Abstract
Hyper-inflammatory responses, lymphopenia, unbalanced immune responses, cytokine storm, large viral replication and massive cell death play fundamental roles in the pathogenesis of COVID-19. Extreme production of many kinds of pro-inflammatory cytokines and chemokines occur in severe COVID-19 that called cytokine storm. Signal transducer and activator of transcription-3 (STAT-3) present in the cytoplasm in an inactive form and can be stimulated by a vast range of cytokines, chemokines and growth factors. Thus, STAT-3 can participate in the induction of inflammatory responses during coronavirus infections. STAT-3 can also suppress anti-virus interferon response and induce unbalanced anti-virus adaptive immune response, through influencing Th17-, Th1-, Treg-, and B cell-mediated functions. Furthermore, STAT-3 can contribute to the M2 macrophage polarization, lung fibrosis and thrombosis. Moreover, STAT-3 may be directly targeted by some virus-derived protein and operate as a pro-viral or anti-viral element in a virus-specific process. Here, the possible contribution of STAT-3 to the pathogenesis of COVID-19 was explained, while providing potential approaches to target this transcription factor in an attempt for COVID-19 treatment.
Keywords: COVID-19, SARS-CoV-2, STAT3, Pathogenesis, Inflammation, Immune response, Treatment
1. Introduction
Both SARS-CoV-2- (such as viral mutations) and human-related factors (such as age, gender, genetic, immune status, comorbidities, obesity and smoking) contribute to the COVID-19 severity and outcomes [1,2]. Massive infiltration of lung alveoli with inflammatory cells such as monocytes, macrophages, lymphocytes, eosinophils and neutrophils occur during COVID-19 [3,4]. The fibroblastic proliferation and bronchiolar epithelial metaplasia also occur in lung alveoli [4]. Clinical exacerbation of COVID-19 accompanies the occurrence of the dysregulation of cytokine production called cytokine storm that is characterized by the dangerous raise of plasma quantities of numerous inflammation-promoting cytokines and chemokines [5,6]. Lung damage is one of the consequences of the cytokine storm which can lead to acute lung injury (ALI) or its more dangerous form called acute respiratory distress syndrome (ARDS) [7]. Overall, hyper-inflammation, lymphopenia, immune dysfunction, cytokine storm, large viral replication, massive cell death, coagulopathy and fibrosis play crucial roles in the multi-organ failure, especially ARDS in COVID-19 [8]. (see Fig. 1, Fig. 2 )
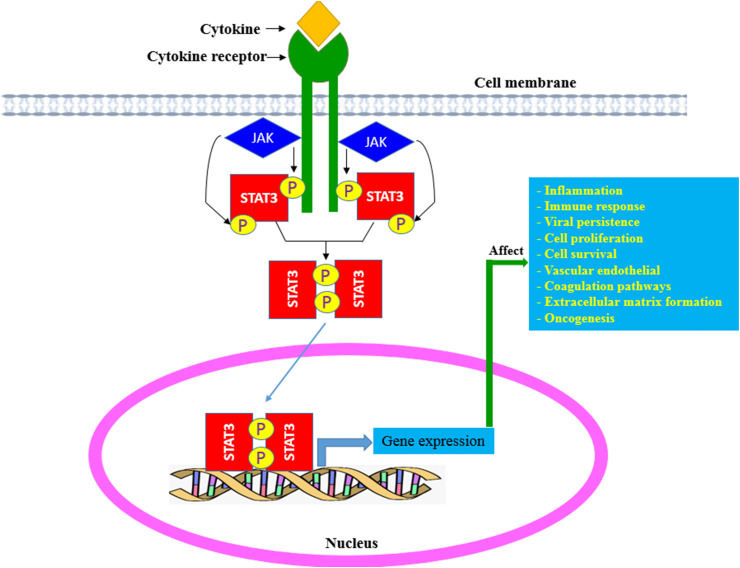
Fig. 1.
Activation of STAT-3: Binding of numerous cytokines or growth factors to their specific receptors induces phosphorylation of the receptor-associated JAK1 and then STAT3 is phosphorylated by JAK1. Phosphorylated STAT3 is dimerized and translocated to the nucleus, which causes the expression of target genes contributing to the immunosuppression, inflammation, angiogenesis, proliferation and survival.
Fig. 2.
Contribution of STAT-3-mediated pathways to COVID-19 pathogenesis. Activated STAT-3 can down-regulate lymphopoiesis, E-cadherin expression by vascular endothelial cells, Treg cell activity and anti-viral immune responses (including type I IFN-mediated signaling, NK cells activity, Th1 cell responses and CD8+ CTLs) supporting the lymphopenia, the vascular leak, hyper-inflammation and virus persistence, respectively. Activated STAT-3 can up-regulate the formation of the extracellular matrix, the expression of plasminogen activator inhibitor-1, the polarization and activation of Th17 cells and the polarization of M2 macrophages promoting lung fibrosis, thrombosis, hyper-inflammation/cytokine storm, and virus persistence/lung fibrosis, respectively.
In mammalian, the signal transducer and activator of transcription factors (STATs) including STAT-1, -2, -3, -4, -5A, -5B and −6 control the expression of genes regulating cell proliferation, cell differentiation, immune responses, inflammation, apoptosis and oncogenesis [9]. Six operative domains of STAT-3 molecule include the N-terminal domain, coiled-coil domain, linker domain, DNA binding domain, SH2 domain and C-terminal transcriptional activation domain [10]. The STAT-3 protein present in the cytoplasm in an inactive form, which is mainly activated by IL-6 upon ligation to its receptor, IL-6Rα. Moreover, STAT-3 can be stimulated by numerous types of cytokines such as TNF-α and IFN-γ, IL-5, IL-9, IL-10, IL-11, IL-12, IL-21, IL-22, IL-27, G-CSF, M-CSF, GM-CSF, monocyte chemotactic protein-1 (MCP-1) and CCL5 [9,10], some of them exist in the COVID-19-related cytokine storm [6].
The binding of the aforementioned cytokines to their corresponding receptors cause conformational alterations in the cytoplasmic domain activating the relevant Janus kinase (JAK), which then phosphorylates the certain tyrosine residues in the intracellular parts of the receptors while attracting and phosphorylating STAT-3. Stimulated STAT-3 releases from the receptor to create homodimers or heterodimers with other STATs via its SH2 domain. Homo- or heterodimerized STAT-3 is then transported from the cytoplasmic region to the cell nucleus and finally attach to the promoter regions of target genes to induce gene expression [10].
IL-6 is a main player among cytokine storm associated with the COVID-19 severity, considering as a predictive marker of COVID-19 fatality [11]. The IL-6/JAK/STAT-3 axis potently contributes to the pathogenesis of COVID-19 [12]. All three components in this axis can be targeted to manage COVID-19. IL-6 blockers target only one cytokine and results using Tocilizumab (an IL-6 blocker) indicate the improvement of the respiratory and laboratory parameters of COVID-19 patients. In addition to IL-6, JAK inhibitors can concurrently interrupt the signal transmission mediated by numerous cytokines [12,13].
The STAT-3 inhibition as a downstream element in the IL-6/JAK/STAT-3 axis may be a more attractive therapeutic strategy to mitigate COVID-19 severity. STAT-3 may have multiple roles during SARS-CoV-2 infection, including induction of pro-inflammatory responses, promotion of cytokine storm, unbalancing of immune responses, impairment of anti-viral immune responses, and exacerbation of lymphopenia. Furthermore, STAT-3 can be accused in the promotion of lung fibrosis, thrombosis and vascular abnormalities. The direct role of STAT-3 in the SARS-CoV-2 replication is also suspicious. Therefore, the targeting of STAT-3 may be better and more efficient than IL-6 and JAK targeting, and thus successive STAT-3 targeting may have therapeutic potentials in COVID-19.
2. STAT-3 contributes to the COVID-19 pathogenesis
2.1. IL-6/STAT-3 axis potentiates inflammatory responses in COVID-19
In SARS-CoV mouse models, high IL-6 levels were associated with severe inflammation and a greater mortality rate [14,15]. In SARS-CoV-infected patients, the elevated IL-6 amounts were correlated with disease severity [15]. A powerful association was also indicated between serum IL-6 levels and the ongoing respiratory failure in COVID-19 patients [16,17]. Raised IL-6 concentrations to levels >80 Pg/mL have been proposed as an indicator to determine the COVID-19 patients with a greater risk of lung dysfunction [18]. The COVID-19-related risk factors such as old ages, hypertension, diabetes, and obesity were associated with greater IL-6 quantities [17,19]. Higher plasma amounts of leptin were detected in COVID‐19 patients [20]. Leptin can synergistically with IL-6 activate STAT-3-mediated signaling in monocytes potentiating cytokine storm, particularly in overweight COVID‐19 patients [20]. Milder disease and lower risk of COVID-19-related mortality in women than in men have been attributed to the regulatory impacts of the estrogen on IL-6 expression [17,19]. The elevated quantities of angiotensin II due to SARS-CoV-2-induced downregulation of ACE2 can also support the IL-6 expression [17]. IL-6 promotes the expression of angiotensin 1 receptor reinforcing the vascular wall response to angiotensin II-mediated signaling such as cytokine secretion [17]. Thus, angiotensin II and IL-6 can induce each other via a positive feedback manner [17].
IL-6 operates via two major signaling pathways named cis or trans. In cis pathway, IL-6 initially creates a complex with the membrane-linked IL-6R and gp130 which then recruits JAKs and STAT-3 [21]. This signaling pathway exerts pleiotropic impacts of IL-6 on the innate- (macrophages, neutrophils and natural killer cells) as well as specific- (T and B cells) immune cells that can support the cytokine storm. In trans pathway, IL-6 firstly attaches to a soluble IL-6 receptor (sIL-6R) and then creates a complex with a gp130 dimer on most types of somatic cells. The IL-6–sIL-6R–JAK-STAT-3 signaling complex is then triggered in membrane-bound IL-6R-negative cells, such as endothelial cells. The trans signaling pathway powerfully exacerbates the cytokine storm via upregulation of the excess IL-6, IL-8, vascular endothelial growth factor (VEGF) and MCP-1, while downregulates the E-cadherin on endothelial cells [21,22]. The VEGF upregulation accompanied by E-cadherin downregulation promotes vascular leakage contributing to the hypotension and ARDS [22].
The presence of SARS-CoV-2 infection or its spike protein promotes IL-6 secretion and induces ADAM-17-mediated release of sIL-6R from epithelial cells [23]. The IL-6 production by SARS-CoV-2-infected epithelial cells cause STAT-3 activation and MCP-1 secretion from endothelial cells (where membrane-bound IL-6R is poorly expressed) via trans-signaling pathway [23]. SARS-CoV-2 infection prevents STAT-3 activation in epithelial cells and inhibits MCP-1 generation in epithelial cells. However, IL-6 production by SARS-CoV-2-infected epithelial cells promote inflammatory responses in endothelial cells through trans-signaling pathway [23].
IL-6/STAT-3 axis can contribute to the COVID-19 pathogenesis via promotion of the Th17 cell development. Th17 cells secrete a large number of cytokines including IL-17A, −17F, −21, −22, −23, −26, TNF-α, GM-CSF and CCL20 [24,25]. Th17 cell hyperactivity has been reported in MERS-CoV and SARS-CoV patients [26,27]. Higher peripheral blood count of CCR6+ Th17 cells were reported in a severe COVID-19 patient [28]. Higher numbers of Th17 cells, elevated plasma levels of Th17-related cytokines (including IL-17, IL-21 and IL-23) and greater expression of Th17-related transcription factor (RORγt) were observed in patients with mild and severe forms of COVID-19 compared with healthy individuals [29]. Some COVID-19-related risk factors such as obesity, hypertension, and chronic kidney disease, increased age (>65 years), diabetes and male gender were associated with elevated Th17 cell activity [30]. The ACE2 down-regulation and hypoxia also promote Th17 cell activity during COVID-19 [30]. Many cytokines among the COVID-19-related cytokine storm directly and/or indirectly derive from Th17 cell responses. Thus, an over-activity of Th17 cells following SARS-CoV-2 infection promotes hyper-inflammation. A potent relation has been indicated between IL-17A levels in the bronchoalveolar lavage fluid (BALF) with lung inflammation and poor outcome in patients with ARDS [31]. IL-22 can also promote the lung edema formation enriched with fibrin and mucins, as observed in patients with SARS-CoV and SARS-CoV-2 infection [32].
Antigenic induction of naïve CD4+ cells accompanied by IL-6-mediated STAT-3 activation in the existence of TGF-β leads to the Th17 cell differentiation [24,25]. IL-23 and IL-21 also reinforce Th17 cell differentiation via STAT-3-mediated signaling pathway. The blockade of STAT-3-mediated signaling interrupts Th17 cell differentiation [33]. Targeting of the STAT-3 axis during the initial stage of COVID-19 can probably attenuate the disease severity such as lung dysfunction via reducing Th17 cell development.
2.2. STAT-3 contributes to the lung fibrosis and injury
Lung fibrosis is one of the serious consequences of respiratory viral infections, such as COVID-19 and SARS [17,34]. Lung fibrosis is due to the large deposition of the extracellular matrix proteins, such as fibronectin impairing lung activity and gas exchange.
TGF-β-associated signaling performs fundamental roles in lung fibrosis, but its contribution to COVID-19-related lung fibrosis remains to be clarified. It has been indicated that SARS-CoV PLpro stimulates the Egr-1-induced TGF-β1 expression through ROS-p38 MAPK-STAT-3 axis promoting the pro-fibrotic reactions [35]. Blood platelets, macrophages, recruited neutrophils and infected type 2 alveolar cells can act as producers of TGF-β during COVID-19 [36,37]. According to lessons from SARS-CoV-1, it was suggested that the N protein of SARS-CoV-2 may support the TGF-β-induced plasminogen activator inhibitor-1 (PAI-1) and collagen I expression, hence promoting lung fibrosis [38].
There is evidence concerning the contribution of the IL-6/STAT-3 axis to lung damage. Elevated levels of the phosphorylated STAT-3 were found in lung biopsies collected from patients with idiopathic lung fibrosis and from mice with bleomycin-induced lung fibrosis [39,40]. STAT-3 inhibition using a small molecule reduces pulmonary fibrosis in bleomycin-induced fibrosis [40]. IL-6 and IL-33 may contribute to the lung fibrosis in mice with Bleomycin-induced through the activation of STAT-3- and AMPK-mediated signals [41].
In a mouse model of pulmonary fibrosis, it was observed that CXCL16 induces the progression of fibrosis via promotion of the STAT-3 phosphorylation in lung fibroblasts. The effect of CXCL16 was remarkably abolished (using a STAT-3 inhibitor) in lung fibroblasts [42]. Targeting of the IL-6-related downstream molecules such as STAT-3 can be considered as an alternative approach to the treatment of fibrosis.
IL-6 overexpression in pulmonary tissues using adenoviral vectors also exacerbated fibrosis, which was related to raised numbers of pro-fibrotic M2-macrophages [43]. Although IL-6 is generally considered as a profibrotic molecule, an experimental study indicated that the IL-6 blockade at the initial inflammatory period (day 2 after bleomycin-induced pulmonary damage) promotes the pneumocyte apoptosis, which more contributed to lung fibrosis, while IL-6 inhibition at the initial fibrotic period (day 8 after induced lung damage) markedly attenuated the fibrosis of the lungs [44].
Using IL-6-defective mice, it has been demonstrated that IL-6 attenuates acute lung damage in the infection of influenza A virus [45]. The proliferation, survival and migration of lung fibroblasts as well as virus-mediated apoptosis of lung epithelial cells were enhanced in IL-6-defective mice infected with influenza A virus [45]. IL-6 can facilitate repair after virus-mediated lung damage by limiting the inflammatory responses and promoting the protective adaptive immune responses [46].
In epithelial cells, excessive IL-6–mediated signaling contributes to the VEGF expression increasing vessel permeability and decreasing myocardial contractility [47]. VEGF secretion and reduced expression of E-cadherin promote vascular permeability and leakage that are implicated in the pathogenesis of hypotension and lung failure in ARDS [48]. IL-6 is also involved in the viral infection progression via activating of the matrix metalloproteinases promoting tissue permeability and edema.
2.3. STAT-3 contributes to the thrombosis
Pulmonary capillary thrombosis may lead to the quick development of hypoxia in COVID-19 patients [49]. There are controversies concerning the IL-6 role in inflammatory hypercoagulation, as it has been stated IL-6 can promote the incidence of fibrotic clot generation in whole blood, but its impact is less prominent than that seen for IL-1 or IL-8 [50]. However, it was also documented that IL-6 is essential for resolution of thrombus as it acts as a regulator of the expression of thrombolysis-related genes such as MMPs in macrophages [51].
In COVID-19 patients, PAI-1 is highly expressed in lungs and plasma [49]. In injured type 2 alveolar cells, the expression of PAI-1 is indirectly upregulated by STAT-3. PAI-1 interacts with TLR4 and triggers IL-6 expression which activates STAT-3 [38]. STAT-3 and PAI-1 reinforce thrombosis and coagulopathy in COVID-19 [49]. STAT-3-mediated PAI-1 upregulation may effectively suppress urokinase-type plasminogen activator and tissue-type plasminogen activator, promoting thrombosis [52]. STAT-3-mediated CRP production can induce tissue factor (TF) which in turn promotes the conversion of prothrombin into thrombin that convert plasma fibrinogen into fibrin causing fibrin-based coagulation [52].
2.4. Contribution of the STAT-3 to the lymphopenia
Lymphopenia may happen in about 1.0%–80.0% of mild/moderate COVID-19 patients and in about 33.0%–96.0% of severe COVID-19 patients [53]. The blood counts of T cells (including total CD3+ cells, CD4+ T- and CD8+ T cells), B lymphocytes and NK cells are diminished in COVID-19 patients, particularly in severe cases [53]. SARS-CoV-2- and immunologic-related mechanisms may lead to the appearance of lymphopenia by affecting the lymphopoiesis, lymphocyte viability, and tissue re-distribution of lymphocytes [53]. Lymphopenia can cause general immune-dysfunction and reinforce cytokine storm, both of them perform a major role in viral spreading and multi-organ dysfunction [53]. Lymphocytopenia exhibits a powerful association with mortality, especially in patients with lower counts of CD3+ T-, CD4+ T-, and CD8+ T cells [54,55].
Using various mouse models, it was indicated that IL-6 inhibits lymphopoiesis by direct impacts on hematopoietic stem cells [56,57]. An inverse relation has been indicated between serum IL-6 amounts and the absolute counts of blood lymphocytes in patients with COVID-19 [58]. The absolute counts of lymphocytes were enhanced within the 24 h after administration of Tocilizumab, a blocking monoclonal antibody against IL-6R [58,59].
In a mouse model of SLE, it has been demonstrated that the uncommitted progenitor cells express IL-6Rα and respond to IL-6 by phosphorylation of STAT-3. Furthermore, high IL-6 levels disrupt lymphopoiesis and promote the production of myeloid cells in a STAT-3-dependent manner [56]. Indeed, higher neutrophil/lymphocyte ratios were indicated in severe COVID-19 patients [53]. IL-6 influences lymphopoiesis through gp130-mediated STAT-3 activation [60]. The involvement of the IL-6/STAT-3 axis in the COVID-19-mediated lymphopenia needs careful dissection. If the role of the IL-6/STAT-3 axis in the COVID-19-mediated lymphopenia proved, then the targeting of STAT-3 can improve the lymphopenia.
3. STAT-3-mediated signaling pathways affect immune responses during COVID-19
3.1. STAT-3 impairs the anti-virus immune responses
In the innate immunity interferons (IFNs) provide the first protective border against viral infections [61]. Type I IFN signaling activates both STAT-1 and STAT-3 factors. However, some SARS-CoV-2 proteins such as ORF6 and NSP1 can inactivate STAT-1 contributing to the inhibition of IFN response, while compensatory leading to the hyper-activation of STAT-3. STAT-3 inhibits the STAT-1-mediated type I IFN response via preventing the creation of the STAT-1 homodimer by generating heterodimers with STAT-1, inhibiting the ISGF3 binding to DNA, reducing the expression of ISGF3 components [62]. Furthermore, during acute lung damage or when STAT-1 is insufficient, the expression of EGFR is upregulated [63,64]. EGFR-mediated signaling also inhibits type I IFN responses through induction of STAT-3 [38]. Thus, STAT-3-mediated inhibition of IFN response, especially during early stages of SARS-CoV-2 infection promotes virus spreading. Due to the importance of the IFN response during the initial phase of infection with SARS-CoV-2 [61], the local targeting of STAT-3 at this stage can improve IFN response and limit the virus spreading.
In specific immunity, CD4+ Th1 cells generate certain cytokines, particularly IL-2, IFN-γ and TNF-α that provide help for CD8+ cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells to destroy the virus infected-cells and reduce viral load [65,66]. Th2 cells release cytokines such as IL-4, -5, -6, and -9 assisting B cells to secrete anti-viral antibody [25,67]. Suitable antibody responses to certain parts of S protein, in particular receptor-binding domain (RBD), can prevent the SARS-CoV-2 attachment to its target cells [68]. Treg cells exhibit immunomodulatory activity through the generation of immunoregulatory cytokines TGF-β, IL-10 and IL-35. However, over-activity of Treg cells assist pathogen persistence, while their poor activity promotes immunopathologic damage [24]. Although the precise role of Th1/Th2 cells during SARS-CoV-2 remains to be clarified, it seems that the balanced Th1/Th2-related responses require for effective control of a viral infection.
Th1 cell responses appear to be protective against SARS-CoV, while Th2 responses are related to SARS progression [69]. It appears that the elimination of SARS-CoV-2 needs the timely and proper activation of CD4+ Th1 cells. In recovered patients with mild COVID-19, all the virus-specific CD4+ T cells were Th1 cells [70]. The CD4+ Th1 cells were reduced in COVID-19 patients who did not respond to antigenic stimulation with SARS-CoV-2-derived major proteins [71].
IL-6/STAT-3 axis inhibits Th1 polarization, promotes Th2 cell responses, suppresses CD8+ CTLs and NK cells, and induces T cell exhaustion, as indicated by the expression of markers including PD-1 and Tim-3 [8,17,72], promoting viral persistence and replication. STAT-3 exerts key modulatory impacts on the T cell-mediated immune responses, as it antagonizes the secretion of Th1 cell-related cytokines such as IFN-γ and IL-12 [[73], [74], [75], [76]].
The immunopathological response can be triggered by inappropriate excessive Th1 cell-induced responses [77,78]. SARS-CoV-2-specific T cells in COVID-19 patients with ARDS largely produced Th1 cell-related cytokines, but the production of Th17- and Th2 cell-linked cytokines were also detected [79]. The raised numbers of Th1 cells were detected in the peripheral lymphoid organs of severe COVID-19 patients which related to the diminished number of follicular helper T (Tfh) cells [80]. In transgenic mice expressing human ACE2, the SARS-CoV-2 infection causes accumulation of macrophage and lymphocyte in the lungs with potent Th1 cell responses and greater amounts of pro-inflammatory cytokines [81]. Raised amounts of IFN-γ, TNF-α, MCP-1 and IP-10 were associated with COVID-19 severity [5,82]. Th1 cell-mediated responses can cause various consequences during different stages of COVID-19.
In COVID-19 patients needing intensive care, the Th2 cell responses-rather than Th1 cell responses-are provoked against SARS-CoV-2 [83]. Large quantities of Th2 cell-related cytokines were measured in fatal COVID-19 cases in comparison with the recovered patients [84]. Some experiments using mouse models indicated that STAT-3 coordinates with STAT-6 in order to support Th2 cell differentiation [85]. STAT-3 regulates the maturation and differentiation and activation of B cells [86]. IL-6 involves in the formation of follicular helper T cells, generation of plasma cells and IgG production [8,17]. Disruption of IL-6 in the very initial phases of virus infection can lead to impaired Tfh cell differentiation and delay in the production of antiviral antibodies [87].
The frequencies of Treg cells were diminished in severe COVID-19 patients [88,89]. Severe COVID‐19 patients display low numbers of Treg cells [90,91] and raised numbers of Th17 cells resulting to the decreased Treg/Th17 cell ratio [28,32]. Organic environmental pollutants lead to the expression of aryl hydrocarbon receptor (AhR) and consequently AhR/IL-6/STAT-3 axis promotes Th17 cell differentiation while suppresses Treg cell polarization causing Treg/Th17 cell imbalance in COVID-19 patients [92]. A deviation in the balance of Th17/Treg cells toward Th17 cells may support the COVID-19 pathogenesis. The Treg/Th17 cell imbalance promotes the severity of lung injury and the development of the ARDS [93,94]. Excessive Th17 cell activities along with weak Treg cell responses may contribute to the massive release of pro‐inflammatory cytokines and chemokines in COVID‐19 patients which reinforce cytokine storm, exacerbating the disease and lead to multi-organ dysfunction and death.
STAT-3-mediated signaling inhibits Treg cell differentiation leading to the promotion of the immunopathologic responses [86]. Critical COVID‐19 patients with poor prognosis displayed greater numbers of Treg cells and Th2 cells than those with a favorite prognosis [95]. Further, primary IL-6 signaling can enhance IL-27-dependent Treg cell differentiation in mouse models of influenza virus and respiratory syncytial virus (RSV) infections preventing immunopathological responses [96].
3.2. STAT-3 can contribute to the development of M2-like macrophages during COVID-19
Activated macrophages were grouped into two distinct subsets, including classical (M1)- and alternative (M2) macrophages. M1 macrophages produce IL-1, IL-6, IL-12, TNF-α, monocyte chemotactic protein 1 (MCP-1), nitric oxide (NO), and reactive oxygen species (ROS) promoting inflammatory responses [6,97]. The lung macrophages display the M1 phenotype during the acute exudative stage of ALI/ARDS [97]. Suppressing the STAT-3 activity has a protective effect against LPS-induced ALI via inhibition of the M1 macrophages [98].
M2 macrophages contribute to the inflammation resolution, angiogenesis, and tissue repair by secretion of IL-1 receptor antagonist, TGF-β, IL-10, arginase 1 and CCL18 [6,97]. The lung macrophages display M2 phenotype and contribute to pulmonary fibrosis during the fibroproliferative stage of ALI/ARDS [97].
Using animal models of RSV infection, it was postulated that the lung macrophage polarization toward an M1 phenotype reduces the virus replication [99]. Depletion of M1 macrophages by apoptosis and necrosis can occur in some viral respiratory infections such as influenza and SARS, facilitating the viral replication [99]. Depending on the type of virus, the polarization of the macrophage to M2 type leads to virus persistence via attenuating the efficient anti-virus immune responses [99]. However, the balanced activation of M2 macrophages needs to prevent the RSV-induced immunopathology [99,100]. Higher numbers of FCN1+- and FCN1lo SPP1+ macrophages (M1-like phenotype) have been observed in the BALF samples collected from patients with severe COVID-19, while the BALF specimens obtained from patients with moderate COVID-19 and healthy individuals exhibited a greater number of FABP4+ macrophages (M2 phenotype) [101].
In addition to antiviral properties, type I IFNs influence the polarization of macrophages [99]. Type I IFNs may lead to the M1 macrophage polarization via induction of the STAT-1 and STAT-2-linked signals, whereas they cause M2 macrophage polarization through stimulation of the STAT-3 and STAT-6-mediated pathways [99,102]. Type I IFN-mediated signaling in the macrophages coincided with the presence of high amounts of STAT-1 and STAT-2 causes their polarization to the M1 phenotype, while high expression of STAT-3 and STAT-6 lead to their polarization to the M2 phenotype [99,102]. An in vitro study using mouse macrophages, it was observed that the suppression of the IL‐6/STAT-3 signaling induced the M1 macrophage polarization, while prevented the polarization of M2 macrophages [103]. IL-6 also induces the polarization of the M2 macrophages via stimulation of STAT-3 correlating with the development of gastric cancer [104].
The mechanisms controlling the macrophage polarization during the different stages of SARS-CoV-2 infection and their orientation in the correct direction need more investigations. If the contribution of the M1 and M2 macrophages in the COVID-19-related pathological events proved, then the targeting of certain STATs such as STAT-3 may be considered as a therapeutic approach.
4. STAT-3 can promote SARS-CoV-2 replication
A number of proteins derived from HBV, HCV, EBV, HSV-1, HCMV, HIV, KSHV, mumps virus, measles virus, Influenza A virus and SARS-CoV can directly interact with STAT-3 and influence viral replication and persistence [10]. STAT-3 can operate as a proviral or antiviral element in a virus-specific process [10]. Concerning the coronaviruses, SARS-CoV infection in Vero E6 cells activates p38 MAPK pathway which reduces cell apoptosis via STAT-3 inactivation [105]. Moreover, the SARS-CoV-2 infection in Vero cells (an epithelial cell line derived from the kidney of African green monkey) leads to the STAT-1 and STAT-3 dephosphorylation, large virus production and apoptosis [106]. However, in Calu-3 cells (an epithelial cell line derived from human airways), SARS-CoV-2 infection induces long-lasting STAT-1 and STAT-3 phosphorylation, small virus production without large apoptosis. Inhibitors that blockade STAT-3 phosphorylation and dimerization reduce SARS-CoV-2 production in the Calu-3 cell line, but not in the Vero cell line [106]. Phosphorylation and dimerization of STAT-3 is essential for the SARS-CoV-2 formation in Calu-3 cells [106]. Thus, the role of STAT-3 in infection with the SARS-CoV-2 can be altered depending on the infected cell type.
5. Targeting of STAT-3
The hyper-activation of STAT-3 can play a pivotal role in the pathogenesis of COVID-19 due to the occurrence of cytokine storm [107]. Thus STAT-3 may be considered as a possible therapeutic target for severe COVID-19 cases. The STAT-3 activity is strongly regulated by the negative controllers in non-activated cells [108]. Post-translational alterations such as methylation, acetylation and sumoylation can control STAT-3 activity via changing the STAT-3 phosphorylation. STAT-3 activity also can be directly and indirectly modulated by noncoding RNAs such as miRNAs and long non-coding RNAs [108].
Different strategies are being developed to suppress STAT-3-related signaling using direct STAT-3 inhibitors or indirectly using the suppressors of upstream kinases [9]. Direct STAT-3 inhibitors bind to different domains of STAT-3 preventing the activation and function of STAT-3. Direct STAT-3 inhibitors including small molecules and oligonucleotides such as siRNA targeting DNA-binding domain, SH2 domain and N-terminal domain [9]. Napabucasin as a small molecule binds to the DNA-binding domain of STAT-3, and Danvatirsen targets the 3′UTR region in the STAT-3 gene and prevents its expression [9]. Metformin prevents STAT-3 activation through decreasing its phosphorylation [38]. Some FDA-approved agents, such as Celecoxib and Pyrimethamine, were considered as STAT-3 suppressors [9]. A number of natural compounds (such as Curcumin, Caffeic acid, Betulinic acid, Capsaicin, Butein, Diosgenin, Celastrol Cucurbitacins, Guggulsterone and Honokiol) targeting STAT-3 was evaluated in preclinical and clinical studies [109].
6. Conclusion
STAT-3 can contribute to the development of several major pathological events during COVID-19. STAT-3 may inhibit the anti-SARS-CoV-2 IFN response and potentiates the unbalanced adaptive immune responses via influencing the differentiation of the effector Th17-, Treg-, Th1- and Th2 cells [[73], [74], [75], [76]], thus promoting the virus persistence and immunopathologic reactions.
The results from an in vitro study using an epithelial cell line indicated that the silencing STAT-3α leads to the upregulation of the ACE2 expression while silencing STAT-3β exerts the opposite impacts [110]. SARS-CoV-2 can utilize STAT-3-induced ACE2 upregulation to promote infection [78]. In alveolar epithelial cells, SARS-CoV-2 infection through ACE2 can activate STAT-3 amplifying the IL-6 expression that triggers a positive feedback process [103]. STAT-3 can also support coagulopathy and lung fibrosis in SARS-CoV-2-infected patients.
Thus, STAT-3 can be considered as an excellent target for COVID-19 therapy. Timely targeting of IL-6/STAT-3 axis can potentiate appropriate immune responses against SARS-CoV-2 while attenuating immunopathology reactions. There are actually some challenges regarding the STAT-3-based therapies in the clinic. Firstly, there are large sequence and conformational similarities between STAT-3 and other STAT members such as STAT-1 and STAT-5 [111]. Biologically, STAT-1 and STAT-3 exert opposite activities. Hence, a direct STAT-3 inhibitor can nonspecifically inhibit STAT-1 causing unwanted adverse effects [111]. Secondly, different STAT molecules can exhibit functional redundancy in their activities. Thus, when a certain STAT is selectively blocked another STAT molecule can exert compensatory impacts. For instance, blockade of STAT-3 which is essential for IL-6-mediated signaling, can allow the cells to react to this cytokine via STAT-1 [9]. Thirdly, there are some functional dualities concerning STAT-3. For example, STAT-3 is involved in both IL-10- and IL-6-mediated signaling pathways. IL-10 promotes Treg cell polarization, whereas IL-6 induces Th17 cell differentiation. The orientation of the functional dualities of STAT-3 in favorite directions need to be clarified in future studies [9]. Fourthly, the main limitations of the inhibitory peptides targeting STAT-3 are low cell permeability, undesirable pharmacokinetic effects, low stability, and suboptimal potency [112,113]. Fifth, there are some concerns about the selectivity, safety and mechanisms of action of small molecules targeting STAT-3 [9]. Sixth, effective delivery of siRNA to a specific cell subset has been considered as a vigorous challenge regarding their clinical utilization [114]. Seventh, the STAT-3 inhibitors that have used in clinical trials can cause adverse reactions such as nausea, hypertension and weakness, limiting their clinical utility [115]. Natural components including butein and curcumin suppress STAT-3 via a number of mechanisms such as affecting the STAT-3 phosphorylation, dimerization and DNA-binding capacity. These compounds exhibit favorable patterns of toxicity, however, they operate nonspecifically and blockade STAT-3 indirectly [113]. Finally, a precise understanding of the role of STAT-3 in different lymphoid- and non-lymphoid cells is essential in order to design novel STAT3-based therapies.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Abbreviations
- ACE2
Angiotensin-converting enzyme 2
- ALI
Acute lung injury
- ARDS
Acute respiratory distress syndrome
- BALF
Bronchoalveolar lavage fluid
- CTL
Cytotoxic T lymphocyte
- DCs
Dendritic cells
- EGFR
Epidermal growth factor receptor
- G-CSF
Granulocyte colony-stimulating factor
- GM-CSF
Granulocyte-macrophage colony-stimulating factor
- JAK
Janus kinase
- IFN
Interferon
- ISGF3
Interferon Stimulated Gene Factor 3
- IRF
Interferon regulatory factor
- MAPK
Mitogen-activated protein kinase
- MCP-1
Monocyte chemoattractant protein-1
- MERS-CoV
Middle East respiratory syndrome-related coronavirus
- ORF
Open reading frame
- PD-1
Programmed death-1
- PAI-1
Plasminogen activator inhibitor-1
- RBD
Receptor-binding domain
- PLpro
Papain-like protease
- SARS-CoV
Severe acute respiratory syndrome-related coronavirus
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- STAT
Signal transducer and activator of transcription factor
- TIM-3
T-cell immunoglobulin and mucin-domain containing-3
- Treg cell
regulatory T cell
- VEGF
Vascular endothelial growth factor
References
- 1.Eaaswarkhanth M., Al Madhoun A., Al-Mulla F. Could the D614G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality? Int. J. Infect. Dis. 2020;96:459–460. doi: 10.1016/j.ijid.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tahaghoghi-Hajghorbani S., Zafari P., Masoumi E., Rajabinejad M., Jafari-Shakib R., Hasani B., et al. The role of dysregulated immune responses in COVID-19 pathogenesis. Virus Res. 2020;290:198197. doi: 10.1016/j.virusres.2020.198197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao X.H., Li T.Y., He Z.C., Ping Y.F., Liu H.W., Yu S.C., et al. [A pathological report of three COVID-19 cases by minimally invasive autopsies] Chin. J. Pathol. 2020;49:E009. doi: 10.3760/cma.j.cn112151-20200312-00193. [DOI] [PubMed] [Google Scholar]
- 4.Wu J.H., Li X., Huang B., Su H., Li Y., Luo D.J., et al. [Pathological changes of fatal coronavirus disease 2019 (COVID-19) in the lungs: report of 10 cases by postmortem needle autopsy] Chin. J. Pathol. 2020;49:568–575. doi: 10.3760/cma.j.cn112151-20200405-00291. [DOI] [PubMed] [Google Scholar]
- 5.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jafarzadeh A., Chauhan P., Saha B., Jafarzadeh S., Nemati M. Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: lessons from SARS and MERS, and potential therapeutic interventions. Life Sci. 2020;257:118102. doi: 10.1016/j.lfs.2020.118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ragab D., Salah Eldin H., Taeimah M., Khattab R., Salem R. The COVID-19 cytokine storm; what we know so far. Front. Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lingeswaran M., Goyal T., Ghosh R., Suri S., Mitra P., Misra S., et al. Inflammation, immunity and immunogenetics in COVID-19: a narrative review. Indian J. Clin. Biochem. 2020;35:260–273. doi: 10.1007/s12291-020-00897-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gharibi T., Babaloo Z., Hosseini A., Abdollahpour-Alitappeh M., Hashemi V., Marofi F., et al. Targeting STAT3 in cancer and autoimmune diseases. Eur. J. Pharmacol. 2020;878:173107. doi: 10.1016/j.ejphar.2020.173107. [DOI] [PubMed] [Google Scholar]
- 10.Chang Z., Wang Y., Zhou X., Long J.E. STAT3 roles in viral infection: antiviral or proviral? Future Virol. 2018;13:557–574. doi: 10.2217/fvl-2018-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X., Xu S., Yu M., Wang K., Tao Y., Zhou Y., et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J. Allergy Clin. Immunol. 2020;146:110–118. doi: 10.1016/j.jaci.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo W., Li Y.X., Jiang L.J., Chen Q., Wang T., Ye D.W. Targeting JAK-STAT signaling to control cytokine release syndrome in COVID-19. Trends Pharmacol. Sci. 2020;41:531–543. doi: 10.1016/j.tips.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz D.M., Kanno Y., Villarino A., Ward M., Gadina M., O'Shea J.J. JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nat. Rev. Drug Discov. 2017;17:78. doi: 10.1038/nrd.2017.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Day C.W., Baric R., Cai S.X., Frieman M., Kumaki Y., Morrey J.D., et al. A new mouse-adapted strain of SARS-CoV as a lethal model for evaluating antiviral agents in vitro and in vivo. Virology. 2009;395:210–222. doi: 10.1016/j.virol.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magro G. SARS-CoV-2 and COVID-19: is interleukin-6 (IL-6) the 'culprit lesion' of ARDS onset? What is there besides Tocilizumab? SGP130Fc. Cytokine X. 2020 doi: 10.1016/j.cytox.2020.100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y., et al. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clin. Infect. Dis. 2020;71(8):1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gubernatorova E.O., Gorshkova E.A., Polinova A.I., Drutskaya M.S. IL-6: relevance for immunopathology of SARS-CoV-2. Cytokine Growth Factor Rev. 2020;53:13–24. doi: 10.1016/j.cytogfr.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herold T., Jurinovic V., Arnreich C., Hellmuth J.C., von Bergwelt-Baildon M., Klein M., Weinberger T., et al. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J. Allergy Clin. Immunol. 2020;146(1):128–136. doi: 10.1016/j.jaci.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strope J.D., Chau C.H., Figg W.D. Are sex discordant outcomes in COVID-19 related to sex hormones? Semin. Oncol. 2020;47:335–340. doi: 10.1053/j.seminoncol.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J., Xu Y., Zhang X., Wang S., Peng Z., Guo J., et al. Leptin correlates with monocytes activation and severe condition in COVID-19 patients. J. Leukoc. Biol. 2021 doi: 10.1002/JLB.5HI1020-704R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett D. In: Cytokine Storm Syndrome. Cron R., Behrens E., editors. Springer; Cham: 2019. IL-6 blockade in cytokine storm syndromes. [DOI] [Google Scholar]
- 22.Narazaki M., Kishimoto T. The two-faced cytokine IL-6 in host defense and diseases. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19113528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patra T., Meyer K., Geerling L., Isbell T.S., Hoft D.F., Brien J., et al. SARS-CoV-2 spike protein promotes IL-6 trans-signaling by activation of angiotensin II receptor signaling in epithelial cells. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jafarzadeh A., Larussa T., Nemati M., Jalapour S. T cell subsets play an important role in the determination of the clinical outcome of Helicobacter pylori infection. Microb. Pathog. 2018;116:227–236. doi: 10.1016/j.micpath.2018.01.040. [DOI] [PubMed] [Google Scholar]
- 25.Jafarzadeh A., Nemati M. Therapeutic potentials of ginger for treatment of Multiple sclerosis: a review with emphasis on its immunomodulatory, anti-inflammatory and anti-oxidative properties. J. Neuroimmunol. 2018;324:54–75. doi: 10.1016/j.jneuroim.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Faure E., Poissy J., Goffard A., Fournier C., Kipnis E., Titecat M., et al. Distinct immune response in two MERS-CoV-infected patients: can we go from bench to bedside? PloS One. 2014;9 doi: 10.1371/journal.pone.0088716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Josset L., Menachery V.D., Gralinski L.E., Agnihothram S., Sova P., Carter V.S., et al. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. mBio. 2013;4:e00165–13. doi: 10.1128/mBio.00165-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tahmasebi S., El-Esawi M.A., Mahmoud Z.H., Timoshin A., Valizadeh H., Roshangar L., et al. Immunomodulatory effects of nanocurcumin on Th17 cell responses in mild and severe COVID-19 patients. J. Cell. Physiol. 2020 doi: 10.1002/jcp.30233. [DOI] [PubMed] [Google Scholar]
- 30.Orlov M., Wander P.L., Morrell E.D., Mikacenic C., Wurfel M.M. A case for targeting Th17 Cells and IL-17A in SARS-CoV-2 infections. J. Immunol. 2020;205:892–898. doi: 10.4049/jimmunol.2000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikacenic C., Hansen E.E., Radella F., Gharib S.A., Stapleton R.D., Wurfel M.M. Interleukin-17A is associated with alveolar inflammation and poor outcomes in acute respiratory distress syndrome. Crit. Care Med. 2016;44:496–502. doi: 10.1097/CCM.0000000000001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu D., Yang X.O. TH17 responses in cytokine storm of COVID-19: an emerging target of JAK2 inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020;53:368–370. doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Min H.K., Choi J., Lee S.Y., Seo H.B., Jung K., Na H.S., et al. Protein inhibitor of activated STAT3 reduces peripheral arthritis and gut inflammation and regulates the Th17/Treg cell imbalance via STAT3 signaling in a mouse model of spondyloarthritis. J. Transl. Med. 2019;17:18. doi: 10.1186/s12967-019-1774-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun P., Qie S., Liu Z., Ren J., Li K., Xi J. Clinical characteristics of hospitalized patients with SARS-CoV-2 infection: a single arm meta-analysis. J. Med. Virol. 2020;92:612–617. doi: 10.1002/jmv.25735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li S.W., Wang C.Y., Jou Y.J., Yang T.C., Huang S.H., Wan L., et al. SARS coronavirus papain-like protease induces Egr-1-dependent up-regulation of TGF-beta1 via ROS/p38 MAPK/STAT3 pathway. Sci. Rep. 2016;6:25754. doi: 10.1038/srep25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen W. A potential treatment of COVID-19 with TGF-beta blockade. Int. J. Biol. Sci. 2020;16:1954–1955. doi: 10.7150/ijbs.46891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lodyga M., Hinz B. TGF-beta1 - a truly transforming growth factor in fibrosis and immunity. Semin. Cell Dev. Biol. 2020;101:123–139. doi: 10.1016/j.semcdb.2019.12.010. [DOI] [PubMed] [Google Scholar]
- 38.Matsuyama T., Kubli S.P., Yoshinaga S.K., Pfeffer K., Mak T.W. An aberrant STAT pathway is central to COVID-19. Cell Death Differ. 2020;27:3209–3225. doi: 10.1038/s41418-020-00633-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pechkovsky D.V., Prêle C.M., Wong J., Hogaboam C.M., McAnulty R.J., Laurent G.J., et al. STAT3-mediated signaling dysregulates lung fibroblast-myofibroblast activation and differentiation in UIP/IPF. Am. J. Pathol. 2012;180:1398–1412. doi: 10.1016/j.ajpath.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 40.Pedroza M., Le T.T., Lewis K., Karmouty-Quintana H., To S., George A.T., et al. STAT-3 contributes to pulmonary fibrosis through epithelial injury and fibroblast-myofibroblast differentiation. Faseb. J. : official publication of the Federation of American Societies for Experimental Biology. 2016;30:129–140. doi: 10.1096/fj.15-273953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shieh J.M., Tseng H.Y., Jung F., Yang S.H., Lin J.C. Elevation of IL-6 and IL-33 levels in serum associated with lung fibrosis and skeletal muscle wasting in a bleomycin-induced lung injury mouse model. Mediat. Inflamm. 2019;2019 doi: 10.1155/2019/7947596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuo S., Zhu Z., Liu Y., Li H., Song S., Yin S. CXCL16 induces the progression of pulmonary fibrosis through promoting the phosphorylation of STAT3. Can. Respir. J. J. Can. Thorac. Soc. 2019;2019 doi: 10.1155/2019/2697376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ayaub E.A., Dubey A., Imani J., Botelho F., Kolb M.R.J., Richards C.D., et al. Overexpression of OSM and IL-6 impacts the polarization of pro-fibrotic macrophages and the development of bleomycin-induced lung fibrosis. Sci. Rep. 2017;7:13281. doi: 10.1038/s41598-017-13511-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kobayashi T., Tanaka K., Fujita T., Umezawa H., Amano H., Yoshioka K., et al. Bidirectional role of IL-6 signal in pathogenesis of lung fibrosis. Respir. Res. 2015;16:99. doi: 10.1186/s12931-015-0261-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang M.L., Wang C.T., Yang S.J., Leu C.H., Chen S.H., Wu C.L., et al. IL-6 ameliorates acute lung injury in influenza virus infection. Sci. Rep. 2017;7:43829. doi: 10.1038/srep43829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lauder S.N., Jones E., Smart K., Bloom A., Williams A.S., Hindley J.P., et al. Interleukin-6 limits influenza-induced inflammation and protects against fatal lung pathology. Eur. J. Immunol. 2013;43:2613–2625. doi: 10.1002/eji.201243018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu B., Li M., Zhou Z., Guan X., Xiang Y. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J. Autoimmun. 2020;111:102452. doi: 10.1016/j.jaut.2020.102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang C., Wu Z., Li J.W., Zhao H., Wang G.Q. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int. J. Antimicrob. Agents. 2020;55:105954. doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuo Y., Warnock M., Harbaugh A., Yalavarthi S., Gockman K., Zuo M., et al. Plasma tissue plasminogen activator and plasminogen activator inhibitor-1 in hospitalized COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.08.29.20184358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bester J., Matshailwe C., Pretorius E. Simultaneous presence of hypercoagulation and increased clot lysis time due to IL-1β, IL-6 and IL-8. Cytokine. 2018;110:237–242. doi: 10.1016/j.cyto.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 51.Nosaka M., Ishida Y., Kimura A., Kuninaka Y., Taruya A., Ozaki M., et al. Crucial involvement of IL-6 in thrombus resolution in mice via macrophage recruitment and the induction of proteolytic enzymes. Front. Immunol. 2019;10:3150. doi: 10.3389/fimmu.2019.03150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cimmino G., Cirillo P. Tissue factor: newer concepts in thrombosis and its role beyond thrombosis and hemostasis. Cardiovasc. Diagn. Ther. 2018;8:581–593. doi: 10.21037/cdt.2018.10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jafarzadeh A., Jafarzadeh S., Nozari P., Mokhtari P., Nemati M. Lymphopenia an important immunological abnormality in patients with COVID-19: possible mechanisms. Scand. J. Immunol. 2020 doi: 10.1111/sji.12967. [DOI] [PubMed] [Google Scholar]
- 54.Zeng Q., Li Y-z, Huang G., Wu W., Dong S-y, Xu Y. Mortality of COVID-19 is associated with cellular immune function compared to immune function in Chinese han population. medRxiv. 2020 doi: 10.1101/2020.03.08.20031229. [DOI] [Google Scholar]
- 55.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maeda K., Malykhin A., Teague-Weber B.N., Sun X.H., Farris A.D., Coggeshall K.M. Interleukin-6 aborts lymphopoiesis and elevates production of myeloid cells in systemic lupus erythematosus-prone B6.Sle1.Yaa animals. Blood. 2009;113:4534–4540. doi: 10.1182/blood-2008-12-192559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maeda K., Baba Y., Nagai Y., Miyazaki K., Malykhin A., Nakamura K., et al. IL-6 blocks a discrete early step in lymphopoiesis. Blood. 2005;106:879–885. doi: 10.1182/blood-2005-02-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000. doi: 10.1016/j.chom.2020.04.009. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang C., Wu Z., Li J.W., Zhao H., Wang G.Q. The cytokine release syndrome (CRS) of severe COVID-19 and Interleukin-6 receptor (IL-6R) antagonist Tocilizumab may be the key to reduce the mortality. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jenkins B.J., Roberts A.W., Greenhill C.J., Najdovska M., Lundgren-May T., Robb L., et al. Pathologic consequences of STAT3 hyperactivation by IL-6 and IL-11 during hematopoiesis and lymphopoiesis. Blood. 2007;109:2380–2388. doi: 10.1182/blood-2006-08-040352. [DOI] [PubMed] [Google Scholar]
- 61.Jafarzadeh A., Nemati M., Saha B., Bansode Y., S J. Protective potentials of type III interferons (IFN-λ) in COVID-19 patients: lessons from different basically and clinically properties attributed to the type I- and III interferons. Viral Immunol. 2020 doi: 10.1089/vim.2020.0076. [DOI] [PubMed] [Google Scholar]
- 62.Tsai M.H., Pai L.M., Lee C.K. Fine-tuning of type I interferon response by STAT3. Front. Immunol. 2019;10:1448. doi: 10.3389/fimmu.2019.01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Finigan J.H., Downey G.P., Kern J.A. Human epidermal growth factor receptor signaling in acute lung injury. Am. J. Respir. Cell Mol. Biol. 2012;47:395–404. doi: 10.1165/rcmb.2012-0100TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Venkataraman T., Coleman C.M., Frieman M.B. Overactive epidermal growth factor receptor signaling leads to increased fibrosis after severe acute respiratory syndrome coronavirus infection. J. Virol. 2017;91 doi: 10.1128/JVI.00182-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miyauchi K. Helper T cell responses to respiratory viruses in the lung: development, virus suppression, and pathogenesis. Viral Immunol. 2017;30:421–430. doi: 10.1089/vim.2017.0018. [DOI] [PubMed] [Google Scholar]
- 66.Frank K., Paust S. Dynamic natural killer cell and T cell responses to influenza infection. Frontiers in cellular and infection microbiology. 2020;10:425. doi: 10.3389/fcimb.2020.00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jafarzadeh A., Shokri F. The antibody response to HBs antigen is regulated by coordinated Th1 and Th2 cytokine production in healthy neonates. Clin. Exp. Immunol. 2003;131:451–456. doi: 10.1046/j.1365-2249.2003.02093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 69.Janice Oh H.L., Ken-En Gan S., Bertoletti A., Tan Y.J. Understanding the T cell immune response in SARS coronavirus infection. Emerg. Microb. Infect. 2012;1:e23. doi: 10.1038/emi.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neidleman J., Luo X., Frouard J., Xie G., Gill G., Stein E.S., et al. SARS-CoV-2-specific T cells exhibit unique features characterized by robust helper function, lack of terminal differentiation, and high proliferative potential. bioRxiv : the preprint server for biology. 2020 doi: 10.1101/2020.06.08.138826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sattler A., Angermair S., Stockmann H., Heim K.M., Khadzhynov D., Treskatsch S., et al. SARS-CoV-2 specific T-cell responses and correlations with COVID-19 patient predisposition. J. Clin. Invest. 2020;130:6477–6489. doi: 10.1172/JCI140965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Velazquez-Salinas L., Verdugo-Rodriguez A., Rodriguez L.L., Borca M.V. The role of interleukin 6 during viral infections. Front. Microbiol. 2019;10:1057. doi: 10.3389/fmicb.2019.01057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kortylewski M., Xin H., Kujawski M., Lee H., Liu Y., Harris T., et al. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Canc. Cell. 2009;15:114–123. doi: 10.1016/j.ccr.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eddahri F., Denanglaire S., Bureau F., Spolski R., Leonard W.J., Leo O., et al. Interleukin-6/STAT3 signaling regulates the ability of naive T cells to acquire B-cell help capacities. Blood. 2009;113:2426–2433. doi: 10.1182/blood-2008-04-154682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nurieva R.I., Chung Y., Hwang D., Yang X.O., Kang H.S., Ma L., et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang X.O., Panopoulos A.D., Nurieva R., Chang S.H., Wang D., Watowich S.S., et al. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 2007;282:9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Y., Zhang Y., Gu W., Sun B. TH1/TH2 cell differentiation and molecular signals. Adv. Exp. Med. Biol. 2014;841:15–44. doi: 10.1007/978-94-017-9487-9_2. [DOI] [PubMed] [Google Scholar]
- 78.Schmitt N., Ueno H. Regulation of human helper T cell subset differentiation by cytokines. Curr. Opin. Immunol. 2015;34:130–136. doi: 10.1016/j.coi.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Science immunology. 2020;5 doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaneko N., Kuo H.H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S., et al. Loss of Bcl-6-expressing t follicular helper cells and germinal centers in COVID-19. Cell. 2020;183:143–157. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yinda C.K., Port J.R., Bushmaker T., Owusu I.O., Avanzato V.A., Fischer R.J., et al. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. PLoS Pathog. 2020;17:e1009195. doi: 10.1371/journal.ppat.1009195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang Y., Shen C., Li J., Yuan J., Wei J., Huang F., et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J. Allergy Clin. Immunol. 2020;146:119–127. doi: 10.1016/j.jaci.2020.04.027. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roncati L., Nasillo V., Lusenti B., Riva G. Signals of T(h)2 immune response from COVID-19 patients requiring intensive care. Ann. Hematol. 2020;99:1419–1420. doi: 10.1007/s00277-020-04066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li C.K., Wu H., Yan H., Ma S., Wang L., Zhang M., et al. T cell responses to whole SARS coronavirus in humans. J. Immunol. 2008;181:5490–5500. doi: 10.4049/jimmunol.181.8.5490. Baltimore, Md : 1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stritesky G.L., Muthukrishnan R., Sehra S., Goswami R., Pham D., Travers J., et al. The transcription factor STAT3 is required for T helper 2 cell development. Immunity. 2011;34:39–49. doi: 10.1016/j.immuni.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Deenick E.K., Pelham S.J., Kane A., Ma C.S. Signal transducer and activator of transcription 3 control of human T and B cell responses. Front. Immunol. 2018;9:168. doi: 10.3389/fimmu.2018.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Karnowski A., Chevrier S., Belz G.T., Mount A., Emslie D., D'Costa K., et al. B and T cells collaborate in antiviral responses via IL-6, IL-21, and transcriptional activator and coactivator, Oct2 and OBF-1. J. Exp. Med. 2012;209:2049–2064. doi: 10.1084/jem.20111504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu Z.X., Ji M.S., Yan J., Cai Y., Liu J., Yang H.F., et al. The ratio of Th17/Treg cells as a risk indicator in early acute respiratory distress syndrome. Crit. Care. 2015;19:82. doi: 10.1186/s13054-015-0811-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang F., Hou H., Luo Y., Tang G., Wu S., Huang M., et al. The laboratory tests and host immunity of COVID-19 patients with different severity of illness. JCI insight. 2020;5 doi: 10.1172/jci.insight.137799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Engin A.B., Engin E.D., Engin A. The effect of environmental pollution on immune evasion checkpoints of SARS-CoV-2. Environ. Toxicol. Pharmacol. 2021;81 doi: 10.1016/j.etap.2020.103520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li G., Cao Y., Sun Y., Xu R., Zheng Z., Song H. Ultrafine particles in the airway aggravated experimental lung injury through impairment in Treg function. Biochem. Biophys. Res. Commun. 2016;478:494–500. doi: 10.1016/j.bbrc.2016.05.059. [DOI] [PubMed] [Google Scholar]
- 94.Lin S., Wu H., Wang C., Xiao Z., Xu F. Regulatory T cells and acute lung injury: cytokines, uncontrolled inflammation, and therapeutic implications. Front. Immunol. 2018;9:1545. doi: 10.3389/fimmu.2018.01545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wei L.-L., Wang W.-J., Chen D.-X., Xu B. Dysregulation of the immune response affects the outcome of critical COVID-19 patients. J. Med. Virol. 2020 doi: 10.1002/jmv.26181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pyle C.J., Uwadiae F.I., Swieboda D.P., Harker J.A. Early IL-6 signalling promotes IL-27 dependent maturation of regulatory T cells in the lungs and resolution of viral immunopathology. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen X., Tang J., Shuai W., Meng J., Feng J., Han Z. Macrophage polarization and its role in the pathogenesis of acute lung injury/acute respiratory distress syndrome. Inflamm. Res. : official journal of the European Histamine Research Society [et al] 2020;69:883–895. doi: 10.1007/s00011-020-01378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao J., Yu H., Liu Y., Gibson S.A., Yan Z., Xu X., et al. Protective effect of suppressing STAT3 activity in LPS-induced acute lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 2016;311 doi: 10.1152/ajplung.00281.2016. L868-l80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sang Y., Miller L.C., Blecha F. Macrophage polarization in virus-host interactions. J. Clin. Cell. Immunol. 2015;6 doi: 10.4172/2155-9899.1000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shirey K.A., Lai W., Pletneva L.M., Karp C.L., Divanovic S., Blanco J.C., et al. Role of the lipoxygenase pathway in RSV-induced alternatively activated macrophages leading to resolution of lung pathology. Mucosal Immunol. 2014;7:549–557. doi: 10.1038/mi.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 102.Drareni K., Gautier J.F., Venteclef N., Alzaid F. Transcriptional control of macrophage polarisation in type 2 diabetes. Semin. Immunopathol. 2019;41:515–529. doi: 10.1007/s00281-019-00748-1. [DOI] [PubMed] [Google Scholar]
- 103.Yin Z., Ma T., Lin Y., Lu X., Zhang C., Chen S., et al. IL-6/STAT3 pathway intermediates M1/M2 macrophage polarization during the development of hepatocellular carcinoma. J. Cell. Biochem. 2018;119:9419–9432. doi: 10.1002/jcb.27259. [DOI] [PubMed] [Google Scholar]
- 104.Fu X.L., Duan W., Su C.Y., Mao F.Y., Lv Y.P., Teng Y.S., et al. Interleukin 6 induces M2 macrophage differentiation by STAT3 activation that correlates with gastric cancer progression. Canc. Immunol. Immunother. : CII. 2017;66:1597–1608. doi: 10.1007/s00262-017-2052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mizutani T., Fukushi S., Murakami M., Hirano T., Saijo M., Kurane I., et al. Tyrosine dephosphorylation of STAT3 in SARS coronavirus-infected Vero E6 cells. FEBS Lett. 2004;577:187–192. doi: 10.1016/j.febslet.2004.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Park B.K., Kim D., Park S., Maharjan S., Kim J., Choi J.K., et al. Differential signaling and virus production in Calu-3 cells and vero cells upon SARS-CoV-2 infection. Biomol. Ther. 2021 doi: 10.4062/biomolther.2020.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hojyo S., Uchida M., Tanaka K., Hasebe R., Tanaka Y., Murakami M., et al. How COVID-19 induces cytokine storm with high mortality. Inflamm. Regen. 2020;40:37. doi: 10.1186/s41232-020-00146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zou S., Tong Q., Liu B., Huang W., Tian Y., Fu X. Targeting STAT3 in cancer immunotherapy. Mol. Canc. 2020;19:145. doi: 10.1186/s12943-020-01258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Siveen K.S., Sikka S., Surana R., Dai X., Zhang J., Kumar A.P., et al. Targeting the STAT3 signaling pathway in cancer: role of synthetic and natural inhibitors. Biochim. Biophys. Acta. 2014;1845:136–154. doi: 10.1016/j.bbcan.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 110.Shamir I., Abutbul-Amitai M., Abbas-Egbariya H., Pasmanik-Chor M., Paret G., Nevo-Caspi Y. STAT3 isoforms differentially affect ACE2 expression: a potential target for COVID-19 therapy. J. Cell Mol. Med. 2020;24:12864–12868. doi: 10.1111/jcmm.15838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thilakasiri P.S., Dmello R.S., Nero T.L., Parker M.W., Ernst M., Chand A.L. Repurposing of drugs as STAT3 inhibitors for cancer therapy. Semin. Canc. Biol. 2021;68:31–46. doi: 10.1016/j.semcancer.2019.09.022. [DOI] [PubMed] [Google Scholar]
- 112.Yue P., Turkson J. Targeting STAT3 in cancer: how successful are we? Expet Opin. Invest. Drugs. 2009;18:45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wong A.L.A., Hirpara J.L., Pervaiz S., Eu J.Q., Sethi G., Goh B.C. Do STAT3 inhibitors have potential in the future for cancer therapy? Expet Opin. Invest. Drugs. 2017;26:883–887. doi: 10.1080/13543784.2017.1351941. [DOI] [PubMed] [Google Scholar]
- 114.Sen M., Grandis J.R. Nucleic acid-based approaches to STAT inhibition. JAK-STAT. 2012;1:285–291. doi: 10.4161/jkst.22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang L., Lin S., Xu L., Lin J., Zhao C., Huang X. Novel activators and small-molecule inhibitors of STAT3 in cancer. Cytokine Growth Factor Rev. 2019;49:10–22. doi: 10.1016/j.cytogfr.2019.10.005. [DOI] [PubMed] [Google Scholar]