Abstract
Recently, we reported that chemokine (C-X-C motif) receptor 4 (CXCR4) heteromerizes with α1-adrenergic receptors (AR) on the cell surface of vascular smooth muscle cells, through which the receptors cross-talk. Direct biophysical evidence for CXCR4:α1-AR heteromers, however, is lacking. Here we utilized bimolecular luminescence/fluorescence complementation (BiLC/BiFC) combined with intermolecular bioluminescence resonance energy transfer (BRET) assays in HEK293T cells to evaluate CXCR4:α1a/b/d-AR heteromerization. Atypical chemokine receptor 3 (ACKR3) and metabotropic glutamate receptor 1 (mGlu1R) were utilized as controls. BRET between CXCR4-RLuc (Renilla reniformis) and enhanced yellow fluorescent protein (EYFP)-tagged ACKR3 or α1a/b/d-ARs fulfilled criteria for constitutive heteromerization. BRET between CXCR4-RLuc and EYFP or mGlu1R-EYFP were nonspecific. BRET50 for CXCR4:ACKR3 and CXCR4:α1a/b/d-AR heteromers were comparable. Stimulation of cells with phenylephrine increased BRETmax of CXCR4:α1a/b/d-AR heteromers without affecting BRET50; stimulation with CXCL12 reduced BRETmax of CXCR4:α1a-AR heteromers, but did not affect BRET50 or BRETmax/50 for CXCR4:α1b/d-AR. A peptide analogue of transmembrane domain (TM) 2 of CXCR4 reduced BRETmax of CXCR4:α1a/b/d-AR heteromers and increased BRET50 of CXCR4:α1a/b-AR interactions. A TM4 analogue of CXCR4 did not alter BRET. We observed CXCR4, α1a-AR and mGlu1R homodimerization by BiFC/BiLC, and heteromerization of homodimeric CXCR4 with proto- and homodimeric α1a-AR by BiFC/BiLC BRET. BiFC/BiLC BRET for interactions between homodimeric CXCR4 and homodimeric mGlu1R was nonspecific. Our findings suggest that the heteromerization affinity of CXCR4 for ACKR3 and α1-ARs is comparable, provide evidence for conformational changes of the receptor complexes upon agonist binding and support the concept that proto- and oligomeric CXCR4 and α1-ARs constitutively form higher-order hetero-oligomeric receptor clusters.
Keywords: G protein-coupled receptor heteromers, homodimer, hetero-oligomer, stromal cell-derived factor 1α, bimolecular luminescence complementation assay, bimolecular fluorescence complementation assay
Introduction
Seven transmembrane (7TM) receptors, of which most are G protein-coupled receptors (GPCRs), form the largest class of membrane proteins in humans [1,2]. 7TM receptors are essential in human physiology, play major roles in a broad variety of disease processes and are the targets of more than one third of all drugs approved by the Federal Drug Administration [1,2].
Accumulating evidence suggests that many 7TM receptors can form homo- and hetero-oligomers, which is thought to alter their pharmacological behavior and affect receptor function [3–7]. While the function of the prototypical GPCR chemokine (C-X-C motif) receptor 4 (CXCR4) appears to depend on the formation of homo-oligomers (nano-clusters) on the cell surface [8,9], CXCR4 has been reported to heteromerize with several other 7TM receptors, such as chemokine (C-C motif) receptor (CCR)2, CCR5, CXCR3, atypical chemokine receptor (ACKR) 3, chemerin receptor 23, β2-adrenergic receptor (AR), δ-opioid receptor or cannabinoid receptor 2 [10–17]. Recently, we provided evidence that heteromeric complexes between CXCR4 and α1-adrenergic receptors (AR) are constitutively expressed in rat and human vascular smooth muscle cells (VSMCs), through which the receptor partners regulate each other [18–21]. In these studies, we employed biochemical methods, such as proximity ligation assays and co-immunoprecipitation experiments, in combination with functional assays to demonstrate heteromerization and functional cross-talk between recombinant and endogenously expressed CXCR4 and α1-ARs. Biophysical evidence for interactions between CXCR4 and α1-ARs, however, is lacking. Furthermore, information on the composition and molecular characteristics of such complexes is not available.
In the present study we utilized bimolecular luminescence and fluorescence complementation (BiLC, BiFC) and intermolecular bioluminescence resonance energy transfer (BRET) assays, which are considered a gold standard for the detection of direct physical interactions between GPCRs [22], to evaluate heteromerization between CXCR4 and α1-ARs. Our findings support the concept that CXCR4 constitutively heteromerizes with α1-ARs in higher-order hetero-oligomeric complexes comprised of proto- and oligomeric receptor partners and provide initial information on the molecular behavior of such complexes.
Materials and methods
Reagents –
Phenylephrine was purchased from Sigma-Aldrich and (C-X-C motif) chemokine ligand 12 (CXCL12) from Protein Foundry. The peptide analogues derived from the transmembrane domains 2 (TM2, LLFVITLPFWAVDAVANWYFGN) and 4 (TM4, VYVGVWIPALLLTIPDFIFAN) of CXCR4 were as described previously [18,20]; their sequences and purity were confirmed by mass spectrometry. Coelenterazine H was from Nanolight Technology.
Plasmids –
The coding sequences of CXCR4, ACKR3, α1a-AR, α1b-AR, α1d-AR and mGlu1R were from Addgene (CXCR4-TANGO, #66262; ACKR3-TANGO, #66265, α1a-AR-TANGO, #66213, α1b-AR-TANGO, #66214, α1d-AR-TANGO, #66215, mGlu1-Tango, #66387). Upper and lower case subscripts are used to denote endogenous and recombinant α1-ARs, respectively [23]. The coding sequence of Renilla luciferase was from CXCR4-hRLuII, which was generously provided by Dr. Michel Bouvier, PCR amplified and ligated at the C-terminus of CXCR4 at the sites of Age I and Xba I (CXCR4-RLuc). To produce ACKR3-enhanced yellow fluorescent protein (EYFP), α1a-AR-EYFP, α1b-AR-EYFP, α1d-AR-EYFP and mGlu1R-EYFP, the cDNA of EYFP was PCR amplified and ligated in-frame with the receptor genes at the C-termini at the sites of Age I and Xba I, respectively. To construct CXCR4 fused with split luciferase, the coding sequence of Renilla luciferase was PCR amplified into two segments, L1 (N-Luc - AA1–229) and L2 (C-Luc - AA230–311) [24], and inserted in pcDNA3.1-CXCR4 at the sites of Age I and Xba I. To construct CXCR4 fused with split-yellow fluorescent protein Venus (V1, N- Venus - AA1–155; V2, C-Venus - AA156–240), V1 and V2 were PCR amplified from D2R-V1 and D2R-V2 (both from Addgene) with a primer that carries the Age I site and matches D2R-V1 and V2 linker sequences and primer sp6 [24]. The amplicons were fused with CXCR4 in-frame at the C-terminus at the Age I and Xba I sites, respectively. α1a-AR and mGlu1R fused with split luciferase fragments (L1 or L2), α1a-AR-L1, α1a-AR-L2, mGlu1-L1 and mGlu1-L2, were generated as for CXCR4-L1 and CXCR4-L2. All plasmids were confirmed by sequencing.
Cell culture –
HEK 293T were as described [21] and cultured in high-glucose Dulbecco’s Modified Eagle’s Medium containing 10 mg/mL sodium pyruvate, 2 mM L-glutamine, 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin in a humidified environment at 37°C, 5% CO2.
BRET assays –
BRET assays were performed as described previously [25,26]. In brief, HEK293T cells were seeded in 12-well plates and transfected with the plasmids indicated using the Lipofectamine 3000 transfection reagent (ThermoScientific). For BRET titration assays, CXCR4-RLuc at a fixed amount of 50 ng was transfected alone or with increasing amounts of EYFP, mGlu1R-EYFP, ACKR3-EYFP or α1a/b/d-AR-EYFP. For BRET assays at a constant energy donor : acceptor ratio [27], increasing amounts of both CXCR4-RLuc and ACKR3-EYFP or α1a/b/d-AR-EYFP were co-transfected at a ratio of 1:10. In all assays, empty vector pcDNA3.1 was added to maintain the total DNA amount for each transfection constant. After an overnight incubation, cells were seeded in poly-L-lysine coated 96-well white plates and incubated again overnight. Cells were then washed with PBS and fluorescence was measured in a Biotek Synergy HT4 plate reader (excitation 485 nm, emission 528 nm). For BRET measurements, coelenterazine H was added at a final concentration of 5 μM. After 10 min incubation at room temperature, luminescence was measured at 460 nm and 528 nm. The BRET signal was calculated as the ratio of the relative luminescence units (RLU) measured at 528 nm over RLU at 460 nm. The net BRET is calculated by subtracting the BRET signal detected when CXCR4-hRLuc was transfected alone. For titration experiments, net BRET ratios are expressed as a function of fluorescence/total luminescence. To test the effects of receptor agonists on BRET, cells were replaced with PBS after 48h of transfection, phenylephrine (final concentration 200 μM) or CXCL12 (final concentration 500 nM) were added and cells were incubated at 37°C for 5 min before the addition of coelenterazine H. To test the effects of TM-derived peptides on BRET, cells were replaced with PBS after 48h of transfection, the TM2 or TM4 peptide were added in a final concentration of 20 μM and cells were incubated at 37°C for 15 min before the addition of coelenterazine H.
Bimolecular luminescence and fluorescence complementation BRET –
For bimolecular luminescence complementation assays (BiLC), CXCR4-L1, α1a-AR-L1 or mGlu1R-L1 at a fixed amount were co-transfected with increasing amounts of CXCR4-L2, α1a-AR-L2 or mGlu1R-L2, respectively. For bimolecular fluorescence complementation (BiFC) assays, CXCR4-V1 at a fixed amount was co-transfected with increasing amounts of CXCR4-V2. After overnight transfection, cells were seeded in poly L-lysine precoated 96 well plates and incubated further overnight before detection of luminescence or fluorescence. For the BiLC BRET assay, CXCR4-L1 and CXCR4-L2 at fixed amounts were co-transfected with increasing amounts of α1a-AR-EYFP. For the BiFC BRET assay, α1a-AR-RLuc or mGlu1R-RLuc at constant amounts were co-transfected with increasing amounts of CXCR4-V1 and CXCR4-V2. For combined BiLC and BiFC BRET, α1a-AR-L1 and α1a-AR-L2 or mGlu1R-L1 and mGlu1R-L2 at constant amounts were transfected with increasing amounts of CXCR4-V1 and CXCR4-V2. BRET was measured as described before.
Data analyses –
Data are expressed as mean ± standard error. Titration curves were analyzed with nonlinear regression analyses. One-way analyses of variance (ANOVA) with Dunnett’s multiple comparison post hoc test for multiple comparisons were used to assess statistical significance. A two-tailed p<0.05 was considered significant. All analyses were calculated with the GraphPad Prism 8.0.2 software.
Results and Discussion
We performed saturation BRET experiments in cells expressing CXCR4-RLuc and α1a/b/d-ARs-EYFP. Saturation BRET experiments in cells expressing CXCR4-RLuc and ACKR3-EYFP were used as a positive control, and in cells expressing CXCR4-hRLuc and EYFP or mGlu1R-EYFP as negative controls [8,11,15,28]. As shown in Fig. 1A/B, the BRET signals between CXCR4-RLuc and ACKR3-EYFP or α1a/b/d-AR-EYFP showed hyperbolic progressions with increasing energy acceptor : donor ratios. Consistent with a non-specific bystander BRET signal (open squares), the BRET signal was low and increased linearly with increasing energy acceptor : donor ratios in cells expressing CXCR4-RLuc and EYFP (Fig. 1A) or CXCR4-RLuc and mGluR1-EYFP (Fig. 1B). The BRET signals for interactions between CXCR4 and ACKR3 or α1a/b/d-ARs were independent of the concentrations of BRET partners when tested at fixed energy acceptor : donor ratios (Fig. 1C). Thus, the observed BRET between CXCR4-RLuc and ACKR3-EYFP or α1a/b/d-AR-EYFP indicate constitutive heteromerization of the receptor partners and support the assumption of direct physical interactions between CXCR4 and α1-ARs [18–21,27]. Comparison of the BRET50 values for each heteromerization pair did not show significant differences (BRET50 (EYFP/Lum): CXCR4:ACKR3 – 0.062 ± 0.004; CXCR4:α1a-AR - 0.055 ± 0.011; CXCR4:α1b-AR - 0.072 ± 0.017; CXCR4:α1d-AR - 0.051 ± 0.018; n=4 for all, p>0.05), suggesting that CXCR4 has a comparable interaction affinity for ACKR3 and α1a/b/d-AR.
Figure 1:
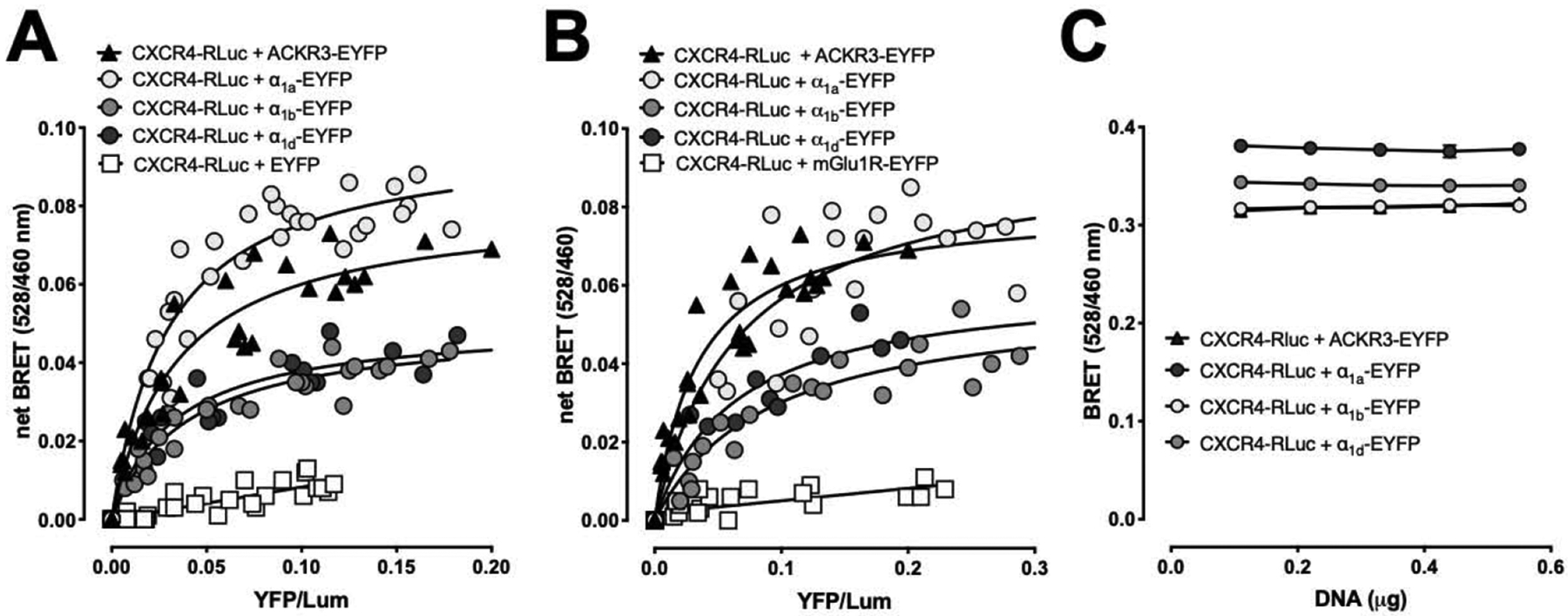
BRET assays indicate that CXCR4 interacts with α1a/b/d-ARs. Graphs are representative of at least three independent experiments. A/B. HEK293T cells were transfected with a fixed amount of CXCR4-Rluc and increasing amounts of ACKR3-EYFP, α1a/b/d-AR-EYFP, EYFP (A) or mGlu1R-EYFP (B). 48 h after transfection, EYFP fluorescence and luminescence were read as described in Methods. Net BRET (528nm/460nm) was plotted against EYFP/luminescence (YFP/Lum). C. HEK293T cells were transfected with increasing amounts of both CXCR4-Rluc and ACKR3-EYFP or :α1a/b/d-AR-EYFP at a fixed ratio (1:10) in quadruplicate. Raw BRET (528nm/460nm) was plotted against total DNA amounts transfected.
Net BRETmax (n=4 for all combinations) was significantly higher for CXCR4:ACKR3 (528nm/460nm: 0.080 ± 0.011) and CXCR4:α1a-AR (528nm/460nm: 0.093 ± 0.006), as compared with net BRETmax for CXCR4:α1b-AR (528nm/460nm: 0.053 ± 0.004, p<0.05 vs. CXCR4:ACKR3 and CXCR4:α1a-AR) and CXCR4:α1d-AR (528nm/460nm: 0.052 ± 0.008, p<0.05 vs. CXCR4:ACKR3 and CXCR4:α1a-AR). These observations, however, are difficult to interpret as the BRETmax signal between CXCR4-RLuc and the EYFP-tagged receptor partners depends on the relative orientation or distance between the energy donor and acceptor, which may differ among the various receptor constructs [27].
Because BRET for various GPCR homo- and heteromers has been reported to be modulated upon agonist binding, we tested whether the selective α1-AR agonist phenylephrine and the cognate CXCR4 agonist CXCL12 affect BRET between CXCR4 and α1a/b/d-AR. Fig. 2 shows typical titration BRET experiments for interactions between CXCR4 and α1a/b/d-ARs (top) and summarizes BRET50 (center) and BRETmax (bottom) values from four independent experiments when cells were exposed to phenylephrine or CXCL12. As compared with untreated cells, phenylephrine significantly increased BRETmax of CXCR4:α1a/b/d-AR heteromers. CXCL12 reduced BRETmax of CXCR4:α1a-AR heteromers but did not affect BRETmax between CXCR4 and α1b/d-AR. Both agonists did not significantly alter BRET50 values. These findings are similar to previous observations on the CXCR4:CCR2 heteromer, which have been interpreted to reflect agonist-induced conformational changes of the receptor complex [25]. Whether the ligand-induced changes in BRETmax that we observed for the CXCR4:α1-AR heteromers also reflect increases in CXCR4:α1a/b/d-AR heteromerization upon binding of phenylephrine to α1-ARs and reduced CXCR4:α1a-AR heteromerization upon binding of CXCL12 to CXCR4, remains unclear because the apparent interaction affinity was not affected by agonist binding. Nevertheless, these observations further support the concept that the BRET signals for CXCR4:α1-AR heteromers are specific because such agonist-induced changes would not be expected for non-specific bystander BRET signals [25].
Figure 2:
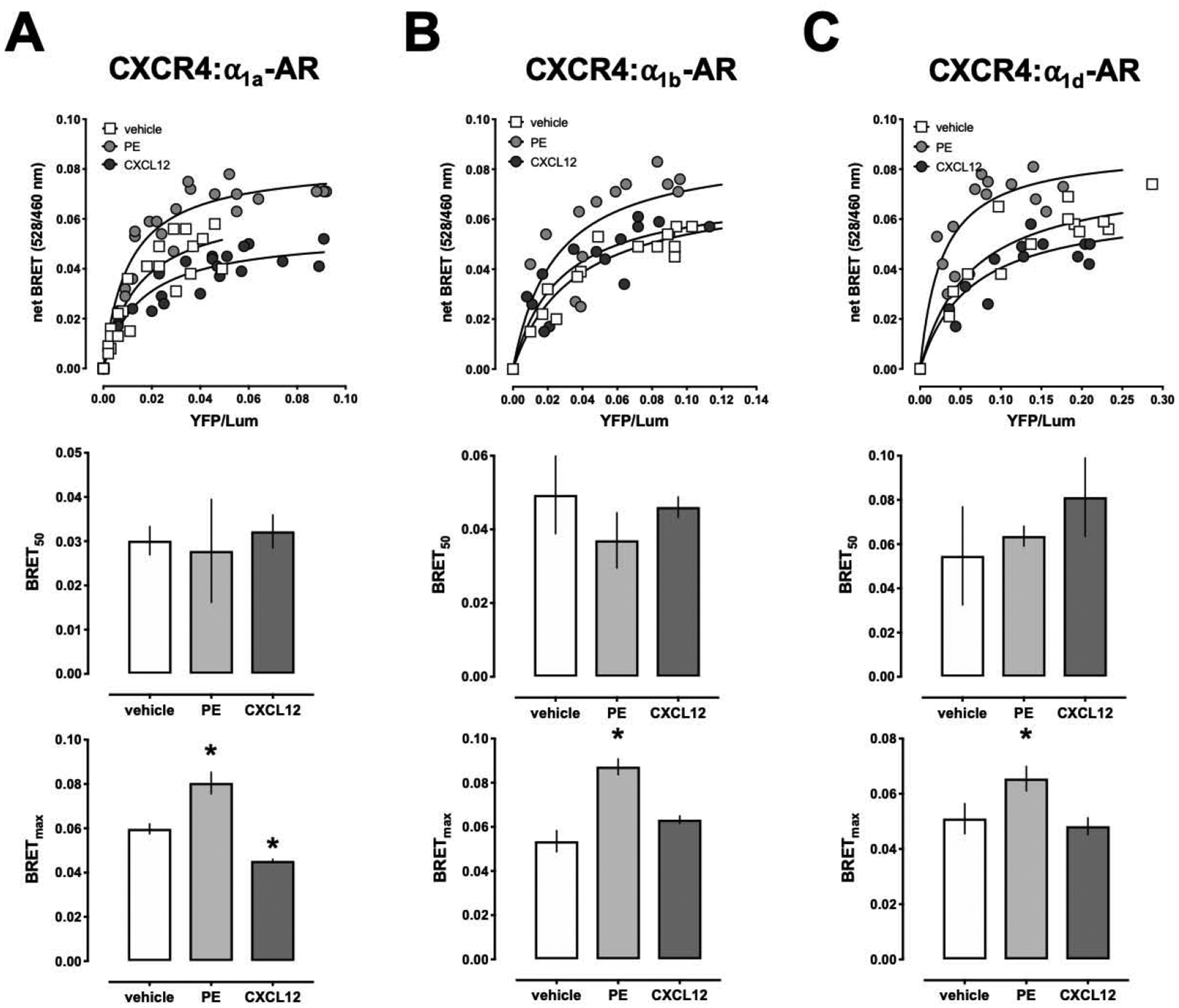
Effects of phenylephrine and CXCL12 on BRET between CXCR4 and α1a/b/d-AR. HEK293T cells were co-transfected with a fixed amount of CXCR4-Rluc and increasing amounts α1a-AR-EYFP (A), α1b-AR-EYFP (B) or α1d-AR-EYFP (C). 48 h after transfection, cells were treated with vehicle (=control), phenylephrine (PE, 200 μM) or CXCL12 (500 nM) for 5 min at 37°C before measuring BRET. Top: Representative measurements from a titration BRET experiment. Center: BRET50 from n=4 independent titration BRET experiments. Bottom: BRETmax from n=4 independent titration BRET experiments. *: p<0.05 vs. control.
We showed previously in proximity ligation assays that a peptide analogue of TM2 of CXCR4 selectively reduces signals corresponding to endogenously expressed CXCR4:α1A/B-AR heteromers in human vascular smooth muscle cells, signals corresponding to recombinant CXCR4:α1b-AR heteromers in HEK293T cells and inhibits β-arrestin cross-recruitment to CXCR4 within the CXCR4:α1b-AR heteromeric complex upon phenylephrine stimulation in Presto-Tango assays in HTLA cells [18,19,21]. Consistent with these observations, the TM2 peptide analogue of CXCR4 significantly increased BRET50 and reduced BRETmax of CXCR4:α1a/b-AR heteromers (Fig. 3A/B), which provides direct evidence that the TM2 peptide analogue interferes with CXCR4:α1a/b-AR heteromerization. While the observation that the TM2 peptide analogue reduced BRETmax but did not increase BRET50 of CXCR4:α1d-AR heteromers implies conformational changes of the heteromeric receptor complex (Fig. 3C), interference with the formation of CXCR4:α1d-AR heteromers cannot be directly inferred from these findings. Furthermore, complimentary data from other assays, such as proximity ligation assays, on the effects of this peptide on CXCR4:α1d-AR heteromers are not available. Thus, we can currently not exclude that CXCR4:α1a/b-AR and CXCR4:α1d-AR heteromerization differs in regard to the underlying structural determinants. Our finding that the TM4 peptide analogue of CXCR4 did not interfere with BRET between CXCR4 and α1a/b/d-ARs is in agreement with the previously described effects of this peptide on CXCR4:α1A-AR heteromers in human vascular smooth muscle cells and indicates selectivity of the effects of the TM2 peptide analogue [19].
Figure 3:
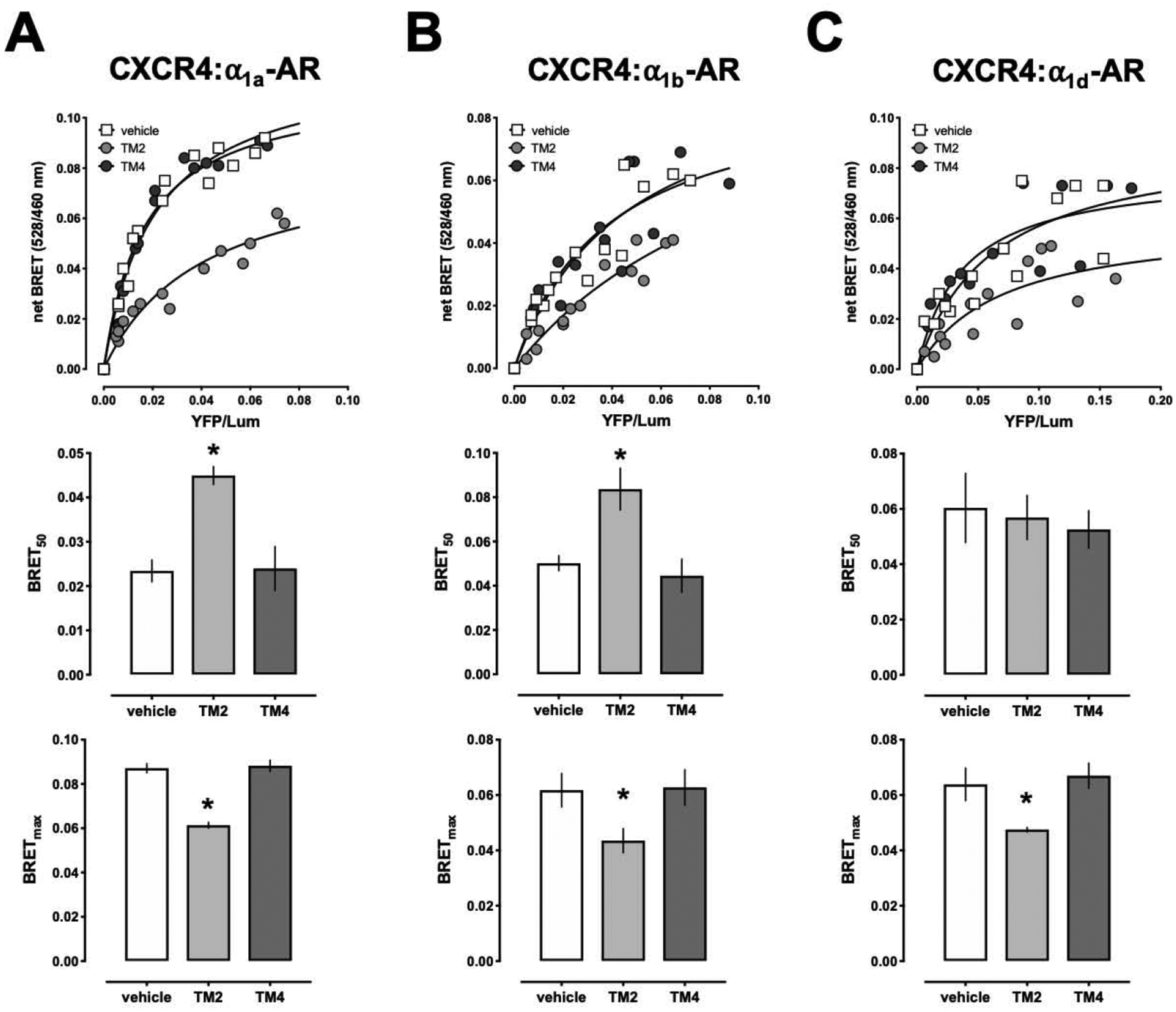
A peptide analogue derived from TM2 of CXCR4 interferes with BRET between CXCR4 and α1a/b/d-ARs. HEK293T cells were co-transfected with a fixed amount of CXCR4-Rluc and increasing amounts of α1a-AR-EYFP (A), α1b-AR-EYFP (B) or α1d-AR-EYFP (C). 48 h after transfection, cells were treated with vehicle (=control), TM2 or TM4 peptides (20 μM) for 15 min at 37°C before measuring BRET. Top: Representative measurements from a titration BRET experiment. Center: BRET50 from n=4 independent titration BRET experiments. *: p<0.05 vs. control. Bottom: BRETmax from n=4 independent titration BRET experiments. *: p<0.05 vs. control.
Because TM-derived peptide analogues of GPCRs can affect receptor dimerization/oligomerization through interference with the correct assembly of the target membrane protein [29,30], the effects of the TM2 peptide analogue that we observed previously and its effects on BRET in the present study may point towards TM2 of CXCR4 as a possible heteromerization interface for α1-ARs [18,19,21]. Crystallographic structures revealed CXCR4 as a homodimer with main interfaces at TM5 and TM6 [31]. Accordingly, we have previously in silico developed a structure-based heteromeric receptor model, in which TM2 of the CXCR4 homodimer serves as heteromerization interface for α1A-AR [19]. In this model heteromerization of CXCR4 with α1A-AR would not interfere with the formation of CXCR4 homodimers.
To test whether the CXCR4 homodimer is able to heteromerize with α1a-AR, we employed BiLC and BiFC assays to detect of CXCR4 homodimerization and tested BiLC and BiFC BRET to assess interactions with α1a-AR. In agreement with homodimerization of CXCR4, we observed strong and robust luminescence and fluorescence signals in cells transfected with CXCR4-L1 and CXCR4-L2 (Fig. 4A) and in cells transfected with CXCR4-V1 and CXCR4-V2, respectively (Fig. 4B). Luminescence and fluorescence signals in cells transfected with CXCR4-L1 and CXCR4-L2 alone or with CXCR4-V1 and CXCR4-V2 alone, respectively, were negligible. As shown in Fig. 4C, in cells transfected with constant amounts of CXCR4-L1 and CXCR4-L2 and with increasing amounts of α1a-AR-EYFP, the BRET signal showed a hyperbolic progression, whereas BRET in cells transfected with constant amounts of CXCR4-L1 and CXCR4-L2 and with increasing amounts of EYFP was low and increased linearly. Similarly, the BRET signal showed hyperbolic progression when cells were transfected with constant amounts of α1a-AR-RLuc and with increasing amounts of CXCR4-V1 and CXCR4-V2 (Fig. 4D). In contrast, the BRET signal in cells expressing constant amounts of mGlu1R-RLuc and increasing amounts of CXCR4-V1 and CXCR4-V2 was low and increased linearly, indicating that neither the CXCR4 protomer nor the CXCR4 homodimer heteromerize with mGlu1R (Fig. 4D). These findings provide evidence that the CXCR4 homodimer heteromerizes with α1a-AR. This implies that α1a-AR interacts with CXCR4 via a heteromerization interface that is different from the homodimerization interface of CXCR4 and experimentally supports our in silico interaction model.
Figure 4:
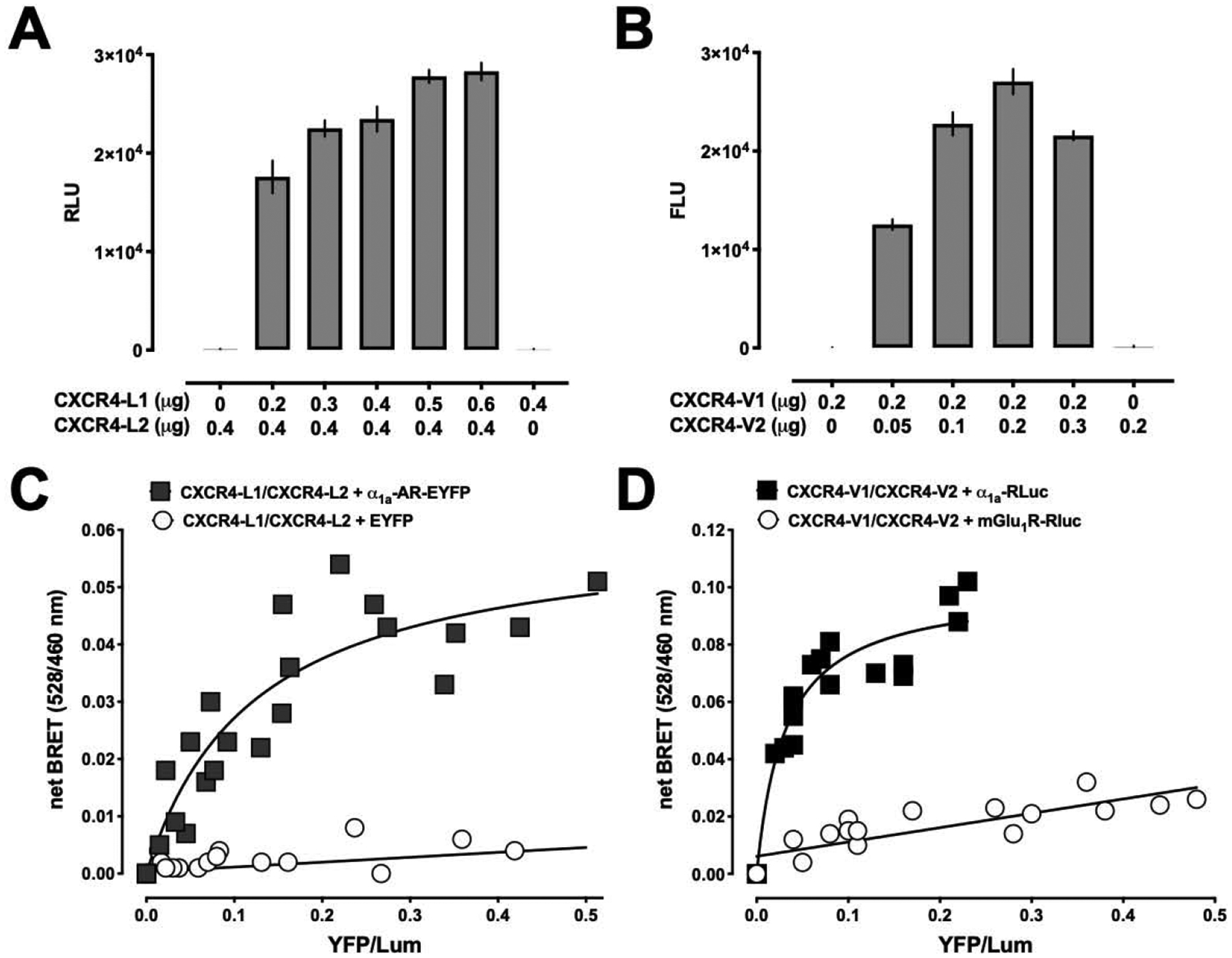
The CXCR4 homodimer interacts with α1a-AR. A/B. Bimolecular luminescence complementation (BiLC, A) and bimolecular fluorescence complementation (BiFC, B) assays to detect dimeric CXCR4. HEK293T cells were co-transfected with CXCR4-L1/2 or CXCR4-V1/2, as indicated. Luminescence was read after the addition of coelenterazine H. N=3. C/D. BiLC and BiFC BRET indicate that the CXCR4 homodimer interacts with α1a-AR. HEK293T cells were co-transfected with a constant amount of CXCR4-L1/2 and with increasing amounts of α1a-AR -EYFP or EYFP (control, C), or with a constant amount of α1a-AR-RLuc or mGlu1R-RLuc and with increasing amounts of CXCR4-V1/2 (control, D). 48 h after transfection, EYFP fluorescence and luminescence were read as described in the Methods. The net BRET (528/460) is plotted against YFP/Lum. The figure is representative of three independent experiments.
Because α1a-AR and mGlu1R are known to form homodimers [32,33], we then tested whether the CXCR4 homodimer may interact with homodimeric α1a-AR or mGlu1R. BiLC assays showed strong luminescence signals in cells co-expressing mGlu1R-L1 and mGlu1R-L2 (Fig. 5A) or α1a-AR-L1 and α1a-AR-L2 (Fig. 5B), respectively, which confirms homodimerization of both receptors. While BiLC and BiFC BRET signals for interactions between homodimeric CXCR4 and homodimeric α1a-AR showed hyperbolic progression with increasing energy acceptor-donor ratios, the signals for interactions between homodimeric CXCR4 and homodimeric mGlu1R were low and increased linearly. These findings suggest that CXCR4 and α1a-AR interact in higher order receptor complexes, whereas oligomeric CXCR4 does not form higher order complexes with oligomeric mGlu1R.
Figure 5:
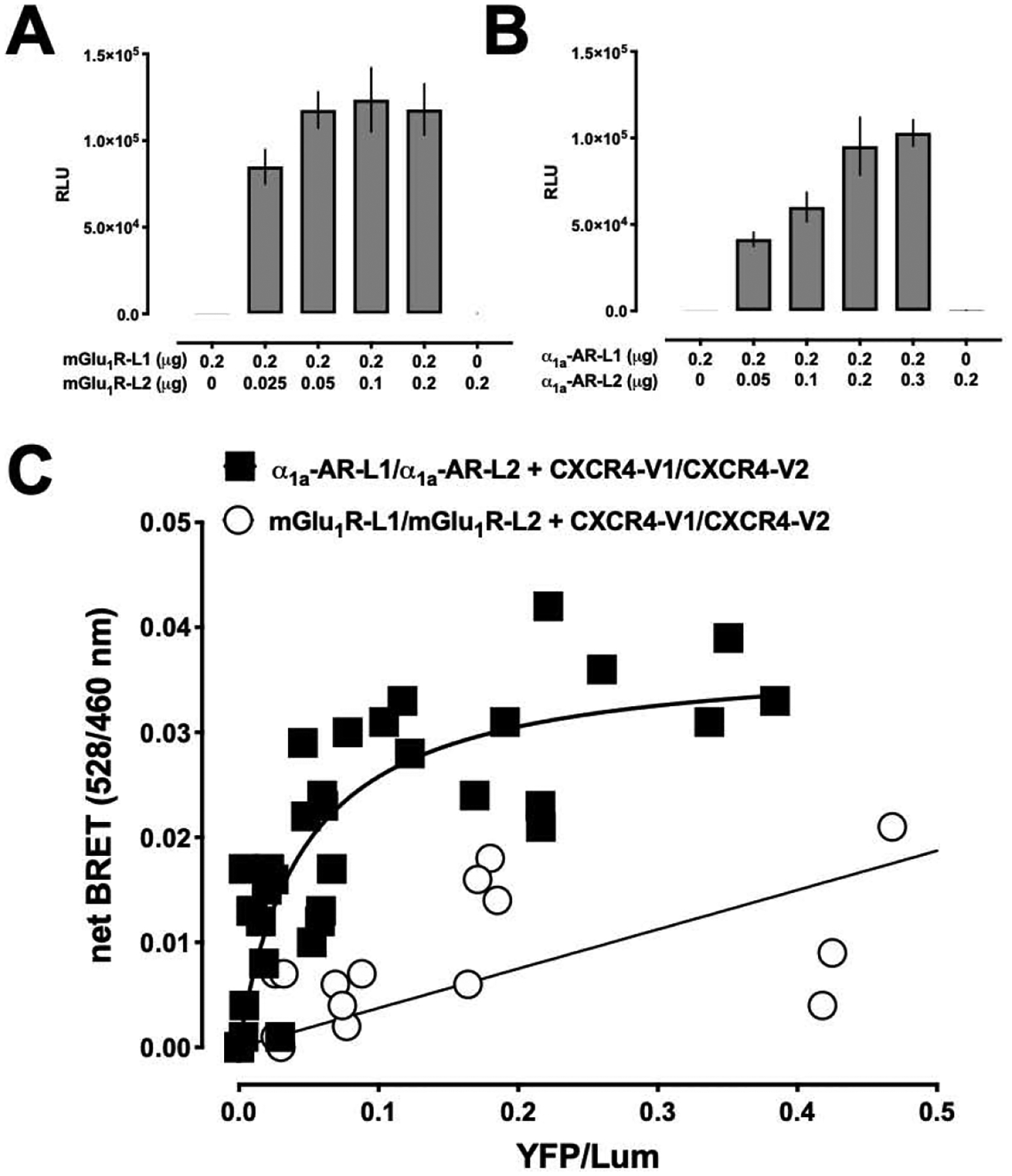
Homodimeric CXCR4 interacts with homodimeric α1a-AR. A/B. Bimolecular luminescence complementation (BiLC) assays to detect dimeric mGLu1R (A) and dimeric α1a-AR (B). HEK293T cells were co-transfected with mGLu1R-L1/2 or α1a-AR-L1/2, as indicated. Luminescence was read after the addition of coelenterazine H. N=3. C. Combined BiLC and BiFC BRET indicates that the CXCR4 homodimer interacts with the α1a-AR homodimer, but not with the mGlu1R homodimer. HEK293T cells were co-transfected with constant amounts of α1a-AR-L1/2 or mGlu1R-L1/L2 and with increasing amounts of CXCR4-V1/V2. 48 h after transfection, EYFP fluorescence and luminescence were read as described in the Methods. The net BRET (528/460) is plotted against YFP/Lum. The figure is representative of three independent experiments.
In conclusion, in the present study we provide biophysical evidence that CXCR4 constitutively heteromerizes with α1-ARs, thus supporting our previous observations from other test systems. Our findings further suggest that the interaction affinities of CXCR4 for ACKR3 and α1-ARs are comparable and that agonist-binding results in conformational changes of the CXCR4:α1-AR complexes. Moreover, we provide evidence that proto- and oligomeric CXCR4 and α1a-AR constitutively form higher-order hetero-oligomeric receptor clusters, which provides initial insights into the molecular composition of these GPCR hetero-oligomers.
We are currently unable to assess interactions of more than four receptor protomers. Nevertheless, previous observations suggest that α1-ARs heterodimerize among each other, that ACKR3 dimerizes with α1a-AR and that the CXCR4:ACKR3 heteromer interacts with α1b/d-ARs [20,32,34]. In combination with the finding from the present study that the interaction affinities of CXCR4 for ACKR3 and α1-ARs are comparable, these data point toward a not anticipated complexity of the organizational structure of these 7TM receptor clusters, which may be comprised of multiple proto- and homo-oligomeric receptors. Further studies will be required to address this possibility.
Supplementary Material
Highlights.
Bimolecular luminescence/fluorescence complementation combined with intermolecular bioluminescence resonance energy transfer assays were utilized
Proto- and oligomeric CXCR4 and α1-adrenoceptors form higher-order hetero-oligomeric receptor complexes
The heteromerization affinity of CXCR4 for ACKR3 and α1-adrenoceptors is comparable
CXCR4:α1-adrenoceptor hetero-oligomers undergo conformational changes upon agonist binding
Funding
Research reported in this publication was supported by the National Institutes of Health under award numbers R01GM107495, R21AA025750 and R21AI139827. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Alexander SP, Christopoulos A, Davenport AP, Kelly E, Marrion NV, Peters JA, Faccenda E, Harding SD, Pawson AJ, Sharman JL, Southan C, Davies JA, Collaborators C, THE CONCISE GUIDE TO PHARMACOLOGY 2017/18: G protein-coupled receptors, Br J Pharmacol 174 Suppl 1 (2017) S17–S129. 10.1111/bph.13878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sharman JL, Benson HE, Pawson AJ, Lukito V, Mpamhanga CP, Bombail V, Davenport AP, Peters JA, Spedding M, Harmar AJ, Nc I, IUPHAR-DB: updated database content and new features, Nucleic Acids Res 41 (2013) D1083–1088. 10.1093/nar/gks960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Quitterer U, AbdAlla S, Discovery of Pathologic GPCR Aggregation, Front Med (Lausanne) 6 (2019) 9. 10.3389/fmed.2019.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gaitonde SA, Gonzalez-Maeso J, Contribution of heteromerization to G protein-coupled receptor function, Curr Opin Pharmacol 32 (2017) 23–31. 10.1016/j.coph.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Franco R, Martinez-Pinilla E, Lanciego JL, Navarro G, Basic Pharmacological and Structural Evidence for Class A G-Protein-Coupled Receptor Heteromerization, Front Pharmacol 7 (2016) 76. 10.3389/fphar.2016.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ferre S, Baler R, Bouvier M, Caron MG, Devi LA, Durroux T, Fuxe K, George SR, Javitch JA, Lohse MJ, Mackie K, Milligan G, Pfleger KD, Pin JP, Volkow ND, Waldhoer M, Woods AS, Franco R, Building a new conceptual framework for receptor heteromers, Nat Chem Biol 5 (2009) 131–134. 10.1038/nchembio0309-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gomes I, Ayoub MA, Fujita W, Jaeger WC, Pfleger KD, Devi LA, G Protein-Coupled Receptor Heteromers, Annu Rev Pharmacol Toxicol 56 (2016) 403–425. 10.1146/annurev-pharmtox-011613-135952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Martinez-Munoz L, Rodriguez-Frade JM, Barroso R, Sorzano COS, Torreno-Pina JA, Santiago CA, Manzo C, Lucas P, Garcia-Cuesta EM, Gutierrez E, Barrio L, Vargas J, Cascio G, Carrasco YR, Sanchez-Madrid F, Garcia-Parajo MF, Mellado M, Separating Actin-Dependent Chemokine Receptor Nanoclustering from Dimerization Indicates a Role for Clustering in CXCR4 Signaling and Function, Mol Cell 70 (2018) 106–119 e110. 10.1016/j.molcel.2018.02.034. [DOI] [PubMed] [Google Scholar]
- [9].Ge B, Lao J, Li J, Chen Y, Song Y, Huang F, Single-molecule imaging reveals dimerization/oligomerization of CXCR4 on plasma membrane closely related to its function, Sci Rep 7 (2017) 16873. 10.1038/s41598-017-16802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Watts AO, van Lipzig MM, Jaeger WC, Seeber RM, van Zwam M, Vinet J, van der Lee MM, Siderius M, Zaman GJ, Boddeke HW, Smit MJ, Pfleger KD, Leurs R, Vischer HF, Identification and profiling of CXCR3-CXCR4 chemokine receptor heteromer complexes, Br J Pharmacol 168 (2013) 1662–1674. 10.1111/bph.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sohy D, Yano H, de Nadai P, Urizar E, Guillabert A, Javitch JA, Parmentier M, Springael JY, Hetero-oligomerization of CCR2, CCR5, and CXCR4 and the protean effects of “selective” antagonists, J Biol Chem 284 (2009) 31270–31279. 10.1074/jbc.M109.054809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].LaRocca TJ, Schwarzkopf M, Altman P, Zhang S, Gupta A, Gomes I, Alvin Z, Champion HC, Haddad G, Hajjar RJ, Devi LA, Schecter AD, Tarzami ST, beta2-Adrenergic receptor signaling in the cardiac myocyte is modulated by interactions with CXCR4, Journal of cardiovascular pharmacology 56 (2010) 548–559. 10.1097/FJC.0b013e3181f713fe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].de Poorter C, Baertsoen K, Lannoy V, Parmentier M, Springael JY, Consequences of ChemR23 heteromerization with the chemokine receptors CXCR4 and CCR7, PLoS One 8 (2013) e58075. 10.1371/journal.pone.0058075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sohy D, Parmentier M, Springael JY, Allosteric transinhibition by specific antagonists in CCR2/CXCR4 heterodimers, J Biol Chem 282 (2007) 30062–30069. 10.1074/jbc.M705302200. [DOI] [PubMed] [Google Scholar]
- [15].Decaillot FM, Kazmi MA, Lin Y, Ray-Saha S, Sakmar TP, Sachdev P, CXCR7/CXCR4 heterodimer constitutively recruits beta-arrestin to enhance cell migration, The Journal of biological chemistry 286 (2011) 32188–32197. 10.1074/jbc.M111.277038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pello OM, Martinez-Munoz L, Parrillas V, Serrano A, Rodriguez-Frade JM, Toro MJ, Lucas P, Monterrubio M, Martinez AC, Mellado M, Ligand stabilization of CXCR4/delta-opioid receptor heterodimers reveals a mechanism for immune response regulation, Eur J Immunol 38 (2008) 537–549. 10.1002/eji.200737630. [DOI] [PubMed] [Google Scholar]
- [17].Coke CJ, Scarlett KA, Chetram MA, Jones KJ, Sandifer BJ, Davis AS, Marcus AI, Hinton CV, Simultaneous Activation of Induced Heterodimerization between CXCR4 Chemokine Receptor and Cannabinoid Receptor 2 (CB2) Reveals a Mechanism for Regulation of Tumor Progression, J Biol Chem 291 (2016) 9991–10005. 10.1074/jbc.M115.712661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tripathi A, Vana PG, Chavan TS, Brueggemann LI, Byron KL, Tarasova NI, Volkman BF, Gaponenko V, Majetschak M, Heteromerization of chemokine (C-X-C motif) receptor 4 with alpha1A/B-adrenergic receptors controls alpha1-adrenergic receptor function, Proc Natl Acad Sci U S A 112 (2015) E1659–1668. 10.1073/pnas.1417564112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Evans AE, Tripathi A, LaPorte HM, Brueggemann LI, Singh AK, Albee LJ, Byron KL, Tarasova NI, Volkman BF, Cho TY, Gaponenko V, Majetschak M, New Insights into Mechanisms and Functions of Chemokine (C-X-C Motif) Receptor 4 Heteromerization in Vascular Smooth Muscle, Int J Mol Sci 17 (2016). 10.3390/ijms17060971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Albee LJ, Eby JM, Tripathi A, LaPorte HM, Gao X, Volkman BF, Gaponenko V, Majetschak M, alpha1-Adrenergic Receptors Function Within Hetero-Oligomeric Complexes With Atypical Chemokine Receptor 3 and Chemokine (C-X-C motif) Receptor 4 in Vascular Smooth Muscle Cells, J Am Heart Assoc 6 (2017). 10.1161/JAHA.117.006575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gao X, Albee LJ, Volkman BF, Gaponenko V, Majetschak M, Asymmetrical ligand-induced cross-regulation of chemokine (C-X-C motif) receptor 4 by alpha1-adrenergic receptors at the heteromeric receptor complex, Sci Rep 8 (2018) 2730. 10.1038/s41598-018-21096-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Szalai B, Hoffmann P, Prokop S, Erdelyi L, Varnai P, Hunyady L, Improved methodical approach for quantitative BRET analysis of G Protein Coupled Receptor dimerization, PLoS One 9 (2014) e109503. 10.1371/journal.pone.0109503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bylund DB, Eikenberg DC, Hieble JP, Langer SZ, Lefkowitz RJ, Minneman KP, Molinoff PB, Ruffolo RR Jr., Trendelenburg U, International Union of Pharmacology nomenclature of adrenoceptors, Pharmacol Rev 46 (1994) 121–136. [PubMed] [Google Scholar]
- [24].Guo W, Urizar E, Kralikova M, Mobarec JC, Shi L, Filizola M, Javitch JA, Dopamine D2 receptors form higher order oligomers at physiological expression levels, EMBO J 27 (2008) 2293–2304. 10.1038/emboj.2008.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Percherancier Y, Berchiche YA, Slight I, Volkmer-Engert R, Tamamura H, Fujii N, Bouvier M, Heveker N, Bioluminescence resonance energy transfer reveals ligand-induced conformational changes in CXCR4 homo- and heterodimers, J Biol Chem 280 (2005) 9895–9903. 10.1074/jbc.M411151200. [DOI] [PubMed] [Google Scholar]
- [26].Albee LJ, LaPorte HM, Gao X, Eby JM, Cheng YH, Nevins AM, Volkman BF, Gaponenko V, Majetschak M, Identification and functional characterization of arginine vasopressin receptor 1A : atypical chemokine receptor 3 heteromers in vascular smooth muscle, Open Biol 8 (2018). 10.1098/rsob.170207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Marullo S, Bouvier M, Resonance energy transfer approaches in molecular pharmacology and beyond, Trends Pharmacol Sci 28 (2007) 362–365. 10.1016/j.tips.2007.06.007. [DOI] [PubMed] [Google Scholar]
- [28].Levoye A, Balabanian K, Baleux F, Bachelerie F, Lagane B, CXCR7 heterodimerizes with CXCR4 and regulates CXCL12-mediated G protein signaling, Blood 113 (2009) 6085–6093. 10.1182/blood-2008-12-196618. [DOI] [PubMed] [Google Scholar]
- [29].Hebert TE, Moffett S, Morello JP, Loisel TP, Bichet DG, Barret C, Bouvier M, A peptide derived from a beta2-adrenergic receptor transmembrane domain inhibits both receptor dimerization and activation, The Journal of biological chemistry 271 (1996) 16384–16392. [DOI] [PubMed] [Google Scholar]
- [30].Tarasov SG, Gaponenko V, Howard OM, Chen Y, Oppenheim JJ, Dyba MA, Subramaniam S, Lee Y, Michejda C, Tarasova NI, Structural plasticity of a transmembrane peptide allows self-assembly into biologically active nanoparticles, Proc Natl Acad Sci U S A 108 (2011) 9798–9803. 10.1073/pnas.1014598108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC, Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists, Science 330 (2010) 1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Stanasila L, Perez JB, Vogel H, Cotecchia S, Oligomerization of the alpha 1a- and alpha 1b-adrenergic receptor subtypes. Potential implications in receptor internalization, The Journal of biological chemistry 278 (2003) 40239–40251. 10.1074/jbc.M306085200. [DOI] [PubMed] [Google Scholar]
- [33].Wu H, Wang C, Gregory KJ, Han GW, Cho HP, Xia Y, Niswender CM, Katritch V, Meiler J, Cherezov V, Conn PJ, Stevens RC, Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator, Science 344 (2014) 58–64. 10.1126/science.1249489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Uberti MA, Hall RA, Minneman KP, Subtype-specific dimerization of alpha 1-adrenoceptors: effects on receptor expression and pharmacological properties, Mol Pharmacol 64 (2003) 1379–1390. 10.1124/mol.64.6.1379. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


