by the Meat and Livestock Australia (MLA) (on behalf of the Australian Lot Feeders’ Association) and CSIRO as part of the MLA mentoring program. The authors acknowledge Angus Australia for facilitating access to progeny from the Angus Sire Benchmarking Program for testing and associated data. We gratefully acknowledge cooperating herd owners, managers, and staff. Special thanks go to Sue Belson, Jim Lea, and Grant Uphill for their technical assistance.
Glossary
Abbreviations
- Ab-IR
antibody-mediated immune response
- Cell-IR
cell-mediated immune response
- IDEX
ImmuneDEX
- ZMEAN
average of standardized Cell-IR and Ab-IR
Conflict of interest statement
The authors declare no real or perceived conflicts of interest.
Literature Cited
- Aleri, J. W., B. C. Hine, M. F. Pyman, P. D. Mansell, W. J. Wales, B. Mallard, M. A. Stevenson, and A. D. Fisher. . 2019. Associations between immune competence, stress responsiveness, and production in Holstein-Friesian and Holstein-Friesian × Jersey heifers reared in a pasture-based production system in Australia. J. Dairy Sci. 102:3282–3294. doi: 10.3168/jds.2018-14578. [DOI] [PubMed] [Google Scholar]
- AUS-MEAT . 2005. Handbook of Australian meat. 7th ed. Brisbane, QLD: AUS-MEAT Ltd. [Google Scholar]
- Crawford, N. F., M. G. Thomas, T. N. Holt, S. E. Speidel, and R. M. Enns. . 2016. Heritabilities and genetic correlations of pulmonary arterial pressure and performance traits in Angus cattle at high altitude. J. Anim. Sci. 94:4483–4490. doi: 10.2527/jas.2016-0703. [DOI] [PubMed] [Google Scholar]
- Dominik, S., L. R. Porto-Neto, C. J. Duff, A. I. Byrne, B. C. Hine, A. Ingham, I. G. Colditz, and A. Reverter. . 2019. A method for developing a breeding objective trait from multiple components using the example of immune competence in Australian Angus cattle. Proc. Assoc. Advmt. Anim. Breed. Genet. 23:402–405. [Google Scholar]
- Freetly, H. C., L. A. Kuehn, R. M. Thallman, and W. M. Snelling. . 2020. Heritability and genetic correlations of feed intake, body weight gain, and residual feed intake of beef cattle as heifers and cows. J. Anim. Sci. 98: skz394. 10.1093/jas/skz394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeneveld, E., M. Kovač, and N. Mielenz. . 2010. VCE user’s guide and reference manual, version 6.0. Neustadt, Germany: Institute of Farm Animal Genetics; 2010. [Google Scholar]
- Gudex, B. W., M. J. McPhee, V. H. Oddy, and B. J. Walmsley. . 2019. Prediction of ossification from live and carcass traits in young beef cattle: model development and evaluation. J. Anim. Sci. 97:144–155. doi: 10.1093/jas/sky422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hine, B. C., A. M. Bell, D. O. D. Niemeyer, C. J. Duff, N. M. Butcher, S. Dominik, A. B. Ingham, and I. Colditz. . 2019. Immune competence traits assessed during the stress of weaning are heritable and favourably genetically correlated with temperament traits in Angus cattle. J. Anim. Sci. 97:4053–4065. doi: 10.1093/jas/skz260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hine, B. C., A. B. Ingham, S. Dominik, and I. G. Colditz. . 2016. Ian Colditz – Mentor for Postdoctoral Fellow. North Sydney (NSW): Meat and Livestock Australia Limited. [Google Scholar]
- McGilchrist, P., R. J. Polkinghorne, A. J. Ball, and J. M. Thompson. . 2019. The meat standards Australia index indicates beef carcass quality. Animal 13:1750–1757. doi: 10.1017/S1751731118003713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumaier, A., and E. Groeneveld. . 1998. Restricted maximum likelihood estimation of covariances in sparse linear models. Genet. Sel. Evol. 30:3–26. doi: 10.1186/1297-9686-30-1-3 [DOI] [Google Scholar]
- Nkrumah, J. D., J. A. Basarab, Z. Wang, C. Li, M. A. Price, E. K. Okine, D. H. Crews, Jr, and S. S. Moore. . 2007. Genetic and phenotypic relationships of feed intake and measures of efficiency with growth and carcass merit of beef cattle. J. Anim. Sci. 85:2711–2720. doi: 10.2527/jas.2006-767. [DOI] [PubMed] [Google Scholar]
- Rauw, W. M., E. Kanis, E. N. Noordhuizen-Stassen, and F. J. Grommers. . 1998. Undesirable side effects of selection for high production efficiency in farm animals: a review. Livest. Prod. Sci. 56:15–33. doi: 10.1016/S0301-6226(98)00147-X [DOI] [Google Scholar]
- Ríos Utrera, A., and L. D. Van Vleck. . 2004. Heritability estimates for carcass traits of cattle: a review. Genet. Mol. Res. 3:380–394. [PubMed] [Google Scholar]
- Thompson-Crispi, K. A., B. Hine, M. Quinton, F. Miglior, and B. A. Mallard. . 2012. Short communication: association of disease incidence and adaptive immune response in Holstein dairy cows. J. Dairy Sci. 95:3888–3893. doi: 10.3168/jds.2011-5201. [DOI] [PubMed] [Google Scholar]
- Torres-Vázquez, J. A., J. H. J. van der Werf, and S. A. Clark. . 2018. Genetic and phenotypic associations of feed intake with growth and carcass traits in Australian Angus cattle. J. Anim. Sci. 96:4521–4531. doi: 10.1093/jas/sky325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, R., R. Polkinghorne, and J. M. Thompson. . 2008. Development of the Meat Standards Australia (MSA) prediction model for beef palatability. Aust. J. Exp. Agric. 48:1368–1379. doi: 10.1071/EA07184 [DOI] [Google Scholar]
- Wilkie, B., and B. Mallard. . 1999. Selection for high immune response: an alternative approach to animal health maintenance? Vet. Immunol. Immunopathol. 72:231–235. doi: 10.1016/s0165-2427(99)00136-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.