Abstract
Research question
Is there a risk of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral exposure and potential cross-contamination from follicular fluid, culture media and vitrification solution within the IVF laboratory using strict patient screening and safety measures?
Design
This was a prospective clinical study. All women undergoing transvaginal oocyte retrieval were required to have a negative SARS-CoV-2 RNA test 3–5 days prior to the procedure. Male partners were not tested. All cases used intracytoplasmic sperm injection (ICSI). The first tube of follicular fluid aspirated during oocyte retrieval, drops of media following removal of the embryos on day 5, and vitrification solution after blastocyst cryopreservation were analysed for SARS-CoV-2 RNA.
Results
In total, medium from 61 patients, vitrification solution from 200 patients and follicular fluid from 300 patients was analysed. All samples were negative for SARS-CoV-2 viral RNA.
Conclusions
With stringent safety protocols in place, including testing of women and symptom-based screening of men, the presence of SARS-CoV-2 RNA was not detected in follicular fluid, medium or vitrification solution. This work demonstrates the possibility of implementing a rapid laboratory screening assay for SARS-CoV-2 and has implications for safe laboratory operations, including cryostorage recommendations.
Keywords: Blastocyst, COVID-19, Cryo-storage, Follicular fluid, Oocyte, SARS-CoV-2
Introduction
Recently, the American Society for Reproductive Medicine (ASRM), European Society for Human Reproduction and Embryology (ESHRE) and International Federation of Fertility Societies (IFFS) released a joint statement affirming that reproduction is an essential human right and that infertility is a time-sensitive although treatable disease affecting 10–12% of couples of reproductive age, the treatment of which should be considered essential care (Veiga et al., 2020). Following the nearly global cessation of infertility treatment at the beginning of the coronavirus disease 2019 (COVID-19) pandemic, restarting the practice of ART within the ongoing pandemic environment is largely an uncharted proposition operating under a ‘proceed-with-caution’ approach. Clinicians and embryologists have implemented best practices as recommended by guidelines from their professional societies. However, hard scientific data are scarce because of the novel nature of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and adjustments to safe practices based on new knowledge and updated recommendations must often be made in real time. Some of the strategies adopted by clinics worldwide include patient and staff questionnaires and testing, adoption of telehealth appointments, use of personal protective equipment and elevated cleaning and disinfection of facilities. Unfortunately, in the embryology laboratory, the risk to gametes and embryos is relatively unknown and management strategies are based upon previous guidelines for known viral pathogens (Simopoulou et al., 2020).
Recent evidence of the presence of the receptor and protease necessary for SARS-CoV-2 infection (ACE2 [angiotensin-converting enzyme 2] and TMPRSS2 [transmembrane serine protease 2]) in human oocytes and embryos (Essahib et al., 2020; Rajput et al., 2020) highlights the potential for infection or cross-contamination in the IVF laboratory and the imperative to implement all possible precautions to keep gametes, embryos, staff and patients safe. To minimize risk of virus exposure in the clinic and laboratory, both the ASRM and ESHRE recommend the use of questionnaire-based triage to identify potentially infected patients and staff, and the use of established infection control procedures. However, the two societies diverge in their recommendations for SARS-CoV-2 testing to mitigate risk in the setting of assisted reproduction. These recommendations continue to change over time. Regardless, the use of these tests without understanding their inherent sensitivity, specificity and temporal limitations may lead to inadvertent infection of patients, staff, physicians, gametes and/or embryos (La Marca and Nelson, 2020).
Although infertility treatment has now resumed, there is still much unknown about the novel coronavirus in the context of the IVF laboratory. it is not known if infection of follicular cells, gametes or embryos occurs in vivo or in vitro, and if so whether the virus may be infectious within the laboratory, resulting in cross-contamination of biological samples and/or staff. There remains concern in the embryology community about how the virus may impact laboratory outcomes as well as staff safety (Anifandis et al., 2020; Arav, 2020). Some suggested precautions for the IVF laboratory until more information becomes available specifically include UV disinfection of liquid nitrogen used for the vitrification and warming of gametes and embryos, extensive washing, closed vitrification systems, reducing the use of embryo micromanipulation techniques that breach the zona pellucida, and eliminating the transfer of any embryos without an intact zona pellucida (Pomeroy and Schiewe, 2020). However, it is difficult to ascertain if these precautions are indeed necessary without further scientific knowledge.
Because the risk of SARS-CoV-2 viral exposure and potential cross-contamination within the IVF laboratory remains largely unclear, the objective of this study was to assess the true risk of exposure to SARS-CoV-2 in an active IVF laboratory when strict patient screening procedures are in place.
Materials and methods
Sample collection
This study included SARS-CoV-2 screening of 300 follicular fluid, 200 vitrification solution (VS) and 61 embryo culture medium samples. Samples were collected during the IVF cycle of patients undergoing fertility treatment at the Colorado Center for Reproductive Medicine. Because transvaginal oocyte retrievals were performed under sedation and under the guidance of the centre's anaesthesia group, each female patient undergoing transvaginal oocyte retrieval (TVOR) was required to have a negative SARS-CoV-2 polymerase chain reaction (PCR) test result 3–5 days before the retrieval procedure. This timing ensured adequate time for the results to be reviewed prior to triggering to avoid having to cancel the retrieval. Due to the lack of anaesthesia use, as well as for cost reasons, male partners were examined using a symptom-based screening approach on the day of collection. All cases examined in this study used standard semen preparation of a double layer gradient followed by sperm swim-up for use during intracytoplasmic sperm injection (ICSI).
The first tube of follicular fluid aspirated during TVOR, which may have involved puncture of more than one follicle, was collected for analysis. Collection entailed pipetting up to 20 µl of the follicular fluid aspirate from the dish into a PCR tube using an RNase/DNAase-free tip. Embryos were cultured in a sequential media system, and samples of the medium were collected on day 5 (following the change from cleavage medium to blastocyst medium on day 3). Care was used to avoid drawing up the mineral oil overlay. Vitrification solution samples were collected after blastocyst exposure to serve as the final dilution step before embryos were placed into storage tanks. Immediately after collection, self-inactivating replication incompetent lentivirus particles (catalogue no. SHC003V; Sigma, Germany) containing the single-stranded viral RNA genome were inoculated into each sample as a positive control for viral RNA stability. This study was conducted under IRB, Fertility Labs of Colorado, IRB no. 20142468, 17 December 2020.
RNA isolation and cDNA synthesis
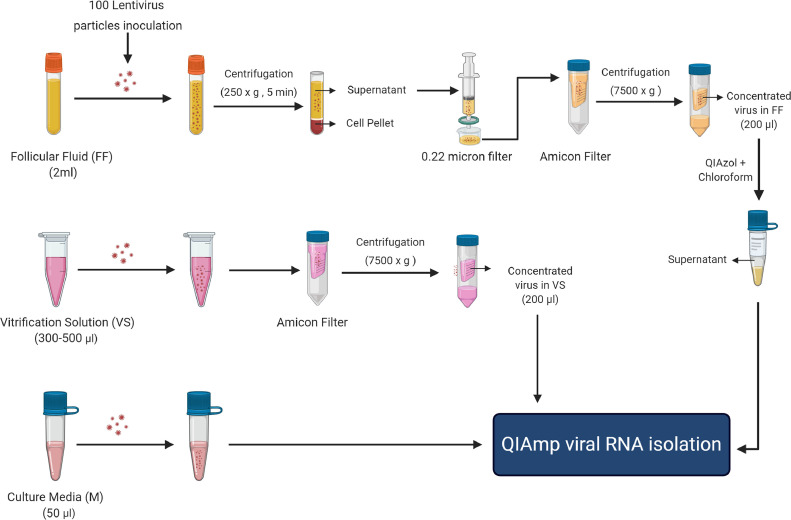
The outline of sample processing before RNA isolation is provided in Figure 1 . Briefly, a total of 100 lentivirus particles were inoculated into approximately 2 ml of follicular fluid, 300–500 µl of vitrification solution and approximately 50 µl of samples of medium collected from patients. After mixing the lentivirus, samples were stored at 4°C and processed for RNA isolation within 5 days of collection. Follicular fluid samples were centrifuged at 250g for 5 min, and the obtained supernatant was passed through a 0.22 µm filter (catalogue no. SLGV033RS; Sigma, Germany) to remove cells and cellular debris. To concentrate the vitrification solution (if it was >200 µl) and filtered follicular fluid (approximately 2 ml), samples were loaded into an Amicon-4 Centrifugal Filter Unit (catalogue no. UFC810024; Sigma) and centrifuged at 4°C in a fixed-angle rotor at 7500g until 200 µl of sample was left in the column. As the concentrated follicular fluid samples were highly viscous due to the high amount of protein present, a protein removal step was performed before RNA isolation. A total of 800 µl of QIAzol (Qiagen, USA) was added to each sample, which was then incubated for 5 min and mixed with 200 µl of chloroform. After centrifugation at 12,000g for 10 min at 4°C, the supernatant (deproteinized follicular fluid) was used for RNA isolation.
Figure 1.
Outline of follicular fluid (FF), vitrification solution (VS) and culture medium (M) sample processing for RNA isolation. After lentivirus inoculation, follicular fluid and vitrification solution samples were centrifuged and passed through a 0.22 µm filter to remove any somatic cells, and then concentrated into a 200 µl volume using an Amicon-4 Centrifugal Filter Unit. As the concentrated follicular fluid samples were highly viscous due to protein enrichment, a protein removal step was performed using QIAzol/chloroform before RNA isolation. Concentrated vitrification solutions and culture media were directly used for RNA isolation.
Samples of approximately 50 µl of medium and approximately 200 µl of concentrated vitrification solution and deproteinized follicular fluid were subjected to RNA isolation using a QIAamp Viral RNA Mini Kit (catalogue no. 52906; Qiagen, USA) following the manufacturer's protocol with some modifications. Briefly, 2 µg of carrier RNA/sample was used for each isolation and RNA was eluted into 16 µl of nuclease-free water. A 260:280 ratio was measured using a NanoDrop One/OneC Microvolume UV-Vis Spectrophotometer (ThermoFisher Scientific, USA). After determining the purity and yield using the NanoDrop, 14 µl of RNA was used for cDNA synthesis using SuperScript IV VILO Master Mix (catalogue no. 11756050; ThermoFisher, USA) on a Applied Biosystems SimpliAmp Thermal Cycler (ThermoFisher Scientific, USA).
Multiplex quantitative PCR
A TaqMan-based multiplex quantitative PCR (qPCR) assay was developed to detect the N1, N2 and ORF1ab loci of the SARS-CoV-2 genome as well as the lentivirus genome (external control) in a single tube reaction. N1 and N2 probes with FAM, ORF1ab with Cy5, and lentivirus probe with SUN labelling were used with their respective primers to prepare the TaqMan assay (TMA) for each locus. Each TMA was tested individually using its specific cDNA template equivalent to 100 copies of lentivirus RNA and SARS-CoV-2 synthetic control RNA in a qPCR. Reaction mixture (20 µl) containing 2X TaqMan (ThermoFisher, USA) Fast Advanced Master Mix (catalogue no. 4444557; ABI), 2 µl of cDNA (100 copies) and 1 µl of TMA was amplified as follows: uracil-N glycosylase incubation at 50°C for 2 min, initial denaturation at 95°C for 2 min and 40 cycles at 95°C for 1 s, 60°C for 20 s. Amplified PCR products were measured based on the fluorescence resulting from TaqMan probe hydrolysis after every cycle. After confirming the specificity for its template, each TMA (N1, N2, ORF1ab and lentivirus) was mixed in equal concentration to develop the multiplex TMA.
To further test the sensitivity and cross-reactivity of the multiplex assay, cDNA equivalent to 100, 20 and 4 copies of the lentivirus and SARS-CoV-2 genome was used to perform the TaqMan-based qPCR as described above. A mixture of SARS-CoV-2 and lentivirus cDNA (equivalent to 100 copies of each) with multiplex TMA was also tested by qPCR to confirm amplification of all the target loci (N1, N2, ORF1ab and lentivirus) in a single tube reaction. All primers and probes used in this assay are provided in Table 1 . The developed multiplex qPCR assay was used to test for the presence of SARS-CoV-2 in all the diagnostic samples. A total of 2 µl of diluted (1:2) cDNA of each diagnostic sample was used to perform the qPCR, along with the positive control (a mixture of SARS-CoV-2 and lentivirus cDNA) and negative control (nuclease-free water) reactions. As lentivirus particles were mixed in each diagnostic sample as an external positive control, samples with no amplification of the lentivirus genome in qPCR were considered to be a false-negative result and were removed from the analysis. Samples with a Ct value above 37 in qPCR amplification in any channel were also considered negative.
Table 1.
Primers used for qRT-PCR
| Target | Accession No / Reference | Primer name | Primer and probe Sequence (5′ → 3′) |
|---|---|---|---|
| N gene | MN908947 | N1 | F: GACCCCAAAATCAGCGAAAT R: TCTGGTTACTGCCAGTTGAATCTG P: FAM-ACCCCGCAT /ZEN/TACGTTTGG TGGACC-3IABkFQ |
| N2 | F: TTACAAACATTGGCCGCAAA R: GCGCGACATTCCGAAGAA P: FAM-ACAATTTGC/ZEN/CCCCAGCGC TTCAG-3IABkF |
||
| ORF1 ab gene | ORF1ab | F: CCCTGTGGGTTTTACACTTAA R: ACGATTGTGCATCAGCTGA R: Cy5- CCG TCT GCG/TAO/GTATGTGGA AAGGT TATGG-3′ IB RQ |
|
| Lentivirus genome | SHC003 (Sigma) | LV | F: TTTCCGTGTCGCCCTTATTC R: CCCAACTGATCTTCAGCATCTT P: SUN/TCACCCAGA/ZEN/AACGCTGGTG AAAGT/3IABkFQ/ |
Results
Validation of TaqMan-based multiplex RT-qPCR assay
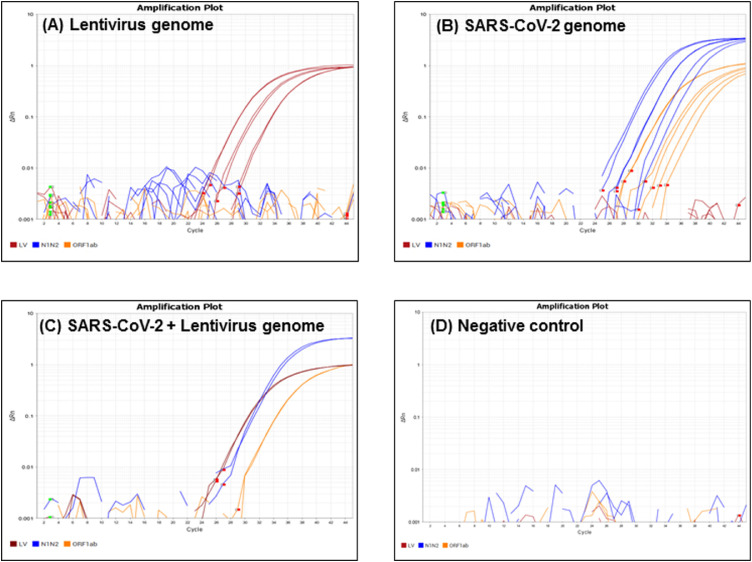
A multiplexed real-time RT-qPCR assay targeting the lentivirus genome (external control) and N1, N2 and ORF1ab loci of the SARS-CoV2 genome was developed and tested with cDNA synthesized from 100, 20 and 4 copies of lentivirus RNA and SARS-CoV-2 synthetic RNA. The results demonstrated specific amplification of SUN-labelled lentivirus TMA (red) with all three concentrations of lentivirus cDNA. In addition, lentivirus cDNA did not show any cross-amplification with SARS-CoV-2 TMA (blue and orange) present in the reaction (Figure 2a ). Similar results were observed with SARS-CoV-2 cDNA. Specific amplification of FAM-labelled N1/N2 (blue) and Cy5-labelled ORF1ab (orange) was detected in qPCR with all three concentrations of SARS-CoV-2 cDNA, but no cross-reactivity was observed with lentivirus TMA (red) present in the reaction (Figure 2b).
Figure 2.
Validation of the multiplex RT-qPCR assay. TaqMan assay containing primers and probes for lentivirus (LV, red), and SARS-CoV-2 (N1N2, blue; ORF1ab, orange) was mixed in an optimized concentration to prepare the multiplex RT quantitative PCR (RT-qPCR) assay. Amplification of 100, 20 and 4 copies of lentivirus genome (A), SARS-CoV-2 genome (B), a mixture of 100 copies of both SARS-CoV-2 and lentivirus genome (C), and the RT-qPCR negative control (D) using multiplex RT-qPCR. Each experiment was repeated three times independently, and the reaction was performed in technical duplicates. Red squares depict the baseline start, and green squares depict the baseline end, of each well.
To test if this assay could successfully amplify all the target loci present in the reaction, a mixture of 100 copies of lentivirus as well as SARS-CoV-2 cDNA was used as a template in a qPCR reaction. A clear amplification of N1/N2, ORF1ab and lentivirus loci was observed (Figure 2c), demonstrating that the multiplex assay can detect both SARS-CoV-2 and lentivirus genomes if they are present in the diagnostic samples. No amplification curve was observed in negative controls that contained all sets of primers and probes with no lentivirus and SARS-CoV-2 genome (Figure 2d), thereby strengthening the validation for primer specificity for the corresponding locus of the genome of interest. These results indicate that N1, N2, ORF1ab and lentivirus primers and their respective probes can be successfully multiplexed in a TaqMan-based RT-qPCR assay.
SARS-CoV-2 screening of follicular fluid, culture medium and vitrification solution using a novel multiplex RT-qPCR assay
Following validation, the TaqMan-based RT-qPCR assay was used to screen SARS-CoV-2 in 300 follicular fluid, 61 medium and 200 vitrification solution samples. These fluids were selected based on the following: follicular fluid was examined to determine if the oocytes might be exposed to the virus in the laboratory, either directly from follicular fluid or via mixture with trace amounts of blood encountered during the normal retrieval process. Samples of medium were examined because this is where the resulting embryos spend the majority of their time and also to determine if exposure to processed spermatozoa from untested males during ICSI or dilution through embryo rinsing/culture might impact the presence or absence of the virus when in the extended presence of the developing embryo. Finally, vitrification solution was examined because it is the final step in the dilution process before embryos are placed into liquid nitrogen tanks and may provide insight into the possible risk of cross-contamination between frozen samples.
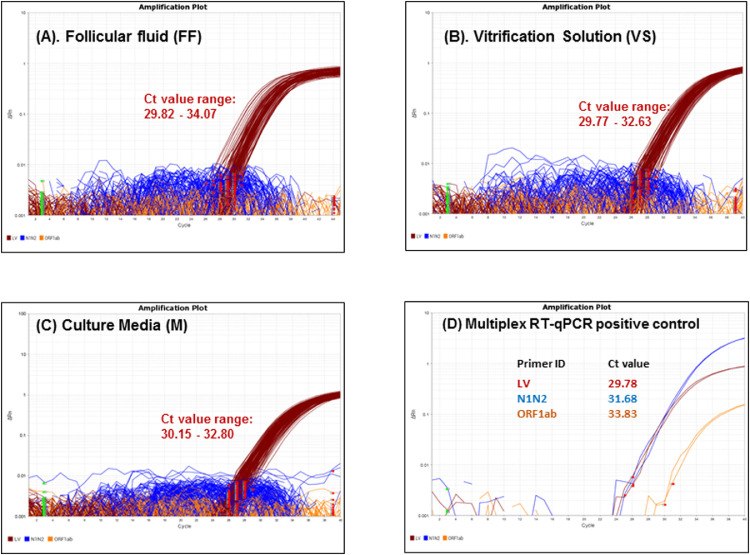
The results showed no amplification of SARS-CoV-2 genome with N1-, N2- and ORF1ab-specific loci during TMA in any of the samples analysed. As expected, a lentiviral amplification signal was observed in all patient samples that were analysed and included in the diagnostic study. Amplification curves for N1, N2, ORF1ab and lentivirus genomes were observed in the positive control (a mixture of lentivirus and SARS-CoV-2 cDNA) included in each RT-qPCR experiment (Figure 3 ). The negative controls in each experiment did not show amplification.
Figure 3.
Multiplex RT quantitative PCR (RT-qPCR)-based SARS-CoV-2 screening of IVF samples. Representative image of multiplex RT-qPCR amplification with RNA isolated from follicular fluid (A), vitrification solution (B), and culture medium (C) collected during patient IVF treatment cycles (n = 50 for each sample category shown). All of the samples showed amplification of the external positive control (lentivirus) and no amplification of N1/N2 (blue) and ORF1ab (orange) of the SARS-CoV-2 loci. (D) Multiplex RT-qPCR of the positive control showed successful amplification of all the target loci. Each reaction was performed in technical duplicates. Red squares depict the baseline start, and green squares depict the baseline end, of each well.
Discussion
The emergence of the respiratory virus, SARS-CoV-2, with possible aerosol transmission, has had tremendous impacts on fertility treatments. Among the various concerns raised from this global pandemic was the possibility of infection of laboratory staff from contaminated patient samples, cross-contamination of samples while inside the laboratory or even reinfection of patients if subsequently using previously infected samples. An abundance of opinions and commentaries on suggested safety precautions and procedural modifications for the IVF laboratory flooded the literature (Alviggi et al., 2020; Anifandis et al., 2020; Arav, 2020; Corona et al., 2020; De Santis et al. 2020; Maggiulli et al., 2020; Perry et al., 2020; Simopoulou et al., 2020; Vaiarelli et al., 2020). With a paucity of data available, these publications were often extremely cautious in their recommendations to avoid unknown or unforeseen issues. This approach was and is prudent to alleviate concerns and permit a safe reopening for essential fertility treatments. However, as new data emerge, an evidenced-based approach to combat infection or cross-contamination risks is feasible, and patient counselling and laboratory procedures can be adjusted accordingly.
Until now, the presence and unknown impact of SARS-CoV-2 in the IVF laboratory has been a concern, but most studies have not examined the laboratory environment for the presence of the virus. Initially viewed as a respiratory virus, as opposed to a sexually transmitted disease, emerging data made it clear that the virus could be detected in various tissues or bodily fluids (Wang et al., 2020). Receptors involved in SARS-CoV-2 signalling were identified in reproductive tissues and cells (Rajput et al., 2020; Stanley et al., 2020). However, whether active virus can be present or whether the virus could act directly upon gametes or embryos to impact development or function is still unknown. As the existing SARS-CoV-2 PCR-based protocol has not been optimized for complex samples such as follicular fluid, culture media and vitrification solutions, a reliable multiplex RT-qPCR protocol for these samples was first developed and this was then used to screen culture medium from 61 patients, vitrification solution from 200 patients and follicular fluid from 300 patients. The data demonstrating the absence of viral particles from these samples demonstrates that the SARS-CoV-2 virus can successfully be excluded from the IVF laboratory, providing reassurance that cross-contamination of the virus between gametes and embryos, as well as exposure of embryologists, is minimal. Importantly, all female patients tested negative with a PCR-based nasal swab test 3–5 days prior to oocyte retrieval and were asymptomatic on the day of the procedure. It is unknown if any of the women had had prior SARS-CoV-2 infections.
That being said, follicular fluid is routinely contaminated with varying amounts of blood. An initial study from China indicated that SARS-CoV-2 was present in less than 1% of blood samples from infected patients (Wang et al., 2020). However, whether infection is possible via blood is unknown. At least one publication describes a platelet transfusion from a SARS-CoV-2-infected individual to an uninfected person with no subsequent infection as a result (Cho et al., 2020). Additionally, because no ovarian follicle aspirates in this study contained SARS-CoV-2 virus, it is unclear if the SARS-CoV-2 virus may be introduced into the oocyte, especially considering the zona pellucida barrier. Importantly, the varying amounts of blood present in the various follicular aspirates could not be quantified. Interestingly, one recent study examined oocytes from two SARS-CoV-2-infected women. The women were diagnosed on the day of their oocyte retrieval as being COVID infected, via PCR assay, which may have implications for the incubation period and the viral presence in blood, follicular fluid or other tissues. However, none of the oocytes examined displayed any viral RNA for the SARS-CoV-2 gene (Barragan et al., 2020).
The current study is the first known report examining embryo culture media within the clinical IVF laboratory for the presence of SARS-CoV-2. No virus was detected in samples obtained from embryo culture media microdroplets. Importantly, several of the embryo culture medium samples that were examined came from patients who did not have prior testing of follicular fluid samples. Additionally, all cycles in this study used ICSI. Sperm preparation for ICSI entailed the routine use of a 45/90 gradient, sperm washing and a final swim-up step prior to the usual dilutions within polyvinyl propylene and other media encountered during routine ICSI procedures.
Interestingly, there are mixed reports on the presence or absence of SARS-CoV-2 in semen. One of the first studies out of Wuhan, China did not detect virus in semen from 34 SARS-CoV-2 seropositive men when tested 8–75 days after diagnosis (Pan et al., 2020). A second study of 38 patients indicated that, in six of the semen samples, virus was detected 6–16 days after symptom onset, although it is unknown whether active virus was present (Li et al., 2020). A third report, from China, of 12 SARS-CoV-2-infected men in the recovery phase demonstrated no virus in semen (Song et al., 2020). Although no symptomatic male patients were knowingly involved in IVF treatments in our study, it is unknown if any of the male patients were asymptomatic and actively infected with SARS-CoV-2. This demonstrates that, with basic, symptom-based screening of men and testing of women undergoing normal laboratory procedures using ICSI, SARS-CoV-2 was not present in the culture media. This has implications for concerns and recommendations regarding zona breaching, future embryo transfer or modes of possible transmission/cross-contamination in the laboratory (Pomeroy and Schiewe, 2020). Importantly, it has been suggested that ICSI may carry the potential to directly place a spermatozoon with SARS-CoV-2 virus directly into the oocyte, which could impact the developing embryo (Perry et al., 2020). It is unknown whether this is the case and it may be a possibility if using semen with active virus.
The current study is also the first known report examining embryo vitrification solutions for the presence of SARS-CoV-2. Similar to embryo culture media, no virus was detected in vitrification solution samples. This is not surprising given the lack of viral detection in follicular fluid and embryo culture media, as well as the additional dilution that occurs during solution exposure. It serves to further reinforce that, with thorough testing and symptom-based screening approaches, as well as the extensive dilution experienced in the IVF laboratory, the presence of the SARS-CoV-2 virus can be avoided. This may have implications for subsequent recommendations regarding cross-contamination and cryostorage procedure modifications for oocytes and preimplantation embryos, such as the use of closed cryo-devices, sterilization of liquid nitrogen, use of separate tanks or other measures (Arav, 2020; Pomeroy and Schiewe, 2020; Yakass and Woodward, 2020). These recommendations may differ from those used for cryopreserving or storing semen (Corona et al., 2020).
Importantly, the multiplex RT-qPCR assay developed in this study targets three loci of the SARS-CoV2 genome to provide better specificity, as well as implementing lentivirus as an external control, which is more similar in size and nature to SARS-CoV-2 compared with the phage RNA used in most of the commercially available kits. The protocol used in this study is sensitive enough to detect 50 viral particles in 2 ml of samples and with four copies of viral genome per qPCR reaction. The multiplex nature of the assay makes it not only cost-effective but also four times less time-consuming than uniplex PCR. Unlike commercially available kits, the current authors have provided complete details of reagents and protocols for RNA isolation and cDNA synthesis, as well as the optimized primers and probes sequences used in the multiplex assay, to aid in direct implementation for SARS-CoV-2 screening in any laboratory. The relatively simple nature and rapid turn-around time of this assay means it could be incorporated into the IVF laboratory during routine embryo culture. If virus is not identified around day 3 of embryo culture, the time when many laboratories change media over in a sequential system, or the day when embryo handling can easily occur for procedures such as zona breaking or embryo grading, there should be confidence that no virus is present during the later stages of culture or after further dilution during cryopreservation. Although virus could be below the detectable limit of the assay, or perhaps inside the cells and not present in media, this is unlikely.
As we continue to learn about the SAR-CoV-2 virus and the impact it may or may not have on gametes and embryos, as well as on any resulting pregnancy and offspring, the field of ART must react accordingly. These data demonstrate that with proper patient screening and testing, and by taking appropriate safety precautions, the virus can be effectively avoided within media in the IVF laboratory. It is important to note this does not mean that SARS-CoV-2 cannot be present inside the laboratory, particularly in the absence of patient testing, as the current study presumably did not include actively infected women or symptomatic men. However, the results, as well as those of other emerging studies, highlight the possibility of continuing to conduct IVF in a safe environment without dramatic changes to existing laboratory protocols for embryos, patients and laboratory staff.
Biography

Jason E. Swain, PhD, HCLD, is the Corporate Laboratory Director of the CCRM Ferility Network, located in North America. His primary research interests include pursuit of methods to improve in-vitro embryo culture conditions through reduction of environmental stressors via modification of both the physical and chemical culture environment.
Key Message.
With proper patient screening, testing and safety precautions, SARS-CoV-2 RNA was not detected in follicular fluid, embryo culture media or vitrification solutions. These findings have implications for safe laboratory practice recommendations in IVF, including cryostorage.
Alt-text: Unlabelled box
Declaration: The authors report no financial or commercial conflicts of interest.
References
- Alviggi C., Esteves S.C., Orvieto R., Conforti A., La Marca A., Fischer R., Andersen C.Y., Buhler K., Sunkara S.K., Polyzos N.P., Strina I., Carbone L., Bento F.C., Galliano D., Yarali H., Vuong L.N., Grynberg M., Drakopoulos P., Xavier P., Llacer J., Neuspiller F., Horton M., Roque M., Papanikolaou E., Banker M., Dahan M.H., Foong S., Tournaye H., Blockeel C., Vaiarelli A., Humaidan P., Ubaldi F.M., P. group COVID-19 and assisted reproductive technology services: repercussions for patients and proposal for individualized clinical management. Reprod. Biol. Endocrinol. 2020;18(1):45. doi: 10.1186/s12958-020-00605-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anifandis G., Messini C.I., Daponte A., Messinis I.E. COVID-19 and fertility: a virtual reality. Reprod. Biomed. Online. 2020;41(2):157–159. doi: 10.1016/j.rbmo.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arav A. A recommendation for IVF lab practice in light of the current COVID-19 pandemic. J. Assist. Reprod. Genet. 2020;37(7):1543. doi: 10.1007/s10815-020-01841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barragan M., Guillen J.J., Martin-Palomino N., Rodriguez A., Vassena R. Undetectable viral RNA in oocytes from SARS-CoV-2 positive women. Hum. Reprod. 2020 doi: 10.1093/humrep/deaa284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.J., Koo J.W., Roh S.K., Kim Y.K., Suh J.S., Moon J.H., Sohn S.K., Baek D.W. COVID-19 transmission and blood transfusion: A case report. J. Infect. Public Health. 2020 doi: 10.1016/j.jiph.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona G., Baldi E., Isidori A.M., Paoli D., Pallotti F., De Santis L., Francavilla F., La Vignera S., Selice R., Caponecchia L., Pivonello R., Ferlin A., Foresta C., Jannini E.A., Lenzi A., Maggi M., Lombardo F. SARS-CoV-2 infection, male fertility and sperm cryopreservation: a position statement of the Italian Society of Andrology and Sexual Medicine (SIAMS) (Societa Italiana di Andrologia e Medicina della Sessualita) J. Endocrinol. Invest. 2020;43(8):1153–1157. doi: 10.1007/s40618-020-01290-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santis L, Anastasi A, Cimadomo, Klinger F.G, Licata E, Pisaturo V, Fernandez L.S, Scarica C. COVID-19: the perspective of Italian embryologists managing the IVF laboratory in pandemic emergency. Hum. Reprod. 2020;35(4):1004–1100. doi: 10.1093/humrep/deaa074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essahib W., Verheyen G., Tournaye H., Van de Velde H. SARS-CoV-2 host receptors ACE2 and CD147 (BSG) are present on human oocytes and blastocysts. J. Assist. Reprod. Genet. 2020 doi: 10.1007/s10815-020-01952-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Marca A, Nelson S.M. SARS-CoV-2 testing in infertile patients: different recommendations in Europe and America. J. Assist. Reprod. Genet. 2020;37(8):1823–1828. doi: 10.1007/s10815-020-01887-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Jin M., Bao P., Zhao W., Zhang S. Clinical Characteristics and Results of Semen Tests Among Men With Coronavirus Disease 2019. JAMA Netw. Open. 2020;3(5) doi: 10.1001/jamanetworkopen.2020.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggiulli R., Giancani A., Fabozzi G., Dovere L., Tacconi L., Amendola M.G., Cimadomo D., Ubaldi F.M., Rienzi L. Assessment and management of the risk of SARS-CoV-2 infection in an IVF laboratory. Reprod. Biomed. Online. 2020;41(3):385–394. doi: 10.1016/j.rbmo.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F., Xiao X., Guo J., Song Y., Li H., Patel D.P., Spivak A.M., Alukal J.P., Zhang X., Xiong C., Li P.S., Hotaling J.M. No evidence of severe acute respiratory syndrome-coronavirus 2 in semen of males recovering from coronavirus disease 2019. Fertil. Steril. 2020;113(6):1135–1139. doi: 10.1016/j.fertnstert.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M.J., Arrington S., Neumann L.M., Carrell D., Mores C.N. It is currently unknown whether SARS-CoV-2 is viable in semen or whether COVID-19 damages spermatozoa. Andrology. 2020 doi: 10.1111/andr.12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeroy K.O., Schiewe M.C. Cryopreservation and IVF in the time of Covid-19: what is the best good tissue practice (GTP)? J. Assist. Reprod. Genet. 2020;37(10):2393–2398. doi: 10.1007/s10815-020-01904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeroy K.O., Schiewe M.C. Cryopreservation and IVF in the time of Covid-19: what is the best good tissue practice (GTP)? J. Assist. Reprod. Genet. 2020 doi: 10.1007/s10815-020-01904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajput S.K., Logsdon D., Khan S.A., Kile R., Engelhorn H.J., Yuan Y., McCormick S., Schoolcraft W.B., Krisher R.L. MATURE HUMAN OOCYTES AND PRE-IMPLANTATION EMBRYOS ARE SUSCEPTIBLE TO SARS-COV-2 INFECTION BASED ON THE PRESENCE OF ACE2 AND TMPRSS2 PROTEINS. ASRM 2020 Virtual Scientific Congress & Expo., ASRM; 2020. [Google Scholar]
- Simopoulou M., Sfakianoudis K., Giannelou P., Rapani A., Siristatidis C., Bakas P., Vlahos N., Pantos K. Navigating assisted reproduction treatment in the time of COVID-19: concerns and considerations. J. Assist. Reprod. Genet. 2020 doi: 10.1007/s10815-020-01942-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Wang Y., Li W., Hu B., Chen G., Xia P., Wang W., Li C., Diao F., Hu Z., Yang X., Yao B., Liu Y. Absence of 2019 novel coronavirus in semen and testes of COVID-19 patientsdagger. Biol. Reprod. 2020;103(1):4–6. doi: 10.1093/biolre/ioaa050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley K.E., Thomas E., Leaver M., Wells D. Coronavirus disease-19 and fertility: viral host entry protein expression in male and female reproductive tissues. Fertil. Steril. 2020;114(1):33–43. doi: 10.1016/j.fertnstert.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaiarelli A., Bulletti C., Cimadomo D., Borini A., Alviggi C., Ajossa S., Anserini P., Gennarelli G., Guido M., Levi-Setti P.E., Palagiano A., Palermo R., Savasi V., Pellicer A., Rienzi L., Ubaldi F.M. COVID-19 and ART: the view of the Italian Society of Fertility and Sterility and Reproductive Medicine. Reprod. Biomed. Online. 2020;40(6):755–759. doi: 10.1016/j.rbmo.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga A., Gianaroli L., Ory S., Horton M., Feinberg E., Penzias A. Assisted reproduction and COVID-19: A joint statement of ASRM, ESHRE and IFFS. Fertil. Steril. 2020;114(3):484–485. doi: 10.1016/j.fertnstert.2020.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakass M.B., Woodward B. COVID-19: should we continue to cryopreserve sperm during the pandemic? Reprod. Biomed. Online. 2020;40(6):905. doi: 10.1016/j.rbmo.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]