Abstract
Abstract
Membrane proteins govern critical cellular processes and are central to human health and associated disease. Understanding of membrane protein function is obscured by the vast ranges of structural dynamics—both in the spatial and time regime—displayed in the protein and surrounding membrane. The membrane lipids have emerged as allosteric modulators of membrane protein function, which further adds to the complexity. In this review, we discuss several examples of membrane dependency. A particular focus is on how molecular dynamics (MD) simulation have aided to map membrane protein dynamics and how enhanced sampling methods can enable observing the otherwise inaccessible biological time scale. Also, time-resolved X-ray scattering in solution is highlighted as a powerful tool to track membrane protein dynamics, in particular when combined with MD simulation to identify transient intermediate states. Finally, we discuss future directions of how to further develop this promising approach to determine structural dynamics of both the protein and the surrounding lipids.
Graphic Abstract
Keywords: Membrane protein dynamics, MD simulation, X-ray solution scattering
The Membrane Protein Influencer
Membrane proteins execute critical cellular processes and enable—with incredible diversity and ingenuity—life as we know it. To appreciate the courage of a membrane protein researcher, it is illustrative to consider the typical spatiotemporal scale and operating conditions of such biological macromolecules. First, the lipid composition varies drastically in membranes across organelles, tissues, and organisms—and also in-between the bilayer leaflets (Harayama and Riezman 2018). In addition, the lipid bilayers display a wide range of structural dynamics from local structural rearrangements (Wiener and White 1992) to transient formation of lipidic micro-assemblies, referred to as lipid rafts (Simons and Sampaio 2011). Second, membrane proteins become inserted into its complex lipid environment by the translocon machinery—itself being a protein—in a process still holding lingering mysteries (Cymer et al. 2015). Third, membrane protein function depends on carefully orchestrated subtle structural rearrangements of amino acid side chains and large-scale conformational changes that span several orders of magnitude on the temporal scale (Fig. 1a). The emerging view is that membrane proteins have coevolved with the membrane lipids to optimize functionality (Lee 2004). Therefore, one of the intricate regulation machineries that control membrane protein activity are the physicochemical properties of the lipids in the surrounding membrane. The endeavor to understand membrane protein function—and associated disease—should therefore ideally monitor the reaction directly in the native membrane on the biological time scale.
Fig. 1.
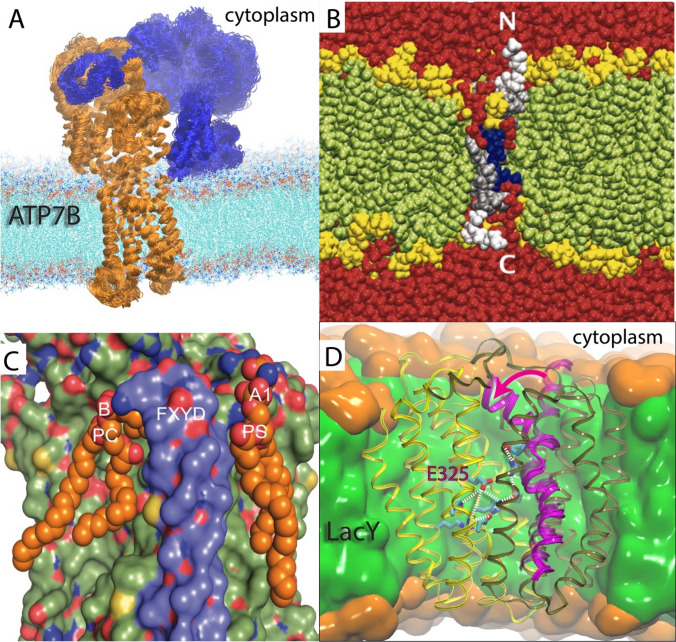
Membrane protein structural rearrangements and lipid regulation. a Homology model of the human Cu+-transporting P-type ATPase (ATP7B) (orange) with associated regulatory domain (blue) inserted into a lipid bilayer. Several simulated overlayed structures visualize thermal fluctuations displayed within a single state of the reaction. b A simulated S4 voltage-sensor peptide (silver with white GGPG flanks) showing bilayer distortion as the charged residues (blue) become solvated by lipid phosphates and water molecules (red). The lipid headgroups and tails are displayed in yellow and green, respectively. Adapted from Freites et al. (2005). c Lipid sites (A1 and B) hosting PC and PS lipids remodeled from a crystal structure at the cytoplasmic side of the Na+, K+ ATPase transporter. Adapted from Cornelius et al. (2015). d Lipid-dependent dynamics (magenta arrow and helix) and internal trigger (protonation state of residue E325) in the Lactose permease (LacY) transporter. The C-terminal and N-terminal domains are colored yellow and tan, respectively. The hydrophobic core of the membrane is colored green, and the polar headgroup region is depicted in orange. Adapted from (Andersson et al. 2012)
Prime examples of the complex protein-lipid interplay are voltage-gated sodium, potassium, and calcium channels that govern nerve impulses. In these proteins, a central ion-conducting pore opens and closes in response to movements of associated voltage-sensing domains induced by changes in the electrical potential (Swartz 2008). The membrane in the vicinity of the voltage-sensing domains deforms locally and enables the formation of a water-filled crevice that hydrates critical acidic and basic amino acid residues, thus allowing them to keep the electrical charge—and hence the capability to respond to changes in the membrane potential (Krepkiy et al. 2009). The water hydration pattern around simulated isolated charge-carrying S4 helices or voltage-sensing domains was shown to depend upon the presence of lipid phosphates in the surrounding membrane (Sands and Sansom 2007; Freites et al. 2005; Andersson et al. 2011) (Fig. 1b), which provides a possible molecular explanation of the lipid head group requirements for optimal functioning of voltage-gated potassium channels (Schmidt et al. 2006; Ramu et al. 2006; Xu et al. 2008). In membranes of heart cells and neurons, polyunsaturated fatty acids (PUFAs) activate voltage-gated potassium channels via electrostatic interaction (Xu et al. 2008; Borjesson et al. 2008) with specific sites of interaction predicted by molecular dynamics (MD) simulation (Yazdi et al. 2016). In addition, phosphatidylinositol 4,5-bisphosphate (PIP2) anionic lipids also regulate voltage-gated potassium channels, in particular the subcategory KCNQ channels (Kruse et al. 2012; Duncan et al. 2020). Regulation includes promoting the coupling between the central ion-conducting pore and the peripheral voltage-sensing domains thus affecting voltage sensitivity (Zhou et al. 2013; Zaydman et al. 2013; Zhang et al. 2013; Kim et al. 2017), which has been located to a linker region connecting the two domains (Rodriguez-Menchaca et al. 2012). A crystal structure of an open-state channel resolved a phosphatidylglycerol (PG) lipid, which is also negatively charged, at the linker region (Long et al. 2007) and several simulations have observed a similar PIP2-interaction site (Kasimova et al. 2014; Kasimova et al. 2015). A second PIP2-interaction site has been proposed to display preference for the closed state of the channel and could therefore possibly be involved in channel deactivation (Zaydman et al. 2013; Zhang et al. 2013; Eckey et al. 2014). Indeed, MD simulations observed PIP2 migration between both sites (Chen et al. 2015). Finally, as an extreme case, PIP2 directly activates so-called inwardly-rectifying potassium channels (Huang et al. 1998).
Pentameric ligand-gated ion channels (pLGICs) bind neurotransmitters and govern fast synaptic transmission (Plested 2016). A common structural basis consists of a central ion-conducting pore forming in-between 4-helical membrane domains from the five subunits. Ligands bind to large extracellular subunits that transmit gating signals to the membrane pore region, which has been observed by e.g. series of crystal structures (Hu et al. 2018) and, at least partially, by MD simulation (Yoluk et al. 2015; Polovinkin et al. 2018; Guros et al. 2020; Damgen and Biggin 2020; Lev and Allen 2020). Several pLGICs show strict requirements for certain membrane lipids (Thompson and Baenziger 2020). Cryo-EM structures in nanodiscs composed of lipids known to disrupt functionality, showed significant structural change between agonist-bound and apo structures in the extracellular domains, but not in the membrane domains (Kumar et al. 2020), thus providing a structural explanation for the lipid dependency. In addition, direct binding has been observed of e.g. phosphatidylglycerol (PG) (2019) and phosphatidylethanolamine (PE) (Henault 2019) lipids and cholesterol (Henin et al. 2014; Brannigan et al. 2008;Zhu et al. 2018; Sharp et al. 2019), that modulates function.
G protein-coupled receptors (GPCRs) induce intracellular signaling in response to external binding of a wide range of stimuli, such as photons and small molecules (Pierce et al. 2002). Ligand binding mediates structural rearrangements in the seven-transmembrane protein that results in G protein dissociation and the intracellular response. Membrane-embedded cholesterol has been identified as critical to GPCR function (Oates and Watts 2011) with proposed effects on ligand binding (Pucadyil and Chattopadhyay 2004), stability (Zocher et al. 2012), and oligomerization (Chakraborty and Chattopadhyay 2015). Advances in GPCR structural biology have generated more than 70 unique receptor structures and signaling complexes (Wang et al. 2020; Congreve et al. 2020), which have resulted in significant insight into protein-lipid interactions by MD simulation approaches. For instance, several cholesterol-binding sites have been identified (Lee and Lyman 2012; Sengupta and Chattopadhyay 2012; Shan et al. 2012), and also have shown that binding alters the conformational state of the receptor (Manna et al. 2016). An observed decrease in the number of hydrogen bonds in the cholesterol-containing protein-lipid interface provides a possible molecular explanation to the enhanced conformational freedom in the presence of cholesterol (Ramirez-Anguita et al. 2018), and the affinities of cholesterol, ganglioside (GM3), and PIP2 lipids were shown to differ between different conformations of GPCR structures (Song et al. 2019). In addition, MD simulations have also shown that the degree of membrane disorder, as affected by lipid composition, facilitate oligomerization and that negatively charged lipids affect activation—but not oligomerization (Marino et al. 2016).
Other examples of membrane proteins with function that depends upon the membrane lipids are ATP-dependent P-type ATPases that transport ions, and also lipids, against their gradients. The Na+,K+ P-type ATPase maintains electrochemical gradients by ATP-dependent transport of three intracellular Na+ ions and two extracellular K+ ions across the membrane and thereby contributes to the membrane resting potential (Nyblom et al. 2013). Several sites of direct lipid-protein interaction have been identified in crystal structures (Cornelius et al. 2015) that have been shown to affect activation, stabilization, and inhibition of transport activity in detergent/lipid micelles (Habeck et al. 2017; Habeck et al. 2015) (Fig. 1c). Such lipid-protein interaction sites have also been identified in crystal structures of Ca2+ P-type ATPase proteins that govern e.g. muscle relaxation (Drachmann et al. 2014). These Ca2+ transporters, or SERCA proteins, have adapted to the thickness of sarcoendoplasmic reticulum membranes (Johannsson et al. 1981) and this adaptation has been assigned to specific steps in the reaction cycle, such as phosphorylation and dephosphorylation (Michelangeli et al. 1991). Due to the unfavorable thermodynamics involved in exposing water to hydrophobic amino acids in the membrane domain of the protein, any mismatching to the surrounding membrane can be expected to be minimized. Both the SERCA protein itself and the inner layer of the surrounding lipids was shown to adapt structurally to different thicknesses of the membrane (Sonntag et al. 2011). Such adaptation has also been shown to induce tilting of the SERCA protein in mixed lipid-detergent micelles (Norimatsu et al. 2017).
The archetypal member of the major facilitator superfamily, the lactose permease of Escherichia coli (LacY), which imports galactopyranoside sugar in a secondary active transport mechanism (Kaback 2015), requires phosphatidylethanolamine (PE) lipids for proper insertion and active transport (Bogdanov et al. 2002). MD simulations identified PE-interaction sites in LacY that were unable to accommodate phosphatidylcholine (PC) lipids, leading to a loss of observed local and global functional dynamics in PC lipid bilayers (Andersson et al. 2012) (Fig. 1d). In addition to such direct lipid-protein interactions, the general physicochemical properties of the membrane also affect LacY structure and function (Bogdanov et al. 2010). While folding, stability, and function of LacY all depend critically on the composition of the lipid bilayer, these properties show significant differences in their lipid dependencies (Findlay and Booth 2017). The LacY transporter is a cardinal example of the complexity of lipid regulation of membrane protein systems. To understand the molecular basis of how lipids can act as allosteric modulators, and how lipid composition can be engineered to curb membrane protein-associated disease, will take persistence, novel methodology, and courage.
The Spatial- Vs. Temporal-Scale Dilemma
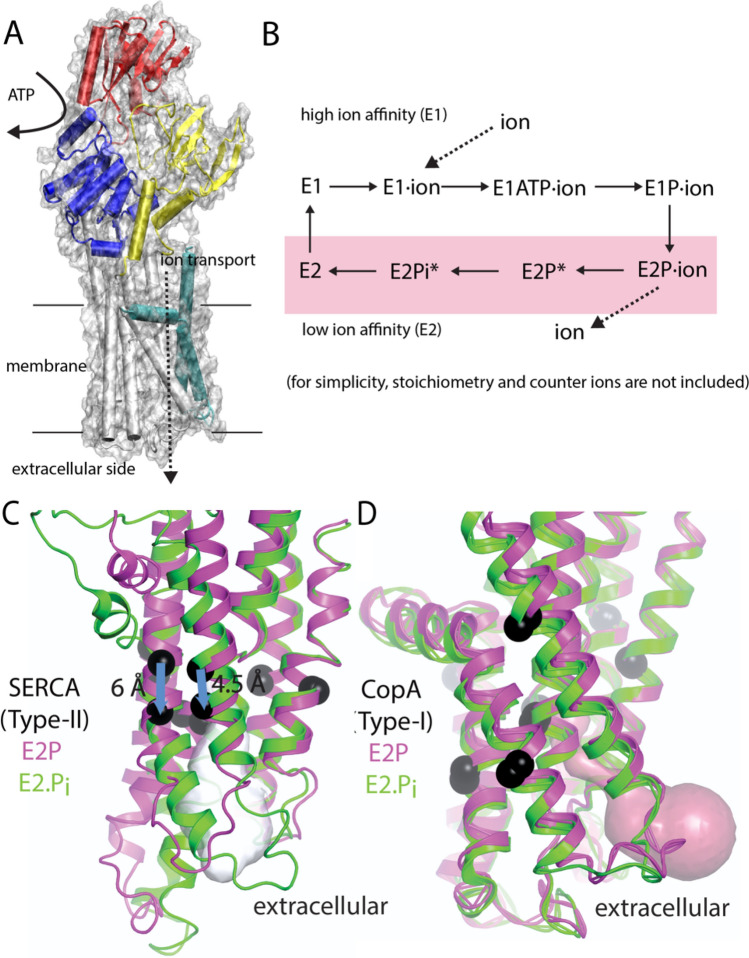
With technical advances and development of novel enhanced sampling algorithms, MD simulation has emerged as a powerful tool for biophysical characterization of complex membrane protein systems (Enkavi et al. 2019). Even though unbiased, atomistic simulations cannot sample the biological time scale, i.e. quite frequently reaction cycles of tens-to-hundreds of milliseconds, certain critical aspects of the membrane protein reactions can be simulated at atomistic detail. For example, the membrane domain of P-type ATPases is attached to large protruding cytoplasmic domains in which the ATP hydrolysis takes place (Fig. 2a). These domains then undergo large-scale conformational changes that drive the transition from so-called E1 states that are open to the cytoplasm, to E2 states with ion-binding sites instead exposed to the extracellular side (Fig. 2b). While crystal structures of the sarcoendoplasmic reticulum calcium ATPase (SERCA) protein, trapped in different modes of action, have contributed enormously to understanding of the transport reaction (Dyla et al. 2019), biophysical characterization, including MD simulation, has added necessary details on the conformational dynamics involved in e.g. activation, inhibition, and regulation (Dyla et al. 2019; Aguayo-Ortiz and Espinoza-Fonseca 2020).
Fig. 2.
P-type ATPase structure, transport reaction, and subtype differences. (a) P-type ATPase architecture, exemplified by a bacterial Cu+-transporting ATPase (CopA), with nucleotide-binding (N), phosphorylation (P), and actuator (A) domains in red, blue, and yellow, respectively. The part of the membrane domain common to all P-type ATPases is shown in white and the CopA-specific helices in cyan. (b) P-type ATPase reaction scheme showing shifts in ion affinity (E1/E2) and accompanying phosphorylation events. Asterisks mark the existing Zn2+ and Cu+ ATPase crystal structures. A side-by-side comparison of the E2P (magenta)-to-E2Pi (green) transition of (c) SERCA and the (d) Cu+ ATPase highlights differences in structural dynamics in the transmembrane domain involved in ion release (black spheres correspond to similar Cα positions in each state). Adapted from Andersson et al. (2014)
Several crystal structures have paved way for a better understanding of P-type ATPases that regulate cellular heavy-metal homeostasis (so-called Type-I ATPases) (Gourdon et al. 2011; Andersson et al. 2014; Wang et al. 2014), but dynamic processes such as entry and release of ions, hydration dynamics, and membrane partitioning remained elusive. Predictions from MD simulations of ion release enabled determination of an ion-release pathway in Cu+-transporting P-type ATPase (CopA) proteins (Andersson et al. 2014). By comparing to the Type-II SERCA transporter, it was clear that copper transport was associated with unique structural changes in the membrane domain (compare Fig. 2c and d). Interestingly, the ion-release mechanism in Zn2+-transporting P-type ATPase (ZntA) proteins, on the other hand, was reminiscent of the prototype SERCA reaction (Wang et al. 2014), which highlights mechanistic variations within the heavy metal-transporting subfamily. Due to the lack of crystal structures trapped in ion-binding states, how heavy-metal ions enter and bind to the internal transport sites remain unknown. A combined modeling-simulation approach predicted membrane partitioning of a protein-associated platform for ion-delivering chaperones and a putative ion-entry mechanism in Cu+-transporting (CopA) P-type ATPases (Gronberg et al. 2016). Hence, atomistic MD simulations can clearly provide critical insight into dynamical processes in membrane proteins that are difficult to assess with experimental methods, but the method is limited by the time scales that can be sampled, which frequently correspond only to a fraction of the full reaction.
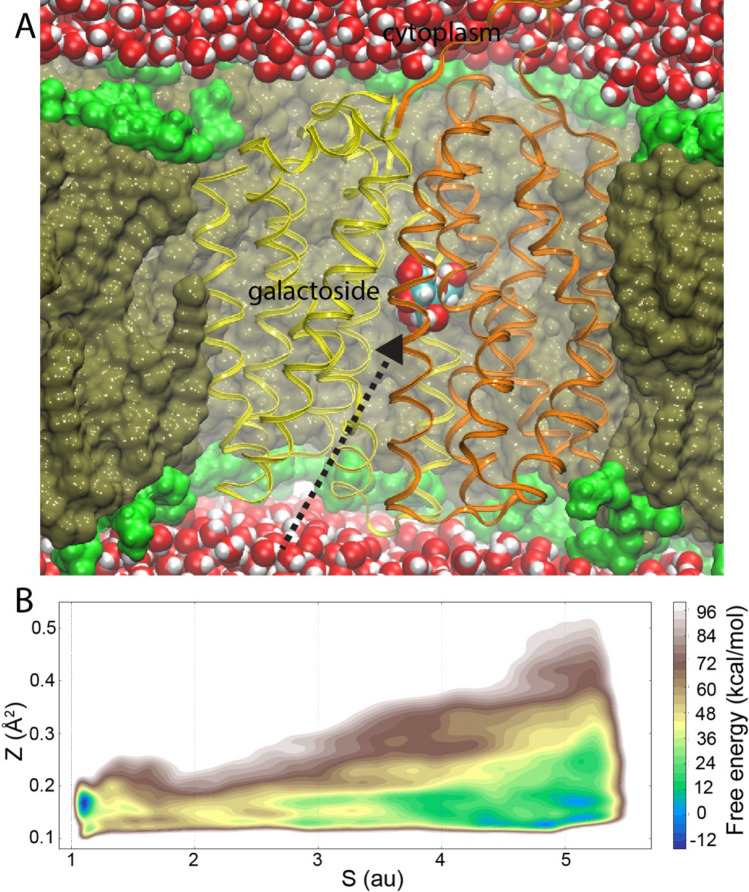
Advancing towards simulations on the biological time scale is a continuous effort. Specialized simulation-dedicated hardware (Shaw et al. 2007; Shaw et al 2014) has enabled tens-to-hundreds microsecond sampling of membrane protein systems. For example, such simulations have provided insight into the energetics of membrane partitioning (Andersson et al. 2013), insertion of outer-membrane proteins (Lundquist et al. 2018), and mechanisms of GPCR receptors (Hollingsworth and Dror 2018), voltage-gated potassium (Jensen et al. 2012) and sodium (Boiteux et al. 2014) channels. Another area of active research is development of enhanced-sampling algorithms. Enhanced sampling methods can enable access to free energy landscapes associated with simulated membrane protein conformational dynamics (Harpole and Delemotte 2018; Howard et al. 2018), and estimate e.g. lipid-binding energetics (Corey et al. 2020). The accelerated weight histogram (AWH) method was used to understand the structural features and thermodynamics underlying ammonia selectivity in aquaporin TIP2;1, a membrane channel permeable to both water and ammonia (Lindahl et al. 2018). A metadynamics approach based on spatial collective variables of a sugar-uptake pathway in the LacY transporter obtained from extended brute-force simulations (Fig. 3a), resulted in a binding free energy that were in excellent agreement with experimental data (Kimanius et al. 2018) (Fig. 3b). Finally, coarse-grained (CG) simulation models enable enhanced sampling by reducing the degrees of freedom in the system, while keeping critical physicochemical properties, although at the cost of atomic resolution (Hedger and Sansom 2016). The CG description has enabled e.g. simulation of multicomponent, asymmetric biological membranes (Marrink et al. 2019) and also containing multiple copies of membrane proteins (Chavent et al. 2016). Despite these impressive advances, simulating membrane proteins in native membranes at the biological time scale of tens-to-hundreds of milliseconds at atomistic detail, does not seem possible within a foreseeable future.
Fig. 3.
Sugar-binding energetics determined by metadynamics-enhanced sampling. a LacY crystal structure open towards the periplasm inserted into a lipid bilayer with hydrophobic core and polar headgroups in brown and green, respectively, with water molecules solvating both sides of the membrane. Extended, unbiased MD simulations identified a sugar-uptake pathway from the periplasm (dashed arrow). b Metadynamics simulations using the location of the uptake pathway (Z and S) as collective variables determined the free energy associated with sugar uptake and binding with excellent agreement to experimental data. Adapted from Kimanius et al. (2018)
From a different perspective, time-resolved X-ray solution scattering (TR-XSS) experiments are ideally suited to provide direct X-ray structural fingerprints on the biological micro-to-milliseconds time scale, albeit at low spatial resolution—in a natural membrane environment. Hence, there is temporal and spatial complementarity between the simulation and TR-XSS methodologies and combining the two should, in principle, allow approaching one of the grand challenges in structural biology, namely to observe proteins operate in single turn-over cycles directly in the natural membrane environment. In TR-XSS experiments, a laser pulse triggers the reaction and intense, synchrotron-generated X-ray pulses monitor the structural changes in the liquid sample (Fig. 4a). The methodology developed from studies tracking the time-dependent formation of transient structural intermediates of photoactive chemicals, such as I2 (Neutze et al. 2001), CH2I2 (Davidsson et al. 2005), and C2H4I2 (Ihee et al. 2005), as well as expansion of the surrounding solvent matrix (Georgiou et al. 2006). Because the heavy atoms are strong X-ray scatterers, relatively subtle structural rearrangements result in prominent difference scattering profiles that extend to the wider angles, i.e. contains structural information of higher resolution. The sensitivity of the TR-XSS method was showcased by resolving high-resolution temporal and spatial differences in the dissociation and recombination following photoactivation of CH2I2 in solvents of varying polarity (Vincent et al. 2009). However, protein large-scale conformational changes involve thousands of atoms and present a very different scenery for TR-XSS visualization. Nevertheless, MD simulations predicted the feasibility of such studies (Andersson et al. 2008).
Fig. 4.
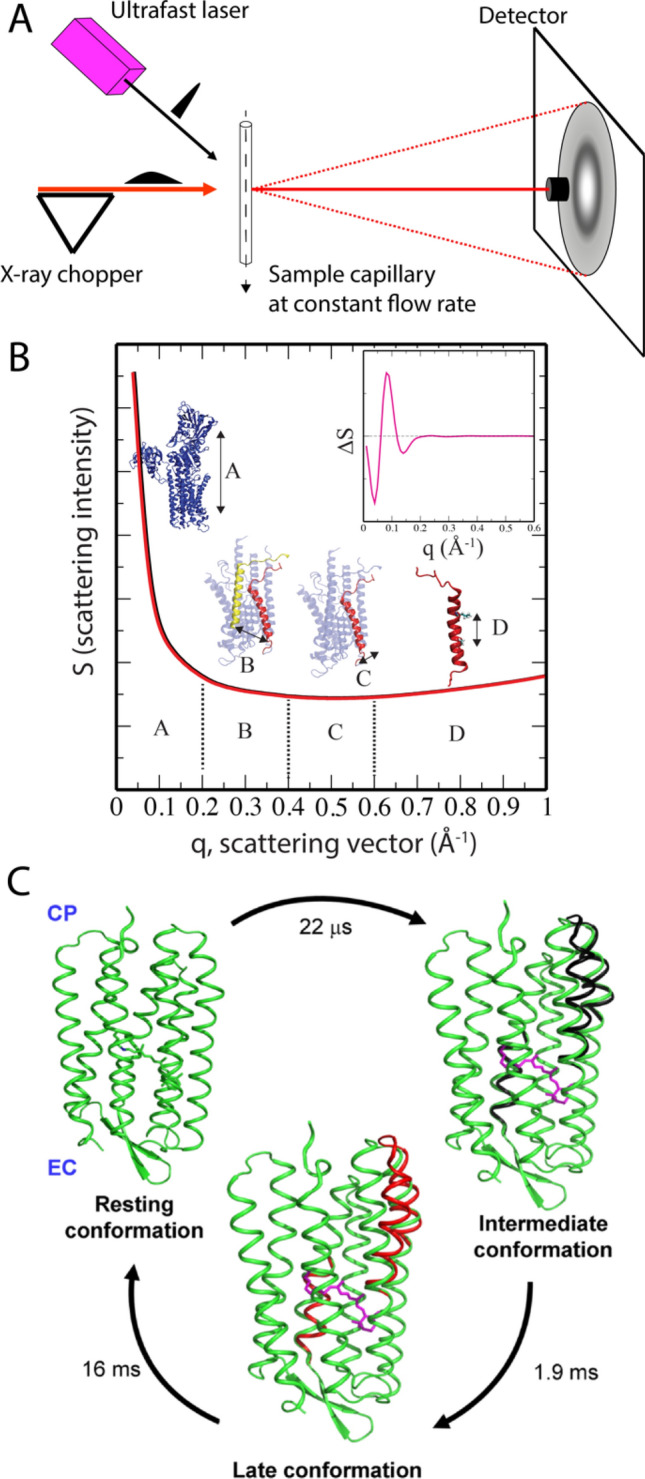
Schematic of the TR-XSS experimental design and resolved membrane protein dynamics. a The pump laser pulse arrives at the sample in the capillary (either static or at a continuous flow rate) before the onset of the X-ray probe pulse, which yields concentric rings on the detector. Subtracting non-activated images from the laser-activated images results in difference images that contain the time-resolved structural data. b Conventional X-ray scattering profiles from two P-type ATPase protein structures (black and red lines), where q = 4π sin(θ)/λ = 4π/2d, where 1/d is the resolution in X-ray crystallography. E.g. q = 0.2 Å−1 corresponds to distances of approximately 30 Å, and q = 1.0 Å−1 to approximately 6 Å. An example of a difference TR-XSS spectrum is shown in the inset. c Bacteriorhodopsin TR-XSS data identified structures and kinetics of two transient intermediates at 22 μs and 1.9 ms with a subsequent relaxation back to the ground state in 16 ms. The structural changes were about twice those observed in crystal structures and revised the bacteriorhodopsin transport mechanism. Adapted from Andersson et al. (2009)
Monitoring Conformational Dynamics in Light-Sensitive Proteins
Conventional, i.e. not resolved in time, small-angle and wide-angle X-ray scattering (SAXS/WAXS) experiments result in data that is a rotational and conformational average of the protein structure, observed as concentric rings on the detector. After radial integration, the structural data are represented as scattering intensity as a function of the scattering vector q, where q acts as an atomic ruler (Fig. 4b). At q < 0.2 Å−1, the global shape and approximate size of the protein is observed. With widening scattering angles, i.e. at higher q, successively more detail is probed in the protein structure: interactions between internal domains (0.2 < q < 0.4 Å−1), interactions between secondary structural elements (0.4 < q < 0.6 Å−1), and between individual side chains (q > 0.6 Å−1). However, because the protein molecules tumble around in the sample, the high-resolution data is inaccessible. In addition, obtaining a unique structural solution of the 1-dimensional scattering spectrum, poses an immense challenge. TR-XSS experiments monitor only the structural differences in the sample, and hence reduce the complexity of the data enormously. Also, the experimental data contain information of the structural changes of all the atoms in the protein molecule, in contrast to traditional spectroscopic methods that register the immediate environment around a certain probe. To obtain the difference scattering profile, a spectrum obtained without laser excitation is subtracted from that registered at a particular point in time after the laser flash. On the micro-to-milliseconds time scale, the obtained difference spectra suffer from low signal-to-noise ratio. Therefore, the TR-XSS experiment needs to be repeated typically hundred to thousands of times, which sets demands on sample size.
Another important prerequisite for a successful TR-XSS experiment is that a large enough population of the species to be studied needs to be activated simultaneously. In this way, synchronized structural dynamics will be performed as the reaction propagates, which directly affects the experimental signal-to-noise. Fast-mixing devices can trigger e.g. protein folding/unfolding (Akiyama et al. 2002), and laser activation can provide an exact trigger—given that the reaction is photosensitive. In the first TR-XSS protein experiment, conformational changes of human hemoglobin were resolved at nanosecond time resolution (Cammarata et al. 2008). Laser-induced release of bound carbon monoxide was used to trigger the reaction, which then was followed from 200 ns to 32 ms. The protein was observed to form the ‘tense’ structure, which is stable in the absence of the ligand, in less than 100 μs. Light-sensitive proteins carry an inherent light trigger and are hence prime candidates for TR-XSS characterization. Such experiments have increased understanding of how bacterial sensor histidine kinases induce signaling networks in bacteria (Takala et al. 2014; Berntsson 2017). In particular, local structural rearrangements were resolved on the μs timescale in the vicinity of the chromophore with a subsequent rotational conformational change occurring within a few milliseconds (Björling 2016).
The first TR-XSS study of a membrane protein focused, not surprisingly, on archaeal rhodopsins including the proton transporter bacteriorhodopsin, well-known for its stability (Andersson et al. 2009). Structural modeling of the time-resolved data involved rigid-body movements based on morphing trajectories extracted from a plethora of available bacteriorhodopsin crystal structures trapped in different states of the photocycle. The resulting structures of transient states at 22 microseconds and 1.9 milliseconds showed that the laser-triggered isomerization of the retinal cofactor induced helical rearrangements that increased over time (Fig. 4c). Helical movements were present already in advance of the so-called primary proton transfer at the retinal, which was in agreement with trapped crystal structure intermediates showing reorganization of the internal H-bond network (Edman et al. 2004; Neutze et al. 2002; Royant et al. 2000). The observed conformational changes in the detergent-micelle solution were about twice the magnitude of those observed in protein crystals, which showcases the restriction posed by crystal lattices. Following the proof-of-principle membrane protein TR-XSS study, conformational changes have also been recorded for proteorhodopsin in detergent-micelles (Malmerberg et al. 2011) and bovine rhodopsin in native rod disc membranes from the bovine retina (Malmerberg et al. 2015).
Indirect Activation of Membrane Protein Dynamics
Transition of the TR-XSS methodology to include membrane proteins not inherently sensitive to light requires finding an indirect means of activating the protein. Caged compounds containing photoremovable groups provide control over release of e.g. ions, neurotransmitter, and ATP molecules (Klan et al. 2013) —and constitutes a possible indirect trigger of protein activity (Fig. 5a). Light-induced ATP release from caged ATP has been used to monitor P-type ATPase activity and time-dependent evolution of structural features with Fourier-Transform Infra-Red (FTIR) spectroscopy (Barth et al. 1996; Ravishankar et al. 2018). Caged ATP activation was also used in early grazing X-ray incidence diffraction studies to trigger synchronization of SERCA reaction cycles and monitoring of intermediate states in partially dehydrated multilayers (Blasie et al. 1985). However, such lamellar diffraction experiments can only resolve profile differences perpendicular to the membrane normal, while solution-scattering studies in principle enable structural determination of 3-dimensional protein envelopes. Nevertheless, the early X-ray studies further showed feasibility for caged ATP triggering of P-type ATPase transport dynamics. Because caged compound activation is an irreversible reaction, the protein solution needs to flow continuously across the focal spots of the laser and X-rays, which sets immense demands on the experimental setup and access to large quantities of the membrane protein.
Fig. 5.
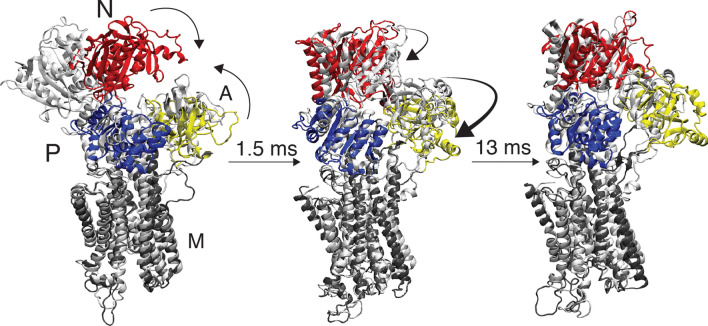
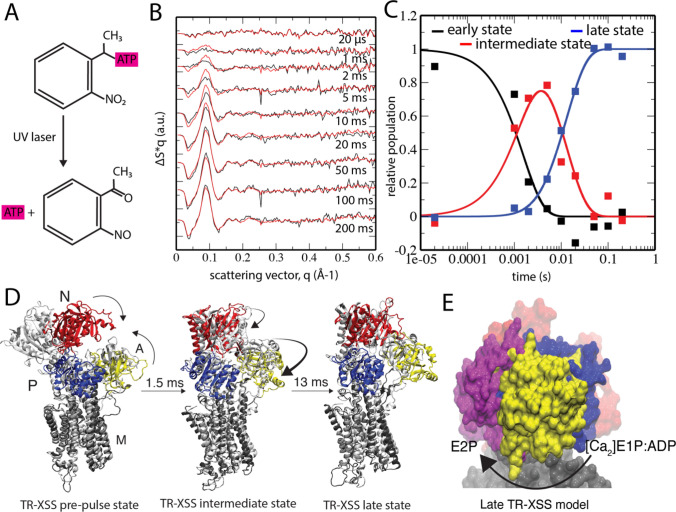
TR-XSS characterization of Ca2+ ATPase (SERCA) kinetics and structural dynamics. a The chemistry of laser-induced release of ATP from caged ATP. b TR-XSS data of SERCA in native membranes (black). The red lines display reconstituted data according to the best-fitting kinetic model. c Temporal shifts in the population densities of the identified transient states; early (black), intermediate (red), and late (blue). d Refined TR-XSS models of prepulse, intermediate, and late states are shown with the closest corresponding crystal structure (white) and rise times from the kinetic analysis. e Structural differences in the A domain between the late TR-XSS model (yellow), the preceding [Ca2]E1P:ADP crystal structure (blue), and the subsequent E2P crystal structure (magenta). The N and M domains are colored red and gray, respectively. The arrow indicates the forward reaction. Adapted from Ravishankar et al. (2020)
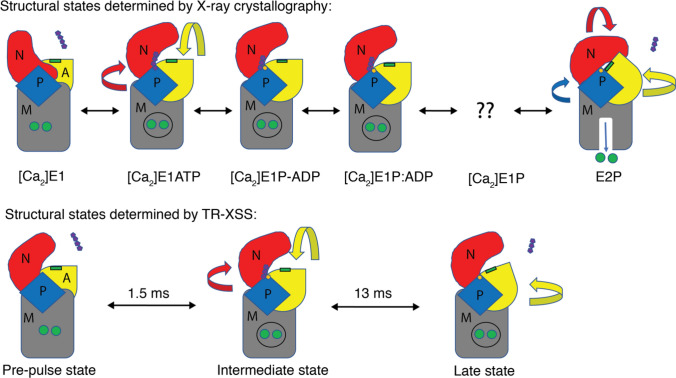
In the first TR-XSS experiment on a non-light sensitive membrane protein, structural dynamics were recorded in sarcoplasmic reticulum (SR) membranes from rabbit skeletal muscle from 20 μs to 200 ms following laser-activation of caged ATP (Ravishankar et al. 2020) (Fig. 5b). The protein content in SR membranes consist to 90% of the SERCA protein (Meissner et al. 1973), which enabled monitoring activity directly in the native membrane. The time-resolved data contained structural fingerprints of two transient intermediate states, at 1.5 ms and 13 ms, respectively (Fig. 5c). Development of a structural refinement protocol based on MD simulation enabled structural interpretation of the identified transient states. In this way, the 1.5-ms intermediate was found to represent a calcium-bound state where the cytoplasmic domains had closed upon the ATP substrate (Fig. 5d). Such domain movements had also been observed in X-ray crystallography (Dyla et al. 2019). In contrast, refinement of the 13-ms intermediate showed a novel arrangement of the cytoplasmic domains (Fig. 5d). In this state, the ADP-binding site was exposed and the so-called actuator (A) domain, which dictates membrane opening and closing, was positioned in-between the principal locations that determine whether the transporter is opened towards the cytoplasm or the SR lumen (Fig. 5e). The existence of such a state had been inferred from biochemical (Danko et al. 2009) and fluorescence microscopy data (Dyla et al. 2017). Hence, the TR-XSS study presented new structural information and kinetics for the decisive “moment of truth” intermediate of SERCA inward-to-outward transport in a native membrane (Fig. 6). This work highlights TR-XSS in combination with MD simulation as a powerful tool to determine structures of transient, high-energy intermediates of membrane proteins in complex lipid settings. The structural refinement relies on existence of high-resolution data, and the TR-XSS methodology should therefore be viewed as a complementary structural-biology technique that is highly timely since it capitalizes on advances in e.g. cryogenic electron microscopy (cryo-EM).
Fig. 6.
Schematic comparison of the principal structural rearrangements in-between crystal structures and TR-XSS models. The pre-pulse state shows reduced opening of the cytoplasmic domains, the intermediate TR-XSS model is similar to a [Ca2]E1ATP state, and displacement of the A domain in the late TR-XSS model exposes the ADP site, but with the N-domain not yet in a E2 position. The ATP and ADP displayed as four and three purple pentagons, the TGES motif is represented by a green rectangle, and phosphorylated aspartic acid in yellow, and the calcium ions are depicted as green circles. Adapted from Ravishankar et al. (2020)
Future Perspective
Identification of structural dynamics of the protein and surrounding membrane is key to understanding membrane protein function. While the TR-XSS methodology holds great promise to contribute to the membrane protein field, the technique is far from standardized and much developmental work is required. For instance, the proof-of-principle TR-XSS characterization of a membrane protein using indirect activation targeted a P-type ATPase protein (Ravishankar et al. 2020). These transporters are exceptional in that the cytoplasmic domains in P-type ATPases protrude significantly from membrane and undergo large-scale conformational change. Therefore, it was possible to model the X-ray scattering data without accounting for dynamics in the membrane. To pave way for similar characterization of less pronounced structural changes in proteins lacking protruding domains, strategies to handle contributions from the surrounding lipids need to be developed. In fact, this presents an opportunity to explore the allosteric nature of membrane lipids. Given the developments within the field of caged-compound chemistry (Klan et al. 2013), electric-field-stimulated protein dynamics in time-resolved crystallography (Hekstra et al. 2016), and synthetic photoswitches (Gorostiza and Isacoff 2008), several possible target proteins can now be envisioned. Also, advances of X-ray free-electron lasers (XFELs) delivering extremely brilliant X-ray pulses have provided access to picosecond structural dynamics, exemplified by myoglobin (Levantino et al. 2015) and a bacterial photosynthetic reaction center (Arnlund 2014). Finally, MD simulations can enable refinement of TR-XSS data. However, such approaches have so-far been case-specific and can potentially suffer from limited sampling, in particular of dynamics of proteins inserted into complex lipid bilayers. While the SERCA MD-based refinement protocol utilized enhanced sampling (Ravishankar et al. 2020), more sophisticated methods have been suggested. For example, energetic restraints can enable driving simulations toward agreement with experimental data using a harmonic biasing potential (Chen and Hub 2015; Björling et al. 2015) or a metadynamics collective variable (Kimanius et al. 2015). Despite the challenges, TR-XSS characterization has emerged as a powerful tool in structural biology to probe protein and lipid dynamics and holds great potential to contribute to understanding of membrane protein functioning and associated disease.
Acknowledgements
Financial support was provided by grants from the Swedish Research Council (2016–03610 and 2020-03840 to MA).
Funding
Open Access funding provided by Umea University.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aguayo-Ortiz R, Espinoza-Fonseca LM (2020) Linking biochemical and structural states of serca: achievements, challenges, and new opportunities. Int J Mol Sci 21 [DOI] [PMC free article] [PubMed]
- Akiyama S, Takahashi S, Kimura T, Ishimori K, Morishima I, Nishikawa Y, Fujisawa T. Conformational landscape of cytochrome c folding studied by microsecond-resolved small-angle x-ray scattering. Proc Natl Acad Sci USA. 2002;99:1329–1334. doi: 10.1073/pnas.012458999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M, Vincent J, van der Spoel D, Davidsson J, Neutze R. A proposed time-resolved X-ray scattering approach to track local and global conformational changes in membrane transport proteins. Structure. 2008;16:21–28. doi: 10.1016/j.str.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Andersson M, Malmerberg E, Westenhoff S, Katona G, Cammarata M, Wohri AB, Johansson LC, Ewald F, Eklund M, Wulff M, Davidsson J, Neutze R. Structural dynamics of light-driven proton pumps. Structure. 2009;17:1265–1275. doi: 10.1016/j.str.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Andersson M, Freites JA, Tobias DJ, White SH. Structural dynamics of the S4 voltage-sensor helix in lipid bilayers lacking phosphate groups. J Phys Chem B. 2011;115:8732–8738. doi: 10.1021/jp2001964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M, Bondar AN, Freites JA, Tobias DJ, Kaback HR, White SH. Proton-coupled dynamics in lactose permease. Structure. 2012;20:1893–1904. doi: 10.1016/j.str.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M, Ulmschneider JP, Ulmschneider MB, White SH. Conformational states of melittin at a bilayer interface. Biophys J. 2013;104:L12–14. doi: 10.1016/j.bpj.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M, Mattle D, Sitsel O, Klymchuk T, Nielsen AM, Moller LB, White SH, Nissen P, Gourdon P. Copper-transporting P-type ATPases use a unique ion-release pathway. Nat Struct Mol Biol. 2014;21:43–48. doi: 10.1038/nsmb.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnlund D, et al. Visualizing a protein quake with time-resolved X-ray scattering at a free-electron laser. Nat Methods. 2014;11:923–926. doi: 10.1038/nmeth.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth A, von Germar F, Kreutz W, Mäntele W. Time-resolved infrared spectroscopy of the Ca2+-ATPase. The enzyme at work. J Biol Chem. 1996;271:30637–30646. doi: 10.1074/jbc.271.48.30637. [DOI] [PubMed] [Google Scholar]
- Berntsson O, et al. Sequential conformational transitions and alpha-helical supercoiling regulate a sensor histidine kinase. Nat Commun. 2017;8:284. doi: 10.1038/s41467-017-00300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björling A, Niebling S, Marcellini M, van der Spoel D, Westenhoff S. Deciphering solution scattering data with experimentally guided molecular dynamics simulations. J Chem Theory Comput. 2015;11:780–787. doi: 10.1021/ct5009735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björling A, et al. Structural photoactivation of a full-length bacterial phytochrome. Sci Adv. 2016;2:e1600920. doi: 10.1126/sciadv.1600920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasie JK, Herbette LG, Pascolini D, Skita V, Pierce DH, Scarpa A. Time-resolved x-ray diffraction studies of the sarcoplasmic reticulum membrane during active transport. Biophys J. 1985;48:9–18. doi: 10.1016/S0006-3495(85)83756-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Heacock PN, Dowhan W. A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J. 2002;21:2107–2116. doi: 10.1093/emboj/21.9.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Heacock P, Guan Z, Dowhan W. Plasticity of lipid-protein interactions in the function and topogenesis of the membrane protein lactose permease from Escherichia coli. Proc Natl Acad Sci USA. 2010;107:15057–15062. doi: 10.1073/pnas.1006286107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux C, Vorobyov I, Allen TW. Ion conduction and conformational flexibility of a bacterial voltage-gated sodium channel. Proc Natl Acad Sci USA. 2014;111:3454–3459. doi: 10.1073/pnas.1320907111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borjesson SI, Hammarstrom S, Elinder F. Lipoelectric modification of ion channel voltage gating by polyunsaturated fatty acids. Biophys J. 2008;95:2242–2253. doi: 10.1529/biophysj.108.130757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannigan G, Henin J, Law R, Eckenhoff R, Klein ML. Embedded cholesterol in the nicotinic acetylcholine receptor. Proc Natl Acad Sci USA. 2008;105:14418–14423. doi: 10.1073/pnas.0803029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarata M, Levantino M, Schotte F, Anfinrud PA, Ewald F, Choi J, Cupane A, Wulff M, Ihee H. Tracking the structural dynamics of proteins in solution using time-resolved wide-angle X-ray scattering. Nat Methods. 2008;5:881–886. doi: 10.1038/nmeth.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty H, Chattopadhyay A. Excitements and challenges in GPCR oligomerization: molecular insight from FRET. ACS Chem Neurosci. 2015;6:199–206. doi: 10.1021/cn500231d. [DOI] [PubMed] [Google Scholar]
- Chavent M, Duncan AL, Sansom MS. Molecular dynamics simulations of membrane proteins and their interactions: from nanoscale to mesoscale. Curr Opin Struct Biol. 2016;40:8–16. doi: 10.1016/j.sbi.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PC, Hub JS. Interpretation of solution x-ray scattering by explicit-solvent molecular dynamics. Biophys J. 2015;108:2573–2584. doi: 10.1016/j.bpj.2015.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang Q, Qiu Y, Li Z, Chen Z, Jiang H, Li Y, Yang H. Migration of PIP2 lipids on voltage-gated potassium channel surface influences channel deactivation. Sci Rep. 2015;5:15079. doi: 10.1038/srep15079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congreve M, de Graaf C, Swain NA, Tate CG. Impact of GPCR structures on drug discovery. Cell. 2020;181:81–91. doi: 10.1016/j.cell.2020.03.003. [DOI] [PubMed] [Google Scholar]
- Corey RA, Stansfeld PJ, Sansom MSP. The energetics of protein-lipid interactions as viewed by molecular simulations. Biochem Soc Trans. 2020;48:25–37. doi: 10.1042/BST20190149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelius F, Habeck M, Kanai R, Toyoshima C, Karlish SJ. General and specific lipid-protein interactions in Na K-ATPase. Biochim Biophys Acta. 2015;1848:1729–1743. doi: 10.1016/j.bbamem.2015.03.012. [DOI] [PubMed] [Google Scholar]
- Cymer F, von Heijne G, White SH. Mechanisms of integral membrane protein insertion and folding. J Mol Biol. 2015;427:999–1022. doi: 10.1016/j.jmb.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgen MA, Biggin PC. A Refined Open State of the Glycine Receptor Obtained via Molecular Dynamics Simulations. Structure. 2020;28(130–139):e132. doi: 10.1016/j.str.2019.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danko S, Daiho T, Yamasaki K, Liu X, Suzuki H. Formation of the stable structural analog of ADP-sensitive phosphoenzyme of Ca2+-ATPase with occluded Ca2+ by beryllium fluoride: structural changes during phosphorylation and isomerization. J Biol Chem. 2009;284:22722–22735. doi: 10.1074/jbc.M109.029702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsson J, Poulsen J, Cammarata M, Georgiou P, Wouts R, Katona G, Jacobson F, Plech A, Wulff M, Nyman G, Neutze R. Structural determination of a transient isomer of CH2I2 by picosecond x-ray diffraction. Phys Rev Lett. 2005;94:245503. [Google Scholar]
- Drachmann ND, Olesen C, Moller JV, Guo Z, Nissen P, Bublitz M. Comparing crystal structures of Ca(2+) -ATPase in the presence of different lipids. FEBS J. 2014;281:4249–4262. doi: 10.1111/febs.12957. [DOI] [PubMed] [Google Scholar]
- Duncan AL, Song W, Sansom MSP. Lipid-dependent regulation of ion channels and g protein-coupled receptors: insights from structures and simulations. Annu Rev Pharmacol Toxicol. 2020;60:31–50. doi: 10.1146/annurev-pharmtox-010919-023411. [DOI] [PubMed] [Google Scholar]
- Dyla M, Terry DS, Kjaergaard M, Sorensen TL, Lauwring Andersen J, Andersen JP, Rohde Knudsen C, Altman RB, Nissen P, Blanchard SC. Dynamics of P-type ATPase transport revealed by single-molecule FRET. Nature. 2017;551:346–351. doi: 10.1038/nature24296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyla M, Kjaergaard M, Poulsen H, Nissen P (2019) Structure and Mechanism of P-Type ATPase Ion pumps. Annu Rev Biochem [DOI] [PubMed]
- Dyla M, Basse Hansen S, Nissen P, Kjaergaard M. Structural dynamics of P-type ATPase ion pumps. Biochem Soc Trans. 2019;47:1247–1257. doi: 10.1042/BST20190124. [DOI] [PubMed] [Google Scholar]
- Eckey K, Wrobel E, Strutz-Seebohm N, Pott L, Schmitt N, Seebohm GN. Kv71-phosphatidylinositol 4,5-bisphosphate interaction sites uncovered by charge neutralization scanning. J Biol Chem. 2014;289:22749–22758. doi: 10.1074/jbc.M114.589796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman K, Royant A, Larsson G, Jacobson F, Taylor T, van der Spoel D, Landau EM, Pebay-Peyroula E, Neutze R. Deformation of helix C in the low temperature L-intermediate of bacteriorhodopsin. J Biol Chem. 2004;279:2147–2158. doi: 10.1074/jbc.M300709200. [DOI] [PubMed] [Google Scholar]
- Enkavi G, Javanainen M, Kulig W, Rog T, Vattulainen I. Multiscale simulations of biological membranes: the challenge to understand biological phenomena in a living substance. Chem Rev. 2019;119:5607–5774. doi: 10.1021/acs.chemrev.8b00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay HE, Booth PJ. The folding, stability and function of lactose permease differ in their dependence on bilayer lipid composition. Sci Rep. 2017;7:13056. doi: 10.1038/s41598-017-13290-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freites JA, Tobias DJ, von Heijne G, White SH. Interface connections of a transmembrane voltage sensor. Proc Natl Acad Sci U S A. 2005;102:15059–15064. doi: 10.1073/pnas.0507618102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou P, Vincent J, Andersson M, Wohri AB, Gourdon P, Poulsen J, Davidsson J, Neutze R. Picosecond calorimetry: time-resolved x-ray diffraction studies of liquid CH2Cl2. J Chem Phys. 2006;124:234507. doi: 10.1063/1.2205365. [DOI] [PubMed] [Google Scholar]
- Gorostiza P, Isacoff EY. Optical switches for remote and noninvasive control of cell signaling. Science. 2008;322:395–399. doi: 10.1126/science.1166022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourdon P, Liu XY, Skjorringe T, Morth JP, Moller LB, Pedersen BP, Nissen P. Crystal structure of a copper-transporting PIB-type ATPase. Nature. 2011;475:59–64. doi: 10.1038/nature10191. [DOI] [PubMed] [Google Scholar]
- Gronberg C, Sitsel O, Lindahl E, Gourdon P, Andersson M. Membrane anchoring and ion-entry dynamics in P-type ATPase copper transport. Biophys J. 2016;111:2417–2429. doi: 10.1016/j.bpj.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guros NB, Balijepalli A, Klauda JB. Microsecond-timescale simulations suggest 5-HT-mediated preactivation of the 5-HT3A serotonin receptor. Proc Natl Acad Sci USA. 2020;117:405–414. doi: 10.1073/pnas.1908848117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habeck M, Haviv H, Katz A, Kapri-Pardes E, Ayciriex S, Shevchenko A, Ogawa H, Toyoshima C, Karlish SJ. Stimulation, inhibition, or stabilization of Na, K-ATPase caused by specific lipid interactions at distinct sites. J Biol Chem. 2015;290:4829–4842. doi: 10.1074/jbc.M114.611384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habeck M, Kapri-Pardes E, Sharon M, Karlish SJ. Specific phospholipid binding to Na, K-ATPase at two distinct sites. Proc Natl Acad Sci USA. 2017;114:2904–2909. doi: 10.1073/pnas.1620799114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama T, Riezman H. Understanding the diversity of membrane lipid composition. Nat Rev Mol Cell Biol. 2018;19:281–296. doi: 10.1038/nrm.2017.138. [DOI] [PubMed] [Google Scholar]
- Harpole TJ, Delemotte L. Conformational landscapes of membrane proteins delineated by enhanced sampling molecular dynamics simulations. Biochim Biophys Acta Biomembr. 2018;1860:909–926. doi: 10.1016/j.bbamem.2017.10.033. [DOI] [PubMed] [Google Scholar]
- Hedger G, Sansom MSP. Lipid interaction sites on channels, transporters and receptors: recent insights from molecular dynamics simulations. Biochim Biophys Acta. 2016;1858:2390–2400. doi: 10.1016/j.bbamem.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekstra DR, White KI, Socolich MA, Henning RW, Srajer V, Ranganathan R. Electric-field-stimulated protein mechanics. Nature. 2016;540:400–405. doi: 10.1038/nature20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henault CM, et al. A lipid site shapes the agonist response of a pentameric ligand-gated ion channel. Nat Chem Biol. 2019;15:1156–1164. doi: 10.1038/s41589-019-0369-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henin J, Salari R, Murlidaran S, Brannigan G. A predicted binding site for cholesterol on the GABAA receptor. Biophys J. 2014;106:1938–1949. doi: 10.1016/j.bpj.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth SA, Dror RO. Molecular dynamics simulation for all. Neuron. 2018;99:1129–1143. doi: 10.1016/j.neuron.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard RJ, Carnevale V, Delemotte L, Hellmich UA, Rothberg BS. Permeating disciplines: overcoming barriers between molecular simulations and classical structure-function approaches in biological ion transport. Biochim Biophys Acta Biomembr. 2018;1860:927–942. doi: 10.1016/j.bbamem.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Ataka K, Menny A, Fourati Z, Sauguet L, Corringer PJ, Koehl P, Heberle J, Delarue M. Electrostatics, proton sensor, and networks governing the gating transition in GLIC, a proton-gated pentameric ion channel. Proc Natl Acad Sci USA. 2018;115:E12172–E12181. doi: 10.1073/pnas.1813378116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CL, Feng S, Hilgemann DW. Direct activation of inward rectifier potassium channels by PIP2 and its stabilization by Gbetagamma. Nature. 1998;391:803–806. doi: 10.1038/35882. [DOI] [PubMed] [Google Scholar]
- Ihee H, Lorenc M, Kim TK, Kong QY, Cammarata M, Lee JH, Bratos S, Wulff M. Ultrafast x-ray diffraction of transient molecular structures in solution. Science. 2005;309:1223–1227. doi: 10.1126/science.1114782. [DOI] [PubMed] [Google Scholar]
- Jensen MO, Jogini V, Borhani DW, Leffler AE, Dror RO, Shaw DE. Mechanism of voltage gating in potassium channels. Science. 2012;336:229–233. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- Johannsson A, Keightley CA, Smith GA, Richards CD, Hesketh TR, Metcalfe JC. The effect of bilayer thickness and n-alkanes on the activity of the (Ca2+ + Mg2+)-dependent ATPase of sarcoplasmic reticulum. J Biol Chem. 1981;256:1643–1650. [PubMed] [Google Scholar]
- Kaback HR. A chemiosmotic mechanism of symport. Proc Natl Acad Sci USA. 2015;112:1259–1264. doi: 10.1073/pnas.1419325112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasimova MA, Tarek M, Shaytan AK, Shaitan KV, Delemotte L. Voltage-gated ion channel modulation by lipids: insights from molecular dynamics simulations. Biochim Biophys Acta. 2014;1838:1322–1331. doi: 10.1016/j.bbamem.2014.01.024. [DOI] [PubMed] [Google Scholar]
- Kasimova MA, Zaydman MA, Cui J, Tarek M. PIP(2)-dependent coupling is prominent in Kv7.1 due to weakened interactions between S4–S5 and S6. Sci Rep. 2015;5:7474. doi: 10.1038/srep07474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RY, Pless SA, Kurata HT. PIP2 mediates functional coupling and pharmacology of neuronal KCNQ channels. Proc Natl Acad Sci USA. 2017;114:E9702–E9711. doi: 10.1073/pnas.1705802114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimanius D, Pettersson I, Schluckebier G, Lindahl E, Andersson M. SAXS-guided metadynamics. J Chem Theory Comput. 2015;11:3491–3498. doi: 10.1021/acs.jctc.5b00299. [DOI] [PubMed] [Google Scholar]
- Kimanius D, Lindahl E, Andersson M. Uptake dynamics in the Lactose permease (LacY) membrane protein transporter. Sci Rep. 2018;8:14324. doi: 10.1038/s41598-018-32624-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klan P, Solomek T, Bochet CG, Blanc A, Givens R, Rubina M, Popik V, Kostikov A, Wirz J. Photoremovable protecting groups in chemistry and biology: reaction mechanisms and efficacy. Chem Rev. 2013;113:119–191. doi: 10.1021/cr300177k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krepkiy D, Mihailescu M, Freites JA, Schow EV, Worcester DL, Gawrisch K, Tobias DJ, White SH, Swartz KJ. Structure and hydration of membranes embedded with voltage-sensing domains. Nature. 2009;462:473–479. doi: 10.1038/nature08542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse M, Hammond GR, Hille B. Regulation of voltage-gated potassium channels by PI(4,5)P2. J Gen Physiol. 2012;140:189–205. doi: 10.1085/jgp.201210806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Wang Y, Zhang Z, Zhao Z, Cymes GD, Tajkhorshid E, Grosman C. Cryo-EM structures of a lipid-sensitive pentameric ligand-gated ion channel embedded in a phosphatidylcholine-only bilayer. Proc Natl Acad Sci USA. 2020;117:1788–1798. doi: 10.1073/pnas.1906823117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AG. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Lee JY, Lyman E. Predictions for cholesterol interaction sites on the A2A adenosine receptor. J Am Chem Soc. 2012;134:16512–16515. doi: 10.1021/ja307532d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev B, Allen TW. Simulating ion channel activation mechanisms using swarms of trajectories. J Comput Chem. 2020;41:387–401. doi: 10.1002/jcc.26102. [DOI] [PubMed] [Google Scholar]
- Levantino M, Schiro G, Lemke HT, Cottone G, Glownia JM, Zhu D, Chollet M, Ihee H, Cupane A, Cammarata M. Ultrafast myoglobin structural dynamics observed with an X-ray free-electron laser. Nat Commun. 2015;6:6772. doi: 10.1038/ncomms7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl V, Gourdon P, Andersson M, Hess B. Permeability and ammonia selectivity in aquaporin TIP2;1: linking structure to function. Sci Rep. 2018;8:2995. doi: 10.1038/s41598-018-21357-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- Lundquist K, Bakelar J, Noinaj N, Gumbart JC. C-terminal kink formation is required for lateral gating in BamA. Proc Natl Acad Sci USA. 2018;115:E7942–E7949. doi: 10.1073/pnas.1722530115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmerberg E, PH MB-G, Katona G, Deupi X, Arnlund D, Wickstrand C, Johansson LC, Westenhoff S, Nazarenko E, Schertler GF, Menzel A, de Grip WJ, Neutze R (2015) Conformational activation of visual rhodopsin in native disc membranes. Sci Signal 8:26 [DOI] [PubMed]
- Malmerberg E, Omran Z, Hub JS, Li X, Katona G, Westenhoff S, Johansson LC, Andersson M, Cammarata M, Wulff M, van der Spoel D, Davidsson J, Specht A, Neutze R. Time-resolved WAXS reveals accelerated conformational changes in iodoretinal-substituted proteorhodopsin. Biophys J. 2011;101:1345–1353. doi: 10.1016/j.bpj.2011.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna M, Niemela M, Tynkkynen J, Javanainen M, Kulig W, Muller DJ, Rog T, Vattulainen I (2016) Mechanism of allosteric regulation of beta2-adrenergic receptor by cholesterol. Elife 5 [DOI] [PMC free article] [PubMed]
- Marino KA, Prada-Gracia D, Provasi D, Filizola M. Impact of lipid composition and receptor conformation on the spatio-temporal organization of mu-opioid receptors in a multi-component plasma membrane model. PLOS Comput Biol. 2016;12:e1005240. doi: 10.1371/journal.pcbi.1005240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrink SJ, Corradi V, Souza PCT, Ingolfsson HI, Tieleman DP, Sansom MSP. Computational modeling of realistic cell membranes. Chem Rev. 2019;119:6184–6226. doi: 10.1021/acs.chemrev.8b00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner G, Conner GE, Fleischer S. Isolation of sarcoplasmic reticulum by zonal centrifugation and purification of Ca2+-pump and Ca2+-binding proteins. Biochim Biophys Acta. 1973;298:246–269. doi: 10.1016/0005-2736(73)90355-6. [DOI] [PubMed] [Google Scholar]
- Michelangeli F, Grimes EA, East JM, Lee AG. Effects of phospholipids on the function of (Ca2(+)-Mg2+)-ATPase. Biochemistry. 1991;30:342–351. doi: 10.1021/bi00216a006. [DOI] [PubMed] [Google Scholar]
- Neutze R, Wouts R, Techert S, Davidsson J, Kocsis M, Kirrander A, Schotte F, Wulff M. Visualizing photochemical dynamics in solution through picosecond x-ray scattering. Phys Rev Lett. 2001;87:195508. doi: 10.1103/PhysRevLett.87.195508. [DOI] [PubMed] [Google Scholar]
- Neutze R, Pebay-Peyroula E, Edman K, Royant A, Navarro J, Landau EM. Bacteriorhodopsin: a high-resolution structural view of vectorial proton transport. Biochim Biophys Acta. 2002;1565:144–167. doi: 10.1016/s0005-2736(02)00566-7. [DOI] [PubMed] [Google Scholar]
- Norimatsu Y, Hasegawa K, Shimizu N, Toyoshima C. Protein-phospholipid interplay revealed with crystals of a calcium pump. Nature. 2017;545:193–198. doi: 10.1038/nature22357. [DOI] [PubMed] [Google Scholar]
- Nyblom M, Poulsen H, Gourdon P, Reinhard L, Andersson M, Lindahl E, Fedosova N, Nissen P. Crystal structure of Na+, K(+)-ATPase in the Na(+)-bound state. Science. 2013;342:123–127. doi: 10.1126/science.1243352. [DOI] [PubMed] [Google Scholar]
- Oates J, Watts A. Uncovering the intimate relationship between lipids, cholesterol and GPCR activation. Curr Opin Struct Biol. 2011;21:802–807. doi: 10.1016/j.sbi.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- Plested AJ. Structural mechanisms of activation and desensitization in neurotransmitter-gated ion channels. Nat Struct Mol Biol. 2016;23:494–502. doi: 10.1038/nsmb.3214. [DOI] [PubMed] [Google Scholar]
- Polovinkin L, Hassaine G, Perot J, Neumann E, Jensen AA, Lefebvre SN, Corringer PJ, Neyton J, Chipot C, Dehez F, Schoehn G, Nury H. Conformational transitions of the serotonin 5-HT3 receptor. Nature. 2018;563:275–279. doi: 10.1038/s41586-018-0672-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucadyil TJ, Chattopadhyay A. Cholesterol modulates ligand binding and G-protein coupling to serotonin(1A) receptors from bovine hippocampus. Biochim Biophys Acta. 2004;1663:188–200. doi: 10.1016/j.bbamem.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Ramirez-Anguita JM, Rodriguez-Espigares I, Guixa-Gonzalez R, Bruno A, Torrens-Fontanals M, Varela-Rial A, Selent J. Membrane cholesterol effect on the 5-HT2A receptor: Insights into the lipid-induced modulation of an antipsychotic drug target. Biotechnol Appl Biochem. 2018;65:29–37. doi: 10.1002/bab.1608. [DOI] [PubMed] [Google Scholar]
- Ramu Y, Xu Y, Lu Z. Enzymatic activation of voltage-gated potassium channels. Nature. 2006;442:696–699. doi: 10.1038/nature04880. [DOI] [PubMed] [Google Scholar]
- Ravishankar H, Barth A, Andersson M (2018) Probing the activity of a recombinant Zn(2+) -transporting P-type ATPase. Biopolymers 109 [DOI] [PubMed]
- Ravishankar H, Pedersen MN, Eklund M, Sitsel A, Li C, Duelli A, Levantino M, Wulff M, Barth A, Olesen C, Nissen P, Andersson M. Tracking Ca(2+) ATPase intermediates in real time by x-ray solution scattering. Sci Adv. 2020;6:0981. doi: 10.1126/sciadv.aaz0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Menchaca AA, Adney SK, Tang QY, Meng XY, Rosenhouse-Dantsker A, Cui M, Logothetis DE. PIP2 controls voltage-sensor movement and pore opening of Kv channels through the S4–S5 linker. Proc Natl Acad Sci USA. 2012;109:E2399–2408. doi: 10.1073/pnas.1207901109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royant A, Edman K, Ursby T, Pebay-Peyroula E, Landau EM, Neutze R. Helix deformation is coupled to vectorial proton transport in the photocycle of bacteriorhodopsin. Nature. 2000;406:645–648. doi: 10.1038/35020599. [DOI] [PubMed] [Google Scholar]
- Sands ZA, Sansom MS. How does a voltage sensor interact with a lipid bilayer? Simulations of a potassium channel domain. Structure. 2007;15:235–244. doi: 10.1016/j.str.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Jiang QX, MacKinnon R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature. 2006;444:775–779. doi: 10.1038/nature05416. [DOI] [PubMed] [Google Scholar]
- Sengupta D, Chattopadhyay A. Identification of cholesterol binding sites in the serotonin1A receptor. J Phys Chem B. 2012;116:12991–12996. doi: 10.1021/jp309888u. [DOI] [PubMed] [Google Scholar]
- Shan J, Khelashvili G, Mondal S, Mehler EL, Weinstein H. Ligand-dependent conformations and dynamics of the serotonin 5-HT(2A) receptor determine its activation and membrane-driven oligomerization properties. PLOS Comput Biol. 2012;8:e1002473. doi: 10.1371/journal.pcbi.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp L, Salari R, Brannigan G. Boundary lipids of the nicotinic acetylcholine receptor: Spontaneous partitioning via coarse-grained molecular dynamics simulation. Biochim Biophys Acta Biomembr. 2019;1861:887–896. doi: 10.1016/j.bbamem.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw D, et al (2007) Anton, a special-purpose machine for molecular dynamics simulation. Proceedings of the 34th Annual International Symposium on Computer Architecture 1-12
- Shaw DE et al (2014) SC '14: proceedings of the international conference for high performance computing, networking, storage and analysis. pp 41-53. 10.1109/SC.2014.9
- Simons K, Sampaio JL. Membrane organization and lipid rafts. CSH Perspect Biol. 2011;3:a004697. doi: 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Yen HY, Robinson CV, Sansom MSP. State-dependent Lipid interactions with the A2a receptor revealed by MD simulations using in vivo-mimetic membranes. Structure. 2019;27(392–403):e393. doi: 10.1016/j.str.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag Y, Musgaard M, Olesen C, Schiott B, Moller JV, Nissen P, Thogersen L. Mutual adaptation of a membrane protein and its lipid bilayer during conformational changes. Nat Commun. 2011;2:304. doi: 10.1038/ncomms1307. [DOI] [PubMed] [Google Scholar]
- Swartz KJ. Sensing voltage across lipid membranes. Nature. 2008;456:891–897. doi: 10.1038/nature07620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala H, Bjorling A, Berntsson O, Lehtivuori H, Niebling S, Hoernke M, Kosheleva I, Henning R, Menzel A, Ihalainen JA, Westenhoff S. Signal amplification and transduction in phytochrome photosensors. Nature. 2014;509:245–248. doi: 10.1038/nature13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MJ, Baenziger JE. Structural basis for the modulation of pentameric ligand-gated ion channel function by lipids. Biochim Biophys Acta Biomembr. 2020;1862:183304. doi: 10.1016/j.bbamem.2020.183304. [DOI] [PubMed] [Google Scholar]
- Tong A, Petroff JT, 2nd, Hsu FF, Schmidpeter PA, Nimigean CM, Sharp L, Brannigan G, Cheng WW (2019) Direct binding of phosphatidylglycerol at specific sites modulates desensitization of a ligand-gated ion channel. Elife 8 [DOI] [PMC free article] [PubMed]
- Vincent J, Andersson M, Eklund M, Wohri AB, Odelius M, Malmerberg E, Kong Q, Wulff M, Neutze R, Davidsson J. Solvent dependent structural perturbations of chemical reaction intermediates visualized by time-resolved x-ray diffraction. J Chem Phys. 2009;130:154502. doi: 10.1063/1.3111401. [DOI] [PubMed] [Google Scholar]
- Wang K, Sitsel O, Meloni G, Autzen HE, Andersson M, Klymchuk T, Nielsen AM, Rees DC, Nissen P, Gourdon P. Structure and mechanism of Zn2+-transporting P-type ATPases. Nature. 2014;514:518–522. doi: 10.1038/nature13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hua T, Liu ZJ. Structural features of activated GPCR signaling complexes. Curr Opin Struct Biol. 2020;63:82–89. doi: 10.1016/j.sbi.2020.04.008. [DOI] [PubMed] [Google Scholar]
- Wiener MC, White SH. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of x-ray and neutron diffraction data III. Complete structure. Biophys J. 1992;61:434–447. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XP, Erichsen D, Borjesson SI, Dahlin M, Amark P, Elinder F. Polyunsaturated fatty acids and cerebrospinal fluid from children on the ketogenic diet open a voltage-gated K channel: a putative mechanism of antiseizure action. Epilepsy Res. 2008;80:57–66. doi: 10.1016/j.eplepsyres.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Xu Y, Ramu Y, Lu Z. Removal of phospho-head groups of membrane lipids immobilizes voltage sensors of K+ channels. Nature. 2008;451:826–829. doi: 10.1038/nature06618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdi S, Stein M, Elinder F, Andersson M, Lindahl E. The Molecular Basis of Polyunsaturated Fatty Acid Interactions with the Shaker Voltage-Gated Potassium Channel. PLOS Comput Biol. 2016;12:e1004704. doi: 10.1371/journal.pcbi.1004704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoluk O, Lindahl E, Andersson M. Conformational gating dynamics in the GluCl anion-selective chloride channel. ACS Chem Neurosci. 2015;6:1459–1467. doi: 10.1021/acschemneuro.5b00111. [DOI] [PubMed] [Google Scholar]
- Zaydman MA, Silva JR, Delaloye K, Li Y, Liang H, Larsson HP, Shi J, Cui J. Kv7.1 ion channels require a lipid to couple voltage sensing to pore opening. Proc Natl Acad Sci USA. 2013;110:13180–13185. doi: 10.1073/pnas.1305167110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhou P, Chen Z, Li M, Jiang H, Gao Z, Yang H. Dynamic PIP2 interactions with voltage sensor elements contribute to KCNQ2 channel gating. Proc Natl Acad Sci USA. 2013;110:20093–20098. doi: 10.1073/pnas.1312483110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yu H, Gu M, Nan FJ, Gao Z, Li M. Phosphatidylinositol 4,5-bisphosphate alters pharmacological selectivity for epilepsy-causing KCNQ potassium channels. Proc Natl Acad Sci USA. 2013;110:8726–8731. doi: 10.1073/pnas.1302167110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Noviello CM, Teng J, Walsh RM, Jr, Kim JJ, Hibbs RE. Structure of a human synaptic GABAA receptor. Nature. 2018;559:67–72. doi: 10.1038/s41586-018-0255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zocher M, Zhang C, Rasmussen SG, Kobilka BK, Muller DJ. Cholesterol increases kinetic, energetic, and mechanical stability of the human beta2-adrenergic receptor. Proc Natl Acad Sci USA. 2012;109:E3463–3472. doi: 10.1073/pnas.1210373109. [DOI] [PMC free article] [PubMed] [Google Scholar]