Abstract
Objective
To identify OA subtypes based on cartilage transcriptomic data in cartilage tissue and characterize their underlying pathophysiological processes and/or clinically relevant characteristics.
Methods
This study includes n = 66 primary OA patients (41 knees and 25 hips), who underwent a joint replacement surgery, from which macroscopically unaffected (preserved, n = 56) and lesioned (n = 45) OA articular cartilage were collected [Research Arthritis and Articular Cartilage (RAAK) study]. Unsupervised hierarchical clustering analysis on preserved cartilage transcriptome followed by clinical data integration was performed. Protein–protein interaction (PPI) followed by pathway enrichment analysis were done for genes significant differentially expressed between subgroups with interactions in the PPI network.
Results
Analysis of preserved samples (n = 56) resulted in two OA subtypes with n = 41 (cluster A) and n = 15 (cluster B) patients. The transcriptomic profile of cluster B cartilage, relative to cluster A (DE-AB genes) showed among others a pronounced upregulation of multiple genes involved in chemokine pathways. Nevertheless, upon investigating the OA pathophysiology in cluster B patients as reflected by differentially expressed genes between preserved and lesioned OA cartilage (DE-OA-B genes), the chemokine genes were significantly downregulated with OA pathophysiology. Upon integrating radiographic OA data, we showed that the OA phenotype among cluster B patients, relative to cluster A, may be characterized by higher joint space narrowing (JSN) scores and low osteophyte (OP) scores.
Conclusion
Based on whole-transcriptome profiling, we identified two robust OA subtypes characterized by unique OA, pathophysiological processes in cartilage as well as a clinical phenotype. We advocate that further characterization, confirmation and clinical data integration is a prerequisite to allow for development of treatments towards personalized care with concurrently more effective treatment response.
Keywords: osteoarthritis, cluster analyses, subtypes, RNA sequencing
Rheumatology key messages
Osteoarthritis subtypes have been identified however poorly characterized.
Whole-transcriptome and clinical data integration characterized the existence of two robust OA patient subgroups.
This study contributes to development of both generic and more personalized OA treatment strategies.
Introduction
OA is the most common degenerative joint disorder affecting over 40% of people >65 years of age [1]. OA is the result of combinatory risk factors, such as genetics, obesity and older age, and mainly affecting the diarthrodial joints (hands, knees and hips). Although several non-drug therapies are available, there is, as of yet, no effective disease-modifying OA treatment [2]. It is generally accepted that lack of insight into diverse underlying OA processes has contributed to this tempered advancement [3]. To identify OA subtypes, cluster analyses have been performed on clinical data [4, 5] radiographic scores or regional quantitative MRI measures of cartilage [6], which indeed discriminated distinct OA phenotypes. Nonetheless, such phenotypes do not necessarily report on the ongoing disease processes in the joint tissues, hence the lack of information on potential targeted treatments.
To discriminate diverse OA pathophysiological processes in cartilage, Soul et al. [7] recently performed a non-negative matrix factorization on RNA sequencing data (RNA-seq) of articular cartilage. Two major subgroups of knee OA patients were identified [7]. These analyses, however, focused on preserved knee OA cartilage only and did not integrate molecular subtypes with clinical data.
In the present study, we used whole-transcriptome RNA-seq data to identify robust OA subgroups, both for knee and for hip OA. Moreover, to identify cluster-specific phenotypes we explored differences in radiographic OA features, joint space narrowing and osteophytosis. Pairwise analysis between preserved and lesioned OA on each cluster highlighted involvement of cluster-specific OA pathways at the molecular level. Together, this study could contribute to the development of both improved generic and more personalized treatment strategies.
Methods
Samples
This study includes a total of n = 66 primary OA patients (41 knees and 25 hips), who underwent a joint replacement surgery, from which macroscopically unaffected (preserved n = 56) and lesioned (n = 45) OA articular cartilage were collected (RAAK study) [8] (Table 1). Ethical approval for the RAAK study was obtained from the medical ethics committee of the Leiden University medical Center (P08.239) and informed consent was obtained from all participants.
Table 1.
Baseline characteristics of RAAK samples with OA phenotypes included in the cluster analyses
| Total (n = 36) | Cluster A (n = 26) | Cluster B (n = 10) | P-valuea | |
|---|---|---|---|---|
| Age (s.d.) | 69.7 (7.8) | 69.7 (8.0) | 69.7 (7.7) | 0.534a |
| Females | 28 (78) | 19 (73) | 9 (90) | 0.492a |
| BMI (s.d.) | 28.2 (2.9) | 28.2 (2.5) | 28.2 (4.0) | 0.518a |
| Knee joints | 25 (69) | 20 (77) | 5 (50) | 0.059a |
| KL score (s.d.) | 2.7 (0.6) | 2.7 (0.7) | 2.7 (0.5) | 0.720a |
| OP score (s.d.)c | 2.7 (2.1) | 2.7 (2.2) | 2.6 (1.7) | 0.931b |
| JSN score (s.d.)c | 4.6 (1.2) | 4.5 (1.2) | 5.0 (1.1) | 0.042b |
P-value by generalized estimation equation (GEE) with cluster-type as dependent variable (Cluster A as reference) and age, sex and joint-type as covariates.
P-value by GEE adjusted for age, sex and KL score as covariates.
The OP score or JSN score is defined as the sum of osteophyte grades (0–3) or JSN grades (0–3), respectively determined at the lateral and medial compartments of the tibio-femoral knee joint or at the inferior and superior compartments of the acetabular-femoral hip joint. JSN: joint space narrowing; OP: osteophyte; KL: Kellgren and Lawrence score.
Clinical data
Pre-operative BMI, age, sex and plain radiographs of the relevant hip or knee were present for n = 36 RAAK subjects included in the RNA-seq and n = 22 RAAK subjects for which expression from microarray data were available [8]. Radiographs scores (Kellgren and Lawrence, osteophytes and joint space narrowing (JSN)), were performed by two experienced readers (J.M., K.H.) to determine OA characteristics prior and independent of cluster analysis. More details are described in Supplementary Materials, available at Rheumatology online.
RNA-seq
Total RNA from cartilage was isolated using Qiagen RNeasy Mini Kit (Qiagen, GmbH, Hilden, Germany). Paired-end 2 × 100 bp RNA sequencing (Illumina TruSeq RNA Library Prep Kit, Illumina HiSeq2000 and Illumina HiSeq4000) was performed. Strand-specific RNA-seq libraries were generated, which yielded a mean of 20 million reads per sample. See Supplementary Materials, available at Rheumatology online for detailed description for alignment, mapping and normalization. Quality control (QC) was performed as previously described [9]. In short, after sequencing QC, principal component analysis (PCA) was applied to remove outliers followed by batch effect correction (details in Supplementary Materials, available at Rheumatology online).
Cluster analysis
To expose subtypes and avoid heterogeneity, we used the preserved cartilage for unsupervised hierarchical clustering. To minimize features (genes), we used a coefficient of variation score [10]. The optimal number of clustering was identified using two approaches: Dynamic Tree Cut [11] and Silhouette width score [12]. To compare similarity between clusters we used the Rand Index, where values ≥0.5 were considered significant [13].
Statistical analyses
Differential expression analysis was performed in two ways: between the different clusters; and between pairs of preserved and lesioned cartilage on each cluster separately. The DESeq2 v1.24 R package [14] was used for normalization and statistical framework. A general linear model assuming negative binomial distribution was applied and age, joint type and gender were used as covariates. For the paired analysis between lesioned and preserved cartilage, we used the paired Wald-test. Genes with False Discovery Rate (FDR) <0.05 were considered significant.
To allow further associations based on OA phenotypes with larger sample size, we integrated microarray data (n = 22) (Illumina HumanHT-12 v3 microarrays) with RNA-seq datasets (n = 36) from preserved and lesioned articular cartilage. A–Z normalization was applied in both data sets, which were further merged to create a unique gene expression dataset with 58 samples with both OA phenotypes, OP score and JSN score (Supplementary Materials, available at Rheumatology online).
Protein–protein interactions and pathway enrichment analysis
Protein–protein interactions (PPI) were performed with STRING v11.0 [15] using the default confidence score >0.4 as a cut-off criterion to evaluate the PPI information. We used four interaction sources: text mining; experiments; curated databases; and co-expression. Moreover, pathway enrichment analysis was performed using Gene Ontology (GO) for biological process and molecular function in all imputed genes with interactions on the PPI network. Pathways with FDR <0.05 were consider significant.
Results
To identify subgroups of OA patients, we integrated whole-transcriptome clustering analysis with clinical data in a step-wise approach (Fig. 1). Hereto RNA-seq dataset from OA articular cartilage samples (56 preserved and 45 lesioned, Supplementary Table S1, available at Rheumatology online) was used. To expose subtypes and avoid heterogeneity, we applied unsupervised hierarchical clustering analysis on OA preserved cartilage transcriptome (n = 56). In order to optimize discriminative power of the RNA-seq data in the cluster analysis, we first determined the CV of the expressed genes and ranked according to highest degree of variation. Subsequently, by comparing the distribution and cluster performance of OA subgroups using different sets of highest ranked genes (Supplementary Fig. S1, available at Rheumatology online), we proceed clustering analyses with the top 1000 genes (Supplementary Table S2, available at Rheumatology online).
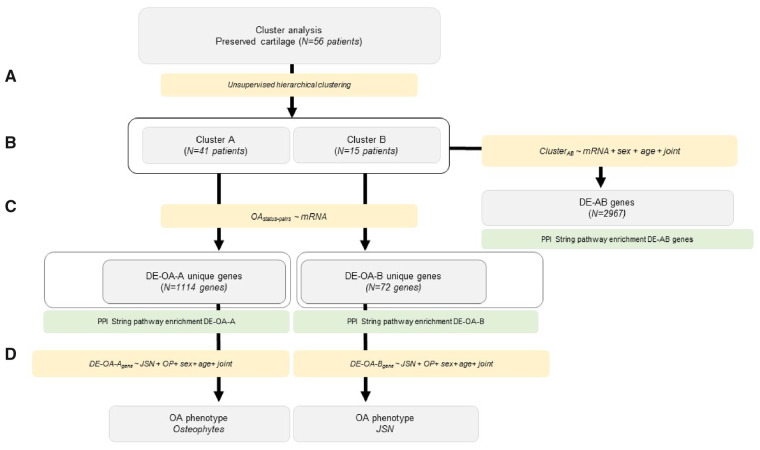
Fig. 1.
Step-wise approach integrating whole-transcriptome clustering analysis with clinical data
(A) Unsupervised hierarchical clustering of the OA preserved cartilage. (B) Differential expression analysis between cluster A and B. (C) PPI and pathway enrichment analysis with differentially expressed genes unique for each cluster. (D) Integration with clinical data.
Unsupervised hierarchical clustering of cartilage samples
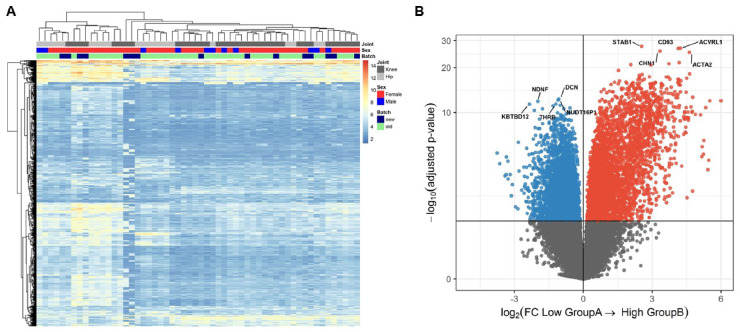
Hierarchical clustering analyses of n = 56 preserved cartilage samples (Fig. 1A) was applied and both the dynamic tree cut and the silhouette width score methods identified two subgroups of respectively, n = 41 (cluster A) and n = 15 (cluster B) OA patients (Fig. 2A). To verify whether our identified clusters were robust to clustering method, we applied the non-negative matrix factorization method adapted from Soul et al. [7] to the RAAK study dataset (Supplementary Materials, available at Rheumatology online). We identified significant comparable clusters in both methods (Rand Index = 0.50).
Fig. 2.
Unsupervised hierarchical clustering of the OA preserved cartilage and volcano plot
(A) Heatmap displaying subgroup A and B. (B) Volcano plot. Cluster B (n = 15) is presented as the effector and cluster A (n = 41) as reference. Blue circles represent genes lower expressed and red circles indicate genes higher expressed in subgroup B relative to subgroup A.
Differential expression analysis between cluster A and cluster B
To investigate whether the preserved cartilage in cluster A or cluster B had a specific transcriptomic profile (Fig. 1B), we performed differential expression analyses between cluster A and cluster B with cluster A as reference (DE-AB genes). We identified 2967 DE-AB genes with FDR < 0.05 (Fig. 2B, Supplementary Table S3, available at Rheumatology online). As shown in Fig. 2B, the most significant DE-AB genes were STAB1, encoding Stabilin-1 (FC = 5.8, FDR = 2.7×10−28) and ACVRL1 encoding Activin A Receptor Like Type 1 (FC = 19, FDR = 1.7×10−27).
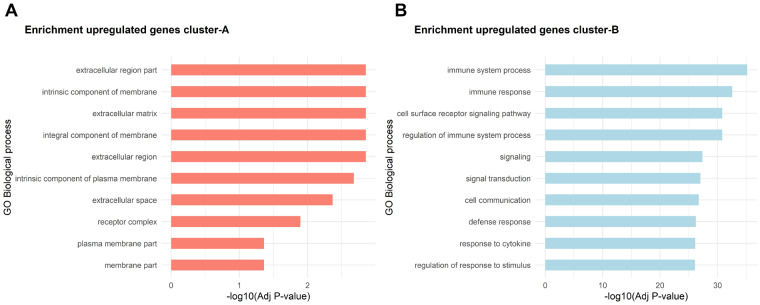
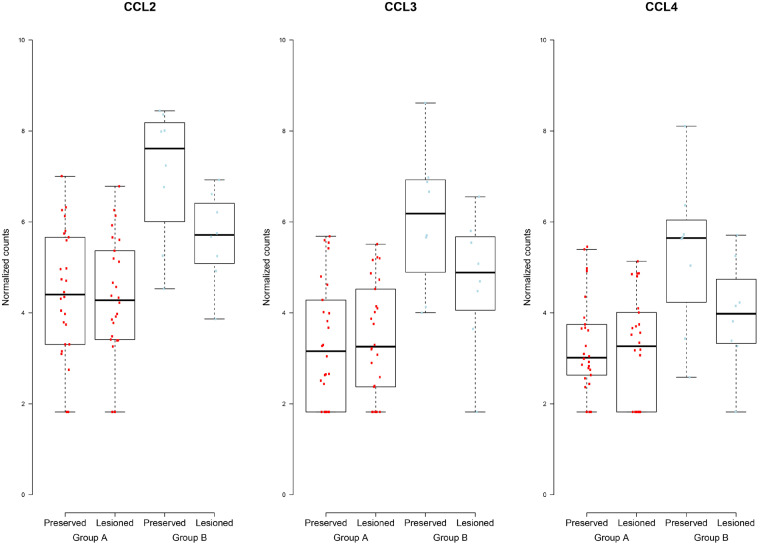
Notably, 66% of DE-AB genes were significantly higher expressed in cluster B relative to cluster A. The DE-AB genes in cluster B relative to cluster A with FC > 5 were enriched for genes involved in immune system process (GO: 0002376, 193 genes, FDR = 6.82×10−36) and cell surface receptor signalling pathway (GO: 0007166, FDR = 1.49×10−31, Fig. 4B, Supplementary Table S4, available at Rheumatology online). Notable is the presence of several chemokine genes that were extremely highly upregulated in cluster B relative to cluster A, such as CCL18 encoding chemokine (C-C motif) ligand 18 (FC = 64, FDR = 9.5×10−13), CXCL1 encoding chemokine (C-X-C motif) ligand 1 (FC = 21, FDR = 1.1×10−10) and CXCL5 encoding chemokine (C-X-C motif) ligand 5 (FC = 38, FDR = 9.3×10−6, see also Fig. 3).
Fig. 4.
Top 10 most significant enriched pathways on each cluster
(A) Bar plot represents significantly enriched GO terms for biological process for genes upregulated in cluster A. (B) Bar plot represents significantly enriched GO terms for biological process with genes upregulated in cluster B.
Fig. 3.
Boxplots showing expression pattern of chemokine activity genes in group A and group B
Conversely, the most significantly upregulated DE-AB genes in cluster A relative to cluster B were DCN encoding decorin (FC = 2.1, FDR = 4.4×10−13), NUDT16P1 encoding nudix hydrolase 16 pseudogene 1 (FC = 2.1, FDR = 7.3×10−13), NDNF encoding neuron derived neurotrophic factor (FC = 4.0, FDR = 1.1×10−12), and THRB encoding thyroid receptor beta (FC = 2.3, FDR = 3.6×10−12). The DE-AB genes in cluster A relative to cluster B with FC >2 in cluster A were enriched for genes involved in extracellular matrix (GO: 0031012, FDR = 1.4×10−3) and integral component of membrane (GO: 0016021, FDR = 1.4×10−3, Fig. 4A, Supplementary Table S5, available at Rheumatology online).
Differential expression (DE) analysis between preserved and lesioned OA cartilage stratified by cluster
To explore the ongoing pathophysiological process in cluster A and cluster B, we performed pairwise DE analyses between preserved and lesioned OA cartilage stratified by cluster (Fig. 1C). These analyses showed n = 3201 significantly DE genes for cluster A (DE-OA-A; Supplementary Fig. S2A and Table S6, available at Rheumatology online), and n = 271 DE genes for cluster B (DE-OA-B; Supplementary Fig. S2B and Table S7, available at Rheumatology online). Of these genes, n = 172 were overlapping in both clusters (Supplementary Fig. S2C and Table S8, available at Rheumatology online).
To explore the biological pathways of these n = 172 overlapping DE genes, we analysed their protein–protein interactions (PPI) using STRING. PPI enrichment showed significantly more interactions between these genes than expected (P<1.0×10−16; Supplementary Fig. S3, available at Rheumatology online), with biologically relevant interplay of genes within GO biological processes such as regulation of response to stimulus (GO: 0018583, FDR = 4.9×10−5), tissue development (GO: 0009888, FDR = 1.6×10−4), and the extracellular region (GO: 0005576, FDR = 9.5×10−12). Moreover, the genes represented in these pathways were previously and consistently reported in OA, such as IL11, FRZB, TNFRSF11B, NGF, WNT16 (Supplementary Table S9, Supplementary Fig. S3, available at Rheumatology online).
Cluster A specific differentially expressed genes with OA pathophysiology (DE-OA-A)
To investigate whether we were able to identify the exclusive pathway in the ongoing pathophysiological process in cluster A patients, we selected for DE-OA-A genes not present among the DE-OA-B genes nor among our previous published DE genes of the overall RNA-seq RAAK dataset (Supplementary Fig. S2C, available at Rheumatology online). This prioritization led to n = 1114 exclusive DE-OA-A genes (Fig. 1C, Supplementary Table S10, available at Rheumatology online).
PPI enrichment analyses of exclusive DE-OA-A genes that are downregulated in lesioned as compared with preserved cartilage (n = 478, P<4.5×10−8, Supplementary Fig. S4, available at Rheumatology online) identified 13 FDR significant GO biological terms and eight GO molecular function terms (Supplementary Table S11, available at Rheumatology online). Of note was enrichment of genes in the chondrocyte hypertrophy pathway (GO-BP: 0003415, FDR = 4.9×10−2) characterized by down regulation of SOX9, RARG and TGFBRP2 and the ion-binding pathway (GO-MF: 0043167; FDR = 2.4×10−2) characterized by genes such as ACACB, COL10A1 (in Supplementary Table S11, available at Rheumatology online).
Conversely, PPI enrichment analyses of exclusive DE-OA-A genes that are upregulated in lesioned as compared with preserved cartilage showed n = 636 highly significantly connected genes (P<1.0×10−16, Supplementary Fig. S5, available at Rheumatology online) in 165 significant GO biological processes such as nucleotide metabolic processes (GO: 0009117, FDR = 1.2×10−7) or cellular component organization – biogenesis (GO: 0071840, FDR = 5.1×10−7, Supplementary Table S12, available at Rheumatology online).
Cluster B specific differentially expressed genes with OA pathophysiology (DE-OA-B)
Similarly to cluster A, we selected for DE-OA-B genes not present among the DE-OA-A genes nor among our previously published DE genes of the overall RNA-seq RAAK dataset (Supplementary Fig. S2C, available at Rheumatology online) and identified n = 72 exclusive DE-OA-B genes (Fig. 1C, Supplementary Table S13, available at Rheumatology online).
PPI enrichment analyses of exclusive DE-OA-B genes that are downregulated in lesioned as compared with preserved cartilage (n = 57) showed highly significant connected genes (P<4.5×10−8) and highlighted 234 FDR significant GO biological terms (Supplementary Table S14 and Fig. S6, available at Rheumatology online) particularly involved in biological immune system process (e.g. GO: 0043567, FDR = 6.1×10−3), among others characterized by downregulation of genes such as HLA-DRB5 and HLA-DPA1. Additionally, notable among the downregulated DE-OA-B genes were genes enriched in the molecular function of chemokine activity (GO: 008009, FDR = 2.7×10−6), characterized by, among others, decreased expression of CCL2, CCL3, CCL4 and CCL4-L1 (Supplementary Table S14, available at Rheumatology online). As shown in Fig. 3, these genes were also among the DE-AB genes significantly upregulated in the preserved OA cartilage in cluster B patients relative to cluster A patients.
Conversely, PPI enrichment on exclusive DE-OA-B genes that were upregulated in lesioned compared with preserved OA cartilage (n = 15) showed eight genes highly significant connected genes (P<3.2×10−5Supplementary Fig. S7, available at Rheumatology online) with enrichment of genes in insulin-like growth factor receptor signalling (GO: 0043567, FDR = 1.0×10−3), characterized by upregulation of IGFBP6, IGFBP5 and BMP2 (Supplementary Table S15, available at Rheumatology online). Another notable upregulated DE-OA-B was DRGX, encoding the dorsal root ganglia homeobox transcription factor. DRGX was highly upregulated in lesioned compared with preserved cartilage in cluster-B (FC = 30, FDR = 4.0×10−4) and is required for the formation of correct projections from nociceptive sensory neurons to the dorsal horn of the spinal cord and normal perception of pain [16].
Characterization of the OA phenotype in cluster A and cluster B patients
To investigate whether patients in cluster A or cluster B could be characterized by specific radiographic OA features (Fig. 1D), pre-operative radiographs were scored for Kellgren and Lawrence scoring (KL score), osteophytes (OPs) and joint space narrowing (JSN). OA phenotypes were available in n = 36 out of n = 56 samples. As shown in Table 1, covariates age, sex and BMI were not significantly different between arthroplasty patients of cluster B relative to cluster A. However, relative to cluster A, we observed more prominent JSN (P=4.0×10−2) in cluster B patients.
To verify our findings, we integrated mRNA expression levels from a previously published microarray mRNA dataset of n = 22 preserved and lesioned cartilage of the RAAK study [8] for which we had also assessed OA phenotypes. Upon merging and normalizing mRNA levels of DE genes of the microarray and RNA-seq (Supplementary methods, available at Rheumatology online), a dataset of 29 DE-AB gene levels in n = 58 patients was established. As shown in Table 2, the majority of the tested DE-AB genes showed significant association between the mRNA expression levels in preserved cartilage and the OA phenotype JSN such as, for example, STAB1 with OR = 1.22 (95% CI 1.01, 1.47, P =0.0350). Among these DE-AB genes was also the NR2F2 showing a highly significant association to higher JSN scores (OR = 1.48, 95%CI 1.26, 1.75, P = 2×10×10−6) in addition to significantly lower OP scores (OR = 0.88, 95%CI 0.81, 0.95, P =1.1×10−3) in cluster B patients relative to cluster A. Moreover, the effect of NR2F2 gene was similar when stratifying for OA status (preserved or lesioned OA cartilage) or for mRNA quantification method (microarray or RNA-seq).
Table 2.
Associations of mRNA expression level of DE cluster A and cluster B genes with OA phenotypes
| Gene | JSN scoreb |
OP scoreb |
||
|---|---|---|---|---|
| OR (95% CI) | P-valuea | OR (95% CI) | P-valuea | |
| NR2F2 | 1.48 (1.26, 1.75) | 0.000002 | 0.88 (0.81, 0.95) | 0.0011 |
| MAP4K4 | 1.37 (1.13, 1.65) | 0.0012 | 1.00 (0.91, 1.1) | 0.9919 |
| TPST2 | 1.30 (1.07, 1.57) | 0.0076 | 0.97 (0.89, 1.06) | 0.4699 |
| HCLS1 | 1.30 (1.05, 1.6) | 0.0155 | 0.92 (0.83, 1.01) | 0.0641 |
| FCER1G | 1.25 (1.03, 1.53) | 0.0276 | 0.91 (0.83, 1.01) | 0.0768 |
| STAB1 | 1.22 (1.01, 1.47) | 0.0350 | 0.92 (0.83, 1.02) | 0.1259 |
| FLRT2 | 1.20 (1.01, 1.43) | 0.0419 | 0.91 (0.83, 1) | 0.0577 |
| CD248 | 1.24 (1.01, 1.53) | 0.0446 | 0.93 (0.85, 1.02) | 0.1043 |
| FYN | 1.19 (0.98, 1.44) | 0.0722 | 0.95 (0.86, 1.05) | 0.3319 |
| TMEM51 | 1.22 (0.98, 1.52) | 0.0729 | 0.92 (0.83, 1.02) | 0.1133 |
| EXT1 | 1.21 (0.98, 1.5) | 0.0757 | 1.00 (0.90, 1.11) | 0.9798 |
| RALA | 1.20 (0.98, 1.47) | 0.0774 | 0.93 (0.85, 1.01) | 0.0675 |
| VWF | 1.22 (0.97, 1.54) | 0.0934 | 0.94 (0.86, 1.03) | 0.1949 |
| CXCL12 | 1.18 (0.97, 1.44) | 0.1059 | 0.94 (0.85, 1.05) | 0.2683 |
| CTSH | 1.15 (0.94, 1.41) | 0.1625 | 0.91 (0.82, 1.00) | 0.0511 |
| SAMHD1 | 1.14 (0.94, 1.38) | 0.1888 | 1.03 (0.92, 1.16) | 0.5804 |
| MMP9 | 1.15 (0.92, 1.43) | 0.2149 | 0.94 (0.85, 1.03) | 0.1728 |
| MMP19 | 1.07 (0.88, 1.3) | 0.4725 | 1.02 (0.92, 1.14) | 0.6726 |
| EMILIN2 | 1.08 (0.86, 1.36) | 0.5190 | 0.94 (0.85, 1.04) | 0.2244 |
| LGALS3BP | 1.05 (0.86, 1.28) | 0.6448 | 1.02 (0.92, 1.13) | 0.7416 |
| PTPN6 | 0.96 (0.78, 1.18) | 0.6890 | 1.00 (0.90, 1.12) | 0.9332 |
| PECAM1 | 1.03 (0.85, 1.26) | 0.7499 | 1.00 (0.91, 1.11) | 0.9475 |
| GIMAP7 | 1.03 (0.84, 1.26) | 0.7564 | 1.02 (0.93, 1.13) | 0.6765 |
| STK17B | 1.03 (0.84, 1.25) | 0.7901 | 1.01 (0.91, 1.12) | 0.8208 |
| FLT4 | 1.03 (0.84, 1.26) | 0.8112 | 1.02 (0.93, 1.13) | 0.6464 |
P-value by generalized estimation equation (GEE) with mRNA expression level in preserved cartilage as dependent variable and age, sex, KL score and joint type as covariates.
The OP score or JSN score is defined as the sum of osteophyte grades (0–3) or JSN grades (0–3), respectively determined at the lateral and medial compartments of the tibio-femoral knee joint or at the inferior and superior compartments of the acetabular-femoral hip joint. JSN: joint space narrowing; OP: osteophyte.
Discussion
By applying hierarchical clustering on transcriptomic RNA-seq dataset of cartilage, two robust OA patient subgroups (A and B) were recognized. The preserved cartilage in cluster A patients, relative to cluster B (DE-AB genes), was enriched for cartilage related pathways, such as extracellular matrix. With respect to preserved cartilage of cluster B, relative to cluster A, we observed pronounced upregulation of genes involved in chemokine activity pathways (Supplementary Table S3, available at Rheumatology online). Nevertheless, upon investigating the OA pathophysiology in cluster B patients (DE-OA-B genes), the chemokine genes were significantly down regulated with OA (Fig. 3). In parallel, the pathophysiological process in cluster A patients (DE-OA-A genes) between preserved and lesioned cartilage was characterized by lower expression of genes involved in ion binding and higher expression of genes involved in small molecular metabolic processes (Supplementary Tables S11 and S12, available at Rheumatology online). Upon integrating radiographic OA data, we showed that among cluster B patients, relative to cluster A, the OA phenotype may be characterized by higher JSN scores and low OP scores or vice versa. This effect was further strengthened and confirmed by compiling our previously generated mRNA datasets (microarray and RNA-seq) of clinically characterized OA patients while using the DE-AB genes in preserved cartilage as a proxy (Table 2). Notably we merged the OP and JSN score of hips and knees, which may not be entirely appropriate given essential differences in these scorings between knee and hip joints. However, prior to the merging OP and JSN sum scores of knee and hip data, we compared the current results to those obtained merging Z scores of OP and JSN of hip and knee, respectively. Because the results appeared almost identical (data not shown) we have provided data of the sum scores as these are easier to interpret.
Among the individual DE-AB genes, we identified STAB1 as the most significant upregulated gene in preserved cartilage of cluster B patients (FC = 5.8, FDR = 2.7×10−28). STAB1 encoding Stabilin-1 is a multifunctional type I transmembrane receptor regulating molecule recycling and cell homeostasis by controlling the intracellular trafficking. Ablation of Stabilin-1 in bone in vivo has been shown to enhance bone resorbing activity in osteoclasts [17]. Moreover, Stabilin-1 has been shown to suppress CCL3 excretion thereby preventing excessive collagen deposition and organ fibrosis [18]. It was subsequently concluded that macrophage Stabilin-1 represents a critical defence against oxidative tissue damage. We found that CCL3 was highly expressed in cluster B relative to cluster A patients while significantly downregulated with OA status (Fig. 3). This could indicate that the expression pattern of STAB1 and CCL3 in cluster B patients reflects a successful defence mechanism against the ongoing OA pathophysiological process. This hypothesis, however, needs to be verified by functional studies, preferably in human in vitro models of OA.
Previously clustering analysis on RNA-seq of knee cartilage was performed by applying the relatively complicated, non-negative matrix factorization method [7]. Here we showed that the unsupervised hierarchical clustering applied to our data showed significant cluster similarity upon applying the network NFM method (Rand Index > 0.5), suggesting that our clusters were identified independent of clustering method. Moreover, we found considerable overlap (45%) in the significant DE genes marking the two clusters including STAB1, NR2F2 and chemokine genes (CCL2, CCL3, CCL4 and CCL4-L1) [7]. This overlap indicates a replication of the existence of two OA subtypes in an independent dataset. Replication of the here-identified associated OA pathophysiological processes and phenotypes is still necessary.
The OA phenotype among patients of cluster B was characterized by pronounced JSN scores. In this respect, two exclusive DE-OA-B genes, DRGX and CCL2 (FC = 30, FDR = 4.0×10−4 and FC = 0.4, FDR = 6.0×10−4, respectively) are worth mentioning. DRGX encoding the dorsal root ganglia homeobox transcription factor is required for normal perception of pain whereas CCL2/CCR2 signalling is involved in neuropathic pain and neuroinflammation [19, 20]. In view of the severe joint space narrowing in cluster B patients at total joint replacement surgery, it is tempting to speculate that the ability to upregulate DRGX and downregulate CCL2 could be particularly relevant to mitigate pain (sensitization) during OA pathology. In parallel, we showed that the expression levels of the DE-AB gene NR2F2 are low in cluster B related to cluster A (FC = 0.2, FDR = 4.1×10−14) and could quantitatively mark OA patients with severe JSN and mild OPs or vice versa. NR2F2 encodes a transcription factor activated by ligands such as high concentrations of 9-cis-retinoic acid and all-trans-retinoic acid, but not by dexamethasone, cortisol or progesterone. NRF2 is activated in response to oxidative stress and induces the expression of its target genes by binding to the antioxidant response element. Oxidative stress has been described as an important factor in OA and its regulation in chondrocytes have been proposed as a potential new target for therapy [21]. The deletion of Nrf2 in a murine post-traumatic model resulted in increased OA severity, while the use of histone deacetylase inhibitors activates Nrf2, repressing IL-1β-induced MMP1, MMP3 and MMP13 gene expression in chondrocytes and in mouse joint tissues [22]. Altogether, this supports that the lower levels of NRF2 gene in cluster B could lead to a more severe form of OA related JSN, as observed in our study.
Finally, we found two HLA genes among the exclusive DE-OA-B genes; HLA-DRB5 and HLA-DPA1, that were significantly downregulated (FC = 0.3, FDR = 1.5×10−2 and FC = 0.3, FDR = 4.5×10−4, respectively). For that matter, HLA-DRB5 has repeatedly been positively associated with RA whereas HLA-DPA1 was recently identified as risk gene in a GWAS on OA phenotypes [23]. In this GWAS it was speculated that the readily available RA drugs could possible function as new candidates to treat OA patients. As these HLA genes are here found to be significantly downregulated in OA lesioned cartilage drug targeting, these genes may likely not have a strong beneficial effect to the ongoing OA pathophysiology. We selected for the exclusive DE-OA-A and DE-OA-B genes (i.e. significantly DE only in either cluster and not significant in the overall dataset) to explore the specific characteristics in the OA pathophysiological process. Nonetheless, due to the relatively large difference in representative patients of cluster A and cluster B, hence, power, we cannot exclude a likely small proportion of false-positive or false-negative selection among these exclusive genes.
Among the most consistently upregulated genes with OA pathophysiology [9] and also reported here, are IL11 and FGF18. Notably, these genes were recently found in large GWAS on OA, also indicating their causal involvement in the OA pathophysiology [23]. Moreover, they were recognized as being targeted by Food and Drug Administration approved drugs [24]. With respect to IL11, the OA risk allele of rs4252548 (missense variant p. Arg112His) acts via lower stability of IL-11, suggesting that the demonstrated upregulation of IL11 expression with OA pathology is a final, yet failing attempt to respond to the OA disease process. Interestingly, recombinant IL-11 molecules are available as therapeutics of thrombocytopenia in cancer patients [25, 26]. As such, this would make IL11 an attractive potential drug for OA patients. Results of the current study suggest, however, that this may be particularly true for cluster A relative to cluster B patients (FC = 19 vs FC = 60, respectively). Similarly, FGF18 (fibroblast growth factor 18) was upregulated in lesioned OA cartilage in both clusters (FC = 1.6 in cluster A and FC = 1.9 in cluster B). FGF18 is a member of the FGF family and participates in several processes, including skeletal growth and development regulation of chondrogenesis, osteogenesis, and bone and mineral homeostasis [27]. Furthermore, intra-articular injections of FGF18 into the knee of OA patients have resulted in a dose-dependent reduction of cartilage loss [28]. More recently, recombinant human FGF18 protein (Sprifermin) is tested for its therapeutic potential as disease-modifying OA drug [29, 30]. As the differential expression of FGF18 showed similar fold change upregulated in lesioned OA cartilage in both clusters, this treatment could affect patients in a similar way.
Taken together, we have here shown the existence of two major OA subtypes independent of joint site. These OA subtypes were characterized by a unique OA, pathophysiological process in cartilage as well as radiographic phenotype. We advocate that further characterization, confirmation, and clinical data integration is a prerequisite to allow for development of treatments towards personalized care with concurrently more effective treatment response.
Supplementary Material
Acknowledgements
We thank all study participants of the RAAK study. The Leiden University Medical Centre have and is supporting the RAAK study and studies are being performed within the scope of the Medical Delta programs Regenerative Medicine 4D: Generating complex tissues with stem cells and printing technology and Improving Mobility with Technology. All authors have made substantial contributions to the completion of this study: Study concept and design: R.C.A., Y.F.M.R., A.M., M.R., I.M.; data analyses: R.C.A., A.M., Y.F.M.R., H.M., J.S.; acquisition of material and data: R.C.A., W.dH., E.H., E.S., N.L., J.M., K.H., R.G.H.H.N., Y.F.M.R., I.M.; preparation of the manuscript: R.C.A., Y.F.M.R., A.M., M.R., I.M.; critical reviewing and approval of the manuscript: all authors.
Funding: This work was supported by Foundation for Research in Rheumatology (FOREUM), Dutch Arthritis Society (DAS-10–1-402), BBMRI-NL complementation project (CP2013-83), Ana Fonds (O2015-27) and the Dutch Scientific Research council NWO/ZonMW VICI scheme (91816631/528).
Disclosure statement: The authors have declared no conflicts of interest.
Supplementary data
Supplementary data are available at Rheumatology online.
References
- 1.GBD 2015 DALYs and HALE Collaborators. Global, regional, and national disability-adjusted life-years (DALYs) for 315 diseases and injuries and healthy life expectancy (HALE), 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016;388:1603–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oo WM, Hunter DJ.. Disease modification in osteoarthritis: are we there yet? Clin Exp Rheumatol 2019;37(Suppl 120):135–40. [PubMed] [Google Scholar]
- 3. Karsdal MA, Michaelis M, Ladel C. et al. Disease-modifying treatments for osteoarthritis (DMOADs) of the knee and hip: lessons learned from failures and opportunities for the future. Osteoarthritis Cartilage 2016;24:2013–21. [DOI] [PubMed] [Google Scholar]
- 4. Knoop J, van der Leeden M, Thorstensson CA. et al. Identification of phenotypes with different clinical outcomes in knee osteoarthritis: data from the Osteoarthritis Initiative. Arthritis Care Res 2011;63:1535–42. [DOI] [PubMed] [Google Scholar]
- 5. Castano-Betancourt MC, Rivadeneira F, Bierma-Zeinstra S. et al. Bone parameters across different types of hip osteoarthritis and their relationship to osteoporotic fracture risk. Arthritis Rheum 2013;65:693–700. [DOI] [PubMed] [Google Scholar]
- 6. Waarsing JH, Bierma-Zeinstra SM, Weinans H.. Distinct subtypes of knee osteoarthritis: data from the osteoarthritis initiative. Rheumatology 2015;54:1650–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Soul J, Dunn SL, Anand S. et al. Stratification of knee osteoarthritis: two major patient subgroups identified by genome-wide expression analysis of articular cartilage. Ann Rheum Dis 2018;77:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ramos YF, den Hollander W, Bovee JV. et al. Genes involved in the osteoarthritis process identified through genome wide expression analysis in articular cartilage; the RAAK study. PLoS One 2014;9:e103056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coutinho de Almeida R, Ramos YFM, Mahfouz A. et al. RNA sequencing data integration reveals an miRNA interactome of osteoarthritis cartilage. Ann Rheum Dis 2019;78:270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li W, Fan M, Xiong M.. SamCluster: an integrated scheme for automatic discovery of sample classes using gene expression profile. Bioinformatics 2003;19:811–7. [DOI] [PubMed] [Google Scholar]
- 11. Langfelder P, Zhang B, Horvath S.. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 2008;24:719–20. [DOI] [PubMed] [Google Scholar]
- 12. Charrad M, Ghazzali N, Boiteau V, Niknafs A.. NbClust: an R package for determining the relevant number of clusters in a data set. Journal of Statistical Software 2014;61:36. [Google Scholar]
- 13. Rand WM. Objective criteria for the evaluation of clustering methods. J Am Stat Assoc 1971;66:846–850. [Google Scholar]
- 14. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Szklarczyk D, Gable AL, Lyon D. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607–d613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Regadas I, Matos MR, Monteiro FA. et al. Several cis-regulatory elements control mRNA stability, translation efficiency, and expression pattern of Prrxl1 (paired related homeobox protein-like 1). J Biol Chem 2013;288:36285–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim SY, Lee EH, Park SY. et al. Ablation of stabilin-1 enhances bone-resorbing activity in osteoclasts in vitro. Calcif Tissue Int 2019;105:205–14. [DOI] [PubMed] [Google Scholar]
- 18. Rantakari P, Patten DA, Valtonen J. et al. Stabilin-1 expression defines a subset of macrophages that mediate tissue homeostasis and prevent fibrosis in chronic liver injury. Proc Natl Acad Sci USA 2016;113:9298–9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ramesh G. Novel therapeutic targets in neuroinflammation and neuropathic pain. Inflamm Cell Signal 2014;1:e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miller RE, Tran PB, Das R. et al. CCR2 chemokine receptor signaling mediates pain in experimental osteoarthritis. Proc Natl Acad Sci USA 2012;109:20602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Poulet B, Beier F.. Targeting oxidative stress to reduce osteoarthritis. Arthritis Res Ther 2016;18:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cai D, Yin S, Yang J, Jiang Q, Cao W.. Histone deacetylase inhibition activates Nrf2 and protects against osteoarthritis. Arthritis Res Ther 2015;17:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Styrkarsdottir U, Lund SH, Thorleifsson G. et al. Meta-analysis of Icelandic and UK data sets identifies missense variants in SMO, IL11, COL11A1 and 13 more new loci associated with osteoarthritis. Nat Genet 2018;50:1681–1687. [DOI] [PubMed] [Google Scholar]
- 24. Tachmazidou I, Hatzikotoulas K, Southam L. et al. Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank data. Nat Genet 2019;51:230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kurzrock R. rhIL-11 for the prevention of dose-limiting chemotherapy-induced thrombocytopenia. Oncology 2000;14(9 Suppl 8):9–11. [PubMed] [Google Scholar]
- 26. Tsimberidou AM, Giles FJ, Khouri I. et al. Low-dose interleukin-11 in patients with bone marrow failure: update of the M. D. Anderson Cancer Center experience. Ann Oncol 2005;16:139–45. [DOI] [PubMed] [Google Scholar]
- 27. Liu Z, Lavine KJ, Hung IH, Ornitz DM.. FGF18 is required for early chondrocyte proliferation, hypertrophy and vascular invasion of the growth plate. Dev Biol 2007;302:80–91. [DOI] [PubMed] [Google Scholar]
- 28. Lohmander LS, Hellot S, Dreher D. et al. Intraarticular sprifermin (recombinant human fibroblast growth factor 18) in knee osteoarthritis: a randomized, double-blind, placebo-controlled trial. Arthritis Rheumatol 2014;66:1820–31. [DOI] [PubMed] [Google Scholar]
- 29. Gigout A, Guehring H, Froemel D. et al. Sprifermin (rhFGF18) enables proliferation of chondrocytes producing a hyaline cartilage matrix. Osteoarthritis Cartilage 2017;25:1858–1867. [DOI] [PubMed] [Google Scholar]
- 30. Reker D, Kjelgaard-Petersen CF, Siebuhr AS. et al. Sprifermin (rhFGF18) modulates extracellular matrix turnover in cartilage explants ex vivo. J Transl Med 2017;15:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.