Abstract
Background
This article presents the results of long-term observations and comparative analysis of genotype–phenotype features in a large group of patients (227 males and one female) with a severe, intermediate and mild form of Hunter syndrome, evaluating the quality and span of their lives, as well as their ability to social adaptation.
Methods
We used electrophoresis of glycosaminoglycans of urine, determination of the activity of lysosomal enzymes in plasma, in dried blood spots according to the generally accepted method and DNA analysis.
Results
The clinical symptomatology of 228 patients with Hunter syndrome was characterized by growth retardation, lesions of the bronchopulmonary, cardiovascular, nervous systems, etc. Thirty-five patients had an attenuated form of the disease. DNA was available from all patients. 19 patients from 10 families had a mild form of the disease. 42 patients from 41 families had an intermediate form of the disease. All other patients had a severe form of the disease. We provide brief clinical examples of some patients with a mild form of Hunter syndrome. Currently, 113 patients with Hunter syndrome receive enzyme replacement therapy (idursulfase or idursulfase beta).
Conclusion
The long-term study of the large number of patients with Hunter syndrome helped identify disease-associated variants leading to severe and mild forms of the disease. The treatment effect and successful social adaptation of patients with a mild form of Hunter syndrome were revealed.
Keywords: Mucopolysaccharidosis, Hunter syndrome, Clinical and genetic analysis, Social adaptation
Background
Hunter syndrome or mucopolysaccharidosis type II (MPS II) is a rare disease with frequency ranges from 1:100,000 to 1:170,000 newborn boys [1–4].
Hunter syndrome is the only type of mucopolysaccharidoses that inherited as X-linked recessive trait. Hence, the majority of patients with Hunter syndrome are male. However, few cases of the disease in girls were described. Most of them are associated with structural abnormalities, inactivation disorders, or monosomy of chromosome X [5–7].
The clinical symptoms of Hunter syndrome usually become noticeable during the first two years of life. The disease is characterized by the progressive course. Clinical manifestations include rough facial features, sunken nose, full lips, hypertelorism, large tongue, corneal clouding, megalocephaly, thick coarse hair, short neck, brachidactyly of the hands and feet, contractures of joints, short stature, diffuse muscle hypotension, hepatosplenomegaly, umbilical and inguinal hernias, cardiomyopathy and gross delay in psycho-speech and motor development for most patients. After the first phenotype descriptions, two clinically different forms of Hunter syndrome were identified: a classical, severe form with severe somatic sings, progressive mental retardation, death at the age of 20 years or earlier; and a mild form characterized by longer life expectancy, fertility and minimal reduced intellect [8]. Later, it was suggested considering MPS II as a continuum between two extremes (severe and attenuated) [9].
The disease is caused by deficiency of the lysosomal enzyme iduronate-2-sulfatase (I2S) [10]. This defect is a result of different nucleotide variants in the IDS gene. The IDS gene is located on the locus Xq28 and consists of 9 exons [11]. At present, over 600 nucleotide variants in the IDS gene have been identified. Most of them are point changes (missense or nonsense variants), 28% are minor deletions and insertions, and 9% are splicing substitutions. Gross rearrangements account for 11%, among which 7% are partial or complete deletions of the IDS gene [12].
The I2S deficiency leads to the accumulation in different tissues of two types of glycosaminoglycans (GAG), i.e. heparan sulfate and dermatan sulfate resulting in the formation of multisystem pathology [13, 14].
The diagnosis of Hunter syndrome in Russia is carried out in several stages. The first stage is based on the identification and assessment of phenotype signs. The second stage consists of determination of urinary GAG level and of their fractions, primarily heparan and dermatan sulfates. At the third stage, the activity of the lysosomal enzyme I2S is measured in plasma or in dry blood spots. The fourth and the final stage is the molecular genetic analysis of the IDS gene.
A prerequisite is also the DNA analysis of the child’s mother to confirm the carrier of the pathogenic variant of the IDS gene. In Russia, the results of the final stage are necessary for the reasonable appointment of enzyme replacement therapy to the patient and successful family genetic counseling.
The purpose of this article is to present results of the comparative analysis of genotype–phenotype features in a large group of patients with Hunter syndrome, to assess the quality and duration of their life, as well as the social adaptability of patients with a mild form of the disease. We focused on describing patients with a mild form of disease, which is important for us since there are not many patients with this form of the disease. Moreover, perhaps physicians should be more thoughtful to identify patients for timely treatment.
Methods
Over the last 30 years (1989–2019), we have observed 228 patients with Hunter syndrome (mucopolysaccharidosis type II): 227 male and one 4-year old girl. The age of patients ranged from two to 65 years. First, all the patients were presumably diagnosed with Hunter syndrome based on the clinical data.
To confirm the disease, we determined the urinary GAG level and measured the I2S activity. As the final step, the DNA analysis of the IDS gene was carried out.
We used the following materials and methods:
Electrophoresis of urinary GAG.
Extraction of GAG from urine and electrophoresis of urinary GAG was carried out exactly according to a standard method that has been described previously [15].
-
2.
Biochemical assay.
The I2S activity was measured in plasma, as was described in the literature [16]. In brief, to 10 μl 5 × diluted in 0.2%BSA plasma 20 μl substrate (1.25 mM MU-alphaIdoA-2S) was added and the mixture was incubated for 4 h at 37 °C. Then 20 μl PiCi buffer and 10 μl LEBT solution (lysosomal enzymes purified from bovine testis) was added with subsequent incubation for 24 h at 37 °C. After adding 200 μl, stop-buffer fluorescence of MU was read. The normal range of the I2S activity was 297–705 nmol/4h/ml. Since 2019, the I2S activity was measured in dried blood spots by MS/MS methods, using the commercial kit [17]. Measuring was performed according to the manufacturer’s protocol.
-
3.
DNA analysis.
The DNA extraction was carried out according to the manufacturer’s protocol using the DIAtomt DNA Prep100 kit (Isogene Lab. Ltd., Russia). The nine exons and exon–intron boundaries of the IDS gene were amplified from DNA samples with primers sets designed according to the reference sequence NC_000023.11. PCR conditions and primers were available upon request. Sequencing was performed according to the manufacturer’s protocol on an ABI Prism 3500XL (Applied Biosystems). To detect gene-pseudogene recombination, two pair primers were used as described in the literature [18]. Gross deletion of the IDS gene was detected only on the genomic DNA level. Break-point of the deletion and inversion IDS/IDS2 were not revealed in this study.
Results
The clinical symptoms of mucopolysaccharidosis type II in the patients observed are presented in Table 1. The symptoms included growth retardation, lesion of the bronchopulmonary system, heart and blood vessels, central nervous system, and hearing organ.
Table 1.
Panel of clinical symptoms in patients with Hunter syndrome (n = 228)
| Clinical symptoms | Number of criteria, % |
|---|---|
| Changes in facial features by «gargoylism» type | 100 |
| Short stature | 95 |
| Skeletal anomalies (dysostosis multiplex) | 100 |
| Pathologies of the cardiovascular system | 100 |
| Cardiomyopathy | 35 |
| Anomalies of the heart valves | 100 |
| Narrowing of coronary arteries | 10 |
| Rhythm disturbance | 20 |
| Obstructive conditions of the respiratory tract | 100 |
| Obstructive sleep apnea | 55 |
| Decrease in lung capacity | 100 |
| Hepatosplenomegaly | 100 |
| Stiffness of major and small joints | 100 |
| Umbilical or inguinal and inguinal-scrotal hernia | 83 |
| Papular eruption on the skin | 5 |
| Retinitis pigmentosa | 5 |
| Progressive conductive or neurosensory hearing loss | 85 |
| Impairments of the nervous system | 85 |
| Mental retardation | 75 |
| Tonic–clonic convulsions | 55 |
Approximately, 1/5 of the patients had a papular rash with papules filled with glycolipid complexes and localized on the lateral and posterior surfaces of the thighs, shoulders and shoulder blades. This symptom is characteristic of Hunter syndrome which does not occur in patients with other types of mucopolysaccharidosis.
All patients demonstrated high excretion of dermatan sulfate and heparan sulfate in urine and low residual IDS activity in plasma or in dried blood spots.
228 patients from 207 families were completely genotyped. 122 different nucleotide variants were detected: 50 missense, 16 nonsense, 11 splicing substitutions, 27 small deletion, 5 small insertions/duplications, 3 small indels, and 10 gross deletions and complex rearrangements. Some nucleotide variants have been found multiple times in different patients from different families (Table 2). Many disease-associated variants found have been previously described (www.hgmd.cf.ac.uk). From 50 missense substitutions, 12 were not detected before. To evaluate their pathogenic influence, three different programs for impact prediction of nucleotide changes were used. All programs revealed probably deleterious effect of nucleotide variants for all except one change found. For nucleotide variant c.103G>C both programs PolyPhen-2 and PMUT Pathogenic mutation prediction revealed a possibly damaging effect. (Table 3). Additional analysis of nucleotide variant c.103G>C with the program Human Splicing Finder interpreted this variant as the most probably affecting splicing due to Broken WT Donor Site (data not showed). Two different nucleotide variants c.1411G>C and c.1418C>T had been detected in one patient (#43). Both nucleotide variants were novel ones. According to the prediction of pathogenicity, both nucleotide substitutions were probably pathogenic (Table 3). Moreover, five new nonsense substitutions, 19 novel small deletions, 2 novel splicing substitutions, 5 novel small insertion/duplications and 3 novel small indels were found. All small deletions, except the two, all small insertions and one small indel were leading to frame shift and premature stop codon. Two other small indels may have affected splicing. As can be seen from Table 2, the largest number of nucleotide variants was registered in exons 3, 5, 7 and 9. The analysis of nucleotide variants showed that the largest share was represented by point changes (missense or nonsense). Gross rearrangements and major deletions accounted for 14.4%, and 9.6% were splicing substitutions. The nucleotide variants c.253G>A, c.257C>T, c.263G>A, c.263G>T, c.514C>T, c.998C>T, c.1327C>T, c.1402C>T; c.1403G>A and the splicing substitution c.1122C>T were detected more than twice in patients from different families. All these point variants involve CpG sites of the IDS gene. The data were consistent with previous studies [19]. The small deletion c.596_599delAACA was detected in five patients from five families. The IDS/IDSP1 inversion has been described in detail [18] and was found in 17 patients from 16 families (see Table 4).
Table 2.
Nucleotide variants found in the IDS gene
| Nucleotide change number | Nucleotide; protein change found | Type of nucleotide change | Exons of IDS gene | HGMD accession | Allele frequency (%) in presented cohort | Comments |
|---|---|---|---|---|---|---|
| 1 | c.103G>C; p.Asp35His | Missense | 1 | None | 0.88 |
Novel (NC_000023.11:g.149505035C>G) ClinVar accession SCV001450592 |
| 2 | c.136G>T; p.Asp46Tyr | Missense | 2 | None | 0.44 |
Novel (NC_000023.11:g.149504261C>A) ClinVar accession SCV001450595 one another described in the same codon |
| 3 | c.136G>A; p.Asp46Asn | Missense | 2 | None | 0.44 |
Novel (NC_000023.11:g.149504261C>T) ClinVar accession SCV001450596 one another described in the same codon |
| 4 | c.160T>G; p.Tyr54Asp | Missense | 2 | CM981010 | 0.44 | |
| 5 | c.187A>G; p.Asn63Asp | Missense | 2 | CM960853 | 1.3 | |
| 6 | c.236C>A; p.Ala79Glu | Missense | 2 | CM981012 | 0.88 | |
| 7 | c.253G>A; p.Ala85Thr | Missense | 3 | CM960855 | 2.2 | |
| 8 | c.253G>T; p.Ala85Ser | Missense | 3 | CM981013 | 0.44 | |
| 9 | c.257C>T; p.Pro86Leu | Missense | 3 | CM950659 | 1.3 | |
| 10 | c.257C>G; p.Pro86Arg | Missense | 3 | CM930414 | 0.44 | |
| 11 | c.263G>A; p.Arg88His | Missense | 3 | CM960857 | 2.65 | |
| 12 | c.262C>T; p.Arg88Cys | Missense | 3 | CM950661 | 1.76 | |
| 13 | c.263G>T; p.Arg88Leu | Missense | 3 | CM981014 | 0.44 | |
| 14 | c.263G>C; p.Arg88Pro | Missense | 3 | CM970749 | 0.44 | |
| 15 | c.283A>T; p.Arg95Trp | Missense | 3 | None | 0.44 |
Novel (NC_000023.11:g.149503447T>A) ClinVar accession SCV001450598 three other described in the same codon |
| 16 | c.305T>G; p.Leu102Arg | Missense | 3 | CM981017 | 0.88 | |
| 17 | c.307T>G; p.Tyr103Asp | Missense | 3 | None | 0.44 |
Novel (NC_000023.11:g.149503423A>C) ClinVar accession SCV001450601 two other described in the same codon |
| 18 | c.325T>C; p.Trp109Arg | Missense | 3 | CM128183 | 0.88 | |
| 19 | c.359C>G; p.Pro120Arg | Missense | 3 | CM930417 | 0.44 | |
| 20 | c.395C>G; p.Ser132Trp | Missense | 3 | CM950663 | 0.88 | |
| 21 | c.403A>G; p.Lys135Glu | Missense | 3 | None | 0.44 |
Novel (NC_000023.11:g.149503327T>C) ClinVar accession SCV001450599 two other described in the same codon |
| 22 | c.476A>C; p.His159Pro | Missense | 4 | CM981026 | 0.44 | |
| 23 | c.512G>A; p.Cys171Tyr | Missense | 5 | None | 0.44 |
Novel (NC_000023.11:g.149498303C>T) ClinVar accession SCV001450602 one another described in the same codon |
| 24 | c.545T>C; p.Leu182Pro | Missense | 5 | CM981027 | 0.44 | |
| 25 | c.551G>T; p.Cys184Phe | Missense | 5 | CM960862 | 0.44 | |
| 26 | c.587T>C; p.Leu196Ser | Missense | 5 | CM981029 | 0.88 | |
| 27 | c.590C>T; p.Pro197Leu | Missense | 5 | None | 1.76 |
Novel (NC_000023.11:g.149498225G>A) ClinVar accession SCV001450603 |
| 28 | c.593A>G; p.Asp198Gly | Missense | 5 | CM981030 | 0.44 | |
| 29 | c.671G>A; p.Gly224Glu | Missense | 5 | CM981031 | 0.44 | |
| 30 | c.697A>G; p.Arg233Gly | Missense | 5 | CM146285 | 1.3 | |
| 31 | c.776T>C; p.Leu259Pro | Missense | 6 | CM030889 | 0.44 | |
| 32 | c.795C>A; p.Asn265Lys | Missense | 6 | CM128190 | 0.88 | |
| 33 | c.795C>G; p.Asn265Lys | Missense | 6 | CM141180 | 0.44 | |
| 34 | c.998C>T; p.Ser333Leu | Missense | 7 | CM920367 | 3.09 | |
| 35 | c.1004A>G; p.His335Arg | Missense | 7 | CM981045 | 0.44 | |
| 36 | c.1006G>C; p.Gly336Arg | Missense | 8 | CM970753 | 0.44 | |
| 37 | c.1019G>A; p.Gly340Asp | Missense | 8 | CM981048 | 1.3 | |
| 38 | c.1028G>A; p.Gly343Glu | Missense | 8 | None | 0.44 |
Novel (NC_000023.11:g.149487077C>T) ClinVar accession SCV001450616 |
| 39 | c.1035G>C, p.Trp345Cys | Missense | 8 | CM950668 | 0.44 | |
| 40 | c.1034G>C; p.Trp345Ser | Missense | 8 | None | 0.44 |
Novel (NC_000023.11:g.149487071C>G) ClinVar accession SCV001450617 four other described in the same codon |
| 41 | p.1037C>T; p.Ala346Val | Missense | 8 | CM950669 | 1.3 | |
| 42 | c.1204G>A; p.Glu402Lys | Missense | 9 | CM167391 | 0.88 | |
| 43 | c.1295G>A; p.Cys432Tyr | Missense | 9 | CM981052 | 0.88 | |
| 44 | c.1402C>T; p.Arg468Trp | Missense | 9 | CM920369 | 2.2 | |
| 45 | c.1403G>A; p.Arg468Gln | Missense | 9 | CM930422 | 4.4 | |
| 46 |
c.1411G>C; p.Asp471His c.1418C>T; p.Pro473Leu |
Missense | 9 | None | 0.44 |
Novel Novel (NC_000023.11:g.[149482988C>G;149482981G>A]) ClinVar accession SCV001450627 |
| 47 | c.1432G>T; p.Asp478Tyr | Missense | 9 | CM981054 | 0.44 | |
| 48 | c.1432G>A; p.Asp478Asn | Missense | 9 | BM1234454 | 0.44 | |
| 49 | c.1454T>G; p.Ileu485Arg | Missense | 9 | CM940967 | 0.88 | |
| 50 | c.1565T>C; p.Leu522Pro | Missense | 9 | HM971766 | 0.44 | |
| 51 | c.196C>T; p.Gln66Term | Nonsense | 2 | CM068304 | 0.44 | |
| 52 | c.361C>T; p.Gln121Term | Nonsense | 3 | CM128174 | 0.44 | |
| 53 | c.514C>T; p.Arg172Term | Nonsense | 5 | CM920366 | 2.65 | |
| 54 | c.598C>T p.Gln200Term | Nonsense | 6 | CM146284 | 0.88 | |
| 55 | c.800G>A; p.Trp267Term | Nonsense | 6 | CM050243 | 0.44 | |
| 56 | c.814C>T; p.Gln272Term | Nonsense | 6 | None | 0.44 |
Novel (NC_000023.11:g.149496411G>A) ClinVar accession SCV001450612 |
| 57 | c.829C>T; p.Gln277Term | Nonsense | 6 | None | 0.44 |
Novel (NC_000023.11:g.149496396G>A) ClinVar accession SCV001450613 |
| 58 | c.998C>A; p.Ser333Term | Nonsense | 7 | None | 0.88 |
Novel (NC_000023.11:g.149490322G>T) ClinVar accession SCV001450615 two other described in the same codon |
| 59 | c.1010G>A; p.Trp337Term | Nonsense | 8 | CM128194 | 0.88 | |
| 60 | c.1234G>T; p.Gly412Term | Nonsense | 9 | None | 0.44 |
Novel (NC_000023.11:g.149483165C>A) ClinVar accession SCV001450623 |
| 61 | c.1288G>T; p.Glu430Term | Nonsense | 9 | CM146287 | 0.44 | |
| 62 | c.1327C>T; p.Arg443Term | Nonsense | 9 | CM920368 | 1.3 | |
| 63 | c.1340T>A; p.Leu447Term | Nonsense | 9 | None | 0.44 |
Novel (NC_000023.11:g.149483059A>T) ClinVar accession SCV001450625 |
| 64 | c.1375G>T; p.Glu459Term | Nonsense | 9 | CM1719915 | 0.44 | |
| 65 | c.1445T>G; p.Leu482Term | Nonsense | 9 | CM981058 | 0.44 | |
| 66 | c.1608T>A; p.Tyr536Term | Nonsense | 9 | CM141189 | 0.44 | |
| 67 | IVS1 as A-G -2; c.104-2A>G | Splicing substitutions | CS982224 | 0.44 | ||
| 68 | IVS2 ds G-C +1; c.240+1G>C | Splicing substitutions | CS050391 | 0.44 | ||
| 69 | IVS2 ds G-T +1; c.240+1G>T | Splicing substitutions | CS982227 | 0.44 | ||
| 70 | IVS2 as C-G -9; c.241-9C>G | Splicing substitutions | None | 0.88 |
Novel (NC_000023.11:g.149503498G>C) ClinVar accession SCV001450634 |
|
| 71 | IVS4 ds G-A +1; c.507+1G>A | Splicing substitutions | CS982228 | 0.88 | ||
| 72 | IVS6 ds G-A +1; c.879+1G>A | Splicing substitutions | CS982229 | 0.44 | ||
| 73 | IVS6 as A-G -2; c.880-2A>G | Splicing substitutions | CS930833 | 0.44 | ||
| 74 | IVS7 ds T-G +2 c.1006+2T>G | Splicing substitutions | None | 0.44 |
Novel (NC_000023.11:g.149490312A>C) ClinVar accession SCV001450635 |
|
| 75 | IVS7 as G-A -1; c.1007-1G>A | Splicing substitutions | CS120471 | 0.44 | ||
| 76 | IVS8 as C-A -15; c.1181-15C>A | Splicing substitutions | CS013824 | 0.44 | ||
| 77 | IVS8 ds C-T -59; c.1122C>T | Splicing substitutions | CS963080 | 5.75 | ||
| 78 | c.118_120delCTT; p.Leu40del | Small deletion | 2 | None | 0.44 |
Novel (NC_000023.11:g.149504277_149504279delAAG) ClinVar accession SCV001450593 |
| 79 | c.121_123delCTC p.Leu41del | Small deletion | 2 | CD941707 | 0.44 | |
| 80 | c.133delG; p.Asp45Metfs | Small deletion | 2 | None | 0.44 |
Novel (NC_000023.11:g.149504264delC) ClinVar accession SCV001450594 |
| 81 | c.248delT; p.Val83Glyfs | Small deletion | 3 | None | 0.44 |
Novel (NC_000023.11:g.149503482delA) ClinVar accession SCV001450597 |
| 82 | c.305delT; p.Leu102Argfs | Small deletion | 3 | None | 0.44 |
Novel (NC_000023.11:g.149503425delA) ClinVar accession SCV001450600 |
| 83 | c.404_405delAA; p.Lys135Serfs | Small deletion | 3 | None | 0.44 |
Described ClinVar accession VCV000499561.1 |
| 84 | c.410_411delTT p.Phe137Serfs | Small deletion | 3 | CD012530 | 0.44 | |
| 85 | c.596_599delAACA; p.Lys199Argfs | Small deletion | 5 | CD941708 | 2.21 | |
| 86 | c.613delG; p.Ala205Profs | Small deletion | 5 | None | 0.44 |
Novel (NC_000023.11:g.149498202delC) ClinVar accession SCV001450604 |
| 87 | c.625_627del TTG; p.Leu209del | Small deletion | 5 | None | 0.44 |
Novel (NC_000023.11:g.149498188_149498190delCAA) ClinVar accession SCV001450605 |
| 88 | c.667_683del17; p.Val223Thrfs | Small deletion | 5 | CD982702 | 0.44 | |
| 89 | c.687delC; p.His229Glnfs | Small deletion | 5 | None | 0.44 |
Novel (NC_000023.11:g.149498128delG) ClinVar accession SCV001450606 |
| 90 | c.715_721del7; p.Gln239Cysfs | Small deletion | 6 | None | 0.44 | Novel (NC_000023.11:g.149496504_149496510delACTTCTG) ClinVar accession SCV001450607 |
| 91 | c.782delC; p.Pro261Leufs | Small deletion | 6 | CD982703 | 0.44 | |
| 92 | c.800_801delGG; p.Trp267Tyrfs | Small deletion | 6 | None | 0.44 |
Novel (NC_000023.11:g.149496424_149496425delCC) ClinVar accession SCV001450611 |
| 93 | c.899_900delAC; p.Tyr300Phefs | Small deletion | 7 | None | 0.44 |
Novel (NC_000023.11:g.149490420_149490421delGT) ClinVar accession SCV001450614 |
| 94 | c.908_909delCT; p.Ser303Cysfs | Small deletion | 7 | CD1412401 | 0.44 | |
| 95 | c.1077delG; p.Ile360Tyrfs | Small deletion | 8 | CD146296 | 0.44 | |
| 96 | c.1129delC; p.Leu377Phefs | Small deletion | 8 | None | 0.44 |
Novel (NC_000023.11:g.149486976delG) ClinVar accession SCV001450618 |
| 97 | c.1191delC; p.Met398Trpfs | Small deletion | 9 | None | 0.44 |
Novel (NC_000023.11:g.149483208delG) ClinVar accession SCV001450620 |
| 98 | c.1214_1220del7; p.Ser405Phefs | Small deletion | 9 | None | 0.44 |
Novel (NC_000023.11:g.149483179_149483185delAAAAGAG) ClinVar accession SCV001450621 |
| 99 | c.1221delT; p.Ser409Argfs | Small deletion | 9 | None | 0.44 |
Novel (NC_000023.11:g.149483178delA) ClinVar accession SCV001450622 |
| 100 | c.1353_1357delGTACC; p.Tyr452Profs | Small deletion | 9 | None | 0.44 |
Novel (NC_000023.11:g. 149483042_149483046delGGTAC) ClinVar accession SCV001450626 |
| 101 | c.1426_1437 del12 p.476_479delAsnSerAspLys | Small deletion | 9 | None | 0.44 |
Novel (NC_000023.11:g.149482962_149482973delCTTGTCAGAATT) ClinVar accession SCV001450629 |
| 102 | c.1431delT; p.Asp478Thrfs | Small deletion | 9 | None | 0.44 |
Novel (NC_000023.11:g.149482968delA) ClinVar accession SCV001450628 |
| 103 | c.1438_1442delCCGAG;p.Pro480Phefs | Small deletion | 9 | None | 0.44 | Novel (NC_000023.11:g.149482957_149482961delCTCGG) ClinVar accession SCV001450630 |
| 104 | c.1466delG p.Val489Alafs | Small deletion | 9 | CD146297 | 0.44 | |
| 105 | c.776_777dupTA; p.Pro260Tyrfs | Small insertions/duplications | 6 | None | 0.44 |
Novel (NC_000023.11:g.149496448_149496449dupTA) ClinVar accession SCV001450608 |
| 106 | c.801_802insG; p.Met268Aspfs | Small insertions/duplications | 6 | None | 0.44 |
Novel (NC_000023.11:g.149496423_149496424insC) ClinVar accession SCV001450610 |
| 107 | c.1151_1152 insTGCGACCCTTT; p.Phe384Leufs | Small insertions/duplications | 8 | None | 0.44 |
Novel (NC_000023.11:g.149486954_149486955 insTGCGACCCTTT) ClinVar accession SCV001450619 |
| 108 | c.1239_c.1240insCT; p.Ala414Leufs | Small insertions/duplications | 9 | None | 0.44 |
Novel (NC_000023.11:g.149483159_149483160insAG) ClinVar accession SCV001450624 |
| 109 | c.1491_1492dupTA; p.Arg498Ileufs | Small insertions/duplications | 9 | None | 0.44 |
Novel (NC_000023.11:g.149482909_149482910dupTA) ClinVar accession SCV001450631 |
| 110 | c.104-1_104delGAinsT | Small indels | 2 | None | 0.44 | Novel (NC_000023.11:g.149504293_149504294delTCinsA) ClinVar accession SCV001450632 |
| 111 | c.240 +2_c.240+3insTCCCAGA | Small indels | Intron 2 | None | 0.44 | Novel (NC_000023.11:g.149504154_149504155insTCCCAGA) ClinVar accession SCV001450633 |
| 112 | c.786_787delGGinsC; p.Ala263Profs | Small indels | 6 | None | 0.44 | Novel (NC_000023.11:g.149496438_149496439delCCinsG) ClinVar accession SCV001450609 |
| 113 | gDNA level exons 1–3 deletion | Gross deletions | 0.88 | |||
| 114 | gDNA level exons 1–4 deletion | Gross deletions | 0.44 | |||
| 115 | gDNA level exons 1–7 deletion | Gross deletions | 0.44 | |||
| 116 | gDNA level exons 1–7 deletion | Gross deletions | 0.44 | |||
| 117 | gDNA level exon 4 deletion | Gross deletions | 0.44 | |||
| 118 | cDNA level del incl. ex 5–6 | Gross deletions | CG984375 | 0.44 | ||
| 119 | gDNA level exon 7 deletion | Gross deletions | 0.44 | |||
| 120 | Complete IDS del | Gross deletions | 3.5 | |||
| 121 | Recomb. between in. 7 and seq. distal of ex. 3 in IDS-2 without exons deletion | Complex rearrangements | CP973598 | 6.19 | ||
| 122 | Recomb. between in. 7 and seq. distal of ex. 3 in IDS-2 with 4–7 exons deletion | Complex rearrangements | 1.32 |
Table 3.
Prediction of functional effects of 12 missense substitutions found
| Nucleotide changes number | Nucleotide/protein change | Programs for prediction of nucleotide changes | ||
|---|---|---|---|---|
| PolyPhen-2 | PMUT pathogenic mutation prediction | Mutation tester | ||
| 1 | c.103G>C; p.Asp35His | Possibly damaging with a score of 0.710 | 0.47 (83%) Neutral | Disease causing |
| 2 | c.136G>T; p.Asp46Tyr | Probably damaging with a score of 1.000 | 0.94 (94%) Disease | Disease causing |
| 3 | c.136G>A; p.Asp46Asn | Probably damaging with a score of 1.000 | 0.89 (92%) Disease | Disease causing |
| 4 | c.283A>T; p.Arg95Trp | Probably damaging with a score of 1.000 | 0.79 (88%) Disease | Disease causing |
| 5 | c.307T>G; Tyr103Asp | Probably damaging with a score of 1.000 | 0.82 (90%) Disease | Disease causing |
| 6 | c.403A>G; p.Lys135Glu | Probably damaging with a score of 1.000 | 0.94 (94%) Disease | Disease causing |
| 7 | c.512G>A; p.Cys171Tyr | Probably damaging with a score of 1.000 | 0.53 (80%) Disease | Disease causing |
| 8 | c.590C>T; p.Pro197Leu | Probably damaging with a score of 0.996 | 0.69 (86%) Disease | Disease causing |
| 9 | c.1028G>A; p.Gly343Glu | Probably damaging with a score of 0.996 | 0.83 (90%) Disease | Disease causing |
| 10 | c.1034G>C; p.Trp345Ser | Probably damaging with a score of 1.000 | 0.89 (92%) Disease | Disease causing |
| 11 | c.1411G>C; p.Asp471His | Probably damaging with a score of 0.999 | 0.81 (89%) Disease | Disease causing |
| 12 | c.1418C>T; p.Pro473Leu | Probably damaging with a score of 0.976 | 0.72 (86%) Disease | Disease causing |
Table 4.
Mutation spectrum in the IDS gene in patients with MPS II
| Family number | Patient number | Age at diagnosis | Enzyme activity in plasma (N = 297–705 nmol/4h/ml) |
Enzyme activity in dried blood spots (N = 2.5–50 μmM/l/h) |
Phenotype | Nucleotide/protein change in IDS gene |
|---|---|---|---|---|---|---|
| 1 | 1 | 7 | 16.2 | Mild | c.187A>G; p.Aspn63Asp | |
| 2 | 2 | 31 | 21.1 | Mild | c.187A>G; p.Aspn63Asp | |
| 3 | 32 | 18.2 | Mild | c.187A>G; p.Aspn63Asp | ||
| 3 | 4 | 8 | 1.2 | Mild | c.236C>A; p.Ala79Glu | |
| 5 | 10 | 8.4 | Mild | c.236C>A; p.Ala79Glu | ||
| 4 | 6 | 11 | 7.4 | Mild | c.305T>G; p.Leu102Arg | |
| 7 | 9 | 8.1 | Mild | c.305T>G; p.Leu102Arg | ||
| 5 | 8 | 55 | – | 0.01 | Mild | c.590C>T; p.Pro197Leu |
| 9 | 12 | – | 0.01 | Mild | c.590C>T; p.Pro197Leu | |
| 10 | 5 | – | 0.01 | Mild | c.590C>T; p.Pro197Leu | |
| 11 | 11 | – | 0.01 | Mild | c.590C>T; p.Pro197Leu | |
| 6 | 12 | 16 | 14.7 | Mild | c.593A>G; p.Asp198Gly | |
| 7 | 13 | 23 | 8.4 | Mild | c.1019G>A; p.Gly340Asp | |
| 14 | 19 | 6.1 | Mild | c.1019G>A; p.Gly340Asp | ||
| 8 | 15 | 33 | 11.7 | Mild | c.1019G>A; p.Gly340Asp | |
| 9 | 16 | 35 | 0.01 | Mild | p.1037C>T; p.Ala346Val | |
| 17 | 23 | 0.05 | Mild | p.1037C>T; p.Ala346Val | ||
| 18 | 18 | 0.07 | Mild | p.1037C>T; p.Ala346Val | ||
| 10 | 19 | 15 | 3.6 | Mild | c.1234G>T; p.Gly412Term | |
| 11 | 20 | 11 | 9.4 | Mild -> intermediate | c.253G>A; p.Ala85Thr | |
| 12 | 21 | 9 | 10.4 | Mild -> intermediate | c.253G>A; p.Ala85Thr | |
| 13 | 22 | 6 | 18.6 | Mild -> intermediate | c.253G>A; p.Ala85Thr | |
| 14 | 23 | 9 | 14.4 | Mild -> intermediate | c.253G>A; p.Ala85Thr | |
| 15 | 24 | 13 | 0.01 | Mild -> intermediate | c.253G>A; p.Ala85Thr | |
| 16 | 25 | 8 | 0.02 | Mild -> intermediate | c.253G>T; p.Ala85Ser | |
| 17 | 26 | 13 | 11.3 | Mild -> intermediate | c.587T>C; p.Leu196Ser | |
| 18 | 27 | 5 | 4.7 | Mild -> intermediate | c.587T>C; p.Leu196Ser | |
| 19 | 28 | 2 | 2.1 | Mild -> intermediate | c.1204G>A; p.Glu402Lys | |
| 20 | 29 | 3 | 1.4 | Mild -> intermediate | c.1204G>A; p.Glu402Lys | |
| 21 | 30 | 4 | – | 0.01 | Mild -> intermediate | c.1438_1442delCCGAG; p.Pro480Phefs |
| 22 | 31 | 28 | 14.8 | Mild -> intermediate | c.1028G>A; p.Gly343Glu | |
| 23 | 32 | 11 | 7.4 | Mild -> intermediate | c.1034G>C; p.Trp345Ser | |
| 24 | 33 | 25 | 4.8 | Mild -> intermediate | c.1035G>C, p.Trp345Cys | |
| 25 | 34 | 8 | 5.1 | Mild -> intermediate | IVS2 as C-G -9; c.241-9C>G | |
| 35 | 10 | 5.2 | Mild -> intermediate | IVS2 as C-G -9; c.241-9C>G | ||
| 26 | 36 | 7 | 0.1 | Intermediate -> severe | c.283A>T; p.Arg95Trp | |
| 27 | 37 | 7 | 0.01 | Intermediate -> severe | c.512G>A; p.Cys171Tyr | |
| 28 | 38 | 5 | 8.4 | Intermediate -> severe | c.545T>C; p.Leu182Pro | |
| 29 | 39 | 6 | 2.3 | Intermediate -> severe | c.551G>T; p.Cys184Phe | |
| 30 | 40 | 6 | 2.7 | Intermediate -> severe | c.1327C>T; p.Arg443Term | |
| 31 | 41 | 7 | 3.18 | Intermediate -> severe | c.1327C>T; p.Arg443Term | |
| 32 | 42 | 2 | – | 0.01 | Intermediate -> severe | c.1327C>T; p.Arg443Term |
| 33 | 43 | 13 | 21.6 | Intermediate -> severe |
c.1411G>C; p.Asp471His c.1418C>T; p.Pro473Leu |
|
| 34 | 44 | 14 | 2.2 | Intermediate -> severe | IVS6 ds G-A +1; c.879+1G>A | |
| 35 | 45 | 11 | 0.01 | Intermediate -> severe | IVS8 ds C-T -59; c.1122C>T | |
| 36 | 46 | 11 | 0.01 | Intermediate -> severe | IVS8 ds C-T -59; c.1122C>T | |
| 37 | 47 | 6 | 0.02 | Intermediate -> severe | IVS8 ds C-T -59; c.1122C>T | |
| 38 | 48 | 8 | 0.01 | Intermediate -> severe | IVS8 ds C-T -59; c.1122C>T | |
| 39 | 49 | 4 | 0.01 | Intermediate -> severe | IVS8 ds C-T -59; c.1122C>T | |
| 40 | 50 | 5 | 2.2 | Intermediate -> severe | IVS8 ds C-T -59; c.1122C>T | |
| 41 | 51 | 3 | 0.01 | Intermediate -> severe | IVS8 ds C-T -59; c.1122C>T | |
| 42 | 52 | 7 | 0.01 | Intermediate -> severe | IVS8 ds C-T -59; c.1122C>T | |
| 43 | 53 | 18 | 0.01 | Intermediate -> severe | IVS8 ds C-T -59; c.1122C>T | |
| 44 | 54 | 4 | 4.9 | Intermediate -> severe | IVS8 ds C-T -59; c.1122C>T | |
| 45 | 55 | 7 | 2.3 | Intermediate -> severe | IVS8 ds C-T -59; c.1122C>T | |
| 46 | 56 | 4 | 7.2 | Intermediate -> severe | IVS8 ds C-T -59; c.1122C>T | |
| 47 | 57 | 5 | Intermediate -> severe | IVS8 ds C-T -59; c.1122C>T | ||
| 48 | 58 | 3 | 4.9 | Intermediate -> severe | c.118_120delCTC; p.Leu40del | |
| 49 | 59 | 4 | 21.4 | Intermediate -> severe | c.121_123delCTC p.Leu41del | |
| 50 | 60 | 6 | 0.01 | Intermediate -> severe | c.625_627del TTG p.Leu209del | |
| 51 | 61 | 9 | 4.4 | Intermediate -> severe | c.1426_1437 del12; p.476_479delAsnSerAspLys | |
| 52 | 62 | 6 | 8.3 | Severe | c.103G>C; p.Asp35His | |
| 63 | 4 | 10.4 | Severe | c.103G>C; p.Asp35His | ||
| 53 | 64 | 3 | 0.35 | Severe | c.136G>T; p.Asp46Tyr | |
| 54 | 65 | 3 | – | 0.01 | Severe | c.136G>A; p.Asp46Asn |
| 55 | 66 | 4 | 0.1 | Severe | c.160T>G; p.Tyr54Asp | |
| 56 | 67 | 4 | 4.6 | Severe | c.257C>T; p. Pro86Leu | |
| 57 | 68 | 5 | 5.1 | Severe | c.257C>T; p. Pro86Leu | |
| 58 | 69 | 2 | 3.5 | Severe | c.257C>T; p. Pro86Leu | |
| 59 | 70 | 10 | 11.8 | Severe | c.257C>G; p. Pro86Arg | |
| 60 | 71 | 6 | 0.01 | Severe | c.263G>A; p.Arg88His | |
| 72 | 3 | 0.01 | Severe | c.263G>A; p.Arg88His | ||
| 61 | 73 | 3 | 0.7 | Severe | c.263G>A; p.Arg88His | |
| 74 | 4 | 0.8 | Severe | c.263G>A; p.Arg88His | ||
| 62 | 75 | 7 | – | 0.01 | Severe | c.263G>A; p.Arg88His |
| 63 | 76 | 3 | 0.01 | Severe | c.263G>A; p.Arg88His | |
| 64 | 77 | 5 | 0.01 | Severe | c.263G>T; p.Arg88Leu | |
| 65 | 78 | 4 | 0.02 | Severe | c.262C>T; p.Arg88Cys | |
| 66 | 79 | 5 | 0.01 | Severe | c.262C>T; p.Arg88Cys | |
| 67 | 80 | 4 | 0.01 | Severe | c.262C>T; p.Arg88Cys | |
| 68 | 81 | 3 | 0.01 | Severe | c.262C>T; p.Arg88Cys | |
| 69 | 82 | 4 | 3.2 | Severe | c.263G>C; p.Arg88Pro | |
| 70 | 83 | 8 | 0.01 | Severe | c.307T>G; p.Tyr103Asp | |
| 71 | 84 | 5 | 4.8 | Severe | c.325T>C; p.Trp109Arg | |
| 72 | 85 | 6 | 1.37 | Severe | c.325T>C; p.Trp109Arg | |
| 73 | 86 | 1 | – | 0.01 | Severe | c.359C>G; p.Pro120Arg |
| 74 | 87 | 4 | 0.01 | Severe | c.395C>G; p.Ser132Trp | |
| 75 | 88 | 9 | 0.01 | Severe | c.395C>G; p.Ser132Trp | |
| 76 | 89 | 3 | 0.01 | Severe | c.403A>G; p.Lys135Glu | |
| 77 | 90 | 10 | 0.1 | Severe | c.476A>C; p.His159Pro | |
| 78 | 91 | 5 | 0.01 | Severe | c.671G>A; p.Gly224Glu | |
| 79 | 92 | 5 | 1.2 | Severe | c.697A>G; p.Arg233Gly | |
| 93 | 2 | 1.59 | Severe | c.697A>G; p.Arg233Gly | ||
| 80 | 94 | 1.5 | 0.01 | Severe | c.697A>G; p.Arg233Gly | |
| 81 | 95 | 15 | 4.1 | Severe | c.776T>C; p.Leu259Pro | |
| 82 | 96 | 3 | 0.15 | Severe | c.795C>A; p.Asn265Lys | |
| 83 | 97 | 3 | 0.49 | Severe | c.795C>A; p.Asn265Lys | |
| 84 | 98 | 3 | 10.8 | Severe | c.795C>G; p.Asn265Lys | |
| 85 | 99 | 10 | 0.01 | Severe | c.998C>T; p.Ser333Leu | |
| 86 | 100 | 7 | 0.01 | Severe | c.998C>T; p.Ser333Leu | |
| 87 | 101 | 6 | 0.01 | Severe | c.998C>T; p.Ser333Leu | |
| 88 | 102 | 7 | 0.01 | Severe | c.998C>T; p.Ser333Leu | |
| 89 | 103 | 5 | 0.01 | Severe | c.998C>T; p.Ser333Leu | |
| 90 | 104 | 7 | 0.01 | Severe | c.998C>T; p.Ser333Leu | |
| 91 | 105 | 1 | 3.5 | Severe | c.998C>T; p.Ser333Leu | |
| 92 | 106 | 2 | 0.01 | Severe | c.1004A>G; p.His335Arg | |
| 93 | 107 | 5 | 2.7 | Severe | c.1006G>C; p.Gly336Arg | |
| 94 | 108 | 10 | 0.1 | Severe | c.1295G>A; p.Cys432Tyr | |
| 95 | 109 | 4 | 0.01 | Severe | c.1295G>A; p.Cys432Tyr | |
| 96 | 110 | 4 | 5.4 | Severe | c.1402C>T; p.Arg468Trp | |
| 97 | 111 | 2 | 0.1 | Severe | c.1402C>T; p.Arg468Trp | |
| 98 | 112 | 3 | 0.01 | Severe | c.1402C>T; p.Arg468Trp | |
| 99 | 113 | 5 | 0.1 | Severe | c.1402C>T; p.Arg468Trp | |
| 100 | 114 | 3 | – | 0.01 | Severe | c.1402C>T; p.Arg468Trp |
| 101 | 115 | 3 | 0.01 | Severe | c.1403G>A; p.Arg468Gln | |
| 102 | 116 | 3 | 0.9 | Severe | c.1403G>A; p.Arg468Gln | |
| 117 | 3 | 0.01 | Severe | c.1403G>A; p.Arg468Gln | ||
| 103 | 118 | 5 | 1.4 | Severe | c.1403G>A; p.Arg468Gln | |
| 104 | 119 | 2 | 0.01 | Severe | c.1403G>A; p.Arg468Gln | |
| 105 | 120 | 2 | 6.3 | Severe | c.1403G>A; p.Arg468Gln | |
| 121 | 4 | 2.8 | Severe | c.1403G>A; p.Arg468Gln | ||
| 106 | 122 | 3 | 10.8 | Severe | c.1403G>A; p.Arg468Gln | |
| 107 | 123 | 4 | 0.01 | Severe | c.1403G>A; p.Arg468Gln | |
| 108 | 124 | 2 | 3.12 | Severe | c.1403G>A; p.Arg468Gln | |
| 109 | 125 | 4 | 0.01 | Severe | c.1432G>T; p.Asp478Tyr | |
| 110 | 126 | 7 | 0.1 | Severe | c.1432G>A; p.Asp478Asn | |
| 111 | 127 | 6 | 0.01 | Severe | c.1454T>G; p.Ileu485Arg | |
| 112 | 128 | 6 | 0.01 | Severe | c.1454T>G; p.Ileu485Arg | |
| 113 | 129 | 8 | 0.01 | Severe | c.1565T>C; p.Leu522Pro | |
| 114 | 130 | 3 | 0.15 | Severe | c.196C>T; p.Gln66Term | |
| 115 | 131 | 2 | 0.01 | Severe | c.361C>T; p.Gln121Term | |
| 116 | 132 | 3 | 4.5 | Severe | c.514C>T; p.Arg172Term | |
| 117 | 133 | 4 | 2.7 | Severe | c.514C>T; p.Arg172Term | |
| 118 | 134 | 2 | 7.8 | Severe | c.514C>T; p.Arg172Term | |
| 119 | 135 | 4 | 4.32 | Severe | c.514C>T; p.Arg172Term | |
| 120 | 136 | 4 | 8.4 | Severe | c.514C>T; p.Arg172Term | |
| 121 | 137 | 2 | – | 0.01 | Severe | c.514C>T; p.Arg172Term |
| 122 | 138 | 6 | 0.01 | Severe | c.598C>T p.Gln200Term | |
| 123 | 139 | 2 | 2.1 | Severe | c.598C>T p.Gln200Term | |
| 124 | 140 | 12 | 0.9 | Severe | c.800G>A; p.Trp267Term | |
| 125 | 141 | 5 | 0.01 | Severe | c.814C>T; p.Gln272Term | |
| 126 | 142 | 4 | 2.1 | Severe | c.829C>T; Gln277Term | |
| 127 | 143 | 8 | 18 | Severe | c.998C>A; p.Ser333Term | |
| 144 | 2 | 10.1 | Severe | c.998C>A; p.Ser333Term | ||
| 128 | 145 | 3 | 0.18 | Severe | c.1010G>A; p.Trp337Term | |
| 129 | 146 | 3 | 4.4 | Severe | c.1010G>A; p.Trp337Term | |
| 130 | 147 | 1 | 0.07 | Severe | c.1288G>T; p.Glu430Term | |
| 131 | 148 | 6 | 0.01 | Severe | c.1340T>A; p.Leu447Term | |
| 132 | 149 | 3 | 0.01 | Severe | c.1375G>T; p.Glu459Term | |
| 133 | 150 | 3.5 | 0.01 | Severe | c.1445T>G; p.Leu482Term | |
| 134 | 151 | 6 | 0.01 | Severe | c.1608T>A;p.Tyr536Term | |
| 135 | 152 | 4 | 0.01 | Severe | IVS1 as A-G -2; c.104-2A>G | |
| 136 | 153 | 4 | 0.01 | Severe | IVS2 ds G-C +1; c.240+1G>C | |
| 137 | 154 | 4 | 4.8 | Severe | IVS2 ds G-T +1; c.240+1G>T | |
| 138 | 155 | 7 | 0.01 | Severe | IVS4 ds G-A +1; c.507+1G>A | |
| 139 | 156 | 16 | 3.96 | Severe | IVS4 ds G-A +1; c.507+1G>A | |
| 140 | 157 | 9 | 3.6 | Severe | IVS6 as A-G -2; c.880-2A>G | |
| 141 | 158 | 4 | 1.14 | Severe | IVS7 ds T-G +2c.1006+2T>G | |
| 142 | 159 | 5 | 0.01 | Severe | IVS7 as G-A -1; c.1007-1G>A | |
| 143 | 160 | 5 | 0.01 | Severe | IVS8 as C-A -15; c.1181-15C>A | |
| 144 | 162 | 6 | 2.1 | Severe | c.133delG p.Asp45Metfs | |
| 145 | 163 | 1.5 | 0.12 | Severe | c.248delT; p.Val83Glyfs | |
| 146 | 164 | 3 | 0.01 | Severe | c.305delT; p.Leu102Argfs | |
| 147 | 165 | 2 | 1.4 | Severe | c.404_405delAA; p.Lys135Serfs | |
| 148 | 166 | 2 | 2.52 | Severe | c.410_411delTT p.Phe137Sfs | |
| 149 | 167 | 6 | 4.2 | Severe | c.596_599delAACA;p.(Lys199Argfs) | |
| 150 | 168 | 9 | 0.58 | Severe | c.596_599delAACA;p.(Lys199Argfs) | |
| 151 | 169 | 7 | 1.2 | Severe | c.596_599delAACA;p.(Lys199Argfs) | |
| 152 | 170 | 5 | 9.1 | Severe | c.596_599delAACA;p.(Lys199Argfs) | |
| 153 | 171 | 4 | 0.7 | Severe | c.596_599delAACA;p.(Lys199Argfs) | |
| 154 | 172 | 3 | 18.1 | Severe | c.613delG; p.Ala205Profs | |
| 155 | 173 | 9 | 0.01 | Severe | c.667-683del17; p.Val223Thrfs | |
| 156 | 174 | 5 | 0.01 | Severe | c.687delC; p.His229Glnfs | |
| 157 | 175 | 6 | 2.3 | Severe | c.715_721del7; p.Gln239Cysfs | |
| 158 | 176 | 2 | 0.01 | Severe | c.782delC p.Pro261Lfs | |
| 159 | 177 | 13 | 0.66 | Severe | c.800_801delGG; p.Trp267Tyrfs | |
| 160 | 178 | 4 | 0.01 | Severe | c.899_900delAC; p.Tyr300Phefs | |
| 161 | 179 | 8 | 8.04 | Severe | c.908_909delCT; p.Ser303Cysfs | |
| 162 | 180 | 1 | 0.43 | Severe | c.1077delG; p.Ile360Tyrfs | |
| 163 | 181 | 12 | 12.6 | Severe | c.1129delC p.Leu377Phefs | |
| 164 | 182 | 4 | 0.01 | Severe | c.1191delC p.Met398Trpfs | |
| 165 | 183 | 6 | 0.01 | Severe | c.1214_1220del7 p.Leu406Hisfs | |
| 166 | 184 | 3 | 2.3 | Severe | c.1221delT p.Cys409Arg fs | |
| 167 | 185 | 16 | 0.79 | Severe | c.1353_1357delGTACCp.Tyr452Profs | |
| 168 | 186 | 3 | 2.4 | Severe | c.1431delT; p.Asp478Thrfs | |
| 169 | 187 | 3 | 1.9 | Severe | c.1466delG p.Val489Alafs | |
| 170 | 188 | 3 | – | 0.01 | Severe | c.776_777dupTA; p.Pro260Tyrfs |
| 171 | 189 | 5 | 0.01 | Severe | c.801_802insG; p.Met268Aspfs | |
| 172 | 190 | 4 | 1.3 | Severe | c.1151_1152 ins11; p.Phe384Leufs | |
| 173 | 191 | 9 | 6.6 | Severe | c.1239_1240insCT; p.Ala414Leufs | |
| 174 | 192 | 5 | 0.01 | Severe | c.1491_1492dupTA; p.Arg498Ileufs | |
| 175 | 193 | 4 | 2.64 | Severe | del c.104-1_104delGAinsT | |
| 176 | 194 | 8 | 0.01 | Severe | c.240+2_240+3 insTCCCAGA (intron 2) | |
| 177 | 195 | 4 | 0.48 | Severe | c.786-787delGGinsC; p.Ala263Profs | |
| 178 | 196 | 2 | – | 0.01 | Severe | gDNA level exons 1–3 deletion |
| 179 | 197 | 3 | – | 0.01 | Severe | gDNA level exons 1–3 deletion |
| 180 | 198 | 7 | 1.9 | Severe | gDNA level exons 1–4 deletion | |
| 181 | 199 | 4 | 0.01 | Severe | gDNA level exons 1–7 deletion | |
| 200 | 1 | 0.01 | Severe | gDNA level exons 1–7 deletion | ||
| 182 | 201 | 2 | 4.2 | Severe | gDNA level exon 4 deletion | |
| 183 | 202 | 5 | 0.01 | Severe | cDNA level del incl. ex 5–6 | |
| 184 | 203 | 2 | 9.6 | Severe | gDNA level exon 7 deletion | |
| 185 | 204 | 5 | 0.01 | Severe | Complete IDS del | |
| 205 | 3 | 0.01 | Severe | Complete IDS del | ||
| 186 | 206 | 2 | 1.3 | Severe | Complete IDS del | |
| 187 | 207 | 3 | 0.01 | Severe | Complete IDS del | |
| 188 | 208 | 7 | 0.01 | Severe | Complete IDS del | |
| 189 | 209 | 10 | 0.01 | Severe | Complete IDS del | |
| 190 | 210 | 3 | 0.01 | Severe | Complete IDS del | |
| 191 | 211 | 2 | 0.01 | Severe | Complete IDS del | |
| 192 | 212 | 5 | 0.01 | Severe | Recomb. between in. 7 and seq. distal of ex. 3 in IDS-2 without exons deletion | |
| 193 | 213 | 2 | 0.01 | Severe | Recomb. between in. 7 and seq. distal of ex. 3 in IDS without exons deletion | |
| 194 | 214 | 3 | 0.01 | Severe | Recomb. between in. 7 and seq. distal of ex. 3 in IDS without exons deletion | |
| 195 | 215 | 6 | 2.5 | Severe | Recomb. between in. 7 and seq. distal of ex. 3 in IDS without exons deletion | |
| 196 | 216 | 6 | 7.2 | Severe | Recomb. between in. 7 and seq. distal of ex. 3 in IDS without exons deletion | |
| 197 | 217 | 6 | 0.43 | Severe | Recomb. between in. 7 and seq. distal of ex. 3 in IDS without exons deletion | |
| 198 | 218 | 3 | 2.4 | Severe | Recomb. between in. 7 and seq. distal of ex. 3 in IDS without exons deletion | |
| 199 | 219 | 3 | 0.01 | Severe | Recomb. between in. 7 and seq. distal of ex. 3 in IDS without exons deletion | |
| 220 | 13 | 3.6 | Severe | Recomb. between in. 7 and seq. distal of ex. 3 in IDS without exons deletion | ||
| 200 | 221 | 5 | 0.01 | Severe | Recomb. between in. 7 and seq. distal of ex. 3 in IDS without exons deletion | |
| 201 | 222 | 2 | 0.01 | Severe | Recomb. between in. 7 and seq. distal of ex. 3 in IDS without exons deletion | |
| 202 | 223 | 1 | 5.1 | Severe | Recomb. between in. 7 and seq. distal of ex. 3 in IDS without exons deletion | |
| 203 | 224 | 5 | 2.3 | Severe | Recomb. between in. 7 and seq. distal of ex. 3 in IDS without exons deletion | |
| 204 | 225 | 1 | 0.01 | Severe | Recomb. between in. 7 and seq. distal of ex. 3 in IDS without exons deletion | |
| 205 | 226 | 3 | 0.01 | Severe | Recomb. between in. 7 and seq. distal of ex. 3 in IDS with 3–7 exons deletion | |
| 206 | 227 | 2 | 0.01 | Severe | Recomb. between in. 7 and seq. distal of ex. 3 in IDS with 3–7 exons deletion | |
| 207 | 228 | 3 | 0.12 | Severe | Recomb. between in. 7 and seq. distal of ex. 3 in IDS with 3–7 exons deletion |
The comparative analysis of the severity of clinical symptoms with the results of nucleotide variants detected in the IDS gene are illustrated in Table 4. It demonstrates that most of nucleotide variants found caused the development of severe forms of the disease characterized by early (from the first months of life) manifestation of the disease, severe damage to vital organs and body systems, severely reduced intelligence and shorter life expectancy.
The protein changes p.Aspn63Asp (three patients; two families), p.Ala79Glu, (two patients; one family), p.Ala85Thr, p.Leu102Arg (two patients; one family), p.Asp198Gly, p.Gly412Term, p.Pro197Leu (four patients; one family), p.Gly340Asp, p.Ala346Val (three patients; one family), p.Arg443Term, the small deletion without frame shift and site-splicing substitution IVS2-9 c->g led to the development of less severe cases with later symptom manifestations, milder symptoms and higher IQ scores. Our data are consistent with the studies of other researchers who analyzed the influence of amino acid substitution on the IDS structure [20, 21].
A brief summary of some patients with a mild form of Hunter syndrome is presented in Table 5.
Table 5.
Summary of some patients with a mild form and a mild -> intermediate form of Hunter syndrome
| Family # | Patient # | Age at diagnosis | Enzyme activity in plasma (N = 297–705 nmol/4h/ml) |
Nucleotide variant in IDS gene | Education | Profession | Date of death | Comment |
|---|---|---|---|---|---|---|---|---|
| 3 | 4 | 10 | 1.2 | c.236C>A (p.Ala79Glu) | University | Teacher | 43 | – |
| 5 | 8 | 8.4 | c.236C>A (p.Ala79Glu) | University |
Economist (38 years old) |
– | Active life | |
| 4 | 6 | 11 | 7.4 | c.305T>G (p.Leu102Pro) | University |
Economist 35 years old) |
– | Active life |
| 7 | 9 | 8.1 | c.305T>G (p.Leu102Pro) | University |
Economist (33 years old) |
– | Active life | |
| 10 | 19 | 15 | 3.6 | c.1234G>T (p.Gly412Term) | University |
Lawyer (32 years old) |
– | Active life |
| 20 | 29 | 3 | 1.4 | c.1204G>A (p.Glu402Lys) | University | Lawyer | 23 | The cause of death is unprofessional tracheal intubation, carried out with the aim of removing the patient from the epileptic status |
| 15 | 24 | 13 | 0.01 | c.253G>A (p.Ala85Thr) | historic | Historian | 29 | Sudden death from acute cardiovascular failure |
Clinical case
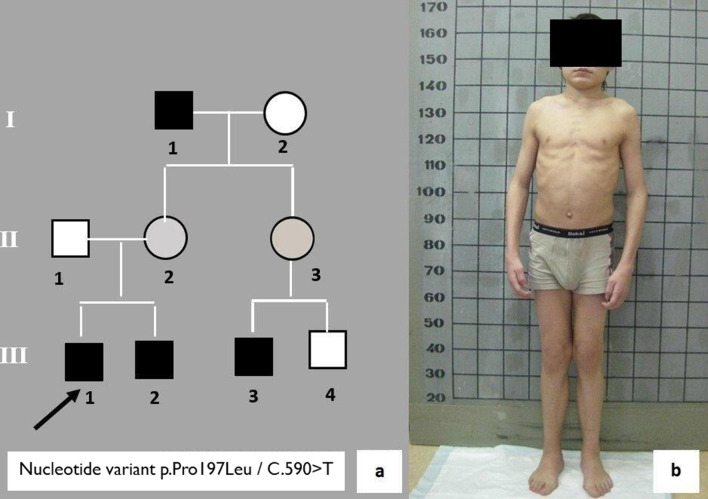
A 12-year old male (#9) was admitted to the genetics department with complaints about rough facial features and stiffness of major and small joints (Fig. 1). While collecting the genealogical history, it was found that the proband’s grandfather on the maternal line exhibits identical symptoms (Fig. 1a—I, 1). The grandfather was 55 years old at the time. The man had a disability and was observed by physicians at the place of residence. The diagnoses he was given were rheumatoid arthritis, hypothyroidism, osteochondrosis, and hypochondroplasia. The proband’s 3-year-old younger brother (Fig. 1a—III, 2) and 4- and 2-year old male cousins (Fig. 1a—III, 3 and III, 4) were considered healthy. Based on the genealogical history and clinical features, the proband was suspected to have mucopolysaccharidosis type II. Examination revealed high rates of renal excretion of heparansulfate and dermatansulfate, a decrease in the activity of I2S in dried blood spots (0.1 μmM/l/h, with the norm being 2.5–50 μmM/l/h) and a missense variant c.590C>T (p.Pro197Leu) in exon 5 of the IDS gene was found; so the diagnosis was confirmed. This nucleotide variant was not described before. The examination of men in this family allowed us to diagnose Hunter syndrome in the grandfather (I, 1), the sibling (III, 2) and the cousin (III, 3) of the proband. Ten years of observation of affected members of this family showed a good social adaptation of patients and long life expectancy of the grandfather. The proband successfully completed a technical college. His sibling and cousin are community college students. All affected family members voluntarily refused to receive enzyme replacement therapy. The clinical observation of the family continues.
Fig. 1.
a A fragment of the pedigree of patient #16 with a mild form of Hunter syndrome; b a 12-year old patient #16 with a mild form of Hunter syndrome
The rare case of Hunter syndrome in girls observed by the authors is due to a disease-associated variant in the IDS gene inherited from the mother and a deletion in the long arm of the X chromosome of paternal origin. The diagnosis was confirmed based on the detection of the absence of enzyme activity of iduronate-2-sulfatase and results of cytogenetic, molecular cytogenetic and molecular genetic examination [22].
Discussion
We present the results of the clinical observation of 228 Russian patients with Hunter syndrome. The diagnosis of mucopolysaccharidosis type II (Hunter syndrome) consisted of four consecutive stages: 1—Assessment of phenotypic characters; 2—Determination of indicators of excretion of urinary glycosaminoglycans and their fractions, primarily heparan and dermatan sulfates; 3—Measurement of the activity of the lysosomal enzyme iduronate-2-sulfatase; 4—DNA diagnostics, including the search for nucleotide substitutions in the IDS gene with an estimate of its frequency and pathogenicity according to the International HMGD Database.
According to the latest concept that MPS II is a continuum between the mild and severe form of the disease, we divided all patients into four groups. Group 1 includes 166 patients with a severe form of the disease, Group 2—patients with a moderate form of the disease, which was subdivided into Group 2a—26 patients with an intermediate -> severe (rather severe) and Group 2b—16 patients with a mild -> intermediate (rather a mild form). Group 3 includes 19 patients with a mild form of Hunter syndrome.
The clinical symptoms of patients with a severe form of mucopolysaccharidosis type II were characterized by an early manifestation of the disease (first months of life), rapid progression of clinical symptoms with the severe damage to the musculoskeletal system, cardiovascular and bronchopulmonary systems, parenchymal organs, hearing, and the formation of rapidly progressing umbilical, inguinal and inguinal-scrotal hernias requiring urgent surgical intervention. These patients completely lacked the ability to move independently and communicate with others; their IQ did not exceed 10–15 points, and life expectancy was short.
The residual activity of I2S in plasma varied from undetectable values to 11.8 nmol/4h/ml. No residual activity in dried blood spots was determined.
Complete deletions of the IDS gene, deletions of several exons, site splicing disease-associated variants, nonsense and missense nucleotide substitutions have been detected in patients with a severe form of the disease (Table 4). Many missense nucleotide variants found in patients with a severe form of the disease have been already described (Table 2). The data of other researchers regarding genotype–phenotype correlations are consistent with our findings [20, 21, 23–27]. Five undescribed missense variants were found in patients with a severe form (c.103G>C; c.136G>T; c.136G>A; c.307T>G; c.403A>G). Some nucleotide substitutions were in the same codon where other changes were found. According to the software analysis for impact prediction of nucleotide changes c.136G>T; c.136G>A; c.307T>G; c.403A>G were probably damaging. For the nucleotide change c.103G>C, PolyPhen-2 and PMUT Pathogenic mutation prediction programs showed a possibly damaging effect. However, this nucleotide variant was predicted as most probably affecting splicing by the Human Splicing Finder software. This seems to us to be a more truthful interpretation since patients had a severe form of the disease. Of course, for complete understanding of the damaging effect of nucleotide variants found additional molecular genetic studies are required. Four novel nonsense variants: c.814C>T; c.829C>T; c.998C>A; c.1340T>A and one new site splicing substitution c.1006+2T>G have been found in patients with a severe form of the disease. All previously undescribed small deletions, small insertions and small indels found in the patients with a severe form resulted in the frame shift and premature stop codon, which is expected to lead to the development of a severe form of the disease. It was established that as a result of extended deletions, the synthesis of the truncated protein occurs, which leads to the violation of its full function, and, thus, to the formation of severe clinical symptoms.
The 2a group of patients with an intermediate -> severe (rather severe) form was characterized by a late formation of the main clinical symptoms of the disease: the first external signs of the disease became evident at 5–6 months of life, heart murmurs were heard at 7–8 months, at the same age hepatosplenomegaly was diagnosed. After six months of life, the child experienced a delay in psychomotor development. However, the children of the second group were capable of independent movement, phrasal speech with limited vocabulary and successful communication. These patients were distinguished by disinhibition, restlessness, quick exhaustion, and lack of concentration. The IQ of the second group of patients did not exceed 45–55 points, and life expectancy, as a rule, was no more than 15–20 years.
The residual activity of I2S in plasma varied from undetectable values to 21.6 nmol/4h/ml. No residual activity in dried blood spots was determined.
The most common nucleotide variants in this group of patients was splicing site substitution c.1122C>T.
Three novel missense variant have been found in these patients: c.283A>T; c.512G>A; [c.1411G>C;1418C>T]. According to the prediction of damaging effect, all these nucleotide variants were disease causing ones (Table 3). Two different nucleotide variants [c.1411G>C;1418C>T] had been detected in one patient. First, it was assumed that one of these nucleotide variants was a nonpathogenic polymorphic one. However, both substitutions had been predicted as nucleotide variants with a potential damaging effect on the protein. For this case, it would be interesting to perform mutagenesis in vitro studies to assess the pathogenic effect of each of the substitutions. Two novel small deletions c.118_120delCTT; p.Leu40del and c.1426_1437 del12 p.476_479delAsnSerAspLys result in only loss of some amino acids without frame shift.
The clinical phenotype of the patients from Group 2b (a mild -> an intermediate form (rather mild)) was between phenotypes of patients from Group 2a and Group 3. The residual activity of I2S in plasma varied from undetectable values to 21.6 nmol/4h/ml. No residual activity in dried blood spots was determined.
The nucleotide variant c.253G>A has been found in five patients from Group 2b. Nucleotide variants c.587T>C and c.1204G>A have been detected twice. Two missense variants c.1028G>A; c.1035G>C, site splicing substitution c.241-9C>G and c.1438_1442delCCGAG; p.Pro480Phefs were novel ones.
The third group of probands with a mild form of the disease was characterized by even a later formation (2–4 years of life) of clinical symptoms. These patients were distinguished by higher body length, less severe changes in the musculoskeletal system and internal organs, a rare formation of hernias or their complete absence, normal intelligence, allowing probands to study in primary, secondary and even professionally oriented schools, successfully graduate and even work in their chosen field, often climbing the corporate ladder to senior level positions. Many of them successfully married and had a healthy offspring. The life expectancy of these patients was the highest and could reach 60 years and above.
The residual activity of I2S in plasma varied from undetectable values to 21.1 nmol/4 h/ml. No residual activity in dried blood spots was determined.
Two variants (c.187A>G, p.1037C>T) were described elsewhere also in a patient with an attenuated form of the disease [24–27]. The four missense variants found in patients with a mild form of Hunter disease (c.236C>A; c.305T>G; c.593A>G; c.1019G>A) were described by authors in their first study of the Russian patient group with MPS II [28]. The nucleotide variants c.590C>T and c.1234G>T were first described in this study.
For all patients the values of the residual activity of I2S in plasma ranged from 0 to 21.6 nmol/4h/ml regardless of age and severity of the disease. For all patients, regardless of age and severity of the disease no residual activity in dried blood spots was determined. Thus, no correlation between the residual activity of enzyme and severity of the disease was observed (Table 4).
Conclusion
In all groups of patients, a different type of nucleotide variant in the IDS gene has been found. It is assumed that the relationship of missense substitution with a severe form of Hunter syndrome, in some cases, can be explained by the pathological role of the replaced amino acid. On the other hand, two nonsense variants: c.1234G>T; p.Gly412Term and c.1438_1442delCCGAG; p.Pro480Phefs, despite the formation of the premature stop codon led to the development of a mild form or a mild -> intermediate form of Hunter syndrome. That suggests that a break in the amino acid chain at a certain position does not always cause several functional damages to the protein. Thus, in authors’ opinion, in order to understand the effect of missense and even nonsense substitutions, specific functional studies are required.
Several point variants (c.253G>A, c.257C>T, c.263G>A, c.263G>T, c.514C>T, c.998C>T, c.1327C>T, c.1402C>T; c.1403G>A, c.1122C>T), a small deletion c.596_599delAACA and recombination between intron 7 in the IDS and exon 3 in IDS2 were detected more than twice in patients from different families. The frequency of these substitutions in the presented cohort of patients varies from 2 to 6%. Thus, these replacements could be relatively common. Probably these point variants that involved CpG sites of the IDS gene, the locus of c.596_599delAACA deletion and recombination region between the IDS and IDS2 are mutagenesis hotspots in the IDS gene [18, 19].
The enzyme replacement therapy with Elaprase has become available in Russia since 2008. According to the Russian public health law, Elaprase was purchased by regional health authorities, which do not always afford the drug due to its high cost. Therefore, not all patients with Hunter syndrome received enzyme replacement therapy. Since 2019, federal authorities started purchasing Elaprase at the expense of the federal budget to provide all patients with mucopolysaccharidosis type II with this enzyme replacing drug.
In 2018, a second enzyme-replacing drug was registered in Russia for the treatment of patients with Hunter syndrome, called idursulfase beta (Hunterase). The drug was developed and successfully tested by Korean researchers.
To date, 113 patients with Hunter syndrome are receiving enzyme replacement therapy in Russia, including 16 adult patients. Enzyme replacement therapy undoubtedly improves the quality and life expectancy of patients with Hunter syndrome, especially patients with a mild form of the disease. In patients with severe (Group 1) and intermediate (Group 2a) forms of the disease, the improvement relates mainly to the state of internal organs (reduction in size liver and spleen, decreased left ventricular mass), increased weight and growth parameters, mainly body length, improved emotional tone, increased step test (if not independent movement of patients), and decreased renal excretion of heparan and dermatan.
No growth in intellectual development (IQ) was observed due to the inability of enzyme replacing drugs to penetrate the blood–brain barrier.
During treatment, patients with a mild form of the disease had more positive changes and a notable increase in tolerance to physical and mental stress.
Acknowledgements
Not applicable
Abbreviations
- DNA
Deoxyribonucleic acid
- GAG
Glycosaminoglycans
- I2S
Iduronate-2-sulfatase
- MPS II
Mucopolysaccharidosis type II
Authors' contributions
AS established a clinical diagnosis, performed a final analysis of the results obtained, designed the article and wrote the first draft of the manuscript. EV performed the DNA diagnostics to search for a disease-associated variant in the IDS gene and interpreted the data received. EN provided a comparative analysis of all clinical data and laboratory results and consulted the patients and their families. EZ determined the activity of lysosomal enzyme iduronate-2-sulfatase in dry blood spots, analyzed all laboratory data obtained and organized a discussion. All authors read and approved the final manuscript.
Funding
This work was financially supported by the Government Assignment of the Russian Ministry of Health, Assignment No. 2 00 079 056. The funders had no role in the study design; in the collection, analysis, or interpretation of the data; or in the preparation, review, or approval of the manuscript.
Availability of data and materials
The datasets generated and analysed during the current study are available in the Human Gene Mutation Database www.hgmd.cf.ac.uk/ (information about nucleotide changes described before) and ClinVar at https://www.ncbi.nlm.nih.gov/clinvar/ (information about novel nucleotide changes), ClinVar accession numbers SCV001450592-SCV001450635 (see also Table 2). The information about the IDS gene sequencing is available at https://www.ncbi.nlm.nih.gov/genome/gdv/ (NC_000023.11).
Declarations
Ethics approval and consent to participate
The study was approved by the Ethics committee of the Research and Clinical Institute of Pediatrics named after Yuri Veltischev of the Pirogov Russian National Research Medical University of the Ministry of Health of the Russian Federation. A written informed consent was obtained from the participants and parents of participants under the age of 18 to take part in the study. The study was done in accordance with the principles outlined in the Helsinki Declaration (1964).
Consent for publication
A written informed consent for the publication of this manuscript including identifying images and other personal and clinical details was obtained from the participants and parents or legal guardians of all participants under the age of 18.
Competing interests
None of the authors has any competing interests in the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Alla Nikolaevna Semyachkina and ElenaYurievna Voskoboeva contributed equally to this study and share first authorship
Contributor Information
Alla Nikolaevna Semyachkina, Email: gokhramenko@pedklin.ru.
Elena Yurievna Voskoboeva, Email: e_voskoboeva@mail.ru.
Ekaterina Alexandrovna Nikolaeva, Email: kate_nikolaeva09@mail.ru.
Ekaterina Yurievna Zakharova, Email: labnbo@yandex.ru.
References
- 1.Poorthuis BJ, Wevers RA, Kleijer WJ, Groener JE, de Jong JG, van Weely S, Niezen-Koning KE, van Diggelen OP. The frequency of lysosomal storage diseases in The Netherlands. Hum Genet. 1999;105(1-2):151–156. doi: 10.1007/s004399900075. [DOI] [PubMed] [Google Scholar]
- 2.Poupetová H, Ledvinová J, Berná L, Dvoráková L, Kozich V. Elleder M The birth prevalence of lysosomal storage disorders in the Czech Republic: comparison with data in different populations. J Inherit Metab Dis. 2010;33(4):387–396. doi: 10.1007/s10545-010-9093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Applegarth DA, Toone JR, Lowry RB. Incidence of inborn errors of metabolism in British Columbia, 1969–1996. Pediatrics. 2000;105(1):e10. doi: 10.1542/peds.105.1.e10. [DOI] [PubMed] [Google Scholar]
- 4.Khan SA, Peracha H, Ballhausen D, Wiesbauer A, Rohrbach M, Gautschi M, Mason RW, Giugliani R, Suzuki Y, Orii KE, Orii T, Tomatsu S. Epidemiology of mucopolysaccharidoses. Mol Genet Metab. 2017;121(3):227–240. doi: 10.1016/j.ymgme.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lonardo F, Di Natale P, Lualdi S, Acquaviva F, Cuoco C, Scarano F, Maioli M, Pavone LM, Di Gregorio G, Filocamo M, Scarano G. Mucopolysaccharidosis type II in a female patient with a reciprocal X;9 translocation and skewed X chromosome inactivation. Am J Med Genet A. 2014;164A(10):2627–2632. doi: 10.1002/ajmg.a.36667. [DOI] [PubMed] [Google Scholar]
- 6.Sukegawa K, Song XQ, Masuno M, Fukao T, Shimozawa N, Fukuda S, Isogai K, Nishio H, Matsuo M, Tomatsu S, Kondo N, Orii T. Hunter disease in a girl caused by R468Q mutation in the iduronate-2-sulfatase gene and skewed inactivation of the X chromosome carrying the normal allele. Hum Mutat. 1997;10(5):361–367. doi: 10.1002/(SICI)1098-1004(1997)10:5<361::AID-HUMU5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 7.Tuschl K, Gal A, Paschke E, Kircher S, Bodamer OA. Mucopolysaccharidosis type II in females: case report and review of literature. Pediatr Neurol. 2005;32(4):270–272. doi: 10.1016/j.pediatrneurol.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 8.McKusick VA. The mucopolysaccharidoses. Heritable disorders of connective tissue. 4. C. V. Mosby (pub.): St. Louis; 1972. pp. 556–574. [Google Scholar]
- 9.Wraith JE, Scarpa M, Beck M, Bodamer OA, De Meirleir L, Guffon N, Lund AM, Malm G, Van der Ploeg AT, Zeman J. Mucopolysaccharidosis type II (Hunter syndrome): a clinical review and recommendations for treatment in the era of enzyme replacement therapy. Europ J Pediat. 2008;167:267–277. doi: 10.1007/s00431-007-0635-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bielicki J, Freeman C, Clements PR, Hopwood JJ. Human liver iduronate-2-sulphatase: purification, characterization and catalytic properties. Biochem J. 1990;271:75–86. doi: 10.1042/bj2710075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flomen RH, Green EP, Green PM, Bentley DR, Giannelli F. Determination of the organisation of coding sequences within the iduronate sulphate sulphatase (IDS) gene. Hum Molec Genet. 1993;2:5–10. doi: 10.1093/hmg/2.1.5. [DOI] [PubMed] [Google Scholar]
- 12.Human Gene Mutation Database. www.hgmd.cf.ac.uk.
- 13.Hunter CA. A rare disease in two brothers. Proc R Soc Med. 1917;10:104–116. [PMC free article] [PubMed] [Google Scholar]
- 14.Young ID, Harper PS. The natural history of severe form of Hunter's disease: a study based on fifty two cases. Dev Med Child Neurol. 1983;25:481–489. doi: 10.1111/j.1469-8749.1983.tb13794.x. [DOI] [PubMed] [Google Scholar]
- 15.Hopwood JJ, Harrison JR. High-resolution electrophoresis of urinary glycosaminoglycans: an improved screening test for the mucopolysaccharidoses. Anal Biochem. 1982;119(1):120–127. doi: 10.1016/0003-2697(82)90674-1. [DOI] [PubMed] [Google Scholar]
- 16.Voznyi YV, Keulemans JL, van Diggelen OP. A fluorimetric enzyme assay for the diagnosis of MPSII (Hunter disease) J Inher Metab Dis. 2001;24(6):675–680. doi: 10.1023/A:1012763026526. [DOI] [PubMed] [Google Scholar]
- 17.Kumar AB, Masi S, Ghomashchi F, et al. Tandem mass spectrometry has a larger analytical range than fluorescence assays of lysosomal enzymes: application to newborn screening and diagnosis of Mucopolysaccharidoses types II, IVA, and VI. Clin Chem. 2015;61:1363–1371. doi: 10.1373/clinchem.2015.242560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagerstedt K, Karsten SL, Carlberg BM, Kleijer WJ, Tönnesen T, Pettersson U, Bondeson ML. Double-strand breaks may initiate the inversion mutation causing the Hunter syndrome. Hum Mol Genet. 1997;6(4):627–633. doi: 10.1093/hmg/6.4.627. [DOI] [PubMed] [Google Scholar]
- 19.Rathmann M, Bunge S, Beck M, Kresse H, Tylki-Szymanska A, Gal A. Mucopolysaccharidosis type II (Hunter syndrome): mutation "hot spots" in the iduronate-2-sulfatase gene. Am J Hum Genet. 1996;59(6):1202–1209. [PMC free article] [PubMed] [Google Scholar]
- 20.Saito S, Ohno K, Okuyama T, Sakuraba H. Structural basis of mucopolysaccharidosis type II and construction of a database of mutant iduronate 2-sulfatases. PLoS ONE. 2016;11(10):e0163964. doi: 10.1371/journal.pone.0163964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zanetti A, D'Avanzo F, Rigon L, Rampazzo A, Concolino D, Barone R, Volpi N, Santoro L, Lualdi S, Bertola F, Scarpa M, Tomanin R. Molecular diagnosis of patients affected by mucopolysaccharidosis: a multicenter study. Eur J Pediatr. 2019;178(5):739–753. doi: 10.1007/s00431-019-03341-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semyachkina AN, Voskoboeva EY, Zakharova EY, Nikolaeva EA, Kanivets IV, Kolotii AD, Baydakova GV, Kharabadze MN, Kuramagomedova RG. Melnikova NV Case report: a rare case of Hunter syndrome (type II mucopolysaccharidosis) in a girl. BMC Med Genet. 2019;20(1):66. doi: 10.1186/s12881-019-0807-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pollard LM, Jones JR, Wood TC. Molecular characterization of 355 mucopolysaccharidosis patients reveals 104 novel mutations. J Inherit Metab Dis. 2013;36(2):179–187. doi: 10.1007/s10545-012-9533-7. [DOI] [PubMed] [Google Scholar]
- 24.Flomen RH, Green PM, Bentley DR, Giannelli F, Green EP. Detection of point mutations and a gross deletion in six Hunter syndrome patients. Genomics. 1992;13(3):543–550. doi: 10.1016/0888-7543(92)90123-A. [DOI] [PubMed] [Google Scholar]
- 25.Goldenfum SL, Young E, Michelakakis H, Tsagarakis S, Winchester B. Mutation analysis in 20 patients with Hunter disease. Hum Mutat. 1996;7(1):76–78. doi: 10.1002/(SICI)1098-1004(1996)7:1<76::AID-HUMU14>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 26.Li P, Huffman P, Thompson JN. Mutations of the iduronate-2-sulfatase gene on a T146T background in three patients with Hunter syndrome. Hum Mutat. 1995;5(3):272–274. doi: 10.1002/humu.1380050314. [DOI] [PubMed] [Google Scholar]
- 27.Froissart R, Da Silva IM, Maire I. Mucopolysaccharidosis type II: an update on mutation spectrum. Acta Paediatr. 2007;96(455):71–77. doi: 10.1111/j.1651-2227.2007.00213.x. [DOI] [PubMed] [Google Scholar]
- 28.Karsten S, Voskoboeva E, Tishkanina S, Pettersson U, Krasnopolskaja X, Bondeson ML. Mutational spectrum of the iduronate-2-sulfatase (IDS) gene in 36 unrelated Russian MPS II patients. Hum Genet. 1998;103(6):732–735. doi: 10.1007/s004390050901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analysed during the current study are available in the Human Gene Mutation Database www.hgmd.cf.ac.uk/ (information about nucleotide changes described before) and ClinVar at https://www.ncbi.nlm.nih.gov/clinvar/ (information about novel nucleotide changes), ClinVar accession numbers SCV001450592-SCV001450635 (see also Table 2). The information about the IDS gene sequencing is available at https://www.ncbi.nlm.nih.gov/genome/gdv/ (NC_000023.11).