Abstract
COVID-19 pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has put the global public health at its highest threat around the world. Previous epidemic caused by the acute respiratory syndrome coronavirus (SARS-CoV) in 2002 is also considered since both the coronaviruses resulted in the similar clinical complications. The outbreak caused by the Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012 had a low rate of disease transmission and death cases. Modes of entry by MERS and SARS coronaviruses are similar to that of SARS-CoV-2, except MERS-CoV utilize different receptor. They all belong to the lineage C of β-coronavirus. Based on the information from the previous reports, the present review is mainly focused on the mechanisms of disease progression by each of these viruses in association to their strategies to escape the host immunity. The viral entry is the first step of pathogenesis associated with attachment of viral spike protein with host receptor help releasing the viral RNA into the host cell. Models of molecular pathogenesis are outlined with virus strategies escaping the host immunity along with the role of various inflammatory cytokines and chemokines in the process. The molecular aspects of pathogenesis have also been discussed.
Keywords: Viral pathogenesis and host immunity, COVID-19 pandemic, Acute respiratory syndrome coronavirus (SARS-CoV-2), SARS-CoV and MERS-CoV
Introduction
Coronaviruses (CoVs) from the family Coronaviridae, and order Nidovirales are the enveloped, non-segmented, positive-sense RNA viruses which are grouped into four genera: alpha (α), beta (β), gamma (γ), and delta (δ).CoVs among which the β-CoVs include the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV) (Rabaan et al. 2020; Fehr and Perlman 2015). All these viruses have the highly conserved genomic organization, consisting of the 5′ cap-leader-UTR (untranslated region)—replicase (around 20 kb)—S (Spike)—E (Envelope)—M (Membrane)—N (Nucleocapsid)—3′ UTR-poly (A) tail scheme, along with the accessory genes interspersed within the structural genes at the 3′ end of the RNA (Fehr and Perlman 2015). The events within the life cycle of these viruses are the same: (1) attachment and entry into the host, (2) expression of the replicase protein, (3) viral replication and transcription, (4) viral assembly and subsequent release (Rabaan et al. 2020; Fehr and Perlman 2015; Noor and Maniha 2020; Chu et al. 2020; Noor 2021). Both SARS-CoV and MERS-CoV may originate from bats; SARS-CoVs were transmitted to humans from market civets; and the MERS-CoVs from the dromedary camels (Huang et al. 2020; Lu et al. 2020). SARS-CoV-2 has the similar transmission pattern like SARS-CoV; i.e., from bats, pangolins, snakes as the intermediate hosts further facilitating the spillover of SARS-CoV-2 to the humans; and its spread is enabled by human-to-human transmission via the respiratory droplets and due to the international traveling (Rabaan et al. 2020; Noor and Maniha 2020). Although the MERS-CoV originated from bats, however, the intermediate host was camel which transferred the infection to humans, and afterwards human-to-human spread triggered the epidemic (Fehr and Perlman 2015). SARS-CoV- 2 infection commonly manifests the onset of pneumonia, fever, dry cough, dyspnea, myalgia, fatigue, and in fatal case, the acute respiratory distress syndrome (ARDS) (Noor and Maniha 2020). Nearly similar clinical manifestations have also been noticed for both SARS-CoV and MERS-CoV (Fehr and Perlman 2015).
SARS-CoV was first isolated from the wild animals sold as food in Guangdong, China in November 2002; and the SARS-CoV epidemic caused around 8000 cases in 26 countries (Rabaan et al. 2020; Chu et al. 2020). The MERS-CoV infection, also known as camel flu was first isolated in the Saudi Arabia in June 2012 with a very limited case of infections (approximately 2500 infected cases) (Chu et al. 2020). At present, the world population is at its highest risk imposed by COVID-19 pandemics (in 223 countries) caused by the SARS-CoV-2 (originated from the Wuhan City of China at the end of December 2019), with 2,360,280 deaths and 107,423,526 infected cases (WHO 2021). Along the last 20 years, the SARS-CoV epidemic took place in 2002; and the onset of the MERS-CoV was noticed in 2012 only in the Middle East (Tay et al. 2020). Thus, SARS-CoV-2 has appeared to be the most dreadful pandemic with a very high transmission dynamics whereas the SARS-CoV or the MERS-CoV had a much lesser impact on the global public health. Study of phylogeny revealed that SARS-CoV-2 is actually quite distant both from the SARS-CoV (79% sequence homology) and MERS-CoV (only 50% homology) (Noor and Maniha 2020).
The extensive study on the pathophysiological impact on the SARS-CoV-2, SARS-CoV and the MERS-CoV concentrated on the alterations within the host immune cells including various pro-inflammatory cytokines and chemokines upon the viral infection (Fehr and Perlman 2015; Lee et al. 2020). The ongoing SARS-CoV-2 and the earlier SARS-CoV have been noticed to share a great deal of resemblances in terms disease progression escaping the host immunity, resulting in ARDS (Rabaan et al. 2020; Fehr and Perlman 2015; Noor 2021). Therefore, figuring out the pathogenic events along with the host immunity avoidance for each of these three viruses would be beneficial for the deeper understanding of the viral infectivity together with the possible scopes of effective drug designing. The information on the viral pathogenesis and the host immunological response for all three viruses has been schematically presented with separate models (Figs. 1, 2, 3) in the current review which may further facilitate to develop anti-viral drugs for mitigating the recurring infection. The key points of the current review include: (1) using the spike (S) protein, both the SARS-CoV-2 and SARS-CoV bind to the angiotensin-converting enzyme 2 (ACE 2) receptor of the host whereas MERS-CoV utilizes the host receptor dipeptidyl peptidase 4 (DPP4) as shown in the models plotted in Figs. 1, 2 and 3 (Rabaan et al. 2020; Fehr and Perlman 2015; Lu et al. 2020; Lee et al. 2020; Zhang et al. 2020; Kaushal 2020); (2) SARS-CoV-2 possesses a longer spike protein than that of SARS-CoV (Rabaan et al. 2020; Fehr and Perlman 2015); (3) antibody-dependent enhancement (ADE) can increase the severity of disease in any respiratory viral infection e.g., respiratory syncytial virus, measles, influenza etc. ADE might be caused by other proteins like cytokines, non-specific antibodies and in case of other viral infections etc.; to exacerbate the infection. ADE has been reported, previously, in SARS-CoV and MERS-CoV (in vivo and in vitro). ADE could occur in COVID-19 too. Recent studies have revealed that S- and RBD-specific antibodies IgG in COVID-19 patients have the lower level of fucosylation within the Fc domain could mediate the antibody-dependent enhancement in cellular cytotoxicity (Fehr and Perlman 2015; Noor 2021; Lee et al. 2020); (4) the regulations of IL-17 are up-regulated and prolonged in MERS-CoV as compared to SARS-CoV, which is believed to induce the immune-mediated pathology contributing to the mortality. Other inflammatory cytokines IL-6, IL-8, IL-1β and anti-viral cytokines TNF-α, IFN-β and IP-10 were also observed as compared to SARS. But, high leucocyte count, C-reactive protein, D-dimer and high expression of IL-2, IL-7, IL-10, G-CSF, IP-10, MCP-1-α, and TNF-α were found in individuals admitted in ICU with COVID-19 disease. This situation is denoted as "cytokine syndrome" or "cytokine storm" contribute to develop lung injury leading to fatal ARDS (Lee et al. 2020; Tay et al. 2020; Lai et al. 2020; Kikkert 2020; Wan et al. 2020; Zhang et al. 2020; Kaushal 2020).
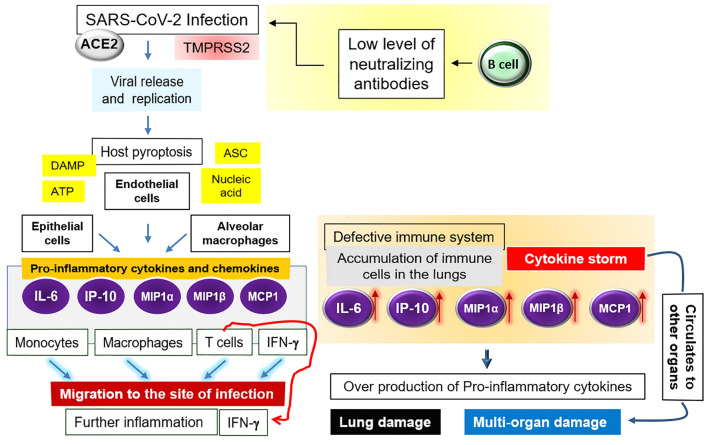
Fig. 1.
Immuno-pathogenicity of SARS-CoV-2. Details are written in the text. Briefly, the SARS-CoV infection triggers the generation of pro-inflammatory cytokines and chemokines (IL-6, IP-10, MIP1α), MIP1β and MCP-1) which attract monocytes, macrophages and T cells (in addition to the IFNγ) to the site of infection, promoting further inflammation. In case of defective immunity, the accumulation of immune cells in the lungs causes the overproduction of pro-inflammatory cytokines, which in turn harms the lung and other organs. The non-neutralizing antibodies further facilitate the viral infection through ADE, and thereby fostering the organ damage
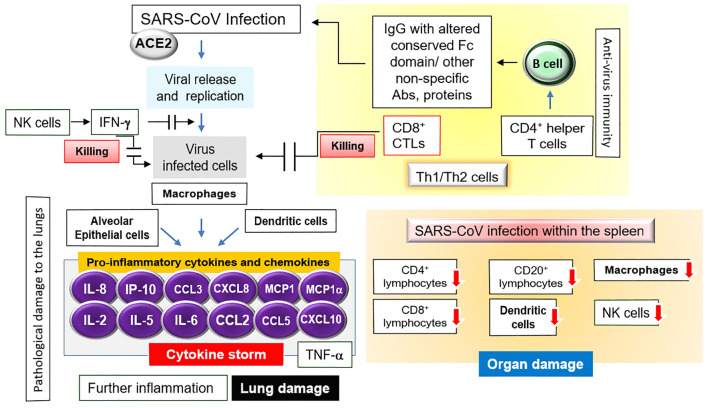
Fig. 2.
Immuno-pathogenicity of SARS-CoV. The interaction between SARS-CoV infection with the subsequent immune response as well as the pathogenesis causing the tissue damage triggering the disease fatality has been shown here. Details are described in the text
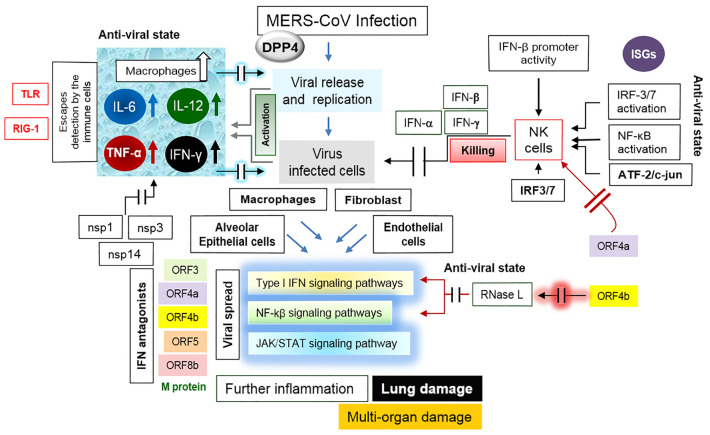
Fig. 3.
Immuno-pathogenicity of MERS-CoV. Details are described in the text. Briefly, upon the viral entry, the type I interferon (IFN)-mediated innate immune response is activated through the production of IFN-α and IFN-β; elevation in macrophages, the tumor necrosis factor α (TNF α), IL-6, IL-12, and IFN-γ. The pathogenesis depends on the IFN antagonists especially ORF4a and ORF4b. Inhibition of the interferon production as well as the type I interferon-induced JAK/STAT and the NF-kβ signaling pathway accelerates the pathogenesis
Proteins of β-coronaviruses (CoVs) required for pathogenesis
In general, all the three viruses consist of large replicase with four structural proteins: S (spike), E (envelope), M (membrane), and N (nucleocapsid) proteins; the N protein holds the RNA genome, and the S, E, and M proteins together create the viral envelope. The elucidation of the genomic organization revealed that the leader sequence and the 5′ UTR consist of multiple stem loop structures helping in the viral RNA replication and transcription while the transcriptional regulatory sequences (TRSs) reside at the beginning of each structural gene to promote their expressions (Fehr and Perlman 2015; Noor and Maniha 2020; Chu et al. 2020; Noor 2021). Usually in CoVs, the S protein is cleaved into two separate polypeptides: the S1 fraction, which makes up the large receptor-binding domain (RBD); and the second part, S2 which forms the stalk of the spike molecule (Fehr and Perlman 2015; Noor 2021). The distinctive attribute of SARS-CoV-2 genome is that the genes orf8 and orf3b encode the spike glycoproteins; and the Orf3b protein encoded by the orf3b gene has been reported to play important role in the SARS-CoV-2 pathogenesis (Noor and Maniha 2020). The N-linked glycosylated S protein utilizes an N-terminal signal sequence to gain access to the ER; and the class I fusion protein of S mediates attachment to the host receptor (Fehr and Perlman 2015). M protein gives the viral shape; and binds to the nucleocapsid (Fehr and Perlman 2015). The E protein mediates the assembly and release of the virus; and its ion channel activity has been shown to be essential for the SARS-CoV pathogenesis (Fehr and Perlman 2015). The N protein has been found with several activities in the viral pathogenesis including the detection of the transcriptional regulatory sequences (TRSs) and the genomic packaging signal; and binding to nsp3, the key component of the replicase complex, and binding to the M protein as well (Fehr and Perlman 2015). Such an interaction facilitates tether the viral RNA to the replicase–transcriptase complex (RTC) trailed by the encapsidation of the viral genome (Rabaan et al. 2020; Fehr and Perlman 2015). The hemagglutinin esterase (HE) can act as a hemagglutinin, binding to the sialic acids on the host surface glycoproteins, and mediating the acetyl-esterase activity which in turn accelerates the S protein-mediated entry of the virus followed by the invasion along the mucosa (Fehr and Perlman 2015).
Pathogenesis mediated by SARS-CoV-2
Like the common traits of β-coronaviruses, the N protein of SARS-CoV-2 is involved in the viral replication, assembly, and budding as well as involved in the host response (Chu et al. 2020). A total of 14 open reading frames (ORFs) within the SARS-CoV-2 have been identified: the first two at 5ʹ UTR encode polyproteins pp1a/ab which mediate the viral replication; and 16 non-structural proteins (nsps, that are needed for the transcription and replication as well (Lai et al. 2020; Kikkert 2020; Wan et al. 2020). Then structural proteins are encoded by the rest of the genome; and at the 3ʹ terminus, the accessory genes: 3a, 3b, p6, 7a, 7b, 8b, 9b and orf14 reside (Noor and Maniha 2020; Chu et al. 2020; Noor 2021). The transmissibility of virus remains undetected/unnoticed during the incubation period after infection, in case of asymptomatic individuals (Rabaan et al. 2020; Noor and Maniha 2020).
Interaction between the spike (S) protein and ACE receptor during the viral entry
As mentioned earlier, the spike (S) protein of the SARS-CoV-2 binds to the angiotensin-converting enzyme 2 (ACE 2) receptor (a metallopeptidase: the key molecule in the renin-angiotensin system) of the human host (Lai et al. 2020). Upon the ACE receptor engagement with the S protein, the cellular surface serine protease TMPRSS2 (the plasma membrane-associated type II transmembrane serine protease) is engaged to prime the spike protein (S) to expedite the membrane fusion that is needed for the release of the viral genome into the host cell cytosol (Noor and Maniha 2020). In addition, the antibody-dependent enhancement (ADE) of viral entry may occur whereby a neutralizing monoclonal antibody (Mab, through the IgG Fc receptor) targets the RBD of the spike, and subsequently allows it to undergo certain conformational changes which permit the viral entry into the IgG Fc receptor-expressing cells (Kikkert 2020; Wan et al. 2020). However, it is to be noted that the ADE phenomenon is noticed with monoclonals in case of SARS-CoV; and normally it does not happen with all coronaviruses. It has also not been noticed in case of SARS-CoV-2 till date (Fehr and Perlman 2015). The phylogenetic analysis revealed that the longer spike protein is encoded by the SARS-CoV-2 in comparison with SARS-CoV and MERS-CoV (Lu et al. 2020). The binding affinity between the SARS-CoV-2 spike ectodomain and human ACE2 was also calculated to be 10- to 20-fold higher than that of the SARS-CoV (Chu et al. 2020). Moreover, the antibody-dependent enhancement (ADE) of viral entry has also been noticed (Wan et al. 2020). Regarding the dreadfulness of SARS-CoV pathogenesis, successive mutations and recombination may largely account; and the evolutionary rate has been calculated up to 10–4 nucleotide substitutions per site per year for RNA viruses like SARS-CoV-2, which in turn may upshot the corresponding mutations at every replication cycle (Lu et al. 2020). Regarding the viral entry, the innate immune signaling is orchestrated through the recognition of the pathogen-associated molecular patterns (PAMPs) by the pattern recognition receptors (PRRs) like the RIG-I-like receptors (RLRs) and the Toll-like receptors (TLRs) from viral components or the replication intermediates (Zhang et al. 2020). Also, this is to be noted that the upregulation of cellular receptor ACE2 by SARS-CoV-2 and SARS-CoV; and the inflammatory cytokines may facilitate their infection (Kaushal 2020; Zhuang et al. 2020).
Pathogenic events after the viral entry and immunological responses
In case of a healthy immune response, the initial inflammation by the SARS-CoV entry attracts the virus-specific T cells to the site of infection as stated earlier (Tay et al. 2020; Kikkert 2020). Indeed, at the time of viral entry, antigen is processed by the antigen presenting cells (APCs) through MHC to activate the T cells; and the antibodies are produced by the B cells thereafter; i.e., IgM within 6–7 days followed by IgG, IgA titer in 3–4 weeks, which are specifically directed to combat the secondary infections (Kaushal 2020). This is to be mentioned that these antibodies usually do not halt the virus at the beginning (entry phase); actually, the viral spread can be arrested if a person is immunized with the virus (Kaushal 2020). However, the recognition of the viruses and apoptotic cells by the alveolar macrophages may eliminate the viral particles by phagocytosis (Tay et al. 2020). As shown in Fig. 1, the pathophysiological impact in the CoV-affected patients largely depends on the alteration of the levels of pro-inflammatory cytokines and chemokines; i.e., the generation of the cytokine storm (Tay et al. 2020; Ralph et al. 2020). Upon the viral entry mediated by ACE2 and TMPRSS2, the active replication with the subsequent release of the virus instigate the host cell to undergo pyroptosis (a condition of cell death and inflammation) together with the release of damage-associated molecular patterns (DAMPs) including ATP, nucleic acids and ASC (apoptosis-associated speck-like protein with a caspase-recruitment domain) oligomers (Tay et al. 2020). The antibodies produced by the B cells may also accelerate the infection through the ADE (Tay et al. 2020). Once the virus gets inside, (1) it travels down the bronchial tubes to the lungs letting the respiratory tree lining to be injured, (2) the nerves of the lining of the airway get irritated, making the mucous membranes of the lungs inflamed and hardened (Lee et al. 2020). As a result, it becomes difficult for lungs to supply oxygen to the blood, resulting in the shortness in breathing or the ARDS (Rabaan et al. 2020; Tay et al. 2020; Kikkert 2020).
As stated earlier, the SARS-CoV-2 first adheres to the ACE2 receptors in nasopharynx and respiratory epithelium; and the host immune response is instigated through the elevated levels of the pro-inflammatory cytokines generated from various lymphocytes, which in turn trigger the lung injury with the subsequent onset of ARDS, a state whereby the alveolar spaces are dominated by the infiltered neutrophils and monocytes which finally lead to hyper cytokinemia. (Kikkert 2020; Kaushal 2020). Besides, the hyper activation of various immune cells uniquely against the nucleocapsid of the virus cause the cytokine storm which is responsible for the ARDS, and the multi-organ malfunction at the later stage (Rabaan et al. 2020; Fehr and Perlman 2015; Lee et al. 2020; Kikkert 2020; Kaushal 2020). This is also worth to mention that the pro-inflammatory macrophages and granulocytes cause the innate inflammation in lungs; i.e., difficulty in breathing as well as cyanosis which in turn require ventilation in the COVID-19 patient. A potential cause of such lung fatality is hyaluronan (HA) which is produced by the HA synthase (HAS2) in CD31-endothelium, EpCAM-lung alveolar epithelial cells, and fibroblasts, triggered by IL-1 and TNF (Kaushal 2020). HA in turn absorbs water in significantly higher amount (approximately 1000-fold than normal), and thereby causing the difficulty in oxygen supply in lungs (Kaushal 2020).
As shown in Fig. 1, the DAMPs are recognized by the neighboring epithelial cells, endothelial cells and the alveolar macrophages, which then trigger the production of the pro-inflammatory cytokines and chemokines including IL-6, IP-10, macrophage inflammatory protein 1α (MIP1α), MIP1β and MCP-1, which entice the monocytes, macrophages, T cells and IFNγ to the site of infection and thus promoting further inflammation (Tay et al. 2020). In case of defective immunity, the above-mentioned cells over accumulate in the lungs resulting in the state of the elevated pro-inflammatory cytokines (cytokine storm), which subsequently impairs the lung infrastructure (Tay et al. 2020; Kikkert 2020). The cytokine storm further circulates to other organs, causing the multi-organ damage (Tay et al. 2020).
Evasion of host immunity by SARS-CoV-2
Dodging of the host protective immunity is the principal strategy to impart the viral pathogenesis (Lee et al. 2020; Kikkert 2020). As the virus spreads along the nasopharyngeal tract, cells of the innate immune defense mechanisms are concomitantly avoided (Kikkert 2020). Indeed, after the viral infection starts, both the adaptive (regulated by the MHC antigens/HLA classes) and innate immunity commence within host (Kikkert 2020; Kaushal 2020). The protective innate immunity is diminished in the severe symptomatic stages of pathogenesis with excessive lymphopenia the NK cells which in turn impart the virus to dodge the host immunity; however, the monocytes and macrophages rush towards the infected site and thus create a hyperinflammatory state (Fehr and Perlman 2015; Noor 2021; Kikkert 2020; Kaushal 2020). Moreover, as the virus skips the NK-cell-mediated protective immunity, eventually the viral spread occurs resulting inflammations of lungs and other major organs (Kaushal 2020). Besides the impairment of innate immunity, the presentation of viral antigens by MHC class I and II can be compromised with the APCs which in turn instigate the weakening of the T-cell response; and thus, the adaptive immunity is also hindered (Noor 2021).
From the molecular point of view, the viral RNA circumvents the innate immune cells by adding a cap-structure (the cap-snatching mechanism, aided by the N protein) to its 5′-end and resulting in the release of the short, 5′-capped transcripts; helping them to be detected by the host translational apparatuses (Kikkert 2020; Vlugt et al. 2018). The 2′-O methylation of the viral cap structures is facilitated by the nsp16, which helps to avoid recognition by the immune sensor MDA5 (melanoma differentiation-associated protein 5) (Kikkert 2020; Menachery et al. 2014). The second mechanism relies on the viral endoribonuclease activity to avoid the protein kinase R (PKR), and the 2′–5′ Oligoadenylate Synthetase (OAS)/RNAse L system (the interferon, IFN effector) (Lee et al. 2020). Thus, the SARS-CoV-2 accelerates its own RNA during the infection processes with the concomitant avoidance of the host immune system (Lee et al. 2020). Thirdly, the N protein reduces the chance of the viral mRNA destruction as well (Kikkert 2020).
Pathogenesis mediated by SARS-CoV and immunological response
Immunological response upon viral infection can either limit virus spreading or surprisingly can cause the pathological damages to the host tissues which mainly depends on the accumulation of pro-inflammatory cytokines (released by activated macrophages) in alveoli as stated earlier (Tay et al. 2020; Kikkert 2020; Ralph et al. 2020; Vlugt et al. 2018; Menachery et al. 2014; Chen et al. 2010; Glass et al. 2003). Like SARS-CoV-2, the prime factor for SARS-CoV progression also depends on the ACE2 receptor although both SARS-CoV and MERS-CoV consists of a relatively shorter spike protein than that of SARS-CoV-2 (Fehr and Perlman 2015; Chu et al. 2020; Ge et al. 2013; Nicholls et al. 2003; Xu and Gao 2004; Gu and Korteweg 2007). As shown in Fig. 2, upon the SARS-CoV infection, the macrophages, dendritic cells, and the alveolar epithelial cells have been shown to trigger a remarkable elevation of the pro-inflammatory chemokines: (1) the macrophage inflammatory protein-1α (MIP-1α), (2) normal T cells; (3) IP-10, (4) interleukin (IL) 2, 5, 6 and IL-8, and (5) monocyte chemoattractant protein-1 (MCP-1), (6) tumor necrosis factor alpha (TNF-α), and (7) chemokines (CXCL10, CCL2, CCL3, and CCL5) correlated with migration of NK cells CXCL8 (Glass et al. 2003; Xu and Gao 2004; Gu and Korteweg 2007). Within the spleen a reduction in the CD4+ lymphocytes, CD8+ lymphocytes, CD20+ lymphocytes, plasmacytoid dendritic cells (pDC), macrophages, and natural killer (NK) cells were observed upon SARS-CoV infection (Xu and Gao 2004; Gu and Korteweg 2007). Additionally, the diminution of the mucosal lymphoid tissue in the small intestines and appendix was noticed along with a succeeding decline in the lymphocytes (Gu and Korteweg 2007). Besides the role of various inflammatory cytokines and chemokines in the pathogenic consequence of the virus as stated above, the molecular pathways recognized by the TLRs (Fehr and Perlman 2015; Kaushal 2020). As the virus enters the host cells; viral RNAs are recognized by the TLRs (7/8,3) to work on three pathways; and the virus turns off two protective immunity intracellular pathways: (1) MyD88 and interferon regulatory transcription factor 7 (IRF7) to produce IFN-1; and (2) tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6) and IRF3 to activate T cells. However, the virus turns on the third intracellular inflammatory pathway; i.e., the p38 MAP Kinase- and AP-1 pathway for the production of inflammatory molecules (Kaushal 2020).
Protective immunity
SARS-CoV-specific serum Abs (specific IgG) become detectable in the late first week after the onset of symptoms and 25 days after the onset of the infection (Xu and Gao 2004). The neutralizing Abs (directed against the S glycoprotein) were noticed to be able to block the viral entry into host cells (Xu and Gao 2004). Using mouse model, a DNA vaccine encoding the S glycoprotein of SARS-CoV was previously noticed to induce T cell and the neutralizing antibody responses (protective immunity) (Xu and Gao 2004; Yang et al. 2004). Other than humoral immunity, the anti-viral CD4+ helper T cells were observed to mediate the production of virus-specific Abs by B cells; and also, the CD8+ cytotoxic T lymphocytes (CTLs) could kill the virus-infected host cells (Fig. 2), and facilitated the lysis of MHC-matched tumor cell lines expressing the S glycoprotein (Xu and Gao 2004). Interestingly, both CD4 and CD8 T-cell responses were found to be induced by immunization with the expression vectors encoding for the S protein of SARS-CoV (Xu and Gao 2004; Yang et al. 2004). Additionally, the earlier evidence showed that the intervention of intra-balance among different cytokines, Th1/Th2 cells, and the innate/adaptive immunity can be an operative way of treating the SARS-CoV infection (Glass et al. 2003).
Pathogenesis mediated by the MERS-CoV
Although the genomic organization is nearly similar to that of SARS-CoV and SARS-CoV-2; however, unlike these two, the MERS-CoV utilizes the host receptor dipeptidyl peptidase 4 (DPP4, which is also known as CD26) to interact with its spike protein during the entry into the host and subsequently to spread the viral infection (Fehr and Perlman 2015; Chafekar and Fielding 2018; Taylor and Mossman 2013; Randall and Goodbourn 2008; Dawson et al. 2019). Besides, to completely understand the MERS-CoV pathogenesis, the role of the MERS-CoV accessory proteins needs to be deciphered for mediating the viral pathogenesis. Like other CoV genomes, the first 5′ two-thirds of the MERS-CoV genome comprises the replicase complex (ORF1a and ORF1b); and the remaining 3′ one-third encodes the structural proteins S, E, M, and N; and the five accessory proteins (ORF3, ORF4a, ORF4b, ORF5 and ORF8b) which are not actually required for the genome replication, but these accessory proteins may facilitate the viral pathogenesis (Chafekar and Fielding 2018; Dawson et al. 2019). Upon entry into the host, the MERS-CoV can infect the alveolar epithelial cells, macrophages, fibroblasts, and the endothelial cells resulting in the alveolar damage; and the virus is capable to replicate within kidney and liver cells (Dawson et al. 2019).
Immunological response and IFN antagonism during MERS-CoV pathogenesis
Upon MERS-CoV entry into the host cell, the type I interferon (IFN)-mediated innate immune response is activated through the production of type I IFNs; i.e., IFN-α and IFN-β (Chafekar and Fielding 2018; Taylor and Mossman 2013; Randall and Goodbourn 2008). An elevation in monocyte-derived macrophages has been noticed (Fig. 3) along with the elevated levels of the tumor necrosis factor α (TNF α), IL-6, IL-12, and IFN-γ (Dawson et al. 2019). MERS-CoV pathogenesis stringently relies on the ORF4a (dsRNA binding protein), which acts as an antagonist of the anti-viral activity of IFN as it (1) inhibits the interferon production (IFN-β promoter activity, the IFN-regulatory transcription factors: IRF3 and IRF7; and the activation of NF-κB (the nuclear factor kappa light chain enhancer of activated B cells); and (2) blocks the ISRE (interferon-stimulated response element) promoter element signaling pathways; i.e., the type I IFN-induced JAK/STAT signaling pathway (Dawson et al. 2019; Yang et al. 2013). As shown in Fig. 3, the MERS-CoV ORF4b inhibits the activation of RNase L, which possesses an IFN-induced potent anti-viral activity and also hinders the type I IFN and NF-kβ signaling pathways, resulting in the viral pathogenesis (Dawson et al. 2019; Thornbrough et al. 2016).
It is clear from the above discussion that escaping the host innate immunity by MERS-CoV largely depends on the IFN antagonism since the MERS-CoV-encoded IFN antagonist proteins are known to block one or more key signaling proteins in the IFN and NF-κB signaling pathways thereby mediating the viral infectivity (Chafekar and Fielding 2018; Taylor and Mossman 2013; Randall and Goodbourn 2008). Impedance of the IFN production by the interference with the global expression of IFN‐stimulated genes (ISGs), and by the degradation of the transcription factors IRF3/7, nuclear factor‐κB or NF‐κB, or ATF‐2/c‐jun (activating transcription factor 2 binding to the cyclic AMP responsive elements or CRE) as well as blocking the JAK/STAT signal transduction pathway or by obstructing the type I IFN receptor are the keys to the MERS-CoV pathogenesis (Taylor and Mossman 2013). MERS-CoV prevents the initial detection of virus by the host immune cells by damaging the Toll‐like receptors (TLR) or the retinoic acid inducible gene I (RIG‐I)‐like receptors, or by masking the receptor ligands (Taylor and Mossman 2013; Yang et al. 2013). In summary, the MERS-CoV pathogenesis has been reported to evolve mechanisms to utilze its M protein, and the ORF4a, ORF4b and ORF5 proteins (all are strong IFN antagonists) to evade the host immune system and thus continuing pathogenesis by inhibiting both type I IFN induction and the NF-kB signaling pathways (Dawson et al. 2019). Besides ORF4a and ORF4b, the MERS-CoV replicase proteins, including nsp1, nsp3, and nsp14 also interfere with the innate immune response signaling pathways (Dawson et al. 2019).
Conclusion
In the current review, the molecular and cellular strategies for the SARS-CoV-2, SARS-CoV, and MERS-CoV to lapse the protective innate immune system have been discussed. Although the genome organization of all these three β-coronaviruses are nearly similar, the priming of pathogenesis differs especially regarding the viral entry; i.e., the SARS-CoV-2 and the SARS-CoV employed their spike protein to the RBD of the host ACE2 receptor whereas the MERS-CoVs used their spike proteins to attach with the host DPP4 receptor. Moreover, the SARS-CoV-2 was found to show stronger affinity for the ACE2 receptor than that of the SARS-CoV. In all three cases, the pathogenesis depends on the escaping performances of the host innate immunity. The pathogenesis of SARS-CoV-2 and SARS-CoV is related to the cytokine storm whereas the MERS-CoV pathogenesis is allied with the host NK cell inactivation by the viral antagonist proteins towards the host IFN. The pathogenesis scheme with such immunological response has been presented as schematic model for each virus. Such a knowledge may aid to further focus on their virulence rate and the transmission dynamics, and the possible anti-viral therapies including the vaccine development strategies.
Acknowledgements
Author is thankful to the group of scientists or associations whose literatures have been extensively used to write this article.
Abbreviations
- ACE 2
Angiotensin-converting enzyme 2 (ACE 2) receptor
- ADE
Antibody-dependent enhancement
- APCs
Antigen presenting cells
- ARDS
Acute respiratory distress syndrome
- CRE
Cyclic AMP responsive elements
- CTLs
Cytotoxic T lymphocytes CTLs
- DPP4
Dipeptidyl peptidase 4
- HA
Hyaluronan
- HAS2
HA synthase
- HE
Hemagglutinin esterase
- IFN
Interferon
- IL
Interleukin
- IRF7
Interferon regulatory transcription factor
- ISGs
IFN‐stimulated genes
- ISRE
Interferon-stimulated response element
- Mab
Monoclonal antibody
- MCP-1
Monocyte chemoattractant protein-1
- MERS-CoV
Middle East respiratory syndrome coronavirus
- MHC
Major histocompatibility complex
- MIP-1α
Macrophage inflammatory protein-1α
- OAS
Oligoadenylate synthetase
- PAMPs
Pathogen-associated molecular patterns
- PRRs
Pattern recognition receptors (PRRs)
- RBD
Receptor-binding domain
- RLR
RIG-I-like receptors (RLRs)
- RTC
Replicase–transcriptase complex
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- TLRs
Toll-like receptors
- TMPRSS2
Plasma membrane-associated type II transmembrane serine protease
- TNF-α
Tumor necrosis factor alpha (TNF-α)
- TRAF6
Tumor necrosis factor receptor (TNFR)-associated factor 6
- TRS
Transcriptional regulatory sequences
- ICU
Intensive care unit
- UTR
Untranslated region
Funding
None.
Declarations
Conflict of interest
Author has declared that he has no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Chafekar A, Fielding BC. MERS-CoV: understanding the latest human coronavirus threat. Viruses. 2018;10(2):93. doi: 10.3390/v10020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lau YF, Lamirande EW, Paddock CD, Bartlett JH, Zaki SR, Subbarao K. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c Mice: CD4+ T cells are important in control of SARS-CoV infection. J Virol. 2010;84(3):1289–1301. doi: 10.1128/JVI.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Chan JF-W, Yuen TT-T, Shuai H, Yuan S, Wang Y, Hu B, Yip CC-Y, Tsang JO-L, Xiner H, Chai Y, Yang D, Hou Y, Chik KK-H, Zhang X, et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1(1):e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson P, Malik MR, Parvez F, Morse SS. What have we learned about middle east respiratory syndrome coronavirus emergence in humans? A systematic literature review. Vector Borne Zoonotic Dis. 2019;19(3):174–192. doi: 10.1089/vbz.2017.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vlugt C, Sikora D, Pelchat M. Insight into influenza: a virus cap-snatching. Viruses. 2018;10(11):641. doi: 10.3390/v10110641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, Zhang YJ, Luo CM, Tan B, Wang N, Zhu Y, Crameri G, Zhang SY, Wang LF, Daszak P, Shi ZL. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass WG, Rosenberg HF, Murphy PM. Chemokine regulation of inflammation during acute viral infection. Curr Opin Allergy Clin Immunol. 2003;3:467–473. doi: 10.1097/00130832-200312000-00008. [DOI] [PubMed] [Google Scholar]
- Gu J, Korteweg C. Pathology and pathogenesis of severe acute respiratory syndrome. Am J Pathol. 2007;170(4):1136–1147. doi: 10.2353/ajpath.2007.061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal A. Immune response and pathogenesis of COVID-19 and The Strategies for developing target drugs. Acta Sci Microbiol. 2020;3(9):92–102. doi: 10.31080/ASMI.2020.03.0682. [DOI] [Google Scholar]
- Kikkert M. Innate immune evasion by human respiratory RNA viruses. J Innate Immun. 2020;12(1):4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WS, Wheatley AK, Kent SJ, DeKosky BJ. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat Microbiol. 2020;5(10):1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery VD, Eisfeld AJ, Schäfer A, Josset L, Sims AC, Proll S, Fan S, Li C, Neumann G, Tilton SC, Chang J, Gralinski LE, Long C, Green R, Williams CM, Weiss J, Matzke MM, Webb-Robertson BJ, Schepmoes AA, Shukla AK, Metz TO, Smith RD, Waters KM, Katze MG, Kawaoka Y, Baric RS. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio. 2014;5(3):e01174-14. doi: 10.1128/mBio.01174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls JM, Poon LL, Lee KC, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor R. Antiviral drugs against severe acute respiratory syndrome coronavirus 2 infection triggering the coronavirus disease-19 pandemic. Tzu Chi Med J. 2021;33(1):7–12. doi: 10.4103/tcmj.tcmj_100_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor R, Maniha SM. A brief outline of respiratory viral disease outbreaks: 1889—till date on the public health perspectives. VirusDisease. 2020;31(4):441–449. doi: 10.1007/s13337-020-00628-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabaan AA, Al-Ahmed SH, Haque S, Sah R, Tiwari R, Malik YS, Dhama K, Yatoo MI, Bonilla-Aldana DK, Rodriguez-Morales AJ. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Infez Med. 2020;28(2):174–184. [PubMed] [Google Scholar]
- Ralph R, Lew J, Zeng T, Francis M, Xue B, Roux M, Toloue Ostadgavahi A, Rubino S, Dawe NJ, Al-Ahdal MN, Kelvin DJ, Richardson CD, Kindrachuk J, Falzarano D, Kelvin AA. 2019-nCoV (Wuhan virus), a novel Coronavirus: human-to-human transmission, travel-related cases, and vaccine readiness. J Infect Dev Ctries. 2020;14:3–17. doi: 10.3855/jidc.12425. [DOI] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89(1):1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KE, Mossman KL. Recent advances in understanding viral evasion of type I interferon. Immunology. 2013;138:190–197. doi: 10.1111/imm.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornbrough JM, Jha BK, Yount B, Goldstein SA, Li Y, Elliott R, Sims AC, Baric RS, Silverman RH, Weiss SR. Middle east respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. mBio. 2016;7(2):e00258. doi: 10.1128/mBio.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Shang J, Sun S, Tai W, Chen J, Geng Q, He L, Chen Y, Wu J, Shi Z, Zhou Y, Du L, Li F (2020) Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J Virol 94(5):e02015–e02019 [DOI] [PMC free article] [PubMed]
- WHO (World Health Organization) (2021) Coronavirus disease (COVID-19) pandemic. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 Last update: 12 Feb 2021, 08:07 pm GMT+6. Accessed 12 Feb 2021
- Xu X, Gao X. Immunological responses against SARS-coronavirus infection in humans. Cell Mol Immunol. 2004;1(2):119–122. [PubMed] [Google Scholar]
- Yang ZY, Kong WP, Huang Y, Roberts A, Murphy BR, Subbarao K, Nabel GJ. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhang L, Geng H, Deng Y, Huang B, Guo Y, Zhao Z, Tan W. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4(12):951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YY, Li BR, Ning BT. The comparative immunological characteristics of SARS-CoV, MERS-CoV, and SARS-CoV-2 coronavirus infections. Front Immunol. 2020;11:2033. doi: 10.3389/fimmu.2020.02033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang MW, Cheng Y, Zhang J, Jiang XM, Wang L, Deng J, Wang PH. Increasing host cellular receptor-angiotensin-converting enzyme 2 expression by coronavirus may facilitate 2019-nCoV (or SARS-CoV-2) infection. J Med Virol. 2020 doi: 10.1002/jmv.26139. [DOI] [PMC free article] [PubMed] [Google Scholar]