Abstract
Background
The outbreak of coronavirus disease 2019 (COVID-19) induced by the novel coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) originated in China and spread to cover the entire world with an ongoing pandemic. The magnitude of the situation and the fast spread of the new and deadly virus, as well as the lack of specific treatment, led to a focus on research to discover new therapeutic agents.
Aim
In this study, we explore the potential inhibitory effects of some active polyphenolic constituents of Rhus spp. (sumac) against the SARS-CoV-2 main protease enzyme (Mpro; 6LU7).
Methods
26 active polyphenolic compounds of Rhus spp. were studied for their antiviral activity by molecular docking, drug likeness, and synthetic accessibility score (SAS) as inhibitors against the SARS-CoV-2 Mpro.
Results
The results show that all tested compounds of sumac provided good interaction with the main active site of SARS-CoV-2 Mpro, with better, lower molecular docking energy (kcal/mol) compared to the well-known drugs chloroquine and favipiravir (Avigan). Only six active polyphenolic compounds of Rhus spp. (sumac), methyl 3,4,5-trihydroxybenzoate, (Z)-1-(2,4-dihydroxyphenyl)-3-(3,4-dihydroxyphenyl)-2-hydroxyprop-2-en-1-one, (Z)-2-(3,4-dihydroxybenzylidene)-6-hydroxybenzofuran-3(2H)-one, 3,5,7-trihydroxy-2-(4-hydroxyphenyl)chroman-4-one, 2-(3,4-dihydroxyphenyl)-3,5-dihydroxy-7-methoxy-4H-chroman-4-one, and 3,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one, were proposed by drug likeness, solubility in water, and SAS analysis as potential inhibitors of Mpro that may be used for the treatment of COVID-19.
Conclusion
Six phenolic compounds of Rhus spp. are proposed for synthesis as potential inhibitors against Mpro and have potential for the treatment of COVID-19. These results encourage further in vitro and in vivo investigations of the proposed ligands and research on the preventive use of Rhus spp. against SARS-CoV-2.
1. Introduction
The coronavirus disease 2019 (COVID-19) crisis started in China in December 2019. By 30 January 2020, about 213 individuals had died and at least 9066 had been infected [1]. It also spread globally, first to a number of Asian countries, as well as to Canada, France, Germany, and the United States. As a result, due to the spread of this new and deadly virus, governments around the world put several major cities on lockdown and put aside all normal plans to deal with the crisis. In addition, on 30 January 2020, the World Health Organization (WHO) declared the COVID-19 outbreak a global health emergency because it could spread to countries that were not prepared [1–4]. Thus, on 11 March 2020, the WHO characterized COVID-19 as a pandemic, which has affected more than 200 countries; by March 2020, there were 30,105 deaths and 638,146 confirmed cases around the world [3], which have increased considerably over time. Genomic and molecular-based analyses show that SARS-CoV-2 is a new type of human-infected β-coronavirus (CoV), which suggested that zoonotic hosts like bats might be the original host of this virus [5]. Situation Reports released by the WHO on 13th April revealed that SARS-CoV-2 infection can induce severe collateral disorders such as severe pneumonia, pulmonary edema, acute respiratory disease syndrome (ARDS), or multiple organ failure (such as shock, acute heart injury, or acute kidney injury), which are ultimately responsible for an increased number of deaths worldwide [6]. This may be also due to the lack of an effective specific antiviral therapy, since SARS-CoV-2 is a novel pathogen. Thus, several drugs such as lopinavir/ritonavir, neuraminidase inhibitors, and other present antiviral drugs have been proposed for the treatment of COVID-19 infection [7, 8]. These drugs produce their antiviral potency via an inhibition process against SARS-CoV-2 proteases [7, 8]. The magnitude of the situation and the lack of a specific therapy for the virus have led to a focus on research to find new therapeutic agents [8]. For the management of COVID-19, preventive and supportive therapies based on herbal plants may complement research on existing antiviral agents.
Medicinal herbs known in ethnopharmacology have been suggested as antiviral agents for the treatment and control of contagious diseases like COVID-19 [9–14]. Most of these studies use molecular docking analysis to identify the potential activities of phytoconstituents present in herbal plants for the treatment of diseases like COVID-19. They focus on the activities of these plant constituents on the main proteases present in CoVs [12–14].
Sumac (Rhus spp.), a flowering plant that grows in temperate and tropical regions, contains over 250 individual species worldwide [15]. It is usually used as spice and a medicinal herb in most of the world, particularly for its antiviral [16, 17], antimicrobial, antibacterial, antioxidant, and wound-healing [18–26] properties.
The antiviral activity of Rhus spp., particularly Rhus chinensis, has been explored in many studies. Different phytochemical fractions of Rhus chinensis showed potent anti-HIV-1 [27–29], anti-herpes simplex virus (HSV) type 1 (HSV-1) [30–32], and anti-HCV activity. This antiviral activity was related to the presence of many active compounds such as phenolics, organic acids, proteins, fibers, volatile oils, fatty acids, vitamins, and minerals [33, 34]. Likewise, severe acute respiratory syndrome coronavirus (SARS-CoV) was significantly inhibited by using a 50% effective concentration (4.5 μm) of tetra-O-galloyl-β-d-glucose isolated from Galla chinensis (Rhus chinensis) [35].
During infection, coronavirus attaches to target cells with the help of angiotensin-converting enzyme 2 (ACE2) present in the spike protein of the virus, which produces a spike protein-host cell protein interaction, whereby the virus genome with its nucleocapsid can easily release into the cytoplasm of the host cells [36, 37]. Sequence analysis of the replicase polyprotein in Avian infectious bronchitis virus, another coronavirus, originally predicted the presence of the coronavirus Mpro protease enzyme [38]. This enzyme was related to chymotrypsin-like cysteine proteases which significantly play a potential role in the replication and transcription of the coronavirus (SARS-CoV). Thus, it is considered a prime target for the discovery of antiviral agents [26, 39–41].
The SARS-CoV genome encodes a number of proteases. The main protease (Mpro) chymotrypsin-like protease (3CLpro) from SARS-CoV-2 (6LU7) has an important role along with other cysteine proteases in the replication of the CoV genome. Thus, synthetic or herbal-based drugs targeting the proteases of SARS-CoV-2 (6LU7) may have a considerable role in the treatment of COVID-19 [38]. Several inhibitors including boceprevir, GC-376, and calpain inhibitors II and XII were identified to have potent activity to inhibit SARS-CoV-2 viral replication in cell culture [39]. The protease enzyme (6LU7) has been successfully crystallized and deposited in the Protein Data Bank (PDB) [40, 41]; thus, it is considered as a potential target for therapeutic strategies, particularly for those who use phytochemicals [7, 42, 43]. It was reported that an evaluation of up-to-date knowledge relating to the characteristics of COVID-19 infection and complications encourages the investigation of the effectiveness of sumac extracts for COVID-19 treatment [44–48]. Recently, active metabolites from 14 cooking seasonings were examined as inhibitors for SARS-CoV-2 main protease (Mpro). A high potency of salvianolic acid A and curcumin as Mpro inhibitors with binding energies of −9.7 and −9.2 kcal/mol, respectively, was identified by in silico molecular docking analysis [47]; the potential activity of salvianolic acid A and curcumin as Mpro inhibitors against the SARS-CoV-2 main protease significantly depends upon forming from nine and six hydrogen bonds, respectively, with amino acids proximal to Mpro's active site [47]. Supporting review study showed that Rhus spp. (sumac) constituents could have a higher potency against the consequences produced by SARS-CoV-2 during human infection. The review study demonstrated that sumac could be useful in COVID-19 infection due to its versatile activities as anti-inflammatory, antimicrobial, antioxidant, and antimalarial effects [48]. In addition, the review article along with others mentioned that use of sumac as syrup or in capsules with different concentrations has no toxicity on human life and could be recommended in treatment protocol for COVID-19 patients [18, 48–51].
Regarding previous molecular docking studies [12, 18, 36, 44–52], we are trying to explore the potential inhibitory effects of some Rhus spp. (sumac) constituents against the SARS-CoV-2 main protease enzyme using molecular docking, drug likeness, and synthetic accessibility score analysis. The study will give more insight on the use of natural products as new therapeutic agents against the pathogenesis of the SARS-CoV-2 (COVID-19).
2. Methodology
2.1. Rhus spp. Phytoconstituents
A total of 26 compounds previously extracted from Rhus spp. [52] were selected to study their potential activity against the SARS-CoV-2 main protease enzyme. These 26 components in Rhus spp. extracts (Figure 1) were selected based on their versatile biological activity, such as antimicrobial, antifungal, antidiabetic, antioxidant, wound-healing, and antiviral activity [26, 36, 52].
Figure 1.
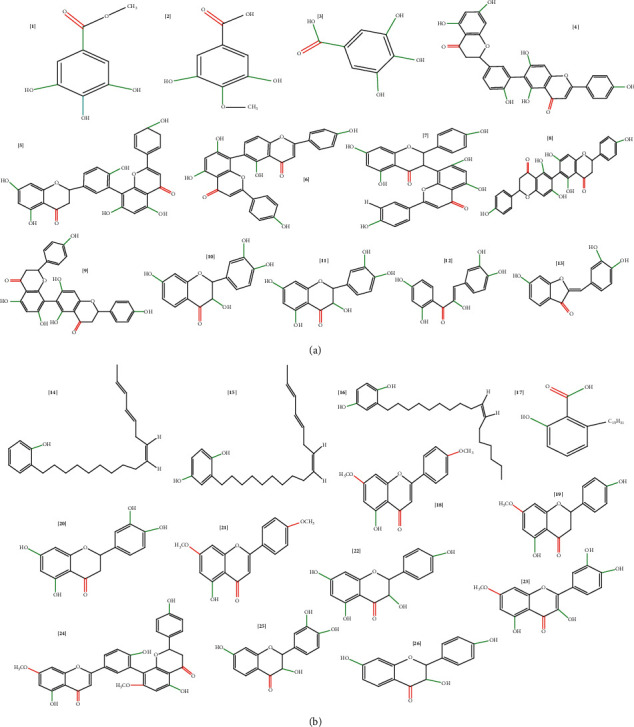
Chemical structures of 26 compounds (a and b) selected from the extract of Rhus spp. [44].
2.2. Molecular Docking Analysis
The crystal structure of the protease enzyme (the molecular target) of SARS-CoV-2 (3clpro/Mpro, PDB ID: 6LU7) was retrieved from RCSB Protein Data Bank (https://www.rcsb.org/) [53]. The proposed natural components in Rhus spp. extracts (Figure 1), in addition, well-known drugs, chloroquine and favipiravir (Avigan), widely tested in clinical trials for the treatment of COVID-19 have been subjected to molecular docking [54]. The structures of the selected sumac compounds along with the proposed drugs were drawn by ChemOffice (2015) freeware and were optimized by HyperChem v.8.1 to have the conformer with minimum energy using AMBER force field [54]. According to previously reported methodology and wording [14, 53–55], for analysis, an open Babel 2.3.2 software was used to convert a molecular format files into pdb format. Then, the ligands under investigations were loaded and their torsion along with rotatable bonds was assigned and saved as ligand PDBQT.
In our experiment by using Molegro Virtual Docker software 6.0, the binding modes of the proposed natural components in Rhus spp. extracts with the protease enzyme (the molecular target) of the SARS-CoV-2 (3clpro/Mpro; PDB ID: 6LU7) were identified [56]. In addition, a blind docking was performed to enumerate the strength of binding interactions between the proposed ligands and the protease enzyme target [14, 56]. In this step, a lowest binding energy (−kcal/mol) represents the best stable conformations of the ligand with the protease enzyme target [56]. Finally, according to the methodology previously reported [14], MGLTools-1.5.6 rc3 were used to analyze all released auto Dock output files (.dlg), whereas the docking parameters were represented as coordinates of the center of binding site with x = 126, y = 126, z = 126, and binding radius = 0.375 Ã. In addition, constituents that may hinder the simulation, such as water molecules, other heteroatoms, and native ligands attached to the target, were removed in the process of accommodating the ligands for the molecular docking study.
2.3. Drug-Likeness Analysis
This analysis qualitatively assesses the chance for a molecule to become an oral drug with respect to bioavailability, as previously described [57]. In this analysis, structural or physicochemical inspections of the proposed herbal compounds and selected drugs chloroquine and favipiravir (Avigan) were used to assess the drug likeness to be considered as an oral drug candidate. The analysis depends on five rule-based filters, with a diverse range of properties, inside of which the molecule is defined as drug-like. The Lipinski (Pfizer) filter is the pioneer rule-of-five implemented from [57]. The original rule-of-five (RO5) deals with orally active compounds and defines four simple physicochemical parameter ranges (MWT ≤ 500, log P ≤ 5, H-bond donors ≤ 5, and H-bond acceptors ≤ 10) associated with 90% of orally active drugs that have achieved phase II clinical status. These physicochemical parameters are associated with acceptable aqueous solubility and intestinal permeability and comprise the first steps in oral bioavailability [57–62]. This analysis is routinely performed to filtrate chemical libraries and exclude compounds with an incompatible or unacceptable pharmacokinetic profile.
In addition, the synthetic accessibility score (SAS) for each molecule extracted from sumac was calculated according to the methodology reported previously [57, 58]. SAS was shown to be essential in both the early drug discovery stage and the drug manufacturing process [59, 60]. To characterize the accessibility of drugs or molecules for synthesis, the synthetic accessibility score was grouped according to previous studies into three groups: easy to synthesize (SAS ≤ 3), moderately easy to synthesize (SAS = 3-4), and difficult to synthesize (>4) [57–63].
3. Results
3.1. Molecular Docking Analysis
The molecular docking parameters, including docking score, ligand-protein interactions, and hydrogen bonds for selected compounds and reference drugs, are provided in Table 1. The possible interactions with the active sites of protease enzyme in this experiment are discussed herein with some details. The reference drugs, Avigan and chloroquine, interact with active sites of protease enzyme 6LU7 with an energy docking score of −2.99 kcal/mol and −6.32 kcal/mol, respectively, as shown in Table 1.
Table 1.
Physicochemical parameters and molecular docking scores of Rhus spp. (sumac) compounds obtained with 6LU7.
| Compounds | Mol. dock scores (kcal/mol) | Protein ligand interactions | Hydrogen bonds energy (kcal/mol) | Properties |
|---|---|---|---|---|
| Favipiravir (Avigan) | −65.45 | −75.47 | −2.99 | MW = 157.104; H-donor = 3; H-acceptor = 3 |
|
| ||||
| Chloroquine | −123.62 | −135.43 | −6.32 | MW = 319.872; H-donor = 1; H-acceptor = 2 |
|
| ||||
| (1) Methyl 3,4,5-trihydroxybenzoate | −81.82 | −72.2 | −22.6 | MW = 184.15; H-donor = 4; H-acceptor = 5 |
|
| ||||
| (2) 3,5-Dihydroxy-4-methoxybenzoic acid | −83.17 | −75.33 | −11.11 | MW = 184.15; H-donor = 2; H-acceptor = 5 |
|
| ||||
| (3) 3,4,5-Trihydroxybenzoic acid | −73.23 | −66.5 | −12.02 | MW = 170.12; H-donor = 3; H-acceptor = 5 |
|
| ||||
| (4) 6-(5-(5,7-Dihydroxy-4-oxochroman-2-yl)-2-hydroxyphenyl)-5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one | −135.36 | −167.5 | −13.5 | MW = 540.47; H-donor = 8; H-acceptor = 10 |
|
| ||||
| (5) 8-(5-(5,7-Dihydroxy-4-oxochroman-2-yl)-2-hydroxyphenyl)-5,7-dihydroxy-2-(4 hydroxycyclohexa-1,5-dien-1-yl)-4H-chromen-4-one | −169.74 | −194.15 | −13.35 | MW = 542.49; H-donor = 8; H-acceptor = 10 |
|
| ||||
| (6) 5,5′,7′-Trihydroxy-2,2′-bis(4-hydroxyphenyl)-4H,4′H-[6,8′-bichromene]-4,4′-dione | −150.98 | −178.50 | −12.79 | MW = 522.46; H-donor = 7; H-acceptor = 9 |
|
| ||||
| (7) 8-(5,7-Dihydroxy-2-(4-hydroxyphenyl)-4-oxochroman-3-yl)-5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one | −155.04 | −180.40 | −13.26 | MW = 540.47; H-donor = 8; H-acceptor = 10 |
|
| ||||
| (8) 5,5′,7,7′-Tetrahydroxy-2,2′-bis(4-hydroxyphenyl)-[6,6′-bichroman]-4,4′-dione | −151.40 | −180.91 | −14.47 | MW = 542.49; H-donor = 8; H-acceptor = 10 |
|
| ||||
| (9) 5,5′,7,7′-Tetrahydroxy-2,2′-bis(4-hydroxyphenyl)-[6,8′-bichroman]-4,4′-dione | −146.88 | −173.17 | −1120 | MW = 542.49; H-donor = 8; H-acceptor = 10 |
|
| ||||
| (10) 2-(3,4-Dihydroxyphenyl)-3,7-dihydroxychroman-4-one | −146.88 | −173.17 | −13.76 | MW = 288.25; H-donor = 5; H-acceptor = 6 |
|
| ||||
| (11) 2-(3,4-Dihydroxyphenyl)-3,5,7-trihydroxychroman-4-one | −97.47 | −121.77 | −11.52 | MW = 304.25; H-donor = 6; H-acceptor = 7 |
|
| ||||
| (12) (Z)-1-(2,4-Dihydroxyphenyl)-3-(3,4-dihydroxyphenyl)-2-hydroxyprop-2-en-1-one | −132.84 | −142.30 | −21.83 | MW = 288.25; H-donor = 6; H-acceptor = 6 |
|
| ||||
| (13) (Z)-2-(3,4-Dihydroxybenzylidene)-6-hydroxybenzofuran-3(2H)-one | −125.38 | −129.38 | −14.31 | MW = 270.24; H-donor = 4; H-acceptor = 5 |
|
| ||||
| (14) 2-((10Z,13E,15E)-Heptadeca-10,13,15-trien-1-yl)phenol | −93.31 | −109.19 | 0.0 | MW = 326.52; H-donor = 1; H-acceptor = 1–22 |
|
| ||||
| (15) 2-((10Z,13E,15E)-Heptadeca-10,13,15-trien-1-yl)benzene-1,4-diol | −141.68 | −137.25 | −5.39 | MW = 342.51; H-donor = 2 H-acceptor = 2 |
|
| ||||
| (16) (Z)-2-(Heptadec-10-en-1-yl)benzene-1,4-diol | −131.14 | −126.86 | −4.54 | MW = 346.55; H-donor = 2; H-acceptor = 2 |
|
| ||||
| (17) 2-Hydroxy-6 pentadecylbenzoic acid | −138.40 | −134.16 | −3.73 | MW = 348.52; H-donor = 1; H-acceptor = 3 |
|
| ||||
| (18) 5-Hydroxy-7-methoxy-2-(4-methoxyphenyl)-4H-chromen-4-one | −116.41 | −130.34 | −5.51 | MW = 298.29; -donor = 2; H-acceptor = 5 |
|
| ||||
| (19) 5-Hydroxy-2-(4-hydroxyphenyl)-7-methoxychroman-4-one | −107.68 | 123.39 | −7.61 | MW = 286.28; H-donor = 3; H-acceptor = 5 |
|
| ||||
| (20) 2-(3,4-Dihydroxyphenyl)-5,7-dihydroxychroman-4-one | −104.17 | −121.38 | −9.6 | MW = 288.25; H-donor = 6; H-acceptor = 6 |
|
| ||||
| (21) 5-Hydroxy-7-methoxy-2-(4-methoxyphenyl)-4H-chromen-4-one | −114.79 | −129.56 | −6.3 | MW = 298.29; H-donor = 2; H-acceptor = 5 |
|
| ||||
| (22) 3,5,7-Trihydroxy-2-(4-hydroxyphenyl)chroman-4-one | −109.41 | −130.17 | −13.34 | MW = 288.25; H-donor = 5; H-acceptor = 6 |
|
| ||||
| (23) 2-(3,4-Dihydroxyphenyl)-3,5-dihydroxy-7-methoxy-4H-chromen-4-one | −105.80 | −125.70 | −15.57 | MW = 316.26; H-donor = 5; H-acceptor = 7 |
|
| ||||
| (24) 5-Hydroxy-2-(4-hydroxy-3-(5-hydroxy-2-(4-hydroxyphenyl)-7-methoxy-4-oxochroman-8-yl)phenyl)-7-methoxy-4H-chromen-4-one | −179.18 | −189.69 | −13.35 | MW = 568.53; H-donor = 6; H-acceptor = 10 |
|
| ||||
| (25) 2-(3,4-Dihydroxyphenyl)-3,7-dihydroxychroman-4-one | −106.43 | −115.60 | −8.57 | MW = 288.25; H-donor = 5; H-acceptor = 6 |
|
| ||||
| (26) 3,7-Dihydroxy-2-(4-hydroxyphenyl)chroman-4-one | −107.14 | −123.05 | −17.21 | MW = 272.25; H-donor = 4; H-acceptor = 5 |
All tested compounds of Rhus spp. have lower energy docking scores compared with the reference drugs Avigan and chloroquine (Table 1), as these compounds are polyphenols and can interact with 6LU7 more strongly through hydrogen bonds, pi-cation interactions, or pi-pi stacking interactions. Compound (14), although it demonstrated a good binding energy, could not form any hydrogen bond with the viral enzyme. Compound (14) showed no perceptible interactions but only electrostatics (Van der Waals) (see Supplementary file).
The obtained results show that the compounds methyl 3,4,5-trihydroxybenzoate (1), (Z)-1-(2,4-dihydroxyphenyl)-3-(3,4-dihydroxyphenyl)-2-hydroxyprop-2-en-1-one (12), (Z)-2-(3,4-dihydroxybenzylidene)-6-hydroxybenzofuran-3(2H)-one (13), 3,5,7-trihydroxy-2-(4-hydroxyphenyl)chroman-4-one (22), 2-(3,4-dihydroxyphenyl)-3,5-dihydroxy-7-methoxy-4H-chroman-4-one (23), and 3,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one (26) give efficient interactions when complexed with the active sites of the protease enzyme 6LU7.
The interaction between these proposed compounds was efficient at lower energy docking scores, –22.6 kcal/mol for compound (1), −21.83 kcal/mol for compound (12), −14.31 kcal/mol for compound (13), −13.34 kcal/mol for compound (22), −15.57 kcal/mol for compound (23), and −17.21 kcal/mol for compound (26), as shown in Table 1. Interactions between proposed compounds (1), (12), (3), (22), (23), and (26) from Rhus spp. and 6LU7 are reported in Figures 2–7 and illustrated in Table 2. Also, the interaction of the proposed drugs, Avigan and chloroquine, with the active sites of protease 6LU7 of SARS-CoV-2 significantly proposed their potency for COVID-19 treatment as shown in Figure 8 and Table 2.
Figure 2.
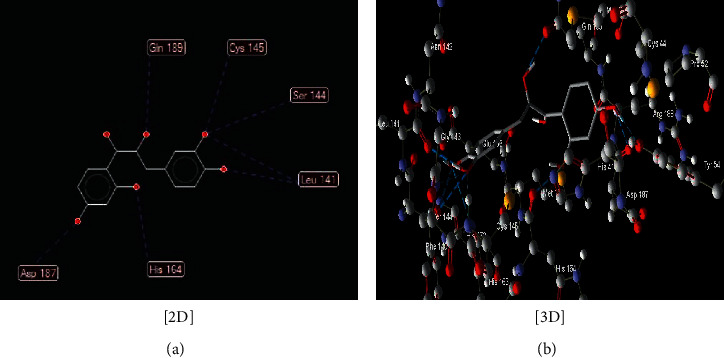
Interaction between compound (12) ((Z)-1-(2,4-dihydroxyphenyl)-3-(3,4-dihydroxyphenyl)-2-hydroxyprop-2-en-1-one) with COVID-19 (6LU7).
Figure 3.
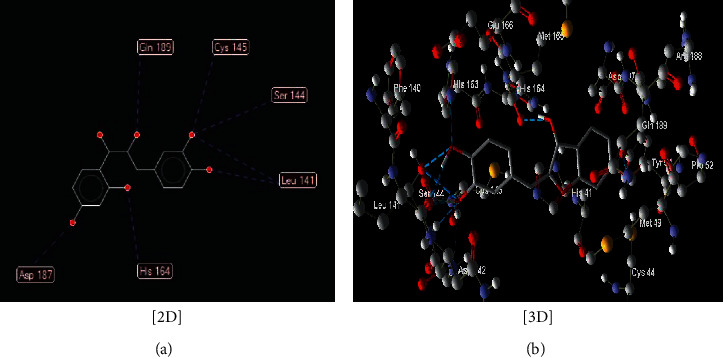
Interaction between compound (13) ((Z)-2-(3,4-dihydroxybenzylidene)-6-hydroxybenzofuran-3(2H)-one) with COVID-19 (6LU7).
Figure 4.
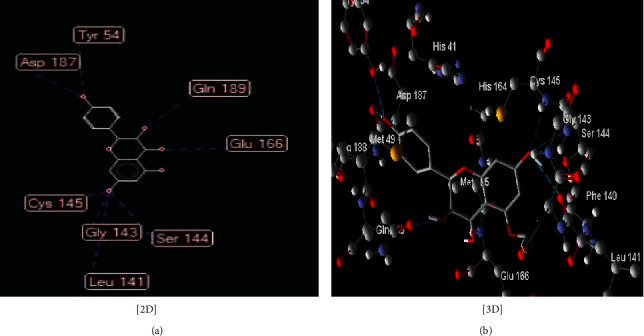
Interaction between compound (22) (3,5,7-trihydroxy-2-(4-hydroxyphenyl)chroman-4-one) with COVID-19 (6LU7).
Figure 5.
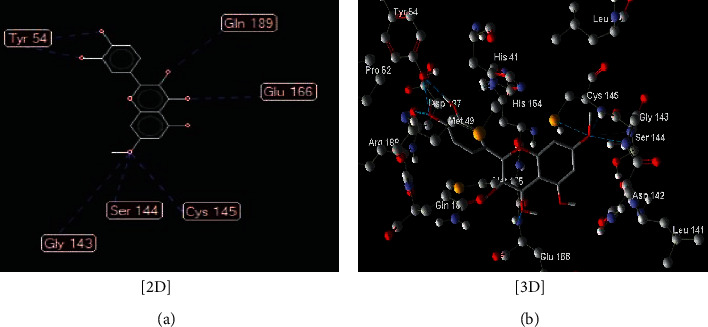
Interaction between compound (23) (2-(3,4-dihydroxyphenyl)-3,5-dihydroxy-7-methoxy-4H-chroman-4-one) with COVID-19 (6LU7).
Figure 6.
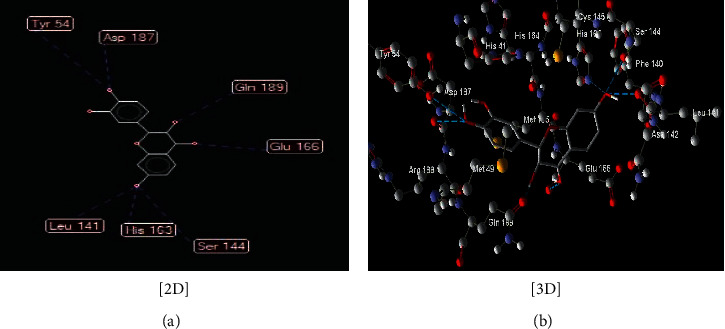
Interaction between compound (26) (3,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one) with COVID-19 (6LU7).
Figure 7.
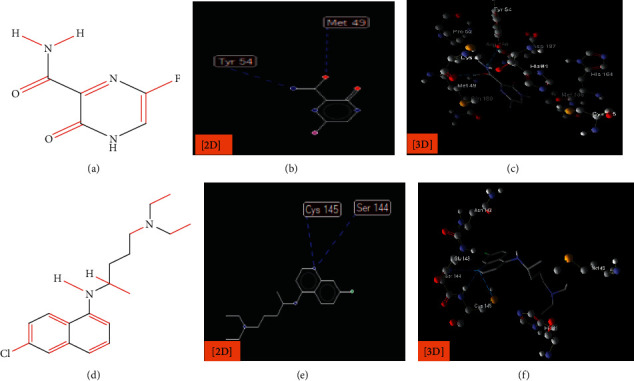
Interaction of the proposed drugs: favipiravir (Avigan): structure (a) and 2D and 3D structures (b and c); chloroquine: structure (d) and 3D structures (e and f) with COVID-19 (6LU7).
Table 2.
Interactions between the sites of 6LU7 in complex with the main proposed Rhus spp. (sumac) compounds for COVID-19 treatment.
| Compound | Name | Type of interactions |
|---|---|---|
| Drugs | Favipiravir (Avigan) (Figure 8) | Hydrogen interactions are possible with the following: |
| Amino acid Tyr 54 (H-donor rotatable) with distance about 2.67 Å and energy of −2.5 kcal/mol | ||
| Amino acid Met 49 (H-donor rotatable) with distance about 2.91 Å and energy of −2.5 kcal/mol | ||
| Chloroquine (Figure 8) | Hydrogen interactions are possible with the following: Amino acid Cys 145 by (two H-donors) with distance about 3.15 and 3.02 Å and energy of −2.24, 1.33 kcal/mol Amino acid Ser 144 (H-donor) with distance about 3.12 Å and energy of −1.13 k cal/mol |
|
|
| ||
| 1 | Methyl 3,4,5-trihydroxybenzoate (Figure 2) | Five hydrogen interactions are possible with the following: |
| Amino acid Phe 140 (H-donor rotatable) with distance about 3.07 Å and energy of −2.50 kcal/mol | ||
| Amino acid Leu 141 (H-donor rotatable) with distance about 3.11 Å and energy of −2.42 kcal/mol | ||
| Amino acid Ser 144 (H-donor rotatable) with distance about 3.13 Å and energy of −2.13 kcal/mol | ||
| Amino acid Cys 145 (H-donor rotatable) with distance about 3.2.96 Å and energy of −1.26 kcal/mol | ||
| Amino acid His 164 (H-donor rotatable) with distance about 3.18 Å and energy of −2.08 kcal/mol | ||
|
| ||
| 12 | (Z)-1-(2,4-Dihydroxyphenyl)-3-(3,4-dihydroxyphenyl)-2-hydroxyprop-2-en-1-one (Figure 3) | Seven hydrogen interactions are possible with the following: |
| Amino acid Gln 189 (H-donor rotatable) with distance 2.71 Å and energy of −2.5 kcal/mol | ||
| Amino acid Cys 145 (H-donor rotatable) with distance 3.08 Å and energy of −2.5 kcal/mol | ||
| Amino acid Leu 141 (H-donor rotatable) with distance 3.02 Å and energy of −2.5 kcal/mol | ||
| Aminoa Leu 141 (H-donor rotatable) with distance 2.93 Å and energy of −2.5 kcal/mol | ||
| Amino acid Ser 144 (H-donor rotatable) with distance 3.11 Å and energy of −2.44 kcal/mol | ||
| Amino acid His 164 (H-donor rotatable) with distance 2.64 Å and energy of −2.50 kcal/mol | ||
| Amino acid Asp 187 (H-donor rotatable) with distance 3.21 Å and energy of −1.97 kcal/mol | ||
|
| ||
| 13 | (Z)-2-(3,4-Dihydroxybenzylidene)-6-hydroxybenzofuran-3(2H)-one (Figure 4) | Seven hydrogen interactions are possible with the following: |
| Amino acid His 164 (H-donor rotatable) with distance 2.88 Å and energy of −2.5 kcal/mol | ||
| Amino acid Leu 141 (H-donor rotatable) with distance 2.87 Å and energy of −2.50 kcal/mol | ||
| Amino acid Leu 141 (H-donor rotatable) with distance 2.62 Å and energy of −2.50 kcal/mol | ||
| Amino acid Cys 145 (H-donor) with distance 3.10 Å and energy of −1.28 kcal/mol | ||
| Amino acid Ser 144 (H-donor rotatable) with distance 2.91 Å and energy of −2.50 kcal/mol | ||
| Amino acid Gln 189 (H-donor rotatable) with distance 2.71 Å and energy of −2.5 kcal/mol | ||
| Amino acid Asp 187 (H-donor rotatable) with distance 3.21 Å and energy of −1.97 kcal/mol | ||
|
| ||
| 22 | 3,5,7-Trihydroxy-2-(4-hydroxyphenyl)chroman-4-one (Figure 5) | Eight hydrogen interactions are possible with the following: |
| Amino acid Asp 187 (H-donor rotatable) with distance 3.17 Å and energy of −2.15 kcal/mol | ||
| Amino acid Tyr 54 (H-donor rotatable) with distance 2.89 Å and energy of −2.50 kcal/mol | ||
| Amino acid Gln 189 (H-donor rotatable) with distance 2.59 Å and energy of −2.39 kcal/mol | ||
| Amino acid Ser 144 (H-acceptor) with distance 2.90 Å and energy of −1.21 kcal/mol | ||
| Amino acid Leu 141 (H-donor rotatable) with distance 3.11 Å and energy of −2.45 kcal/mol | ||
| Amino acid Gly 143 (H-acceptor) with distance 3.16 Å and energy of −0.83 kcal/mol | ||
| Amino acid Cys 145 (H-donor rotatable) with distance 3.00 Å and energy of −1.47 kcal/mol | ||
| Amino acid Glu 166 (H-acceptor) with distance 2.96 Å and energy of −2.33 kcal/mol | ||
|
| ||
| 23 | 2-(3,4-Dihydroxyphenyl)-3,5-dihydroxy-7-methoxy-4H-chroman-4-one (Figure 6) | Eight hydrogen interactions are possible with the following: |
| Amino acid Tyr 54 (H-donor rotatable) with distance 2.66 Å and energy of −2.5 kcal/mol | ||
| Amino acid Tyr 54 (H-donor rotatable) with distance 2.93 Å and energy of −2.5 kcal/mol | ||
| Amino acid Cys 145 (H-donor rotatable) with distance 3.30 Å and energy of −1.50 kcal/mol | ||
| Amino acid Cys 145 (H-donor rotatable) with distance 3.08 Å and energy of −2.41 kcal/mol | ||
| Amino acid Gln 189 (H-donor rotatable) with distance 3.29 Å and energy of −1.53 kcal/mol | ||
| Amino acid Glu 166 (H-acceptor) with distance 2.96 Å and energy of −2.33 kcal/mol Amino acid Ser 144 (H-donor rotatable) with distance 3.09 Å and energy of −0.92 kcal/mol | ||
| Amino acid Gly 143 (H-donor rotatable) with distance 2.86 Å and energy of −1.38 kcal/mol | ||
|
| ||
| 26 | 3,7-Dihydroxy-2-(4-hydroxyphenyl)chroman-4-one (Figure 7) | Seven hydrogen interactions are possible with the following: |
| Amino acid Tyr 54 (H-donor rotatable) with distance 2.67 Å and energy of −2.5 kcal/mol | ||
| Amino acid Asp 187 (H-donor rotatable) with distance 2.79 Å and energy of −2.5 kcal/mol | ||
| Amino acid Gln 189 (H-donor rotatable) with distance 2.5897 Å and energy of −2.5 kcal/mol | ||
| Amino acid Glu 166 (H-donor rotatable) with distance 3.11 Å and energy of −2.40 kcal/mol | ||
| Amino acid Ser 144 (H-donor rotatable) with distance 2.64 Å and energy of −2.50 kcal/mol | ||
| Amino acid His 163 (H-donor rotatable) with distance 3.15 Å and energy of −2.59 kcal/mol | ||
| Amino acid Leu 141 (H-donor rotatable) with distance 2. 93 Å and energy of −2.50 kcal/mol | ||
Figure 8.
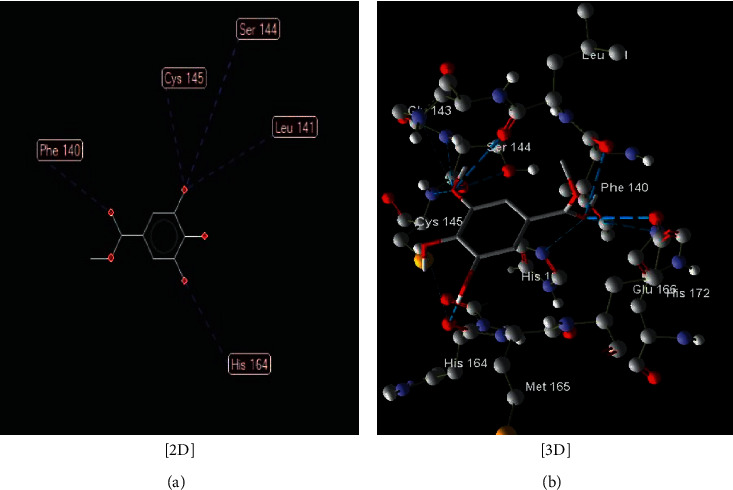
Interaction between compound (1) (methyl 3,4,5-trihydroxybenzoate) with COVID-19 (6LU7).
Due to the varied biological activity of the extracted polyphenolic compounds of Rhus spp. as potential active materials in the treatment of several diseases, several compounds are proposed: (1), (12), (13), (22), (23), and (26). These compounds showed the lowest docking energy with efficient interaction with the active sites of protease 6LU7 of SARS-CoV-2 through many hydrogen bonds compared with Avigan and chloroquine (Tables 1 and 2 and Figures 2–8).
3.2. Drug-Likeness Analysis
To study the availability of the proposed sumac compounds for synthesis, drug likeness, physicochemical properties, and synthetic accessibility score (SAS) were assessed according to the Lipinski rule-of-five. The drug likeness, water solubility, bioavailability scores, and SAS scores for all proposed active polyphenolic compounds of sumac and proposed Avigan and chloroquine are reported in Table 3.
Table 3.
Physicochemical parameters and drug-likeness scores of Rhus spp. (sumac) compounds obtained with 6LU7.
| Compounds | Mol. w. (g/mol) | Water solubility | Drug likeness | Bio. score | Synthetic accessibility score |
|---|---|---|---|---|---|
| Drugs | |||||
| Favipiravir (Avigan) | 157.104 | Very soluble | Yes | 0.55 | 2.03 |
| Chloroquine | 319.872 | Moderately soluble | 0.55 | 2.76 | |
| (1) Methyl 3,4,5-trihydroxybenzoate | 184.15 | Very soluble | Yes | 0.55 | 1.5 |
| (2) 3,5-Dihydroxy-4-methoxybenzoic acid | 184.15 | Very soluble | Yes | 0.56 | 1.38 |
| (3) 3,4,5-trihydroxybenzoic acid | 170.12 | Very soluble | Yes | 0.56 | 1.22 |
| (4) 6-(5-(5,7-Dihydroxy-4-oxochroman-2-yl)-2-hydroxyphenyl)-5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one | 540.47 | Poorly soluble | No | 0.17 | 4.60 |
| (5) 8-(5-(5,7-Dihydroxy-4-oxochroman-2-yl)-2-hydroxyphenyl)-5,7-dihydroxy-2-(4 hydroxycyclohexa-1,5-dien-1-yl)-4H-chromen-4-one | 542.49 | Moderately soluble | No | 0.17 | 5.33 |
| (6) 5,5′,7′-Trihydroxy-2,2′-bis(4-hydroxyphenyl)-4H,4′H-[6,8′-bichromene]-4,4′-dione | 522.46 | Poorly soluble | No | 0.17 | 4.13 |
| (7) 8-(5,7-Dihydroxy-2-(4-hydroxyphenyl)-4-oxochroman-3-yl)-5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one | 540.47 | Poorly soluble | No | 0.17 | 4.98 |
| (8) 5,5′,7,7′-Tetrahydroxy-2,2′-bis(4-hydroxyphenyl)-[6,6′-bichroman]-4,4′-dione | 542.49 | Poorly soluble | No | 0.17 | 4.58 |
| (9) 5,5′,7,7′-Tetrahydroxy-2,2′-bis(4-hydroxyphenyl)-[6,8′-bichroman]-4,4′-dione | 542.49 | Poorly soluble | No | 0.17 | 4.66 |
| (10) 2-(3,4-Dihydroxyphenyl)-3,7-dihydroxychroman-4-one | 288.25 | Soluble | Yes | 0.55 | 3.46 |
| (11. 2-(3,4-Dihydroxyphenyl)-3,5,7-trihydroxychroman-4-one | 304.25 | Soluble | Yes | 0.55 | 3.51 |
| (12) (Z)-1-(2,4-Dihydroxyphenyl)-3-(3,4-dihydroxyphenyl)-2-hydroxyprop-2-en-1-one | 288.25 | Moderately soluble | Yes | 0.55 | 2.78 |
| (13) (Z)-2-(3,4-Dihydroxybenzylidene)-6-hydroxybenzofuran-3(2H)-one | 270.24 | Moderately soluble | Yes | 0.55 | 2.89 |
| (14) 2-((10Z,13E,15E)-Heptadeca-10,13,15-trien-1-yl)phenol | 326.52 | Poorly soluble | Yes | 0.56 | 3.35 |
| (15) 2-((10Z,13E,15E)-Heptadeca-10,13,15-trien-1-yl)benzene-1,4-diol | 342.51 | Poorly soluble | Yes | 0.55 | 3.46 |
| (16) (Z)-2-(Heptadec-10-en-1-yl)benzene-1,4-diol | 346.55 | Poorly soluble | Yes | 0.55 | 3.23 |
| (17) 2-Hydroxy-6 pentadecylbenzoic acid | 348.52 | Poorly soluble | Yes | 0.56 | 2.97 |
| (18) 5-Hydroxy-7-methoxy-2-(4-methoxyphenyl)-4H-chromen-4-one | 298.29 | Moderately soluble | Yes | 0.55 | 3.14 |
| (19) 5-Hydroxy-2-(4-hydroxyphenyl)-7-methoxychroman-4-one | 286.28 | Soluble | Yes | 0.55 | 3.11 |
| (20) 2-(3,4-Dihydroxyphenyl)-5,7-dihydroxychroman-4-one | 288.25 | Soluble | Yes | 0.55 | 3.11 |
| (21) 5-Hydroxy-7-methoxy-2-(4-methoxyphenyl)-4H-chromen-4-one | 298.29 | Moderately soluble | Yes | 0.55 | 3.14 |
| (22) 3,5,7-Trihydroxy-2-(4-hydroxyphenyl)chroman-4-one | 288.25 | Soluble | Yes | 0.55 | 3.42 |
| (23) 2-(3,4-Dihydroxyphenyl)-3,5-dihydroxy-7-methoxy-4H-chromen-4-one | 316.26 | Soluble | Yes | 0.55 | 3.30 |
| (24) 5-Hydroxy-2-(4-hydroxy-3-(5-hydroxy-2-(4-hydroxyphenyl)-7-methoxy-4-oxochroman-8-yl)phenyl)-7-methoxy-4H-chromen-4-one | 568.53 g/mol | Poorly soluble | No | 0.17 | 4.93 |
| (25) 2-(3,4-Dihydroxyphenyl)-3,7-dihydroxychroman-4-one | 288.25 | Soluble | Yes | 0.55 | 3.46 |
| (26) 3,7-Dihydroxy-2-(4-hydroxyphenyl)chroman-4-one | 272.25 | Soluble | Yes | 0.55 | 3.36 |
The obtained results showed that six compounds (methyl 3,4,5-trihydroxybenzoate, (Z)-1-(2,4-dihydroxyphenyl)-3-(3,4-dihydroxyphenyl)-2-hydroxyprop-2-en-1-one, (Z)-2-(3,4-dihydroxybenzylidene)-6 hydroxybenzofuran-3(2H)-one, 3,5,7-trihydroxy-2-(4-hydroxyphenyl)chroman-4-one, 2-(3,4-dihydroxyphenyl)-3,5-dihydroxy-7-methoxy-4H-chroman-4-one, and 3,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one) showed drug likeness with good solubility in water and higher bioavailability scores (0.55–0.56), and lower SAS range (1.5 to 3.42), as shown in Tables 3 and 4. These scores were efficient and optimal when compared to other docked compounds (4, 5, 6, 7, 8, 9, and 24) that showed no drug likeness (Table 3). This was due to the higher molecular weight of these compounds, which affects biological interactions and solubility in water, and consequently gave a lower bioavailability score (0.17) with a higher range of SAS (4.13 to 5.33), leading to difficulty in synthesizing them as drugs, as shown in Table 3. However, only six compounds (methyl 3,4,5-trihydroxybenzoate, (Z)-1-(2,4-dihydroxyphenyl)-3-(3,4-dihydroxyphenyl)-2-hydroxyprop-2-en-1-one, (Z)-2-(3,4-dihydroxybenzylidene)-6 hydroxybenzofuran-3(2H)-one, 3,5,7-trihydroxy-2-(4-hydroxyphenyl)chroman-4-one, 2-(3,4-dihydroxyphenyl)-3,5-dihydroxy-7-methoxy-4H-chroman-4-one, and 3,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one) showed drug likeness with good solubility in water and higher bioavailability scores (0.55–0.56) and lower SAS range (1.5 to 3.42), as shown in Tables 3 and 4. The obtained physicochemical parameters, particularly higher bioavailability scores, good solubility, lower SAS, and drug likeness, in addition to the lowest energy docking score (kcal/mol), as shown in Table 4, argue for the use of these six compounds as potential inhibitors against the SARS-CoV-2 main protease enzyme.
Table 4.
Chemical structures, physicochemical properties, and energy docking scores (kcal/mol) of the main proposed Rhus spp. (sumac) compounds for COVID-19 treatment.
| Compound | Name | Hydrogen bond energy (kcal/mol) | Drug-likeness properties | |||
|---|---|---|---|---|---|---|
| Drug likeness | Water solubility | Bio. score | Synthetic accessibility score | |||
| 1 | Methyl 3,4,5-trihydroxybenzoate | −22.6 | Yes | Very soluble | 0.56 | 1.5 |
| 12 | Z)-1-(2,4-Dihydroxyphenyl)-3-(3,4-dihydroxyphenyl)-2-hydroxyprop-2-en-1-one | −21.83 | Yes | Moderately soluble | 0.55 | 2.78 |
| 13 | (Z)-2-(3,4-Dihydroxybenzylidene)-6-hydroxybenzofuran-3(2H)-one | −14.31 | Yes | Moderately soluble | 0.55 | 2.89 |
| 22 | 3,5,7-Trihydroxy-2-(4-hydroxyphenyl)chroman-4-one | −13.34 | Yes | Soluble | 0.55 | 3.42 |
| 23 | 2-(3,4-Dihydroxyphenyl)-3,5-dihydroxy-7-methoxy-4H- chroman-4-one | −15.57 | Yes | Soluble | 0.55 | 3.30 |
| 26 | 3,7-Dihydroxy-2-(4-hydroxyphenyl)chroman-4-one | −17.21 | Yes | Soluble | 0.56 | 3.36 |
4. Discussion
In this study, we are trying to decipher the proposed mechanism of the most potent active polyphenolic compounds extracted from Rhus spp. (sumac) in terms of binding affinity, necessary hydrogen bond formation [18, 54, 56], drug likeness, physicochemical properties, bioavailability, and synthetic accessibility score (SAS) [56–62], responsible for inhibition of target enzyme of the SARS-CoV-2. This study might lead to clinical trials for the treatment of infections caused by coronaviruses. Thus, a total of 26 known active polyphenolic compounds extracted from Rhus spp. (sumac) were measured for their activity as potential inhibitors against the SARS-CoV-2 main protease enzyme.
Molecular docking, drug-likeness analysis, and synthetic accessibility score (SAS) were performed for each active polyphenolic compound and compared with the results of Avigan and chloroquine as reference control drugs. All studied active polyphenolic compounds showed interactions with more hydrogen atoms to the main active site of the SARS-CoV-2 protease enzyme. This fits the complexation or interaction performed at lower energy docking score (kcal/mol) compared to that when Avigan and chloroquine interacted with the same active site. The polyphenolic nature of the extracted sumac compounds plays a pivotal role in their versatile biological activity, including antioxidant, antimicrobial, wound-healing, and antiviral activity [16–35, 64].
The presence of polyphenolic active groups with more hydrogen atoms in sumac compounds [57–60] easily facilitates the orientation and tight interaction with different amino acids present in the core of the main active site of the SARS-CoV-2 protease enzyme. In addition, it was reported previously that the antiviral activity of most herbal plants, including sumac, is related to the presence of active compounds such as phenolics, organic acids, proteins, fibers, volatile oils, fatty acids, vitamins, and minerals [33, 34].
To study the accessibility of the extracted active compounds of sumac for drug synthesis, drug-likeness analysis and SAS were performed. Only six active compounds (methyl 3,4,5-trihydroxybenzoate (1), (Z)-1-(2,4-dihydroxyphenyl)-3-(3,4-dihydroxyphenyl)-2-hydroxyprop-2-en-1-one (12), (Z)-2-(3,4-dihydroxybenzylidene)-6-hydroxybenzofuran-3(2H)-one (13), 3,5,7-trihydroxy-2-(4-hydroxyphenyl)chroman-4-one (22), 2-(3,4-dihydroxyphenyl)-3,5-dihydroxy-7-methoxy-4H-chroman-4-one (23), and 3,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one (26)) showed a lower energy docking score (kcal/mol), acceptable with the Lipinski rule-of-five (drug likeness), good water solubility, high bioavailability (0.55–0.56), and lower range of SAS (1.5–3.42). These physicochemical parameters suggest the potential for synthesis as inhibitors against the main active site of the SARS-CoV-2 protease enzyme 6LU7. In our data, most of the proposed compounds, particularly, 12, 13, 23, and 26, along with Avigan and chloroquine, have hydrogen bonds with different lengths but identical bond energies, as shown in Table 2. This may be due to the fact that, in most proteins, almost all distances between two atoms are longer than the covalent bond length, so they tend to cause steric hindrance which may affect hydrogen bond energy during interaction with the main active site of the SARS-CoV-2 protease. In addition, all H atoms are donor rotatable, which could be sterically hindered by the surrounding atoms or molecules, thus resulting in the minimization of bond energies of H-bonds with longer lengths.
In most studies, the calculated SAS, drug likeness, bioavailability, water solubility, and lower energy docking scores (kcal/mol) could be used to support various drug discovery processes, particularly for herbal plants [54, 56–63]. It was reported previously that the drug discovery process is easier for ligands or compounds with lower SAS than higher SAS [38, 63, 65]. Like our results, ligands or compounds with the lowest energy docking scores, such as Nigellidine compound extracted from Nigella sativa L, when docked into the active site of 6LU7, produced lower energy with good orientation, which argues for its use as a potential drug candidate against COVID-19 [44]. Also, several molecular docking studies that used plant-based ligands such as kaempferol, quercetin, luteolin-7-glucoside, oleuropein, curcumin, catechin, epicatechin-gallate, caffeine, capsaicin, and hypericin show potential inhibitory activity against COVID-19 [9, 14, 38, 63, 65–68]. In addition, a high potency of salvianolic acid A and curcumin as herbal compounds were reported as Mpro inhibitors by using docking calculations [44].
5. Conclusion
The antiviral activity of 26 active polyphenolic compounds of Rhus spp. (sumac) was analyzed by molecular docking, drug likeness, and synthetic accessibility score (SAS). The results showed that all tested compounds provided good interaction in the main active site of the SARS-CoV-2 protease enzyme 6LU7 with lower molecular docking energy (kcal/mol) compared to two drugs already under clinical tests. Together, the molecular docking, drug likeness, and synthetic accessibility score (SAS) data proposed six active polyphenolic compounds of Rhus spp. for the synthesis of potential inhibitors against the main active site of the SARS-CoV-2 protease enzyme 6LU7, which may be candidates for the treatment of COVID-19. The data herein provide new directions for in vitro and in vivo investigations of the proposed ligands to develop new inhibitors as SARS-CoV-2 antivirals.
Acknowledgments
The authors are grateful to the Deanship of Scientific Research, King Saud University, for funding through Vice Deanship of Scientific Research Chairs.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Supplementary Materials
Interaction between compound (14) (2-((10Z,13 E,15E)-heptadeca-10,13,15-trien-1-yl)) phenol with COVID-19 (6LU7). The 2- and 3-dimensional (2D and 3D) structures of compound (14) showed no hydrogen bonding and no perceptible interactions, only electrostatics exist (Van der Waals).
References
- 1.Pollard C. A., Morran M. P., Nestor-Kalinoski A. L. The COVID-19 pandemic: a global health crisis. Physiological Genomics. 2020;52(11):549–557. doi: 10.1152/physiolgenomics.00089.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. World Health Organization. 2020. https://www.who.int/home.
- 3.Hui D. S., I Azhar E., Madani T. A., et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health-the latest 2019 novel coronavirus outbreak in Wuhan, China. International Journal of Infectious Diseases. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh Malik Y. Emerging novel coronavirus (2019-NCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments. Veterinary Quarterly. 2020;40 doi: 10.1080/01652176.2020.1727993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu R., Zhao X., Li J., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020;395(10224):565–574. doi: 10.1016/s0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D., Hu B., Hu C., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):p. 1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) BioScience Trends. 2020;14(1):p. 69. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- 8.Xu Z., Peng C., Shi Y., et al. Nelfinavir Was Predicted to Be a Potential Inhibitor of 2019-nCoV Main Protease by an Integrative Approach Combining Homology Modelling, Molecular Docking and Binding Free Energy Calculation. Cold Spring Harbor, NY, USA: bioRxiv; 2020. [DOI] [Google Scholar]
- 9.Khaerunnisa S., Kurniawan H., Awaluddin R., Suhartati S., Soetjipto S. Potential inhibitor of COVID-19 main protease (Mpro) from several medicinal plant compounds by molecular docking study. Preprints. 2020 doi: 10.20944/preprints202003.0226.v1.2020030226 [DOI] [Google Scholar]
- 10.Liu Q., Lu L., Hua M., et al. Jiawei-yupingfeng-tang, a Chinese herbal formula, inhibits respiratory viral infections in vitro and in vivo. Journal of Ethnopharmacology. 2013;150(2):521–528. doi: 10.1016/j.jep.2013.08.056. [DOI] [PubMed] [Google Scholar]
- 11.Nadia B., Meriem B., Meriem K. Ethnobotanical study of medicinal plants used by herbalists for the treatment of respiratory diseases in the region of Oran. Algeria. 2020;2:p. 6. [Google Scholar]
- 12.Ton A. T., Gentile F., Hsing M., Ban F., Cherkasov A. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Molecular Informatics. 2020;39(8) doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amin S., Mir S. R., Kohli K., Ali B., Ali M. A study of the chemical composition of black cumin oil and its effect on penetration enhancement from transdermal formulations. Natural Product Research. 2010;24(12):1151–1157. doi: 10.1080/14786410902940909. [DOI] [PubMed] [Google Scholar]
- 14.Agrawal A., Jain N. K., Kumar N., Kulkarni G. T. Molecular Docking Study to Identify Potential Inhibitor of COVID-19 Main Protease Enzyme: An In-Silico Approach. Washington, DC, USA: ChemRxiv; 2020. [DOI] [Google Scholar]
- 15.USDA. Germplasm Resources Information Network. Beltsville, MD, USA: United States Department of Agriculture; 2007. http://www.ars-grin.gov/npgs/aboutgrin.html. [Google Scholar]
- 16.Monavari H. R., Hamkar R., Norooz-Babaei Z., Adibi L., Noroozi M., Ziaei A. Antiviral effect assay of twenty five species of various medicinal plants families in Iran. IJMM. 2007;1(2):49–59. [Google Scholar]
- 17.Lin Y.-M., Anderson H., Flavin M. T., et al. In vitro anti-HIV activity of biflavonoids isolated from Rhus succedanea and Garcinia multiflora. Journal of Natural Products. 1997;60(9):884–888. doi: 10.1021/np9700275. [DOI] [PubMed] [Google Scholar]
- 18.Alghadir A., Gabr S. Efficacy of Rhus coriaria (sumac) juice in reducing muscle pain during aerobic exercise. Acta Physiologica Hungarica. 2016;103(2):p. 231. doi: 10.1556/036.103.2016.2.10. [DOI] [PubMed] [Google Scholar]
- 19.Gabr S. A., El-Metwall M. M., Al-Ghadir A. H. Antioxidant and antibacterial active constituents of Rhus coriaria. Biotechnology (Faisalabad) 2014;13(2):37–45. doi: 10.3923/biotech.2014.37.45. [DOI] [Google Scholar]
- 20.Gabr S. A., Alghadir AH H. Phytochemical analysis and in vitro antifungal activities of bioactive fractions from leaves of Rhus coriaria (SUMAC) Journal of Pure and Applied Microbiology. 2015;9(1):559–565. [Google Scholar]
- 21.Saxena G., McCutcheon A. R., Farmer S., Hancock R. E. W. Antimicrobial constituents of Rhus glabra. Journal of Ethnopharmacology. 1994;42(2):95–99. doi: 10.1016/0378-8741(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 22.Towers S. M., Halkman A. K. Inhibition of some foodborne bacteria by alcohol extract of sumac (Rhus coriaria L.) Journal of Food Safety. 2004;24(4):257–267. doi: 10.1111/j.1745-4565.2004.00506.x. [DOI] [Google Scholar]
- 23.ÖzcanAl-HAQ M. Antioxidant activities of rosemary, sage, and sumac extracts and their combinations on stability of natural peanut oil. Journal of Medicinal Food. 2003;6(3):267–270. doi: 10.1089/10966200360716698. [DOI] [PubMed] [Google Scholar]
- 24.Kossah R., Nsabimana C., Zhang H., Chen W. Evaluation of antimicrobial and antioxidant activities of Syrian Sumac fruit extract. Journal of Natural Products. 2013;6:96–102. [Google Scholar]
- 25.Onkar S., Mohammed A., Nida A. New antifungal aromatic compounds from the seeds of Rhus coriaria L. International Research Journal of Pharmacy. 2011;2:188–194. [Google Scholar]
- 26.Gabr S. A., Alghadir A. H. Evaluation of the biological effects of lyophilized hydrophilic extract of Rhus coriaria on myeloperoxidase (MPO) activity, wound healing, and microbial infections of skin wound tissues. Evidence-Based Complementary and Alternative Medicine. 2019;2019:14. doi: 10.1155/2019/5861537.5861537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang R.-R., Gu Q., Yang L.-M., Chen J.-J., Li S.-Y., Zheng Y.-T. Anti-HIV-1 activities of extracts from the medicinal plant Rhus chinensis. Journal of Ethnopharmacology. 2006;105(1-2):269–273. doi: 10.1016/j.jep.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Wang R.-R., Gu Q., Wang Y.-H., et al. Anti-HIV-1 activities of compounds isolated from the medicinal plant Rhus chinensis. Journal of Ethnopharmacology. 2008;117(2):249–256. doi: 10.1016/j.jep.2008.01.037. [DOI] [PubMed] [Google Scholar]
- 29.Gu Q., Wang R.-R., Zhang X.-M., et al. A new benzofuranone and anti-HIV constituents from the stems of Rhus chinensis. Planta Medica. 2007;73(3):279–282. doi: 10.1055/s-2007-967113. [DOI] [PubMed] [Google Scholar]
- 30.Kurokawa M., Hozumi T., Basnet P., et al. Purification and characterization of eugeniin as an anti-herpesvirus compound from Geum japonicum and Syzygium aromaticum. The Journal of Pharmacology and Experimental Therapeutics. 1998;284(2):728–735. [PubMed] [Google Scholar]
- 31.Kurokawa M., Nagasaka K., Hirabayashi T., et al. Efficacy of traditional herbal medicines in combination with acyclovir against herpes simplex virus type 1 infection in vitro and in vivo. Antiviral Research. 1995;27(1-2):19–37. doi: 10.1016/0166-3542(94)00076-k. [DOI] [PubMed] [Google Scholar]
- 32.Nakano M., Kurokawa M., Hozumi T., et al. Suppression of recurrent genital herpes simplex virus type 2 infection by Rhus javanica in Guinea pigs. Antiviral Research. 1998;39(1):25–33. doi: 10.1016/s0166-3542(98)00023-0. [DOI] [PubMed] [Google Scholar]
- 33.Anwer T., Sharma M., Khan G., et al. Rhus coriaria ameliorates insulin resistance in non-insulin-dependent diabetes mellitus (Niddm) rats. Acta Poloniae Pharmaceutica-Drug Research. 2013;70:861–867. [PubMed] [Google Scholar]
- 34.Özcan M., Haciseferogullari H. A condiment [sumac (Rhus coriaria L.) fruits]: some physico-chemical properties. Bulgarian Journal of Plant Physiology. 2004;30:74–84. [Google Scholar]
- 35.Yi L., Li Z., Yuan K., et al. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. Journal of Virology. 2004;78(20):11334–11339. doi: 10.1128/jvi.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W., Moore M. J., Vasilieva N., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mubarak A., Alturaiki W., Hemida M. G. Middle East respiratory syndrome coronavirus (MERS-CoV): infection, immunological response, and vaccine development. Journal of Immunology Research. 2019;2019:11. doi: 10.1155/2019/6491738.6491738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ertl P., Schuffenhauer A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. Journal of Cheminformatics. 2009;1:p. 8. doi: 10.1186/1758-2946-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim J., Jeon S., Shin H.-Y., et al. The author’s response: case of the index patient who caused tertiary transmission of coronavirus disease 2019 in Korea: the application of lopinavir/ritonavir for the treatment of COVID-19 pneumonia monitored by quantitative RT-PCR. Journal of Korean Medical Science. 2020;35(7):p. e79. doi: 10.3346/jkms.2020.35.e89. [DOI] [PubMed] [Google Scholar]
- 40.Boceprevir W. J. GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Research. 2020;30(8):678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L., Lin D., Sun X., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sacco M. D., Ma C., Lagarias P., et al. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L. Science Advances. 2020;6(50) doi: 10.1126/sciadv.abe0751.eabe0751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.RCSB PDB-6LU7. The crystal structure of COVID-19 main protease in complex with an inhibitor N3’. 2020.
- 44.Bouchentouf S., Missoum N. Washington, DC, USA: ChemRxiv; 2020. Identification of compounds from Nigella sativa as new potential inhibitors of 2019 novel coronavirus (Covid-19): molecular docking study. [DOI] [Google Scholar]
- 45.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. Journal of Medical Virology. 2020;92(4):p. 418. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dasgupta Y., Golovine K., Nieborowska-Skorska M., et al. Drugging DNA repair to target T-ALL cells. Leukemia & Lymphoma. 2018;59(7):1746–1749. doi: 10.1080/10428194.2017.1397662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ibrahim M. A. A., Abdelrahman A. H. M., Hussien T. A., et al. In silico drug discovery of major metabolites from spices as SARS-CoV-2 main protease inhibitors. Computers in Biology and Medicine. 2020;126:p. 104046. doi: 10.1016/j.compbiomed.2020.104046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korkmaz H. Could sumac be effective on COVID-19 treatment? Journal of Medicinal Food. 2020. pp. 1–6. [DOI] [PubMed]
- 49.Wu Z., Ma Y., Zhao L., Cai S., Cheng G. Acute and subchronic toxicities of the ethanol and hot-water extracts from Chinese sumac (Rhus chinensis Mill.) fruits by oral administration in rats. Food and Chemical Toxicology. 2018;119:14–23. doi: 10.1016/j.fct.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 50.Rahideh S. T., Shidfar F., Khandozi N., Rajab A., Hosseini S. P., Mirtaher S. M. The effect of sumac (Rhus coriaria L.) powder on insulin resistance, malondialdehyde, high sensitive C-reactive protein and paraoxonase 1 activity in type 2 diabetic patients. Journal of Research in Medical Sciences. 2014;19:933–938. [PMC free article] [PubMed] [Google Scholar]
- 51.Alsamri H., Athamneh K., Pintus G., Eid A. H., Iratni R. Pharmacological and antioxidant activities of Rhus coriaria L. (Sumac) Antioxidants. 2021;10(1):p. 73. doi: 10.3390/antiox10010073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chemical Computing Group Inc. Molecular Operating Environment Version 2014. Montreal, Canada: Chemical Computing Group Inc.; 2014. [Google Scholar]
- 53.Abu-Reida I. M., Jamous R. M., Ali-Shtayeh M. S. Phytochemistry , pharmacological properties and industrial applications of Rhus coriaria L. ( sumac ) Jordan Journal of Biological Sciences. 2014;7(4):p. 233. doi: 10.12816/0008245. [DOI] [Google Scholar]
- 54. https://www.rcsb.org.
- 55.Rayne S., Mazza G. Biological activities of extracts from sumac (Rhus spp.): a review. Plant Foods for Human Nutrition. 2007;62(4):165–175. doi: 10.1007/s11130-007-0058-4. [DOI] [PubMed] [Google Scholar]
- 56.Molegro virtual docker 6.0. 2013.
- 57.Hypercube Inc. HyperChem Program Version 8.1. Gainesville, FL, USA: Hypercube Inc.; 2002. [Google Scholar]
- 58.Lipinski C. A., Lombardo F., Dominy B. W., Feeney P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings 1PII of original article: S0169-409X(96)00423-1. The article was originally published in Advanced Drug Delivery Reviews 23 (1997) 3-25. 1. Advanced Drug Delivery Reviews. 2001;46(1–3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 59.Bertelli F. Management of Chemical and Biological Samples for Screening Applications. New York, NY, USA: Wiley; 2012. What does an HTS file of the future look like? pp. 275–304. [Google Scholar]
- 60.Lipinski C. A., December Lead- and drug-like compounds: the rule-of-five revolution. Drug Discovery Today: Technologies. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 61.Jayaram B., Singh T., Mukherjee G., Mathur A., Shekhar S., Shekhar V. Sanjeevini: a freely accessible web-server for target directed lead molecule discovery. BMC Bioinformatics. 2012;13(S17):p. S7. doi: 10.1186/1471-2105-13-s17-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports. 2017;7(1):p. 42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonnet P. Is chemical synthetic accessibility computationally predictable for drug and lead-like molecules? A comparative assessment between medicinal and computational chemists. European Journal of Medicinal Chemistry. 2012;54:679–689. doi: 10.1016/j.ejmech.2012.06.024. [DOI] [PubMed] [Google Scholar]
- 64.Duan D., Li Z., Luo H., Zhang W., Chen L., Xu X. Antiviral compounds from traditional Chinese medicines Galla Chinese as inhibitors of HCV NS3 protease. Bioorganic & Medicinal Chemistry Letters. 2004;14(24):6041–6044. doi: 10.1016/j.bmcl.2004.09.067. [DOI] [PubMed] [Google Scholar]
- 65.Abu-Reidah I. M., Ali-Shtayeh M. S., Jamous R. M., Arráez-Román D., Segura-Carretero A. HPLC-DAD-ESI-MS/MS screening of bioactive components from Rhus coriaria L. (Sumac) fruits. Food Chemistry. 2015;166:179–191. doi: 10.1016/j.foodchem.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 66.Yang H., Bartlam M., Rao Z. Drug design targeting the main protease, the achilles heel of coronaviruses. Current Pharmaceutical Design. 2006;12(35):4573–4590. doi: 10.2174/138161206779010369. [DOI] [PubMed] [Google Scholar]
- 67.Shabana M. M., El Sayed A. M., Yousif M. F., El Sayed A. M., Sleem A. A. Bioactive constituents from Harpephyllum caffrum Bernh. and Rhus coriaria L. Pharmacognosy Magazine. 2011;7(28):298–306. doi: 10.4103/0973-1296.90410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shabir A. Rhus coriaria Linn, a plant of medicinal, nutritional and industrial importance: a review. Journal of Animal and Plant Sciences. 2012;2(2):505–512. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Interaction between compound (14) (2-((10Z,13 E,15E)-heptadeca-10,13,15-trien-1-yl)) phenol with COVID-19 (6LU7). The 2- and 3-dimensional (2D and 3D) structures of compound (14) showed no hydrogen bonding and no perceptible interactions, only electrostatics exist (Van der Waals).
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


